Abstract
Peniciadametizine A (1); a new dithiodiketopiperazine derivative possessing a unique spiro[furan-2,7'-pyrazino[1,2-b][1,2]oxazine] skeleton, together with a highly oxygenated new analogue, peniciadametizine B (2); as well as two known compounds, brasiliamide A (3); and viridicatumtoxin (4), were isolated and identified from Penicillium adametzioides AS-53, a fungus obtained from an unidentified marine sponge. The unambiguous assignment of the relative and absolute configuration for the spiro center C-2 of compound 1 was solved by the combination of NMR and ECD measurements with Density-Functional Theory (DFT) conformational analysis and Time-Dependent Density-Functional Theory-Electronic Circular Dichroism (TDDFT-ECD) calculations. The spiro[furan-2,7'-pyrazino[1,2-b][1,2]oxazine] skeleton of 1 has not been reported yet among natural products and the biosynthetic pathway for 1 and 2 was discussed. Compounds 1 and 2 showed inhibitory activity against the pathogenic fungus Alternaria brassicae.
Keywords: marine sponge, endophytic fungus, Penicillium adametzioides, secondary metabolites, antifungal activity
1. Introduction
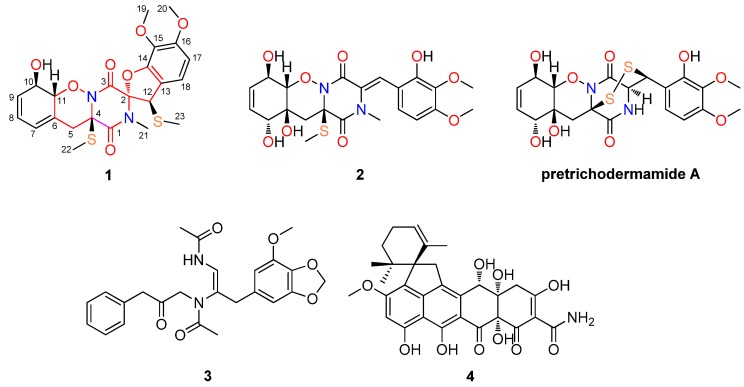
The dithiodiketopiperazine derivatives represent a unique class of fungal metabolites generally possessing a disulfide bridge or having two methylthio groups, with the sulfur-atom usually attached to α-positions of the cyclic dipeptide [1]. In contrast, dithiodiketopiperazines with the disulfide functionality attached to both α- and β-positions of the amino acid residues, such as gliovirin [1] and aspirochlorine [2], are reported very rarely. A peculiar structural feature of these two compounds is the presence of an O-alkyl-oxime or a spiro[furan-pyrazino] functionality, respectively [1,2,3]. In the last two decades, marine filamentous fungi have been recognized as an important source of structurally unusual and biologically active natural products [4]. During our ongoing search for bioactive metabolites from marine-derived fungi [5,6,7,8,9], we have recently reported a new spiroquinazoline derivative, N-formyllapatin A, from Penicillium adametzioides AS-53, a fungus isolated from an unidentified sponge collected at the Hainan Island of China [7]. Futher exploration of the fungal extract resulted in the isolation and characterization of two new diketopiperazine analogues, peniciadametizines A and B (1 and 2); as well as two known compounds, brasiliamide A (3) [10]; and viridicatumtoxin (4) [11] (Figure 1). Among them, compound 1 is a dithiodiketopiperazine derivative possessing a unique spiro[furan-2,7'-pyrazino[1,2-b][1,2]oxazine] skeleton and the two sulfur atoms are attached to the α- and β-positions of the amino acid residues. This paper describes the isolation, structural elucidation, and antimicrobial activity of compounds 1–4.
Figure 1.
Structures of the isolated compounds 1–4 and reference compound pretrichodermamide A.
2. Results and Discussion
Peniciadametizine A (1), obtained as a colorless amorphous powder, had the molecular formula C23H26N2O7S2 on the basis of the positive HR-ESI-MS adduct ion [M + Na]+ at m/z 529.1060, implying the presence of 12 degrees of unsaturation. The IR spectrum exhibited broad absorption for OH groups (3334 cm−1) as well as for amide chromophores (1679 and 1602 cm−1). Inspection of the 13C NMR and DEPT spectroscopic data of 1 (Table 1) revealed the presence of five methyls (with two oxygenated, one nitrogenated, and two sulfurated), one methylene, eight methines (with five aromatic/olefinic and three sulfur/oxygenated sp3), and nine quaternary (with two amide carbonyl, five aromatic/olefinic, and two nitrogen/oxygenated/thiogenated sp3) carbons. The 1H NMR and HSQC data (Table 1) indicated the presence of 26 protons, attributable to five methyl singlets (H3-19 to H3-23), one aliphatic methylene (H2-5), and eight methines, with two aromatic (H-17 and H-18), three olefinic (H-7 to H-9), and three sulfur/oxygenated (H-10 to H-12), as well as an exchangeable OH group (10-OH).
Table 1.
1H-, 13C-NMR and HMBC data of compounds 1 and 2.
