Abstract
The transcription factor brachyury (T, BRA) is one of the first markers of gastrulation and lineage specification in vertebrates. Despite its wide use and importance in stem cell and developmental biology, its functional genomic targets in human cells are largely unknown. Here, we use differentiating human embryonic stem cells to study the role of BRA in activin A-induced endoderm and BMP4-induced mesoderm progenitors. We show that BRA has distinct genome-wide binding landscapes in these two cell populations, and that BRA interacts and collaborates with SMAD1 or SMAD2/3 signalling to regulate the expression of its target genes in a cell-specific manner. Importantly, by manipulating the levels of BRA in cells exposed to different signalling environments, we demonstrate that BRA is essential for mesoderm but not for endoderm formation. Together, our data illuminate the function of BRA in the context of human embryonic development and show that the regulatory role of BRA is context dependent. Our study reinforces the importance of analysing the functions of a transcription factor in different cellular and signalling environments.
KEY WORDS: SMAD, T-BOX, Embryonic stem cells, Gastrulation, Gene regulatory networks, Human
Summary: In differentiating hESCs, BRACHYURY has cell type-specific targets and functions: it acts with SMAD1 to promote mesoderm specification, while in the endoderm it interacts with SMAD2/3.
INTRODUCTION
The three primary germ layers (ectoderm, mesoderm and endoderm) arise from the pluripotent epiblast during gastrulation in the amniote embryo (Arnold and Robertson, 2009; Tam and Loebel, 2007); this can be modelled in vitro using pluripotent stem cells (Murry and Keller, 2008). The brachyury gene (T, BRA) encodes a T-box transcription factor that plays an essential role in mesoderm formation (Papaioannou, 2014; Showell et al., 2004). Brachyury is expressed first in the primitive streak during gastrulation, and later in the notochord and tailbud (Herrmann et al., 1990; Wilkinson et al., 1990). Mice lacking one copy of the gene have a short tail, while homozygous embryos die around embryonic day (E) 9-10 (Chesley, 1935). The latter develop only the first seven somites and lack a proper notochord; in addition, they display defects in left-right asymmetry and in cell migration (Showell et al., 2004). Importantly, the mutant phenotype also includes severe defects in cardiovascular and placental development (David et al., 2011; Inman and Downs, 2006; King et al., 1998). Brachyury is widely used as the earliest marker of mesodermal and also endodermal differentiation in embryonic stem cell (ESC) studies and during gastrulation, because both these cell lineages derive from the primitive streak (Murry and Keller, 2008). Indeed, definitive endoderm progenitors co-express Bra, Sox17, Foxa2, Gsc and other endoderm markers (D'Amour et al., 2005; Kubo et al., 2004; Tada et al., 2005). Based on morphological analyses, mouse Bra homozygotes seem to form a normal foregut but an abnormal hindgut (Chesley, 1935). However, on a molecular level, the role of BRA in endoderm formation remains poorly understood.
Genomic targets of BRA orthologues have previously been identified in zebrafish (Morley et al., 2009) and Xenopus embryos (Gentsch et al., 2013), and in mouse embryoid bodies (Evans et al., 2012; Lolas et al., 2014) using ChIP-chip or ChIP-seq approaches. Recently, BRA target genes were identified in differentiating hESCs (Tsankov et al., 2015), but the regulatory impact of BRA was not functionally characterized in these experiments. Thus, despite its important role in vertebrate development and its widespread use in stem cell biology (Murry and Keller, 2008; Papaioannou, 2014), the precise regulatory role of BRA and its genome-wide functional role are poorly understood in humans.
Human embryonic stem cells (hESCs) represent the best available system in which to study the molecular mechanisms underpinning human embryonic development (Murry and Keller, 2008). We have developed two in vitro protocols (activin or BMP4 based) (Bernardo et al., 2011) that can induce hESCs to differentiate into distinct cell populations, both expressing BRA. Strikingly, these populations have the characteristics of the anterior and posterior regions of the early primitive streak (Alev et al., 2010) from which, respectively, endoderm or mesoderm cells arise in vivo (Lawson et al., 1991; Parameswaran and Tam, 1995).
Here, we perform chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) to identify genomic targets of BRA in these two cell populations. Interestingly, these genome-wide binding events differ in activin-treated or BMP4-treated cells, suggesting that BRA interacts with the genome depending on the signalling environment and cell identity. We provide functional validation of these genomic binding events by modulating levels of BRA within different signalling environments in differentiating hESCs and by analysing the expression pattern of BRA target genes in mouse embryos that lack brachyury.
Importantly, our experiments also indicate that BRA physically interacts with downstream effectors of activin or BMP4 signalling: SMAD2/3 in endoderm progenitors and SMAD1 in mesoderm progenitors. We conclude that BMP4-SMAD1 signalling and BRA action are essential for proper mesodermal differentiation, while simultaneously repressing endodermal fates. By contrast, in partnership with eomesodermine (EOMES) and activin-SMAD2/3 signalling, BRA proves to be sufficient, but not necessary, to activate endodermal gene expression.
RESULTS
An in vitro differentiation system to study the role of BRA in human gastrulation
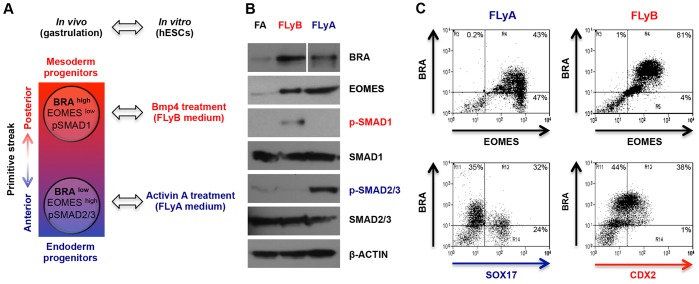
We have previously optimised chemically defined conditions that cause hESCs to differentiate as progenitors of endoderm or of mesoderm (Bernardo et al., 2011). When hESCs are induced to differentiate (36 h of treatment with Fgf2 and the PI3 kinase inhibitor Ly294002) in an activin A-based medium (called FLyA), they upregulate endoderm markers such as CER1, FOXA2, GSC and SOX17. By contrast, hESCs similarly induced to differentiate in a BMP4-based medium (called FLyB) express mesodermal genes such as CDX2, TBX6, FOXF1 and BMP4 (Bernardo et al., 2011). Thus, FLyA-treated hESCs resemble the anterior region of the early primitive streak, whereas FLyB-treated hESCs resemble the posterior region (Fig. 1A) (Alev et al., 2010; Arnold and Robertson, 2009).
Fig. 1.
An in vitro differentiation system to study the role of BRA in human gastrulation. (A) hESCs differentiated in FLyA (blue) or FLyB (red) media for 36 h resemble the anterior (endoderm progenitors) or posterior (mesoderm progenitors) regions of the early primitive streak. (B) Western blots showing the expression of BRA, EOMES, phospho-SMAD1, total SMAD1, phospho-SMAD2/3, total SMAD2/3 and β-actin during pluripotency (FA), and in FLyA and FLyB conditions. (C) Flow cytometry analysis of hESCs differentiated in FLyA or FLyB media for 36 h. FLyA-treated cells were co-immunostained for BRA and EOMES (upper left), or for BRA and SOX17 (lower left). FLyB-treated cells were co-immunostained for BRA and EOMES (upper right), or for BRA and CDX2 (lower right).
Significantly, these two differentiated populations express different levels of BRA and EOMES, two T-BOX transcription factors that are key regulators of gastrulation (Herrmann et al., 1990; Russ et al., 2000; Arnold et al., 2008). FLyA-treated cells are predominantly BRAlow/EOMEShigh and upregulate SOX17 (Fig. 1B,C; supplementary material Fig. S1A,B), whereas FLyB-treated cells are mainly BRAhigh/EOMESlow and upregulate CDX2 (Fig. 1B,C; supplementary material Fig. S1A,B) (Bernardo et al., 2011; Mendjan et al., 2014). As expected, activin A or BMP4 treatment triggers the phosphorylation of their downstream targets, SMAD2/3 or SMAD1, respectively (Fig. 1B). This in vitro differentiation system allows us to capture and analyse the transient progenitor populations that give rise to two primary embryonic tissue lineages: endoderm and mesoderm.
Distinct genome-wide BRA-binding landscapes
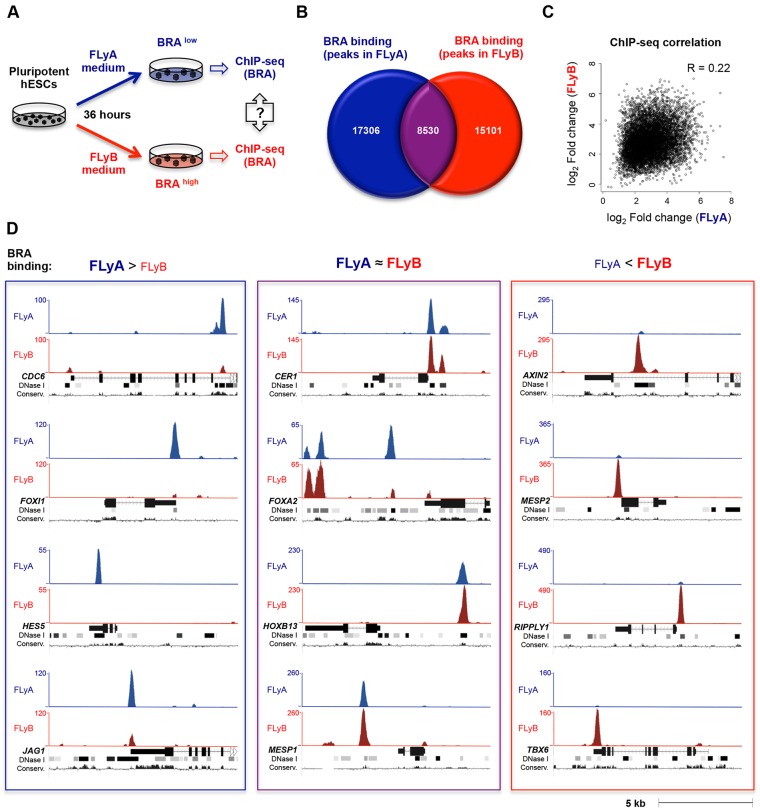
FLyA- and FLyB-treated hESCs express, respectively, genes that are characteristic of anterior and posterior regions of the primitive streak. They also express different levels of BRA, which reaches its peak at around 36 h of differentiation (Bernardo et al., 2011). We used this differentiation system to identify genomic targets of BRA by ChIP-seq and to ask whether these differ between endoderm and mesoderm progenitors (Fig. 2A).