| No. | 1 (Acquired in DMSO-d6) | 2 (Acquired in Acetone-d6) | ||||
|---|---|---|---|---|---|---|
| δC a, Type | δH (J in Hz) b | HMBC | δC a, Type | δH (J in Hz) b | HMBC | |
| 1 | 161.7, C | 164.9, C | ||||
| 2 | 100.8, C | 129.0, C | ||||
| 3 | 158.7, C | 160.9, C | ||||
| 4 | 70.4, C | 68.1, C | ||||
| 5 | 36.8, CH2 | 3.58, d (14.0) | 1, 4, 6, 7, 11 | 33.0, CH2 | 2.09, d (15.0) | 6, 7 |
| 2.98, d (14.0) | 2.54, d (15.0) | |||||
| 6 | 129.7, C | 74.0, C | ||||
| 7 | 122.2, CH | 5.82, d (2.5) | 6, 8 | 75.2, CH | 4.40, br s | 6, 8 |
| 8 | 122.4, CH | 5.76, dd (10.0, 2.5) | 6, 10 | 130.3, CH | 5.54, br d (10.4) | 6, 10 |
| 9 | 132.2, CH | 5.69, br d (10.0) | 7, 11 | 128.9, CH | 5.60, br d (10.4) | 7, 11 |
| 10 | 69.5, CH | 4.54, br dd (13.5, 4.5) | 65.9, CH | 4.65, br s | 9 | |
| 11 | 91.4, CH | 4.92, d (13.5) | 10 | 89.8, CH | 4.00, d (8.2) | 6, 10 |
| 12 | 53.5, CH | 4.95, br s | 2, 3, 13, 14, 23 | 118.1, CH | 7.25, s | 3, 14, 18 |
| 13 | 118.3, C | 115.6, C | ||||
| 14 | 149.7, C | 150.1, C | ||||
| 15 | 132.3, C | 137.1, C | ||||
| 16 | 153.1, C | 154.8, C | ||||
| 17 | 107.0, CH | 6.76, d (8.4) | 13, 15, 16 | 104.8, CH | 6.66, d (8.7) | 13, 15 |
| 18 | 119.2, CH | 6.94, d (8.4) | 12, 14, 16 | 126.0, CH | 6.90, d (8.7) | 14, 16 |
| 19 | 60.2, CH3 | 3.73, s | 15 | 61.0, CH3 | 3.79, s | 15 |
| 20 | 56.2, CH3 | 3.80, s | 16 | 56.4, CH3 | 3.89, s | 16 |
| 21 | 28.2, CH3 | 2.78, s | 1, 2 | 35.6, CH3 | 2.91, s | 1, 2 |
| 22 | 13.2, CH3 | 2.36, s | 4 | 13.9, CH3 | 2.26, s | 4 |
| 23 | 13.0, CH3 | 1.78, s | 12 | |||
| 10-OH | 5.29, d (4.5) | |||||
a Measured at 125 MHz. b Measured at 500 MHz. Multiplicities were determined by DEPT and HSQC experiments.
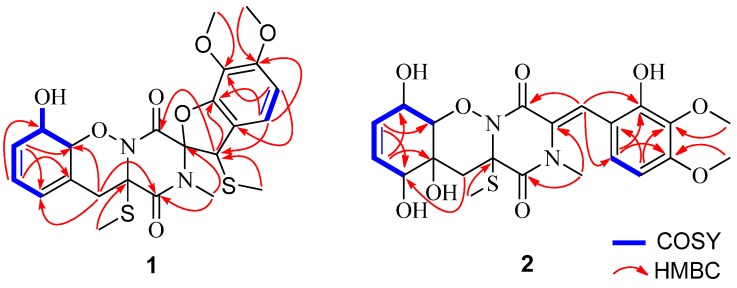
Detailed interpretation of the COSY and HSQC spectra of 1 resulted in the elucidation of two discrete proton-proton spin-coupling systems corresponding to a =CH–CH=CH–CH(OH)–CH– unit (I, C-7 through C-11) and an aromatic –CH=CH– residue (II, C-17 to C-18) (Figure 2). HMBC correlations from H-7, H-8 to C-6, from H2-5 to C-1, C-4, C-6, C-7, and C-11, as well as from H3-22 to C-4 indicated the involvement of an O-alkyl-oxime motif, thus affording half of the ketopiperazine ring in the molecule of 1. The structure of the other half of the ketopiperazine ring was deduced by the HMBC correlations from H-12 to C-2, C-3, C-13, and C-14, from H-17 to C-13, C-15, and C-16, and from H-18 to C-12, C-14, and C-16. Furthermore, HMBC correlations from H3-19 to C-15, from H3-20 to C-16, from H3-21 to C-1 and C-2, and from H3-23 to C-12 verified the position of these methyl groups. Finally, the connection of C-2 and C-14 through an oxygen atom allowed the construction of the spiro[furan-pyrazino] functionality to match the molecular formula and the required degrees of unsaturation.
Figure 2.
Key 1H–1H COSY and HMBC correlations of compounds 1 and 2.
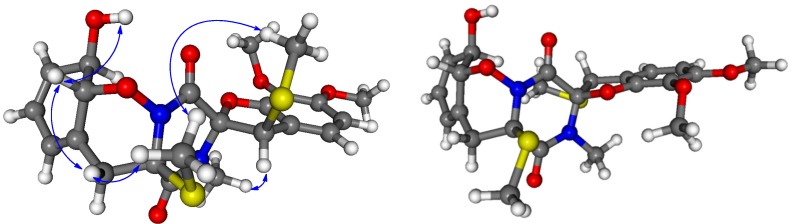
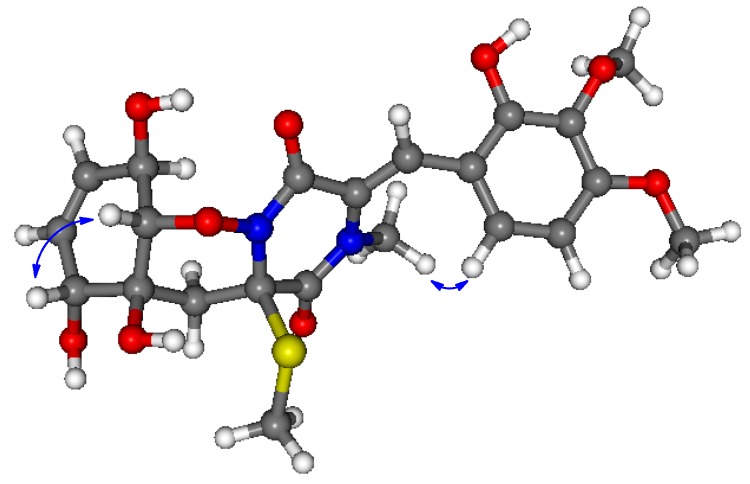
Analysis of vicinal proton-proton coupling constant and NOESY data enabled assignment of the relative configuration of 1. The large 3J coupling constant of H-10 and H-11 (13.5 Hz) and the observed NOE correlation from the proton of 10-OH to H-11 established the trans-diaxial orientation of H-10 and H-11, whereas the observed NOE correlations from Ha-5 to H-11 and H3-22 and from H3-22 to H3-23 placed these groups on the same side of the molecule (Figure 3). However, the NOESY spectrum lacked diagnostic correlations to unambiguously determine the relative configuration of the spiro carbon (C-2), although the NOE correlation from H-12 to H3-21 was observed in the NOESY spectrum. To solve this problem, conformational analyses were carried out on the diastereomeric (2R,4R,10R,11R,12R)-1 and (2S,4R,10R,11R,12R)-1. In the B97D/TZVP (triple-zeta valence polarization) computed conformers of (2R,4R,10R,11R,12R)-1, differing mainly in the orientation of the methylthio and methoxy groups, the 1,2-oxazine ring adopted a boat conformation and the N-methyl protons and H-12 were on the same face at a distance of 2.68 Å (Figure 3). Similarly, the two methylthio groups were also on the same side in accordance with their observed NOE correlation. In contrast, in the computed conformers of (2S,4R,10R,11R,12R)-1, the N-methyl protons and the H-12 were pointing towards opposite directions (distance 4.59 Å) such as the two methylthio groups (Figure 3). Thus pronounced NOE correlation of H-12 and H3-21 verified the (2R*,4R*,10R*,11R*,12R*) relative configurations of 1 as shown in Figure 3.
Figure 3.
Measured NOE correlations of 1 shown on the lowest-energy B97D/TZVP computed conformer of (2R,4R,10R,11R,12R)-1 and lowest-energy conformer of (2S,4R,10R,11R,12R)-1.
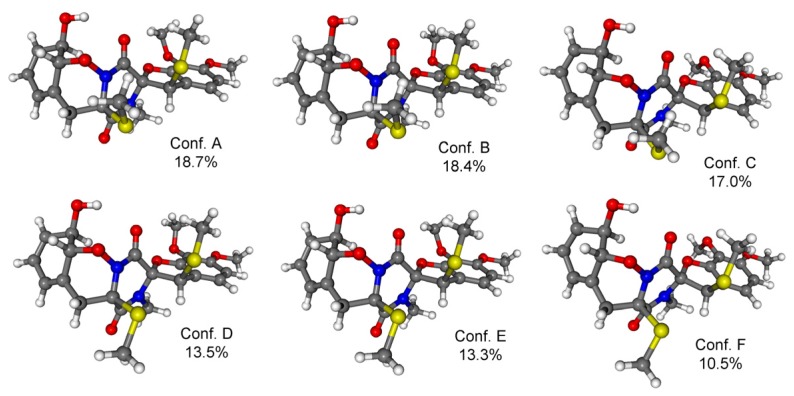
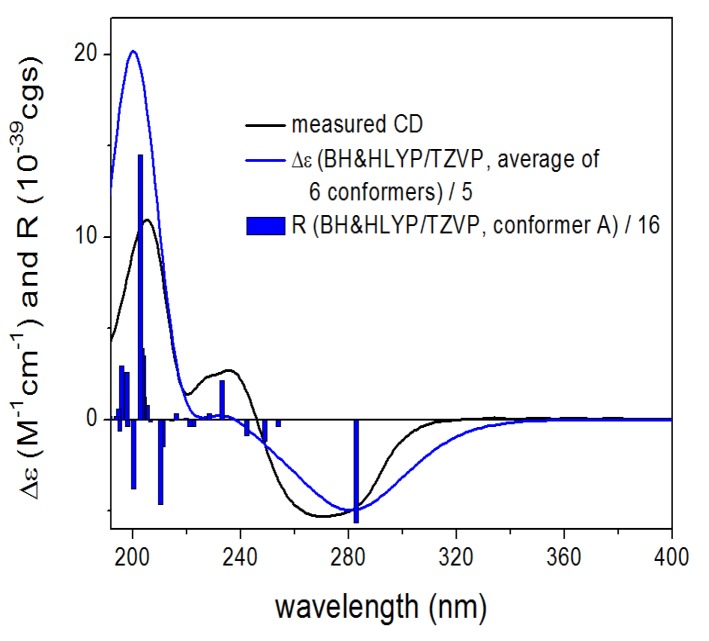
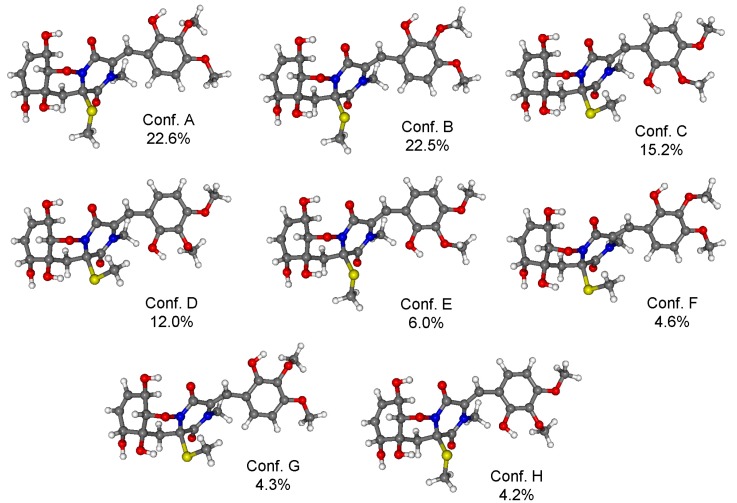
In order to determine the absolute configuration of 1, Time-Dependent Density Functional Theory-Electronic Circular Dichroism (TDDFT-ECD) calculations were carried out on the B97D/TZVP conformers of (2R,4R,10R,11R,12R)-1 with functionals (B3LYP, BH & HLYP, PBE0) and TZVP basis set (Figure 4). The ECD spectrum of 1 showed a broad negative transition (CE) at 269 nm and positive Cotton effects (CEs) at 237, 226, and 205 nm. The computed TDDFT-ECD spectra of (2R,4R,10R,11R,12R)-1 reproduced well the experimental curve with the BH & HLYP/TZVP method giving the best agreement (Figure 5), which allowed determining the absolute configuration as (−)-(2R,4R,10R,11R,12R) for 1.