Fig. 2.
BRA exhibits distinct genomic binding profiles in FLyA- and in FLyB-differentiated hESCs. (A) hESCs treated with FLyA (BRAlow) or FLyB (BRAhigh) media for 36 h were used to analyse and compare the genome-wide binding of BRA (ChIP-seq). (B) Venn diagram showing the detectable overlap between BRA binding (ChIP-seq peaks) in FLyA- and FLyB-treated hESCs. (C) Dot plot of ChIP-seq fold enrichment values (normalised to Input samples) of common BRA peaks in FLyA- and FLyB-treated hESCs. R, correlation coefficient. (D) Examples of ChIP-seq peaks depicting BRA-binding profiles in hESCs treated with FLyA (blue track) or FLyB (red track) media: stronger peaks in FLyA (left); peaks detected in both FLyA and FLyB (centre); stronger peaks in FLyB (right). Tracks under ChIP-seq peaks: gene locus (exons depicted as full rectangles, introns depicted as lines with chevrons), DNase I-hypersensitive clusters (ENCODE project) and mammalian conservation profiles (UCSC genome browser). The y axis shows the number of normalised unique reads.
Two replicate ChIP-seq experiments, both with FLyA- and FLyB-treated cells, were carried out using two different anti-BRA antibodies, one from R&D Systems and the other from Santa Cruz Biotechnology. These replicates showed high correlation coefficients for BRA binding in both datasets (R=0.72 for FLyA conditions and R=0.73 for FLyB conditions) (supplementary material Fig. S2A). Overlapping ChIP-seq peaks (present using either antibody) are shown in supplementary material Table S1. However, the ChIP-seq experiments using the Santa Cruz antibody had lower signal-to-noise ratio and yielded lower ‘unique read’ counts. We therefore used the datasets generated with the R&D Systems antibody to perform all subsequent analyses.
ChIP-seq analysis indicated that there are 25,836 BRA-binding events in FLyA-treated hESCs and 23,631 in FLyB-treated hESCs (Fig. 2B). Although there is considerable overlap between these two datasets (Fig. 2B; supplementary material Fig. S2B), a large proportion of peaks are unique to each cell population: 68% in the FLyA condition and 64% in the FLyB condition (Fig. 2B). For example, BRA binds in the vicinity of genes such as: (1) CDC6, FOXI1, HES5 and JAG1 when cultured in FLyA but not so strongly in FLyB (Fig. 2D; left blue rectangle, FLyA>FLyB); (2) AXIN2, MESP2, RIPPLY1 and TBX6 when cultured in FLyB but not so strongly in FLyA (Fig. 2D; right red rectangle, FLyA<FLyB); and (3) CER1, FOXA2, HOXB13 and MESP1 when hESCs are cultured in FLyA or FLyB (Fig. 2D; central purple rectangle, FLyA≈FLyB). Moreover, the ‘fold enrichment’ correlation coefficient for peaks detected in both FLyA and FLyB conditions was low (R=0.22) (Fig. 2C), further underscoring the differences in BRA binding between FLyA- and FLyB-treated hESCs. Importantly, these differences in BRA binding in FLyA or FLyB conditions were confirmed by ChIP-qPCR on a set of selected targets (supplementary material Fig. S2C,D). These observations reveal that BRA has distinct genome-wide binding landscapes in hESC-derived endoderm and mesoderm progenitors.
Developmental significance of cell type-specific BRA binding: different target genes in endoderm and mesoderm progenitors
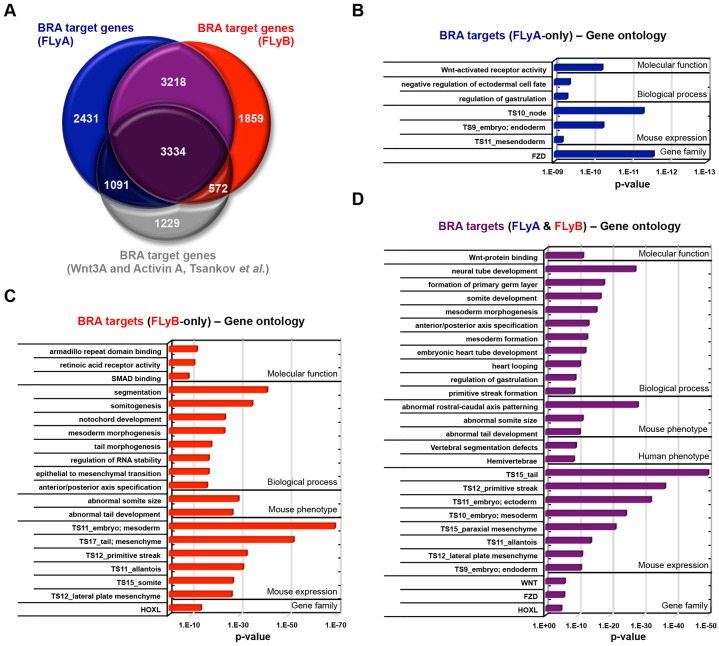
Using a ‘nearby gene’ peak annotation approach (up to 50 kb on either side of a gene), BRA binding was detected in the vicinity of 10,074 genes in FLyA-treated hESCs and 8983 genes in FLyB-treated hESCs (Fig. 3A; supplementary material Table S1). As suggested by the overlap analysis (Fig. 2A), BRA binds to many of its target genes in a cell type-specific manner. Thus, in the FLyA dataset, 35% of targets were unique to this condition and 27% were unique to the FLyB dataset (Fig. 3A). These percentages are lower than those concerning the binding overlap (Fig. 2B) because several peaks are often located around a single locus (in both promoter and enhancer regions) and because we excluded the most distal intergenic peaks, the assignment of which to the nearest gene can lead to a higher false discovery rate (data not shown).
Fig. 3.
BRA has different sets of target genes in FLyA- and FLyB-treated hESCs. (A) Venn diagram showing the overlap of BRA putative target genes between FLyA-treated hESCs (blue), FLyB-treated hESCs (red) and WNT3A/activin- treated hESCs (grey; Tsankov et al., 2015). (B-D) Gene ontology analyses (GREAT algorithm; McLean et al., 2010) of BRA-binding regions detected only in FLyA (B), only in FLyB (C) and in both FLyA and FLyB (D). Ontology terms are ranked according to their enrichment P-values: ‘Gene family’ terms (P-value <1×10−5), all other terms (P-value <1×10−9). TS, Theiler stage of mouse development.
Recently, BRA genomic binding was investigated by ChIP-seq in hESC-derived ‘mesendoderm’ progenitors (12 h treatment with WNT3A and activin A) (Tsankov et al., 2015). This dataset shows a substantial overlap with ours both in terms of binding regions (supplementary material Fig. S3A-C) and target genes (Fig. 3A). However, many unique binding events are detected in each dataset, further suggesting that the regulatory role of BRA is context dependent.
To investigate the developmental significance of our findings, we performed gene ontology analyses using GREAT (McLean et al., 2010) (supplementary material Table S2), distinguishing between FLyA-only putative targets (Fig. 3B), FLyB-only putative targets (Fig. 3C), and targets common to both FLyA and FLyB (Fig. 3D). In all subsets, there was clear enrichment for genes that are co-expressed with Bra during mouse embryogenesis and involved in all aspects of its complex mutant phenotype (Fig. 3, Table 1; supplementary material Table S2).
Table 1.
Examples of BRA target genes in differentiating hESCs*

The gene ontology analysis of the recent human dataset (Tsankov et al., 2015) included few highly significant terms (P-value <1×10−9) (supplementary material Table S2). However, the top term in the ‘mouse expression’ category was ‘TS9_primitive streak’ (P=4.92×10−9) and we note that several BRA targets that are common to the Tsankov et al. dataset and our dataset make biological sense (Table 1).
Strikingly, whereas FLyA-only BRA targets were enriched for genes expressed in anterior primitive streak derivatives, such as the node and endoderm (Fig. 3B), FLyB-only BRA targets were enriched for genes expressed in mid/posterior primitive streak derivatives, such as tail mesoderm, lateral plate mesoderm, extraembryonic mesoderm, allantois and somitic/paraxial mesoderm (Fig. 3C). BRA targets that were common to both FLyA and FLyB conditions are expressed in all germ layers (Fig. 3D).
Overall, our results show that in FLyA- and FLyB-treated cells, BRA binds to the genome in a manner that is correlated with the transcriptional and developmental identity of each cell population, endoderm and mesoderm, respectively.