Figure 4.
Structures and Boltzmann populations of B97D/TZVP conformers Polarizable Continuum Model (PCM) solvent model for MeCN) of (2R,4R,10R,11R,12R)-1.
Figure 5.
Experimental ECD (black) spectrum of 1 compared with BH & HLYP/TZVP ECD spectra (blue) of (2R,4R,10R,11R,12R)-1 computed for the B97D/TZVP conformers (PCM/MeCN). Bars represent the computed rotational strengths of the lowest-energy conformer.
Compound 2 was also obtained as a colorless amorphous powder. It has the molecular formula C22H26N2O9S as determined by HR-ESI-MS data. The IR spectrum exhibited broad absorption for OH groups (3275 cm−1) as well as for amide chromophores (1678 and 1608 cm−1). The 13C NMR data (Table 1) exhibited the presence of 22 carbon signals, which were assigned by DEPT and HSQC experiments as four methyls (with two oxygenated, one nitrogenated, and one thiogenated), one methylene, eight methines (with five sp2 and three oxygenated sp3), and nine quaternary (with two amide carbonyl, five aromatic/olefinic, and two nitrogenated/oxygenated/thiogenated sp3) carbons. The 1H NMR data (Table 1) along with HSQC experiment displayed resonances characteristic for four methyls (H3-19 to H3-22), one methylene (H2-5), three oxymethines (H-7, H-10, and H-11), and five aromatic/olefinic methines (H-8, H-9, H-12, H-17, and H-18) (Table 1). Comprehensive analysis of the NMR data revealed that the structure of 2 was similar to pretrichodermamide A [1] (Figure 1). The main differences were the presence of signals for additional N-methyl (CH3-21, δC/H 35.6/2.91) and S-methyl (CH3-22, δC/H 13.9/2.26) in the NMR spectra of 2, which was proved by the HMBC correlations from H3-21 to C-1 and C-2 and from H3-22 to C-4 (Figure 2). In addition, two methines (CH-2, δC/H 59.0/4.41) and (CH-12, δC/H 45.0/4.49) in the NMR spectra of pretrichodermamide A [1] disappeared in those of 2, and instead signals (C-2, δC 129.0) and (CH-12, δC/H 118.1/7.25) corresponding to a double bond were observed in the NMR spectra of 2. These changes were supported by the observed HMBC correlations from H-12 to C-3, C-14, and C-18 (Figure 2). Thus the planar structure of 2 could be assigned as shown in Figure 1.
The (4R*,6S*,7R*,10R*,11S*) relative configuration of 2 was determined by the analysis of vicinal proton-proton coupling constants and NOESY experiments. The coupling constant between H-10 and H-11 (8.2 Hz) suggested that the two protons had trans-diaxial arrangement, and the NOE correlation between H-7 and H-11 indicated the same orientation of these protons (Figure 6). The chemical shift value of H-12 (δH 7.25), which is downfield-shifted by the deshielding effect of the C=O of C-3, suggested the (Z)-configuration of the C=C double bond at C-2(12), which was further confirmed by the NOE correlation between H3-21 and H-18 (Figure 6).
Figure 6.
Structure of the lowest-energy B97D/TZVP conformer (PCM solvent model for MeCN) of (4R,6S,7R,10R,11S)-2 with the measured NOE correlations.
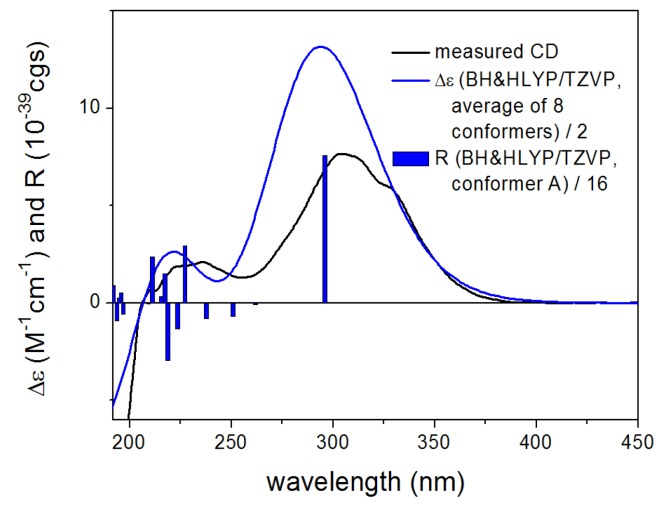
For the configurational assignment of 2, conformational analysis and TDDFT-ECD calculations of the solution conformers were carried out. The B97D/TZVP reoptimization of the initial Merck Molecular Force Field (MMFF) conformers of (4R,6S,7R,10R,11S)-2 afforded eight slightly different conformers, in which the equatorial 10-OH was hydrogen-bonded to the C-3 carbonyl oxygen, while 6-OH to the 4-SMe group fixing the chair conformation of the 1,2-oxazine ring (Figure 7). The ECD spectrum of 2 was dominated by the intense positive CE at 304 nm accompanied with shoulders and weaker positive CEs at 236, 220, and 207 nm. The TDDFT-ECD spectra of (4R,6S,7R,10R,11S)-2 reproduced well the experimental ECD transitions (Figure 8) and thus the absolute configuration was determined as (+)-(4R,6S,7R,10R,11S) and the trivial name 6,7-dihydroxy-seco-peniciadametizine A was assigned to 2.