BRA genomic binding overlaps with EOMES and SMAD2/3 in endoderm progenitors
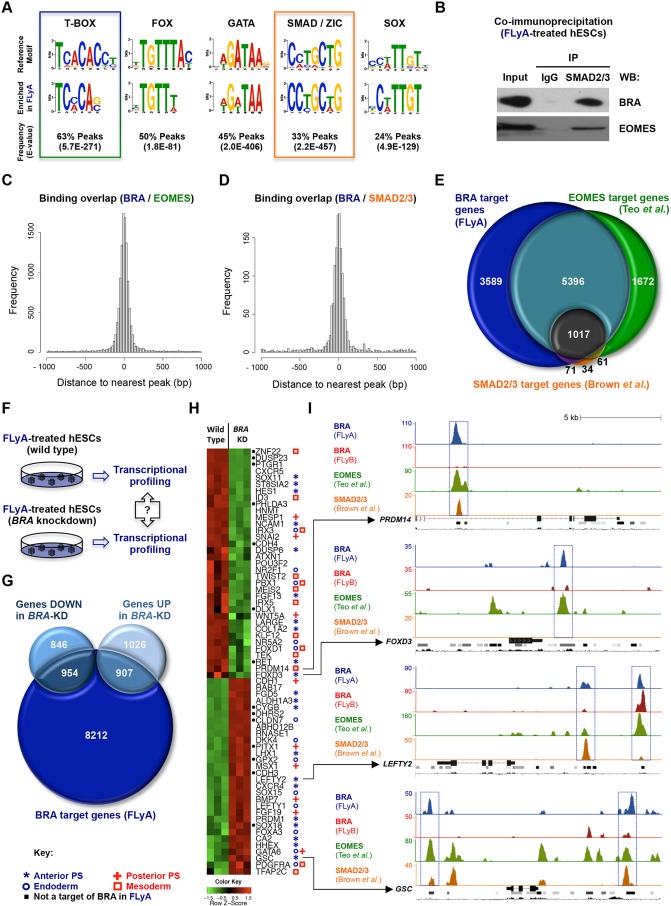
Having identified genomic targets of BRA in differentiating hESCs, we asked whether BRA-binding regions (200 bp sequences centred on ChIP-seq peaks) were enriched for specific DNA sequences. To this end, we performed de novo DNA motif analyses using the MEME suite (Bailey et al., 2009). For FLyA-treated cells, these revealed enrichment of a motif resembling the T-BOX consensus-binding sequence (Kispert and Herrmann, 1993) in 63% of peaks (Fig. 4A). Motifs for other protein families were also enriched in BRA FLyA peaks (Fig. 4A; supplementary material Fig. S4A), including FOX, GATA/GSC, SMAD/ZIC, SOX, POU and STAT, suggesting that BRA might interact with members of these families. Likely candidates, based on their expression patterns in vivo and in FLyA-treated cells (Alev et al., 2010; Pfister et al., 2007; Bernardo et al., 2011), include FOXA2, GATA4/6, GSC, SMAD2/3, SOX17 and POU5F1/OCT4 (supplementary material Fig. S4C,D) (Mullen et al., 2011). Significantly, we were able to show by co-immunoprecipitation that both BRA and EOMES interact with SMAD2/3 (Fig. 4B), suggesting cooperation with activin signalling. BRA-binding peaks show a remarkable overlap with those of EOMES (Teo et al., 2011) and of SMAD2/3 (Brown et al., 2011) in hESC-derived mesendodermal progenitors (Fig. 4C,D), and share with BRA many putative common target genes (Fig. 4E). Importantly, the genomic binding of EOMES and SMAD2 in FLyA-treated cells was confirmed by ChIP-qPCR on a set of selected BRA target regions (supplementary material Fig. S4B).
Fig. 4.
BRA in the context of activin A signalling. (A) Comparison of DNA recognition sites of five protein families (row above) and DNA motifs enriched at BRA FLyA ChIP-seq peaks (row below). (B) Co-immunoprecipitation of SMAD2/3 (pulldown) with BRA and EOMES (WB, western blot) in FLyA-treated hESCs; IgG (negative control immunoglobulin). (C,D) Histograms showing the distance between BRA-binding peaks in FLyA-treated hESCs and EOMES binding (Teo et al., 2011) or SMAD2/3 binding (Brown et al., 2011) in FLyAB-treated hESCs. (E) Venn diagram showing the overlap of putative target genes between BRA in FLyA-treated hESCs (FLyA, blue), EOMES (green) and SMAD2/3 (orange). (F) Wild-type (control) and BRA knockdown hESCs were differentiated for 36 h in FLyA and profiled for transcriptome-wide (microarray) differential expression analysis. (G) Venn diagram showing the overlap between BRA putative target genes (FLyA, dark blue) and genes that were either up- or downregulated (FDR <0.05) in BRA knockdown hESCs when compared with wild-type hESCs. (H) Microarray gene expression heat-map of wild-type versus BRA knockdown (KD) hESCs grown in FLyA for 36 h. Green indicates downregulation and red indicates upregulation. Symbols after gene names indicate expression pattern in vivo (Mouse Genome Informatics; Alev et al., 2010). (I) ChIP-seq peaks depicting BRA binding in hESCs treated with FLyA (blue) or FLyB (red), and EOMES binding (green) or SMAD2/3 binding (orange). Tracks under ChIP-seq peaks: gene locus (exons depicted as full rectangles, introns depicted as lines with chevrons), DNase I-hypersensitive clusters (ENCODE project) and mammalian conservation profiles (UCSC genome browser). The y axis shows the number of normalised unique reads. Blue boxes highlight FLyA-specific BRA binding peaks.
BRA is largely dispensable for the expression of key endoderm markers
Both EOMES and SMAD2/3 are essential for proper expression of endoderm markers in differentiating hESCs (Brown et al., 2011; Teo et al., 2011), but little is known about the role of BRA in this context. We therefore sought to discover which putative BRA targets require normal BRA levels for their correct expression in FLyA-treated hESCs. BRA knockdown (shRNA KD) hESCs were compared with their wild-type counterparts by transcriptional profiling at 36 (Fig. 4F; supplementary material Table S3) and 72 h of differentiation, in more mature endoderm cells (supplementary material Fig. S4F and Table S3). At the protein level, BRA was virtually absent in BRA knockdown cells (supplementary material Fig. S4E). Misregulated transcripts in BRA knockdown cells were compared with BRA-bound genes in the FLyA condition (Fig. 4G; supplementary material Fig. S4G). Up- and downregulated genes, both at 36 and at 72 h, showed enrichment for BRA targets (P<0.01, Pearson's Chi-squared).
We proceeded to perform gene ontology analysis of misregulated BRA targets using GREAT (McLean et al., 2010) (supplementary material Table S4). Although BRA was necessary for the upregulation of many target genes that are developmentally important and expressed under FLyA conditions (Fig. 4H; supplementary material Fig. S4H), this subset appears to be enriched for genes expressed in neurectoderm derivatives post-gastrulation (supplementary material Table S4), which are not gene ontology categories that are developmentally relevant to FLyA-treated cells. Interestingly, however, key genes involved in epithelial-to-mesenchymal transition (EMT) were downregulated at 36 h (SNAI2, TWIST2 and FOXD3), together with the upregulation of CDH1/E-cadherin, a classical hallmark of impaired EMT (Lamouille et al., 2014).
Strikingly, the expression of many genes expressed in the anterior primitive streak and involved in endoderm formation was either unaltered or even upregulated in BRA knockdown cells (P=3.49×10−11 at 36 h, P=1.48×10−11 at 72 h; supplementary material Table S4). The latter included genes such as GSC, GATA6, HHEX, LEFTY1/2, CXCR4, OTX2 and LHX1 (Fig. 4H; supplementary material Fig. S4H). These observations were confirmed by qRT-PCR in another BRA knockdown clone (81% BRA knockdown efficiency in FLyA treatment at 36 h) when compared with a scrambled (mock shRNA) control line (supplementary material Fig. S4I). Under FLyA conditions, many of these genes were not only bound by BRA but also by EOMES and SMAD2/3 (Fig. 4I).
Together, these data show that the genome-wide binding of BRA in endoderm progenitors broadly overlaps with that of EOMES and SMAD2/3, both of which are essential regulators of endoderm formation. However, BRA is not necessary for the expression of most key endoderm markers.
BRA genomic binding overlaps with EOMES and SMAD1 in mesoderm progenitors
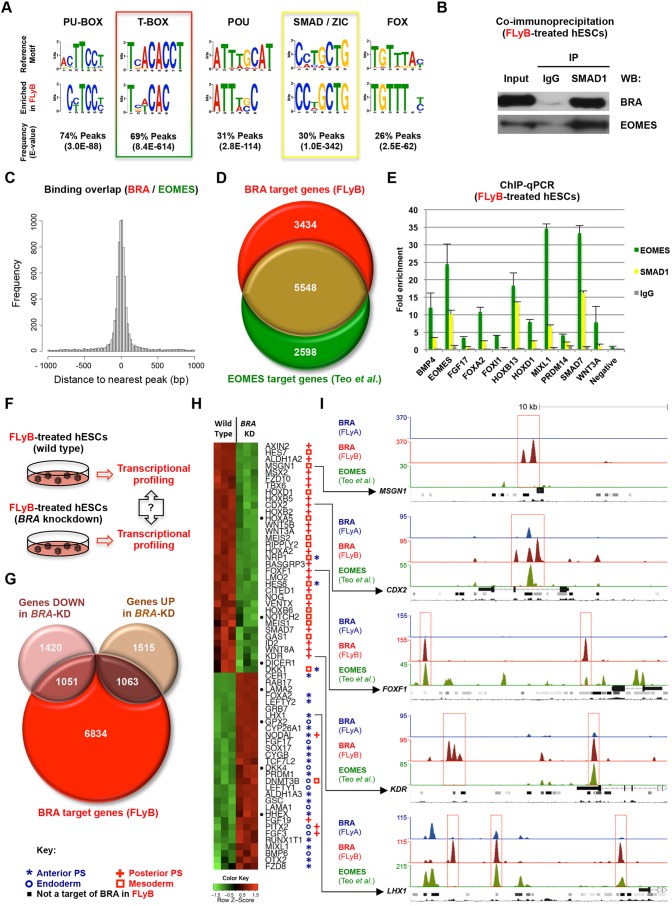
We also performed de novo DNA motif analysis (Bailey et al., 2009) using the FLyB BRA peak dataset. As observed with the FLyA dataset, this analysis revealed enrichment (69% of peaks) of the T-BOX consensus binding sequence (Kispert and Herrmann, 1993) (Fig. 5A). Motifs characteristic of other protein families were also found in BRA FLyB peaks (Fig. 5A; supplementary material Fig. S5A), including PU-BOX, POU, SMAD/ZIC, FOX, KLF and GATA, again suggesting that BRA might interact with members of these families. Likely candidates, based on their expression patterns in vivo and in FLyB-treated cells (Alev et al., 2010; Pfister et al., 2007; Bernardo et al., 2011), include SMAD1, FOXF1, GATA2/3 and POU5F1/OCT4 (supplementary material Fig. S5B,C) (Mullen et al., 2011). Previous work has shown that a BRA orthologue, Xbra, directly interacts with Smad1 in Xenopus embryos (Messenger et al., 2005). Significantly, we were able to show by co-immunoprecipitation that both BRA and EOMES interact with SMAD1 (Fig. 5B), suggesting cooperation with BMP4 signalling. BRA binding peaks also showed close proximity to those of EOMES in hESC-derived mesendodermal progenitors (Teo et al., 2011) (Fig. 5C), with a substantial number of common putative target genes (Fig. 5D). Importantly, the genomic binding of EOMES and SMAD1 in FLyB-treated cells was confirmed by ChIP-qPCR on a set of selected BRA target regions (Fig. 5E).
Fig. 5.
BRA in the context of BMP4 signalling. (A) Comparison of DNA recognition sites of five protein families (row above) and DNA motifs enriched in BRA FLyB ChIP-seq peaks (below). (B) Co-immunoprecipitation of SMAD1 (pulldown) with BRA and EOMES (western blot) in FLyB-treated hESCs; IgG (negative control immunoglobulin). (C) Histogram showing the distance between BRA-binding peaks in FLyB-treated hESCs and EOMES binding (Teo et al., 2011) in FLyAB-treated hESCs. (D) Venn diagram showing the overlap of putative target genes between BRA in FLyB-treated hESCs (FLyB, red) and EOMES (green). (E) Graph with fold enrichment values (ChIP over input) for EOMES binding (green), SMAD1 binding (yellow) and control IgG binding (grey) to BRA target regions in FLyB-treated hESCs (36 h). Error bars correspond to s.d. (n=3). ChIP-qPCR values were normalised to the highest control IgG value (PRDM14). (F) Wild-type (control) and BRA knockdown hESCs were differentiated for 36 h in FLyB and profiled for transcriptome-wide (microarray) differential expression analysis. (G) Venn diagram showing the overlap between BRA putative target genes (FLyB, red) and genes that were either up- or downregulated (FDR <0.05) in BRA knockdown hESCs when compared with wild-type hESCs. (H) Microarray gene expression heat-map of wild-type versus BRA knockdown (KD) hESCs grown in FLyB for 36 h. Green indicates downregulation and red indicates upregulation. Symbols after gene names indicate expression pattern in vivo (Mouse Genome Informatics; Alev et al., 2010). (I) ChIP-seq peaks depicting BRA binding in hESCs treated with FLyA (blue) or FLyB (red), and EOMES binding (green). Tracks under ChIP-seq peaks: gene locus (exons depicted as full rectangles, introns depicted as lines with chevrons), DNase I-hypersensitive clusters (ENCODE project), mammalian conservation profiles (UCSC genome browser). The y axis shows the number of normalised unique reads. Red boxes highlight FLyB-specific BRA binding peaks.
BRA is necessary for mesoderm gene expression
We next asked which putative BRA targets require normal BRA levels for their correct expression in BMP4-treated cells by comparing the transcriptomes of BRA knockdown (shRNA KD) hESCs with their wild-type counterparts at 36 h of FLyB differentiation (Fig. 5F; supplementary material Table S3) and at 72 h of FLyB/FB differentiation (supplementary material Fig. S5D and Table S3), when cells resemble extraembryonic and lateral plate mesoderm (Bernardo et al., 2011; Cheung et al., 2012; Mendjan et al., 2014). Misregulated transcripts in BRA knockdown cells were compared with BRA-bound genes in the FLyB condition (Fig. 5G; supplementary material Fig. S5E). Up- and downregulated genes, both at 36 and 72 h, showed enrichment for BRA targets (P<0.01, Pearson's Chi-squared).
We then proceeded to perform gene ontology analysis of misregulated BRA targets using GREAT (McLean et al., 2010) (supplementary material Table S4). Significantly, BRA was necessary for the normal upregulation of many genes expressed in BMP4-treated cells (at 36 and 72 h) that are essential for mesoderm development. These include ALDH1A2 (RALDH2), AXIN2, CDX2, FOXF1, KDR, LMO2, MSGN1, MEIS1, TBX6 and WNT3A (Fig. 5H; supplementary material Fig. S5F). Indeed, gene ontology analysis shows enrichment for phenotypes that are reminiscent of BRA mutant embryos, such as ‘abnormal somite size’ (P=9.26×10−24), ‘abnormal tail development’ (P=1.90×10−22), ‘abnormal gastrulation’ (P=1.56×10−17) and ‘abnormal vascular development’ (P=3.29×10−16) (supplementary material Table S4). Interestingly, several HOX genes were also downregulated upon BRA knockdown at 36 h (Fig. 5H) (P=1.30×10−12; supplementary material Table S4), an effect that was even more marked at 72 h of differentiation (supplementary material Fig. S5F) (P=3.26×10−6; supplementary material Table S4).
Noticeably, the expression of many endoderm regulators was upregulated in BRA knockdown cells grown in FLyB conditions (P=3.87×10−19 at 36 h, P=8.36×10−16 at 72 h; supplementary material Table S4). These include CER1, CYP26A1, EOMES, FOXA2, GSC, GATA6, HHEX, LEFTY1/2, LHX1, MIXL1, OTX2 and SOX17 (Fig. 5H; supplementary material Fig. S5F). These observations were confirmed by qRT-PCR in another BRA knockdown clone (89% BRA knockdown efficiency in FLyB treatment at 36 h) when compared with a scrambled (mock shRNA) control line (supplementary material Fig. S5G). Strikingly, BRA was bound in the vicinity of some of these genes in regions only detected in the FLyB condition (e.g. LHX1, Fig. 5I).
Together, these data show that the genome-wide binding of BRA in mesoderm progenitors overlaps with that of EOMES. However, unlike EOMES (Teo et al., 2011), BRA was necessary for the expression of many genes involved in mesoderm formation, while simultaneously repressing the expression of endoderm markers.
BRA cooperates with activin or BMP4 signalling to upregulate endoderm or mesoderm markers
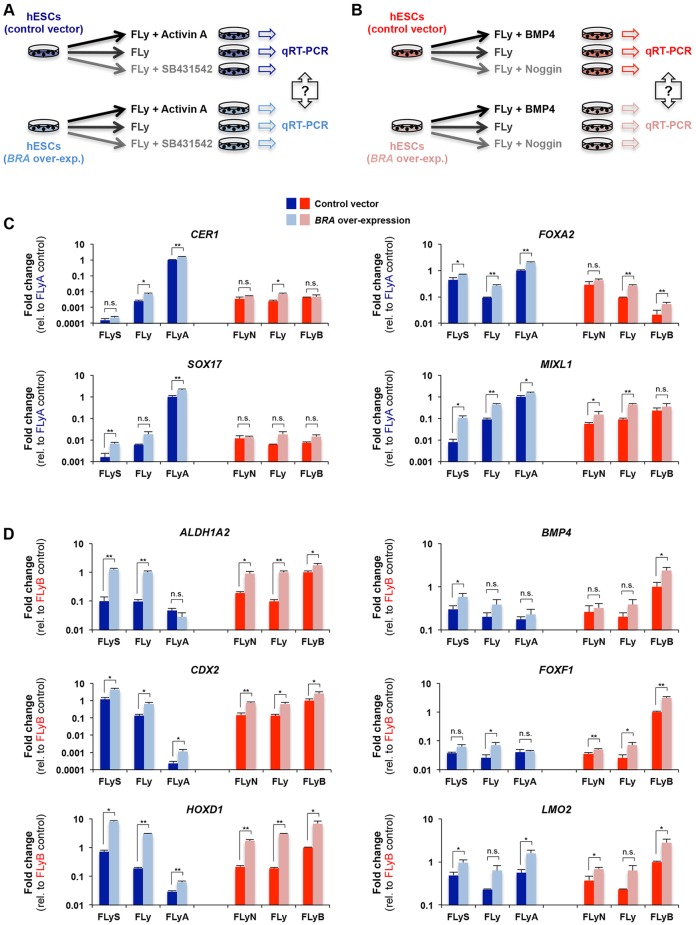
Having established that BRA is required for the expression of many mesodermal but not endodermal genes, we asked whether BRA overexpression (BRA OE) (supplementary material Fig. S6A,B) in hESCs was sufficient to up- or downregulate its genomic targets. Bearing in mind that BRA has different targets in activin- or BMP4-treated cells, we analysed the phenotype of control and BRA OE hESC subclones in different signalling environments. All cells were grown in the presence of Fgf2 and Ly294002 as a basal differentiation medium (FLy) with the addition of activin (FLyA), BMP4 (FLyB), SB431542 (FLyS) to block activin-SMAD2/3 signalling or noggin (NOG) (FLyN) to block BMP4-SMAD1 signalling (Fig. 6A,B).
Fig. 6.
BRA depends on the signalling environment to regulate key developmental genes. (A,B) hESCs transfected with either an empty/control vector or a BRA over-expression vector were differentiated for 36 h, as indicated and samples were collected to perform comparative gene expression analysis by qRT- PCR. (C,D) qRT-PCR of BRA target genes expressed in endoderm/anterior primitive streak (blue) or in mesoderm/posterior primitive streak (red), respectively. hESCs transfected with either an empty vector (dark colours) or a BRA overexpression vector (light colours) were differentiated as indicated in A,B. For each group of germ layer markers, gene expression is presented as fold change over the wild type reference sample (control vector): FLyA for endoderm genes (C) and FLyB for mesoderm genes (D), as indicated on the y axis. Bars indicate s.d. (n=3) (Student's two-tailed t-test: *P<0.05; **P<0.01; n.s., not significant). F, FGF2; Ly, Ly294002; B, BMP4; A, activin A; N, noggin (N); S, SB431542.