Figure 7.
Structures and Boltzmann populations of B97D/TZVP conformers (PCM solvent model for MeCN) of (4R, 6S, 7R, 10R,11S)-2.
Figure 8.
Experimental ECD (black) spectrum of 2 compared with BH & HLYP/TZVP ECD spectra (blue) of (4R,6S,7R,10R,11S)-2 computed for the B97D/TZVP conformers (PCM/MeCN). Bars represent the computed rotational strengths of the lowest-energy conformer.
In addition to the new compounds 1 and 2, two known secondary metabolites, brasiliamide A (3) [10] and viridicatumtoxin (4) [11] (Figure 1), were also isolated, and their structures were elucidated by comparing the NMR data with those of literature reports.
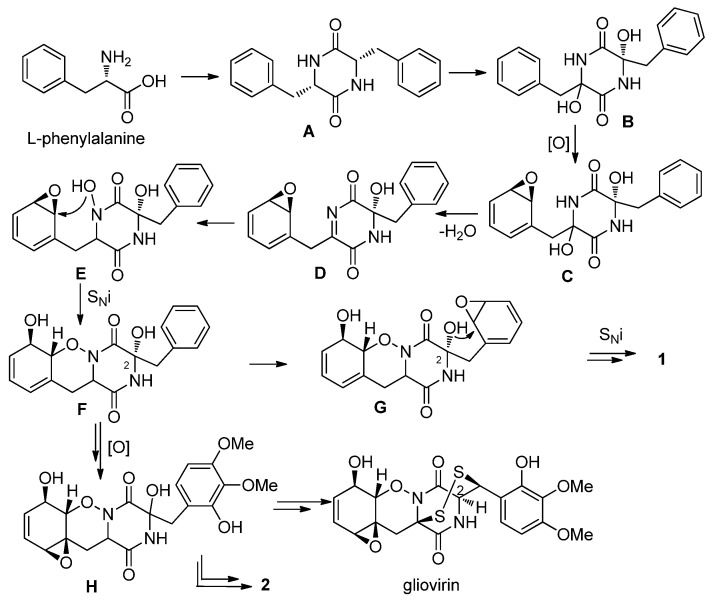
Peniciadametizine A (1) represents a new member of the gliovirin family whose sulfide functionalities are attached to the α- and β-positions of the degraded amino acid residues. Particularly, the spiro[furan-2,7'-pyrazino[1,2-b][1,2]oxazine] skeleton of 1 has not been reported yet among natural products. The isolation of compounds 1 and 2 provided further insight into the biosynthesis of thiodiketopiperazine derivatives, which has been recently studied by several research groups [12]. The biosynthesis of these derivatives starts with the dimerization of L-phenylalanine followed by bishydroxylation to afford intermediate B, the hydroxyl groups of which are usually replaced in the following step(s) by thiol groups (Scheme 1). However, the structures of 1 and 2 suggest a different pathway, in which the two halves of the diketopiperazine moiety follow different sequence of reactions. Upon epoxidation of the phenyl ring one of the hydroxyl groups is lost by dehydration to produce an imine group, the nitrogen of which is then hydroxylated (Scheme 1). The isolation of a related N-hydroxyl intermediate was reported earlier in targeted gene-deletion study [13]. The N-hydroxyl group opens the epoxide ring forming a 1,2-oxazine ring (E→F), while the epoxidation of the Δ6 double bond provides intermediate H, the precursor of both gliovirin and compound 2. If the hydroxyl group at C-2 of intermediate F is involved in an epoxide ring-opening reaction, a dihydrobenzo[b]furan moiety is produced (F→G→1), the further transformation of which affords compound 1. It is noteworthy that gliovirin has (2S) absolute configuration as confirmed by X-ray analysis [14], which means that the original (S) configuration of intermediate A was inverted during the biosynthetic scheme.
Scheme 1.
Proposed biosynthetic pathway for 1 and 2.
Compounds 1 and 2 were tested for cytotoxic activities against nine human tumor cell lines (DU145, HeLa, HepG2, MA, MCF-7, SGC-7901, SW1990, SW480, and U251). None of them showed potent activity (IC50 > 10 μM). However, they exhibited weak brine shrimp lethality against Artemia salina, with lethal ratio of 45.5 and 62.4% at the concentration of 100 μg/mL (Table S1 in the Supplementary file). Compounds 1 and 2 were also assayed for antimicrobial activities against human- and aqua-pathogenic bacteria (Aeromonas hydrophilia, Edwardsiella tarda, Escherichia coli, Staphyloccocus aureus, Vibrio alginolyticus, V. anguillarum, V. parahaemolyticus, and V. harveyi), and plant pathogenic fungi (Alternaria brassicae, Colletotrichum gloeosporioides, Fusarium graminearum, and Gaeumannomyces graminis), and they exhibited selective activities against A. brassicae with MIC values of 4.0 and 32.0 μg/mL, respectively, compared to amphotericin B with an MIC value of 1.0 μg/mL.