Overexpression of BRA caused the upregulation of both endodermal (CER1, FOXA2, SOX17, MIXL1) and mesodermal (ALDH1A2, BMP4, CDX2, FOXF1, HOXD1, LMO2) target genes. Strikingly, however, strong upregulation of genes characteristic of endoderm/anterior primitive streak required activin signalling (Fig. 6C). Genes characteristic of mesoderm/posterior primitive streak either required or benefitted from BMP4 signalling and, interestingly, in some cases, activin antagonism (Fig. 6D). These experiments reveal that BRA expression alone is not sufficient to fully upregulate some of its target genes to wild-type levels and that it requires appropriate signalling cues to activate the expression of its genomic targets.
BRA target genes in the mouse
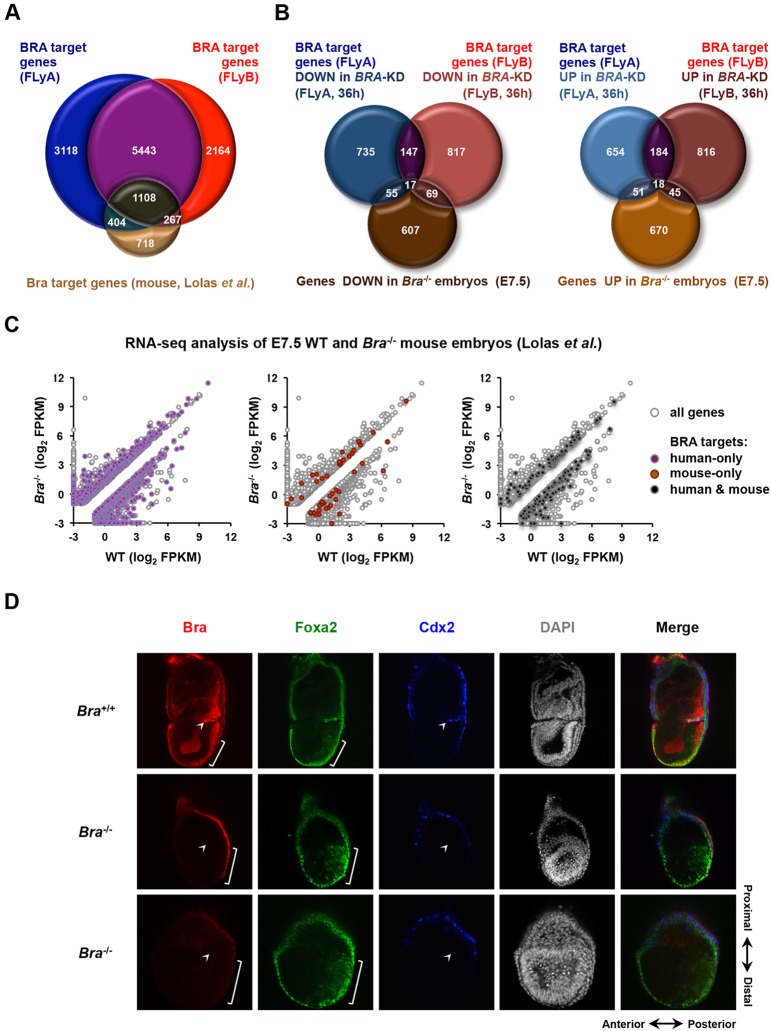
BRA putative target genes have been identified by ChIP-seq in activin-treated mouse embryoid bodies (Lolas et al., 2014). Our analysis reveals a considerable overlap between our datasets and that of Lolas et al., as 71% of the mouse targets are included in the human datasets (Fig. 7A). Indeed, many of these conserved targets make biological sense (Table 1). Importantly, our datasets also reveal the identity of BRA targets not identified in mouse (Table 1), including ALDH1A2, HES7, KDR, LMO2 and MEOX1.
Fig. 7.
BRA target gene expression in mouse embryos. (A) Venn diagram showing the overlap between BRA putative target genes identified in FLyA-treated hESCs (blue), FLyB-treated hESCs (red) and activin-treated mouse embryoid bodies (light brown, Lolas et al., 2014). (B) Venn diagrams showing the overlap between BRA putative target genes that are misregulated in FLyA- or FLyB-treated BRA knockdown hESCs at 36 h and genes misregulated in E7.5 Bra−/− mouse embryos (Lolas et al., 2014). Left diagram, downregulated genes; right diagram, upregulated genes. (C) RNA-seq analysis of E7.5 wild-type and Bra−/− mouse embryos (Lolas et al., 2014); coloured dots indicate BRA target genes identified only in human (FLyA and FLyB datasets, purple), only in mouse (Lolas et al., brown) or in both human and mouse (black). Scale represents log2 FPKM (fragments per kilobase of exon per million fragments mapped). Only differentially expressed (fold change >2) genes are shown. (D) Confocal microscopy analysis (middle embryo stack) of mouse gastrulae (E7.0) immunostained for Bra (red), Foxa2 (green) and Cdx2 (blue). Nuclei were stained with DAPI. Upper row shows a wild-type embryo (Bra+/+). Bottom rows show Bra mutant embryos (Bra−/−). Spatial orientation of the embryos is shown in the lower right corner.
In order to investigate whether our observations made with BRA knockdown hESCs in vitro are also relevant in vivo, we analysed the expression of BRA targets in mouse Bra mutants compared with wild-type embryos at E7.5-8.0 (Fig. 7B; supplementary material Table S5) (fold change>2; RNA-seq data from Lolas et al., 2014). E7.5 is a comparable developmental stage to our 36 h hESC differentiation protocol. These analyses show that orthologues of key BRA mesodermal targets such as Aldh1a2, Cdh2 (N-cadherin), Foxd3, Hes7, several Hox genes, Msgn1, Tbx4, Tbx6 and Wnt5b are downregulated in Bra−/− mouse embryos (supplementary material Table S5). In parallel, targets such as Cdh1, Cer1 and Eomes are upregulated in Bra−/− mouse embryos (supplementary material Table S5). Other anterior/endoderm markers, including Cyp26a1, Foxa2, Gata6, Otx2 and Sox17, are also upregulated in Bra−/− mouse embryos but to a lesser extent (fold change <2). More broadly, 823 putative BRA targets exclusively found in the human ChIP-seq datasets show misregulation in mouse mutant embryos (Fig. 7C), underscoring the value of our new data.
However, RNA-seq data analysis does not provide information on spatial expression patterns in vivo. Thus, we analysed the expression pattern of two key BRA target genes at the protein level in both wild-type and Bra mutant mouse embryos: the mesoderm regulator Cdx2 (Chawengsaksophak et al., 2004) and the endoderm regulator Foxa2 (Ang and Rossant, 1994; Weinstein et al., 1994; Burtscher and Lickert, 2009). In Bra-null embryos, Cdx2 mRNA is downregulated 1.22 fold, while Foxa2 mRNA is upregulated 1.30 fold when compared with wild type (Lolas et al., 2014).
In agreement with our prediction, Cdx2 nuclear expression, which is visible in a small group of cells in the proximal primitive streak where extraembryonic mesoderm is being formed (Bra+/+, white arrowhead, Fig. 7D) (Beck et al., 1995) is completely lacking in Bra mouse mutants (Bra−/−, white arrowheads, Fig. 7D). Foxa2 expression, which marks the distal (future anterior) region of the primitive streak (Bra+/+, white bracket, Fig. 7D) and is also expressed in extraembryonic endoderm (Burtscher and Lickert, 2009) is clearly present in Bra-null embryos (Bra−/−, white brackets, Fig. 7D; supplementary material Fig. S7A), and is seemingly upregulated in the mid/proximal primitive streak. Together, the results obtained with mouse embryos that lack Bra are consistent with our findings in differentiating hESCs: BRA upregulates key mesoderm genes and is dispensable for the expression of several endoderm markers.
DISCUSSION
BRA participates in distinct gene regulatory networks in different cell lineages and species
BRA ChIP-seq analysis of hESCs differentiating in FLyA-, FLyB- or WNT3A- and activin-containing media (Tsankov et al., 2015) showed similar binding sites but also significantly different DNA occupancy levels of BRA. Chromatin accessibility is suggested to be a prerequisite for many sequence-specific transcription factors such as BRA to directly bind DNA (Biggin, 2011; Voss and Hager, 2014). Thus, differences in BRA binding may arise from differential nucleosome occupancy, distinct histone modifications, or the presence or absence of specific protein partners (Biggin, 2011; Spitz and Furlong, 2012; Voss and Hager, 2014). It may be that activin, BMP4 and WNT3A signalling cascades interact with chromatin remodelling complexes (van Grunsven et al., 2005) so as to affect DNA accessibility in a cell type-specific manner.
It is unlikely that the distinct BRA binding in FLyA or FLyB occurs solely because BRA protein levels are lower in hESCs cultured in FLyA rather than FLyB, as there are many binding sites in FLyA-cultured cells with equal or even higher occupancy levels than in FLyB-culture cells. Furthermore, the strong correlation of BRA differential binding with the distinct transcriptional identities of FLyA- or FLyB-treated cells suggests that these events are biologically meaningful. For example, in the FLyB condition (BRAhigh), BRA binds close to many genes involved in posterior mesoderm development, consistent with fate-mapping studies showing that this tissue emerges from the posterior primitive streak, where Bra levels are higher (Lawson et al., 1991; Wilson and Beddington, 1997). Gene ontology analysis reveals that BRA binds in the vicinity of genes involved in specific developmental contexts and that are expressed in tissues where BRA function is essential. These contexts include: epithelial-to-mesenchymal transition; node and notochord formation; the establishment of left-right asymmetry (including heart looping), axial extension and somitogenesis; and cardiovascular development (Fig. 3).
Interestingly, the most highly enriched category in FLyA and FLyB common peaks was ‘neural tube development’. Indeed, BRA seems to repress neurectoderm genes in the tail bud ‘stem cell’ pool that gives rise to posterior structures such as somites, the notochord and the neural tube (Martin and Kimelman, 2010; Gentsch et al., 2013; Tzouanacou et al., 2009; Wilson et al., 2009).
We observed a substantial overlap between BRA targets identified in FLyA- or FLyB-treated hESCs and those identified in WNT3A/activin-treated hESCs (Tsankov et al., 2015) or identified in activin-treated mouse embryoid bodies (Lolas et al., 2014) – the latter suggesting a considerable degree of evolutionary conservation. The fact that the BRA targets we uncovered differ from those of Tsankov et al. could be due to distinct signalling environments, developmental stages, ChIP protocols, antibodies used or other technical issues. We note that both in Tsankov et al. and in Lolas et al. the authors used activin treatment to induce ESC differentiation, thus probably enriching for endoderm progenitors. Indeed, their datasets show a greater overlap with FLyA treatment, rather than with FLyB. Possibly due to this, Tsankov et al. and Lolas et al. did not detect BRA binding in the vicinity of many important mesodermal loci (Table 1). These include key genes in posterior mesoderm and cardiovascular development such as ALDH1A2, HAND1, HES7, KDR, LMO2 and MEOX1, which thus seem to be human specific and unique to our datasets. In summary, our ChIP-seq datasets, together with those recently generated (Lolas et al., 2014; Tsankov et al., 2015), have greatly expanded our knowledge of BRA targets in mammals and now provide a rich resource for future studies to discover new regulators of human gastrulation.
BRA and its potential protein interactors
In addition to the T-BOX consensus binding motif (T-site), DNA motif analyses revealed enrichment for binding sites for protein families with important roles during embryonic development such as FOX, POU, SOX and GATA. Of the many motifs enriched in BRA-binding sites, we focused on the SMAD/ZIC motif (Yoon et al., 2011). SMAD proteins are frequently recruited by master transcription factors to regulate cell type-specific gene regulatory programs (Mullen et al., 2011). Indeed, we found that BRA interacts with SMAD2/3 in FLyA-treated cells and with SMAD1 in FLyB-treated cells. Overexpression of BRA in hESCs upregulates several mesodermal and endodermal target genes, but this effect is particularly potent in the presence of either BMP4 or activin in the culture medium, thus reinforcing the idea that BRA collaborates with these SMAD signalling cascades to regulate its targets in a cell type-specific manner.
BRA binding in FLyA-treated hESCs overlaps very significantly with EOMES binding (Teo et al., 2011; Tsankov et al., 2015). Like other T-BOX proteins in zebrafish (Bra, Ntl, Spt and Tbx6) (Wardle and Papaioannou, 2008), in Xenopus (Xbra, Xbra3, VegT and Eomes) (Gentsch et al., 2013) or in the mouse (Bra and Eomes) (Costello et al., 2011, David et al., 2011), human EOMES and BRA might be functionally interconnected in distinct developmental contexts.
Interestingly, the POU core motif was also enriched in BRA peaks. Although OCT4 is traditionally regarded as a pluripotency regulator, it also acts as lineage specifier (Loh and Lim, 2011; Thomson et al., 2011; Wang et al., 2012). Indeed, OCT4 is expressed throughout the primitive streak during gastrulation (Downs, 2008), together with Bra. Although we (data not shown) and others (Pereira et al., 2011) have been unable to detect a direct physical interaction between BRA and OCT4, it is tempting to speculate that BRA might cooperate with OCT4 and/or other pluripotency factors during the early stages of gastrulation by co-regulating some common target genes, possibly through dynamic sequential binding to these loci (Spitz and Furlong, 2012; Voss and Hager, 2014). Recent ChIP-seq data analysis seems to corroborate this hypothesis (Tsankov et al., 2015).
Searching for functional targets of BRA
Comparison of our ChIP-seq data with results obtained from transcriptional profiling of BRA knockdown cells showed that, although significantly enriched for BRA targets, around half of up- and downregulated transcripts do not appear to be direct targets of BRA and, conversely, that the expression of most putative direct targets is not affected by loss of BRA function. This outcome may derive in part from the inefficiency of our shRNA approach to completely eliminate BRA (supplementary material Fig. S4E), but it is also likely to reflect the functional complexity of transcriptional networks (Biggin, 2011; Spitz and Furlong, 2012). For example, BRA shares a vast number of targets with EOMES. This complicates the analysis of BRA-driven gene regulatory networks because a redundancy with EOMES might mask the number of functional target genes of BRA, as seen in Xenopus embryos (Gentsch et al., 2013).
The number of functional targets of BRA might well be extended if BRA knockdown hESCs were tested in other differentiation protocols where BRA has proposed functions, including primordial germ cells (Aramaki et al., 2013), axial mesoderm (Winzi et al., 2011), and paraxial and cardiac mesoderm (Mendjan et al., 2014). Indeed, our BRA ChIP-seq data revealed that BRA binds in the vicinity of genes involved in the formation of these lineages (Fig. 3).
Importantly, the comparison of our human datasets with mouse Bra ChIP-seq data and RNA-seq from Bra-null gastrulae (Lolas et al., 2014) provides in vivo validation for key BRA targets and underscores the novelty of our work, because around 800 BRA putative targets exclusively identified in our ChIP-seq datasets are either down- or upregulated in Bra mutant embryos (Fig. 7C).
BRA as an essential mesoderm inducer and as an apparent endoderm repressor
Gene ontology analysis of transcriptional changes for BRA target genes observed in BMP4-treated BRA knockdown cells revealed a clear phenotypic scenario: while several BRA target genes involved in mesoderm formation were markedly downregulated, many genes important for endoderm development were upregulated. In agreement with our results, it has been shown that Bra, in collaboration with Mixl1, is able to repress endoderm or anterior primitive streak markers such as Gsc and Pdgfra (Pereira et al., 2011). However, the mechanism by which BRA regulates endoderm differentiation is likely to be complex, because elevated levels of BRA in FLyA-treated cells cause an increase in expression of endodermal marker genes (Fig. 6). Similar results have been obtained by Kalisz et al. (2012), who showed that BRA knockdown and BRA overexpression both cause upregulation of the endodermal markers GSC and SOX17 in hESCs.
Interestingly, the forced expression of several BRA orthologues in Xenopus animal caps leads to mesoderm but not to endoderm formation, except when using the Drosophila and Ciona orthologues (Marcellini et al., 2003). In these two organisms, the N-terminal domain of Bra, which is essential for the interaction with SMAD1 (Messenger et al., 2005), is not conserved. Indeed, a truncated form of Xbra, which can bind DNA but is unable to interact with SMAD1, can induce the expression of Gsc, an anterior/endoderm marker (Messenger et al., 2005). These observations suggest that Bra induces mesoderm formation when cooperating with SMAD1, and endoderm formation when the interaction with SMAD1 is absent (Marcellini, 2006). This may account for the aforementioned paradox that BRA can both repress and activate endoderm gene expression.
By contrast, gene ontology analysis of downregulated transcripts in activin-treated BRA knockdown cells at either 36 or 72 h of differentiation did not reveal any significant biological insights into the role of BRA as an activator in FLyA-treated cells. However, one important aspect that emerged from this profiling was the observation that genes classically involved in EMT were misregulated in FLyA-treated BRA knockdown cells and also in vivo (Lolas et al., 2014). This provides a new molecular insight into the known migratory defects of Bra knockout cells (Yanagisawa et al., 1981) and agrees with the EMT-promoting role of BRA in human cancer samples (Fernando et al., 2010). Indeed, the focus on BRA as an important player in cancer biology, particularly in chordoma, has been growing over the past decade (Papaioannou, 2014; Nelson et al., 2012).
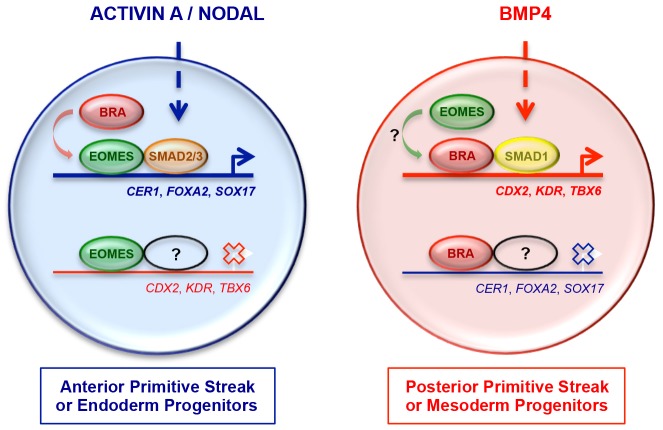
A model for BRA-mediated cell fate decisions in the primitive streak
Our results provide the basis for the following model explaining the establishment of the gene expression patterns in the primitive streak that result in cells acquiring different fates (Fig. 8). Anterior early primitive streak cells, which give rise to definitive endoderm and axial mesoderm (Lawson et al., 1991), experience high levels of NODAL signalling (Tam and Loebel, 2007; Arnold and Robertson, 2009). This induces the expression of high levels of EOMES (Arnold et al., 2008; Teo et al., 2011) and low levels of BRA (Bernardo et al., 2011; Huber et al., 2004) (Fig. 1). EOMES then cooperates with NODAL-SMAD2/3 signalling to induce the expression of anterior markers such as CER1, FOXA2 and SOX17, while repressing posteriorly expressed genes (Brown et al., 2011; Teo et al., 2011). Interestingly, by targeting the same genomic locations as EOMES, BRA is also able to activate the expression of endodermal genes, by cooperating with SMAD2/3 (Fig. 4). However, BRA is not required for anterior primitive streak gene expression (Fig. 4) where EOMES is the main player (Arnold et al., 2008; Teo et al., 2011).
Fig. 8.
BRA, EOMES and SMAD signalling mediate mesoderm or endoderm cell fate choice during gastrulation. Simplified model of gene regulatory mechanisms operating in cells of the anterior (blue) or posterior (red) early primitive streak. Dashed arrows indicate activin A or NODAL as upstream activators of SMAD2/3, and BMP4 as the upstream activator of SMAD1. Crosses over white arrows indicate transcriptional silencing, whereas coloured arrows indicate transcriptional activation.