3. Experimental Section
3.1. General
Optical rotations were measured on an Optical Activity AA-55 polarimeter (Optical Activity Ltd., Cambridgeshire, UK). UV spectra were measured on a PuXi TU-1810 UV-visible spectrophotometer (PuXi Analytical Instrument Co. Ltd, Beijing, China). CD spectra were acquired on a J-810 spectropolarimeter (JASCO International Co. Ltd, Tokyo, Japan). IR spectra were obtained on a Thermo Scientific Nicolet iS10 spectrophotometer (Thermo Fisher, Waltham, MA, USA). The 1H, 13C, and 2D NMR spectra were acquired using a Bruker Avance 500 spectrometers (Bruker Biospin Group, Karlsruhe, Germany). Low- and high-resolution MS data were obtained on a Q-TOF Ultima Global GAA076 LC mass spectrometer (Waters Asia, Ltd., Singapore). Semi-preparative HPLC was performed using a Dionex UltiMate U3000 system (Dionex Corporation, Sunnyvale, CA, USA) with an Agilent Prep RP-18 column (21.2 × 250 mm, 10 μm) under UV detection. Column chromatography (CC) was performed with silica gel (200–300 mesh, Qingdao Haiyang Chemical Factory, Qingdao, China), Lobar LiChroprep RP-18 (40–60 μm, Merck, Darmstadt, Germany), and Sephadex LH-20 (18–110 μm, Merck, Darmstadt, Germany). Solvents for extraction and purification were distilled prior to use.
3.2. Fungal Material
The fungus Penicillium adametzioides AS-53 was isolated from the fresh tissue of an unidentified marine sponge that was collected from Wenchang, Hainan, China, in September 2011. The fungus was identified by analysis of its Internal Transcribed Spacer (ITS) region of the rDNA, as described in our previous report [15]. The sequence data of 5.8S rDNA and ITS regions derived from the fungal strain have been submitted to and deposited at GenBank with accession no. KJ906543. The nucleotide BLAST search result showed that the sequence was the most similar (99%) to the sequence of P. adametzioides strain DTO115I8 (compared to accession no. KC773824.1). The strain is preserved at the Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences.
3.3. Fermentation
For chemical investigation, the fresh mycelia of P. adametzioides AS-53 were grown on potato dextrose agar (PDA) medium at 28 °C for 5 days and were then inoculated into 1 L conical flask containing 300 mL of potato-dextrose broth (PDB) medium (1000 mL sea water, 20 g glucose, 5 g peptone, and 3 g yeast extract, pH (6.5–7.0)) for 30 days at room temperature.
3.4. Extraction and Isolation
All of the fermented cultures (60 flasks, 18 L) were filtered to separate the broth from the mycelia. The former was extracted four times with ethyl acetate (EtOAc), while the latter was extracted four times with mixture of 80% acetone and 20% H2O. The acetone solution was evaporated under reduced pressure to afford an aqueous solution, which was then extracted four times with EtOAc. Because the TLC and HPLC profiles of the two EtOAc solutions were almost identical, they were combined and concentrated under reduced pressure to give an extract (37 g), which was fractionated by silica gel vacuum liquid chromatography (VLC) using different solvents of increasing polarity from petroleum ether (PE) to MeOH to yield 8 fractions (Frs. 1–8) based on TLC analysis. Fr. 4 (3.6 g) was further purified by reversed-phase column chromatography (CC) over Lobar LiChroprep RP-18 with a MeOH–H2O gradient (from 10:90 to 100:0) to yield 10 subfractions. Fr. 4-5 (MeOH–H2O 1:1, 480 mg) was further fractionated by CC on silica gel eluting with CHCl3–MeOH (from 100:1 to 10:1) to afford five subfractions (from Fr. 4-5-1 to Fr. 4-5-5). Fr. 4-5-1 (45 mg) was then purified by preparative TLC (CHCl3–acetone, 2:1) to give compound 1 (4.5 mg, Rf = 0.5). Fr. 4-8 (MeOH–H2O 4:1, 180 mg) was further fractionated by CC on silica gel eluting with CHCl3–MeOH (from 150:1 to 20:1) to afford compound 4 (8.3 mg). Fr. 5 (4.4 g) was further purified by reversed-phase CC over Lobar LiChroprep RP-18 (Merck, Darmstadt, Germany) with a MeOH–H2O gradient (from 10:90 to 100:0) to yield 10 subfractions. Fr. 5-6 (MeOH–H2O 3:2, 350 mg) was further fractionated by CC on silica gel eluting with CHCl3–MeOH (from 100:1 to 10:1) to afford five subfrations, one of which (Fr. 5-6-4, 45 mg) was purified by CC on Sephadex LH-20 (MeOH), to yield compound 3 (8.5 mg). Fr. 6 (4.7 g) was further purified by reversed-phase CC over Lobar LiChroprep RP-18 with a MeOH–H2O gradient (from 10:90 to 100:0) to yield 10 subfractions. Fr. 6-4 (MeOH–H2O 2:3, 850 mg) was further fractionated by CC on silica gel eluting with CHCl3–MeOH (from 100:1 to 10:1) to afford four subfractions (from Fr. 6-4-1 to Fr. 6-4-4). Fr. 6-4-3 (140 mg) was purified by semi-preparative HPLC eluting with 45% aqueous MeOH to afford 2 (6.2 mg, tR = 25.155 min).
Peniciadametizine A (1): colorless amorphous powder; −25.0 (c 0.16, MeOH); UV (MeOH) λmax (log ε) 206 (4.14), 276 (3.14) nm; ECD (MeCN) λmax (Äε) 205 (11.42), 226sh (2.37), 237 (2.86), 269 (−5.31) nm; IR vmax 3334, 2918, 2849, 1679, 1602, 1502, 1462, 1365, 1286, 1091, 1053 cm−1; 1H and 13C NMR data, see Table 1; TOF-ESI-MS m/z 545 [M + K]+, 529 [M + Na]+; HR-ESI-MS m/z 529.1060 [M + Na]+ (calcd for C23H26N2O7S2Na, 529.1074).
Peniciadametizine B (2): colorless solid; +150.0 (c 0.06, MeOH); UV (MeOH) λmax (log ε) 202 (4.17), 256 (3.36), 322 (3.63) nm; ECD (MeCN) λmax (Äε) negative below 206 nm, 207 (1.25), 220 (1.98), 236 (2.23), 304 (7.74), 312 sh (7.51), 328sh (6.04) nm; IR vmax 3275, 2919, 2850, 1678, 1608, 1504, 1463, 1361, 1288, 1098, 1042 cm−1; 1H and 13C NMR data, see Table 1; TOF-ESI-MS m/z 989 [2M + H]+, 533 [M + K]+, 517 [M + Na]+, 495 [M + H]+; HR-ESI-MS m/z 495.1428 [M + H]+ (calcd for C22H27O9N2S+, 495.1432).