At the opposite end of the early primitive streak, which gives rise to extraembryonic and posterior mesoderm (Lawson et al., 1991; Parameswaran and Tam, 1995), cells experience high levels of BMP4 signalling (Tam and Loebel, 2007; Arnold and Robertson, 2009). This induces high levels of BRA and low levels of EOMES (Bernardo et al., 2011; Huber et al., 2004) (Fig. 1). BRA, in cooperation with BMP4-SMAD1 signalling (Marcellini, 2006; Messenger et al., 2005), then induces the characteristic expression of posterior markers such as HOX genes (Wacker et al., 2004), CDX2, TBX6 and KDR (Huber et al., 2004), while repressing the expression of anterior genes (Fig. 5). Whether EOMES can activate mesodermal gene expression in collaboration with BMP4-SMAD1 signalling remains an unresolved issue.
Together, our findings illuminate the function of BRA in the human species. We have shown that BRA is indeed both necessary and sufficient to regulate the transcription of many of its putative targets, which are key players during mesoderm or endoderm development. Moreover, our study shows that the regulatory role of BRA is context dependent, thus establishing an intimate collaboration of BRA with SMAD2/3 in an activin/NODAL-dominated context and with SMAD1 in a BMP4-dominated context.
Our study thus reinforces the value of hESCs as tools to model human embryonic development (Murry and Keller, 2008) and emphasizes the importance of analysing the functions of individual members of complex transcription factor networks in distinct cellular and signalling contexts (Spitz and Furlong, 2012).
MATERIALS AND METHODS
Human ESC culture in chemically defined conditions
Human ESCs (H9 line, WiCell) were grown in a chemically defined medium (CDM) as previously described (Bernardo et al., 2011; Vallier and Pedersen, 2008). For differentiation, cells were grown in CDM containing PVA instead of BSA and supplemented as described in the text and in the methods in the supplementary material. Transfection and selection of stable knockdown or overexpression clones were carried out as described in the methods in the supplementary material.
RNA extraction, cDNA synthesis and quantitative PCR
Total RNA was extracted using the RNeasy Mini kit (Qiagen). Half a microgram of RNA was reverse transcribed using the Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Scientific). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) mixtures were prepared using Fast SYBR Green Master Mix (Applied Biosystems). PCR reactions were performed in a 7500 Fast Real-Time PCR System (Applied Biosystems). All procedures followed manufacturer's instructions. See also methods and Table S6 in the supplementary material.
Co-immunoprecipitation and western blotting
Nuclear extract preparation, co-immunoprecipitation and western blotting are described in the methods in the supplementary material.
Immunofluorescence
Cells were fixed for 10 min at room temperature in 4% paraformaldehyde and immunostained as described in the methods in the supplementary material. Fluorescent images were taken using an Olympus IX71 microscope.
Flow cytometry of intracellular proteins
Cells were fixed and immunostained using the Cytofix-Cytoperm kit (BD Biosciences) as described in the methods in the supplementary material. Cells were analysed using a Beckman Coulter CyAnADP flow cytometer and FlowJo software (Becton Dickinson).
ChIP-seq analysis
Chromatin immunoprecipitation was performed as previously described (Brown et al., 2011), with some modifications. Sequencing libraries were prepared using the ChIP-seq DNA sample kit (Illumina, IP-102-1001) with some modifications and sequenced with a Genome Analyzer II (Illumina). Data were further processed using the MACS peak finder (Zhang et al., 2008). DNA motifs were analysed using the MEME suite (Bailey et al., 2009). ChIP-seq data were visualised using the UCSC Genome Browser (Kent et al., 2002). Gene ontology analysis was performed using GREAT (McLean et al., 2010) with default parameters. Data are available in the GEO database (www.ncbi.nlm.nih.gov/geo) under accession number GSE60606. See methods in the supplementary material for further details.
Microarray analysis
Sample preparation was performed according to manufacturer's instructions (Illumina). Labelled extracts were hybridised to whole-genome bead array (HumanWG-6 v3.0 Expression BeadChip) on an Illumina BeadArray reader. Gene expression heat-maps were generated by importing subsets of processed microarray data as described in the methods in the supplementary material. Data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-2912 and E-MTAB-464.
Mouse embryo work
T/+×T/+ mice (King et al., 1998) were mated for embryo collections. Late gastrulae were dissected (E6.75-7.0) for further analysis. All mouse studies were performed under a UK Home Office project license and complied fully with the UK Animals (Scientific Procedures) Act 1986 as implemented by the University of Cambridge and the Medical Research Council. Embryos were fixed for 20-30 min at room temperature in 4% paraformaldehyde (PFA) supplemented with 0.1% Tween-20 (Sigma) and 0.01% Triton-X100 (Sigma). Immunostaining was performed following standard procedures as described in the methods in the supplementary material. Fluorescent images were captured using a Zeiss LSM 710 microscope.
Supplementary Material
Acknowledgements
We thank M. Alexander for assisting with hESC culture, A. Sesay for processing Illumina libraries, A. Kuony for technical support in optimizing embryo immunostaining, T. Spruce for advice on embryo work, A. Evans for sharing unpublished data, R&D Systems for the SOX17-APC antibody, and members of the Pedersen and Smith laboratories for comments and suggestions. T.F. thanks J. Wysocka for support and advice during the revision of this manuscript.
Footnotes
Competing interests
After participating in the work described, M.W.B.T. has become an employee of Celgene Research, part of Celgene.
Funding
This work was supported by: Fundação para a Ciência e a Tecnologia, Portugal [SFRH/BD/39758/2007] (T.F.); the British Heart Foundation (A.S.B., D.O. and V.L.M.); a UK Medical Research Council Programme grant, The Evelyn Trust and Cambridge Hospitals National Institute for Health Research Biomedical Research Centre funding (R.A.P.); and the UK Medical Research Council [U117597140] and Wellcome Trust (G.E.G., J.C.S.). During the last phases of the work, G.E.G. and J.C.S. were supported by the Francis Crick Institute, which receives its core funding principally from Cancer Research UK, the UK Medical Research Council and the Wellcome Trust. Deposited in PMC for immediate release.
Author contributions
T.F. designed, performed and analysed most experiments, carried out gene ontology, DNA motif, and mouse data analyses, and wrote the manuscript with input from all authors; A.S.B. collected, immunostained and imaged mouse embryos, immunostained and performed flow cytometry analysis of hESCs, and contributed to experimental design; S.M. performed western blot analysis and co-immunoprecipitations, assisted by V.L.M.; E.D. and M.W.B.T. analysed ChIP-seq data; T.F. and G.E.G. prepared sequencing libraries; T.F., A.S.B. and D.O. generated BRA knockdown clones; T.F. and A.S.B. prepared samples for microarray analysis; M.W.B.T. analysed microarray data; J.C.S. and R.A.P. co-supervised the study and co-edited the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.117838/-/DC1
References
- Alev C., Wu Y., Kasukawa T., Jakt L. M., Ueda H. R. and Sheng G. (2010). Transcriptomic landscape of the primitive streak. Development 137, 2863-2874. 10.1242/dev.053462 [DOI] [PubMed] [Google Scholar]
- Ang S.-L. and Rossant J. (1994). HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78, 561-574. 10.1016/0092-8674(94)90522-3 [DOI] [PubMed] [Google Scholar]
- Aramaki S., Hayashi K., Kurimoto K., Ohta H., Yabuta Y., Iwanari H., Mochizuki Y., Hamakubo T., Kato Y., Shirahige K. et al. (2013). A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev. Cell 27, 516-529. 10.1016/j.devcel.2013.11.001 [DOI] [PubMed] [Google Scholar]
- Arnold S. J. and Robertson E. J. (2009). Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91-103. 10.1038/nrm2618 [DOI] [PubMed] [Google Scholar]
- Arnold S. J., Hofmann U. K., Bikoff E. K. and Robertson E. J. (2008). Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development 135, 501-511. 10.1242/dev.014357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W. and Noble W. S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202-W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck F., Erler T., Russell A. and James R. (1995). Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 204, 219-227. 10.1002/aja.1002040302 [DOI] [PubMed] [Google Scholar]
- Bernardo A. S., Faial T., Gardner L., Niakan K. K., Ortmann D., Senner C. E., Callery E. M., Trotter M. W., Hemberger M., Smith J. C. et al. (2011). BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9, 144-155. 10.1016/j.stem.2011.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D. (2011). Animal transcription networks as highly connected, quantitative continua. Dev. Cell 21, 611-626. 10.1016/j.devcel.2011.09.008 [DOI] [PubMed] [Google Scholar]
- Brown S., Teo A., Pauklin S., Hannan N., Cho C. H.-H., Lim B., Vardy L., Dunn N. R., Trotter M., Pedersen R. et al. (2011). Activin/Nodal signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells 29, 1176-1185. 10.1002/stem.666 [DOI] [PubMed] [Google Scholar]
- Burtscher I. and Lickert H. (2009). Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo. Development 136, 1029-1038. 10.1242/dev.028415 [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K., de Graaff W., Rossant J., Deschamps J. and Beck F. (2004). Cdx2 is essential for axial elongation in mouse development. Proc. Natl. Acad. Sci. USA 101, 7641-7645. 10.1073/pnas.0401654101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesley P. (1935). Development of the short-tailed mutant in the house mouse. J. Exp. Zool. 70, 429-459. 10.1002/jez.1400700306 [DOI] [Google Scholar]