3.5. Computational Section
Mixed torsional/low mode conformational searches were carried out by means of the Macromodel 9.9.223 [16] software using Merck Molecular Force Field (MMFF) (Schrödinger, LLC, Portland, OR, USA) with implicit solvent model for chloroform applying a 21 kJ/mol energy window. Geometry reoptimizations of the resultant conformers (B3LYP/6-31G(d) level in vacuo and B97D/TZVP [17,18] level with PCM solvent model for MeCN) and TDDFT calculations were performed with Gaussian 09 [19] using various functionals (B3LYP, BH & HLYP, PBE0) and TZVP basis set. ECD spectra were generated as the sum of Gaussians [20] with 3600 and 4200 cm−1 half-height width (corresponding to ca. 28 and 33 nm at 280 nm), using dipole-velocity computed rotational strengths. Boltzmann distributions were estimated from the Zero Point Vibrational Energy ZPVE corrected B3LYP/6-31G(d) energies in the gas-phase calculations and B97D/TZVP energies in the solvent model calculations. The MOLEKEL [21] software package was used for visualization of the results.
3.6. Brine Shrimp Assay
Evaluations of the isolated compounds for brine shrimp (A. salina) lethality were determined as described previously [22]. Colchicine was used as a positive control.
3.7. Antimicrobial Assays
The antimicrobial activities against human- and aqua-pathogenic bacteria (A. hydrophilia, E. coli, E. tarda, S. aureus, V. alginolyticus, V. anguillarum, V. harveyi, and V. parahaemolyticus) as well as four plant pathogenic fungi (A. brassicae, C. gloeosprioides, F. graminearum, and G. graminis) of the new compounds 1 and 2 were evaluated using the methods described previously [22]. Chloramphenicol and amphotericin B were used as positive controls against bacteria and fungi, respectively.
3.8. Cytotoxicity Assays
The cytotoxic activities of the new compounds 1 and 2 against 9 tumor cell lines including human prostate carcinoma cell line (Du145), human cervixl carcinoma cell line (HeLa), human liver hepatocellular cells (HepG2), mouse Leydig tumor cell line (MA), human breast carcinoma cell line (MCF-7), human gastric carcinoma cell line (SGC-7901), human pancreatic cancer cell line (SW1990), human colon carcinoma cancer (SW480), and human glioma cells (U251) were determined according to previously reported methods [23,24].
4. Conclusions
In conclusion, two new diketopiperazines, peniciadametizines A and B (1 and 2), along with two known secondary metabolites, brasiliamide A (3) and viridicatumtoxin (4) were isolated and characterized from P. adametzioides AS-53, a fungal strain obtained from an unidentified marine sponge. To our knowledge, compound 1 possesses a spiro[furan-2,7'-pyrazino[1,2-b][1,2]oxazine] skeleton, which has not been described yet among natural products. Both compounds exhibited selective activity against the pathogenic fungus A. brassicae.
Acknowledgments
This work was financially supported by the Natural Science Foundation of China (31270403 and U1406402) and from the Ministry of Science and Technology of China (2013AA092901). T.K. thanks the Hungarian National Research Foundation (OTKA K105871) for financial support and the National Information Infrastructure Development Institute (NIIFI 10038) for CPU time.
Supplementary Files
Author Contributions
Yang Liu performed the experiments for the isolation, structure elucidation, and bioassay of the compounds, and prepared the manuscript; Xiaoming Li performed the 1D and 2D NMR measurements; Linghong Meng performed part of the isolation and structure elucidation of the compounds. Attila Mándi performed the computational work; Tibor Kurtán and Bingui Wang supervised the research work and revised the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Seephonkai P., Kongsaeree P., Prabpai S., Isaka M., Thebtaranonth Y. Transformation of an irregularly bridged epidithiodiketopiperazine to trichodermamide A. Org. Lett. 2006;8:3073–3075. doi: 10.1021/ol061046l. [DOI] [PubMed] [Google Scholar]
- 2.Magnus P., Mitchell I.S. Hemi-thioacetal pummerer reaction for the synthesis of gliovirin benzylic sulfide models. Tetrahedron Lett. 1998;39:9131–9134. doi: 10.1016/S0040-4039(98)02064-4. [DOI] [Google Scholar]
- 3.Klausmeyer P., McCloud T.G., Tucker K.D., Cardellina J.H., II, Shoemaker R.H. Aspirochlorine class compounds from Aspergillus flavus inhibit azole-resistant Candida albicans. J. Nat. Prod. 2005;68:1300–1302. doi: 10.1021/np050141k. [DOI] [PubMed] [Google Scholar]
- 4.Blunt J.W., Copp B.R., Keyzers R.A., Munro M.H.G., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2014;31:160–258. doi: 10.1039/c3np70117d. [DOI] [PubMed] [Google Scholar]
- 5.Meng L.H., Li X.M., Liu Y., Wang B.G. Penicibilaenes A and B, sesquiterpenes with a tricyclo[6.3.1.01,5]dodecane skeleton from the marine isolate of Penicillium bilaiae MA-267. Org. Lett. 2014;16:6052–6055. doi: 10.1021/ol503046u. [DOI] [PubMed] [Google Scholar]
- 6.Zhang P., Mándi A., Li X.M., Du F.Y., Wang J.N., Li X., Kurtán T., Wang B.G. Varioxepine A, a 3H-oxepine-containing alkaloid with a new oxacage from the marine algal-derived endophytic fungus Paecilomyces variotii. Org. Lett. 2014;16:4834–4837. doi: 10.1021/ol502329k. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Li X.M., Meng L.H., Wang B.G. N-Formyllapatin A, a new N-formylspiroquinazoline derivative from the marine-derived fungus Penicillium adametzioides AS-53. Phytochem. Lett. 2014;10:145–148. doi: 10.1016/j.phytol.2014.08.018. [DOI] [Google Scholar]
- 8.Zhang P., Meng L.H., Mándi A., Kurtán T., Li X.M., Liu Y., Li X., Li C.S., Wang B.G. Brocaeloids A-C, 4-oxoquinoline and indole alkaloids with C-2 reversed prenylation from the mangrove-derived endophytic fungus Penicillium brocae. Eur. J. Org. Chem. 2014;2014:4029–4036. doi: 10.1002/ejoc.201400067. [DOI] [Google Scholar]
- 9.Meng L.H., Li X.M., Lv C.T., Li C.S., Xu G.M., Huang C.G., Wang B.G. Sulfur-containing cytotoxic curvularin macrolides from Penicillium sumatrense MA-92, a fungus obtained from the rhizosphere of the mangrove Lumnitzera racemosa. J. Nat. Prod. 2013;76:2145–2149. doi: 10.1021/np400614f. [DOI] [PubMed] [Google Scholar]
- 10.Fujita T., Makishima D., Akiyama K., Hayashi H. New convulsive compounds, brasiliamides A and B, from Penicillium brasilianum Batista JV-379. Biosci. Biotechnol. Biochem. 2002;66:1697–1705. doi: 10.1271/bbb.66.1697. [DOI] [PubMed] [Google Scholar]
- 11.Silverton J.V., Kabuto C., Akiyama T. The structure and absolute configuration of viridicatumtoxin: 2'S-(2'á,7'â,11'aâ,12'â)-7',7'a,8',11',11'a,12'-hexahydro-5',6',7'a,10',11'a,12'-hexahydroxy-3'-methoxy-2,6,6-trimethyl-7',8'-dioxospiro[2-cyelohexene-1,2'(l'H)-cyelopenta[de]naphthaeene]-9'-carboxamide methanolate. Acta Cryst. 1982;38:3032–3037. [Google Scholar]
- 12.Guo C.J., Yeh H.H., Chiang Y.M., Sanchez J.F., Chang S.L., Bruno K.S., Wang C.C.C. Biosynthetic pathway for the epipolythiodioxopiperazine acetylaranotin in Aspergillus terreus revealed by genome-based deletion analysis. J. Am. Chem. Soc. 2013;135:7205–7213. doi: 10.1021/ja3123653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chankhamjon P., Boettger-Schmidt D., Scherlach K., Urbansky B., Lackner G., Kalb D., Dahse H.M., Hoffmeister D., Hertweck C. Biosynthesis of the halogenated mycotoxin sspirochlorine in koji mold involves a cryptic amino acid conversion. Angew. Chem. Int. Ed. 2014;53:13409–13413. doi: 10.1002/anie.201407624. [DOI] [PubMed] [Google Scholar]
- 14.Stipanovic R.D., Howell C.R. The structure of gliovirin, a new antibiotic from Gliocladium virens. J. Antibiotics. 1982;35:1326–1330. doi: 10.7164/antibiotics.35.1326. [DOI] [PubMed] [Google Scholar]
- 15.Wang S., Li X.M., Teuscher F., Li D.L., Diesel A., Ebel R., Proksch P., Wang B.G. Chaetopyranin, a benzaldehyde derivative, and other related metabolites from Chaetomium globosum, an endophytic fungus derived from the marine red alga Polysiphonia urceolata. J. Nat. Prod. 2006;69:1622–1625. doi: 10.1021/np060248n. [DOI] [PubMed] [Google Scholar]
- 16.Schrödinger LLC; [(accessed on 31 December 2013)]. MacroModel: Versatile, full-featured program for molecular modeling. Available online: http://www.schrodinger.com/productpage/14/11/ [Google Scholar]
- 17.Grimme S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006;27:1787–1799. doi: 10.1002/jcc.20495. [DOI] [PubMed] [Google Scholar]
- 18.Sun P., Xu D.X., Mándi A., Kurtán T., Li T.J., Schulz B., Zhang W. Structure, absolute configuration, and conformational study of 12-membered macrolides from the fungus Dendrodochium sp. associated with the sea cucumber Holothuria nobilis Selenka. J. Org. Chem. 2013;78:7030–7047. doi: 10.1021/jo400861j. [DOI] [PubMed] [Google Scholar]
- 19.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., et al. Gaussian 09, Revision B.01. Gaussian Inc.; Wallingford, CT, USA: 2010. [Google Scholar]
- 20.Stephens P.J., Harada N. ECD cotton effect approximated by the Gaussian curve and other methods. Chirality. 2010;22:229–233. doi: 10.1002/chir.20733. [DOI] [PubMed] [Google Scholar]
- 21.Varetto U. MOLEKEL 5.4. Swiss National Supercomputing Centre; Manno, Switzerland: 2009. [Google Scholar]
- 22.An C.Y., Li X.M., Luo H., Li C.S., Wang M.H., Xu G.M., Wang B.G. 4-Phenyl-3,4-dihydroquinolone derivatives from Aspergillus nidulans MA-143, an endophytic fungus Isolated from the mangrove plant Rhizophora stylosa. J. Nat. Prod. 2014;76:1896–1901. doi: 10.1021/np4004646. [DOI] [PubMed] [Google Scholar]
- 23.Bergeron R.I., Cavanaugh P.F., Jr., Kline S.J., Hughes R.G., Jr., Elliott G.T., Porter C.W. Antineoplastic and antiherpetic activity of spermidine catecholamide iron chelators. Biochem. Biophys. Res. Commun. 1984;121:848–854. doi: 10.1016/0006-291X(84)90755-1. [DOI] [PubMed] [Google Scholar]
- 24.Mosmann T.J. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.