- Cheung C., Bernardo A. S., Trotter M. W. B., Pedersen R. A. and Sinha S. (2012). Generation of human vascular smooth muscle subtypes provides insight into embryological origin–dependent disease susceptibility. Nat. Biotechnol. 30, 165-173. 10.1038/nbt.2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello I., Pimeisl I.-M., Dräger S., Bikoff E. K., Robertson E. J. and Arnold S. J. (2011). The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 13, 1084-1091. 10.1038/ncb2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour K. A., Agulnick A. D., Eliazer S., Kelly O. G., Kroon E. and Baetge E. E. (2005). Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23, 1534-1541. 10.1038/nbt1163 [DOI] [PubMed] [Google Scholar]
- David R., Jarsch V. B., Schwarz F., Nathan P., Gegg M., Lickert H. and Franz W.-M. (2011). Induction of MesP1 by Brachyury(T) generates the common multipotent cardiovascular stem cell. Cardiovasc. Res. 92, 115-122. 10.1093/cvr/cvr158 [DOI] [PubMed] [Google Scholar]
- Downs K. M. (2008). Systematic localization of Oct-3/4 to the gastrulating mouse conceptus suggests manifold roles in mammalian development. Dev. Dyn. 237, 464-475. 10.1002/dvdy.21438 [DOI] [PubMed] [Google Scholar]
- Evans A. L., Faial T., Gilchrist M. J., Down T., Vallier L., Pedersen R. A., Wardle F. C. and Smith J. C. (2012). Genomic targets of Brachyury (T) in differentiating mouse embryonic stem cells. PLoS ONE 7, e33346 10.1371/journal.pone.0033346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando R. I., Litzinger M., Trono P., Hamilton D. H., Schlom J. and Palena C. (2010). The T-box transcription factor Brachyury promotes epithelial-mesenchymal transition in human tumor cells. J. Clin. Invest. 120, 533-544. 10.1172/JCI38379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch G. E., Owens N. D. L., Martin S. R., Piccinelli P., Faial T., Trotter M. W. B., Gilchrist M. J. and Smith J. C. (2013). In vivo T-box transcription factor profiling reveals joint regulation of embryonic neuromesodermal bipotency. Cell Rep. 4, 1185-1196. 10.1016/j.celrep.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann B. G., Labeit S., Poustka A., King T. R. and Lehrach H. (1990). Cloning of the T gene required in mesoderm formation in the mouse. Nature 343, 617-622. 10.1038/343617a0 [DOI] [PubMed] [Google Scholar]
- Huber T. L., Kouskoff V., Fehling H. J., Palis J. and Keller G. (2004). Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 432, 625-630. 10.1038/nature03122 [DOI] [PubMed] [Google Scholar]
- Inman K. E. and Downs K. M. (2006). Brachyury is required for elongation and vasculogenesis in the murine allantois. Development 133, 2947-2959. 10.1242/dev.02454 [DOI] [PubMed] [Google Scholar]
- Kalisz M., Winzi M., Bisgaard H. C. and Serup P. (2012). EVEN-SKIPPED HOMEOBOX 1 controls human ES cell differentiation by directly repressing GOOSECOID expression. Dev. Biol. 362, 94-103. 10.1016/j.ydbio.2011.11.017 [DOI] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M. and Haussler D. (2002). The human genome browser at UCSC. Genome Res. 12, 996-1006. 10.1101/gr.229102.ArticlepublishedonlinebeforeprintinMay2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T., Beddington R. S. P. and Brown N. A. (1998). The role of the brachyury gene in heart development and left–right specification in the mouse. Mech. Dev. 79, 29-37. 10.1016/S0925-4773(98)00166-X [DOI] [PubMed] [Google Scholar]
- Kispert A. and Herrmann B. G. (1993). The Brachyury gene encodes a novel DNA binding protein. EMBO J. 12, 3211-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Shinozaki K., Shannon J. M., Kouskoff V., Kennedy M., Woo S., Fehling H. J. and Keller G. (2004). Development of definitive endoderm from embryonic stem cells in culture. Development 131, 1651-1662. 10.1242/dev.01044 [DOI] [PubMed] [Google Scholar]
- Lamouille S., Xu J. and Derynck R. (2014). Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178-196. 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson K. A., Meneses J. J. and Pedersen R. A. (1991). Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113, 891-911. [DOI] [PubMed] [Google Scholar]
- Loh K. M. and Lim B. (2011). A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell 8, 363-369. 10.1016/j.stem.2011.03.013 [DOI] [PubMed] [Google Scholar]
- Lolas M., Valenzuela P. D. T., Tjian R. and Liu Z. (2014). Charting Brachyury-mediated developmental pathways during early mouse embryogenesis. Proc. Natl. Acad. Sci. USA 111, 4478-4483. 10.1073/pnas.1402612111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcellini S. (2006). When Brachyury meets Smad1: the evolution of bilateral symmetry during gastrulation. Bioessays 28, 413-420. 10.1002/bies.20387 [DOI] [PubMed] [Google Scholar]
- Marcellini S., Technau U., Smith J. C. and Lemaire P. (2003). Evolution of Brachyury proteins: identification of a novel regulatory domain conserved within Bilateria. Dev. Biol. 260, 352-361. 10.1016/S0012-1606(03)00244-6 [DOI] [PubMed] [Google Scholar]
- Martin B. L. and Kimelman D. (2010). Brachyury establishes the embryonic mesodermal progenitor niche. Genes Dev. 24, 2778-2783. 10.1101/gad.1962910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M. and Bejerano G. (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495-501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S., Mascetti V. L., Ortmann D., Ortiz M., Karjosukarso D. W., Ng Y., Moreau T. and Pedersen R. A. (2014). NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell 15, 310-325. 10.1016/j.stem.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Messenger N. J., Kabitschke C., Andrews R., Grimmer D., Núñez Miguel R., Blundell T. L., Smith J. C. and Wardle F. C. (2005). Functional specificity of the Xenopus T-domain protein Brachyury is conferred by its ability to interact with Smad1. Dev. Cell 8, 599-610. 10.1016/j.devcel.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Morley R. H., Lachani K., Keefe D., Gilchrist M. J., Flicek P., Smith J. C. and Wardle F. C. (2009). A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc. Natl. Acad. Sci. USA 106, 3829-3834. 10.1073/pnas.0808382106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A. C., Orlando D. A., Newman J. J., Lovén J., Kumar R. M., Bilodeau S., Reddy J., Guenther M. G., DeKoter R. P. and Young R. A. (2011). Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147, 565-576. 10.1016/j.cell.2011.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry C. E. and Keller G. (2008). Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661-680. 10.1016/j.cell.2008.02.008 [DOI] [PubMed] [Google Scholar]
- Nelson A. C., Pillay N., Henderson S., Presneau N., Tirabosco R., Halai D., Berisha F., Flicek P., Stemple D. L., Stern C. D. et al. (2012). An integrated functional genomics approach identifies the regulatory network directed by brachyury (T) in chordoma. J. Pathol. 228, 274-285. 10.1002/path.4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou V. E. (2014). The T-box gene family: emerging roles in development, stem cells and cancer. Development 141, 3819-3833. 10.1242/dev.104471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran M. and Tam P. P. L. (1995). Regionalisation of cell fate and morphogenetic movement of the mesoderm during mouse gastrulation. Dev. Genet. 17, 16-28. 10.1002/dvg.1020170104 [DOI] [PubMed] [Google Scholar]
- Pereira L. A., Wong M. S., Lim S. M., Sides A., Stanley E. G. and Elefanty A. G. (2011). Brachyury and related Tbx proteins interact with the Mixl1 homeodomain protein and negatively regulate Mixl1 transcriptional activity. PLoS ONE 6, e28394 10.1371/journal.pone.0028394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S., Steiner K. A. and Tam P. P. L. (2007). Gene expression pattern and progression of embryogenesis in the immediate post-implantation period of mouse development. Gene Expr. Patterns 7, 558-573. 10.1016/j.modgep.2007.01.005 [DOI] [PubMed] [Google Scholar]
- Russ A. P., Wattler S., Colledge W. H., Aparicio S. A. J. R., Carlton M. B. L., Pearce J. J., Barton S. C., Surani M. A., Ryan K., Nehls M. C. et al. (2000). Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404, 95-99. 10.1038/35003601 [DOI] [PubMed] [Google Scholar]
- Showell C., Binder O. and Conlon F. L. (2004). T-box genes in early embryogenesis. Dev. Dyn. 229, 201-218. 10.1002/dvdy.10480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F. and Furlong E. E. M. (2012). Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613-626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- Tada S., Era T., Furusawa C., Sakurai H., Nishikawa S., Kinoshita M., Nakao K., Chiba T. and Nishikawa S.-I. (2005). Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development 132, 4363-4374. 10.1242/dev.02005 [DOI] [PubMed] [Google Scholar]
- Tam P. P. L. and Loebel D. A. F. (2007). Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 8, 368-381. 10.1038/nrg2084 [DOI] [PubMed] [Google Scholar]
- Teo A. K. K., Arnold S. J., Trotter M. W. B., Brown S., Ang L. T., Chng Z., Robertson E. J., Dunn N. R. and Vallier L. (2011). Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 25, 238-250. 10.1101/gad.607311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson M., Liu S. J., Zou L.-N., Smith Z., Meissner A. and Ramanathan S. (2011). Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 145, 875-889. 10.1016/j.cell.2011.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov A. M., Gu H., Akopian V., Ziller M. J., Donaghey J., Amit I., Gnirke A. and Meissner A. (2015). Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344-349. 10.1038/nature14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouanacou E., Wegener A., Wymeersch F. J., Wilson V. and Nicolas J.-F. (2009). Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell 17, 365-376. 10.1016/j.devcel.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Vallier L. and Pedersen R. (2008). Differentiation of human embryonic stem cells in adherent and in chemically defined culture conditions. Curr. Protoc. Stem Cell Biol. Chapter 1, Unit 1D.4.1-1D.4.7 10.1002/9780470151808.sc01d04s4 [DOI] [PubMed] [Google Scholar]
- van Grunsven L. A., Verstappen G., Huylebroeck D. and Verschueren K. (2005). Smads and chromatin modulation. Cytokine Growth Factor Rev. 16, 495-512. 10.1016/j.cytogfr.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Voss T. C. and Hager G. L. (2014). Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 15, 69-81. 10.1038/nrg3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker S. A., McNulty C. L. and Durston A. J. (2004). The initiation of Hox gene expression in Xenopus laevis is controlled by Brachyury and BMP-4. Dev. Biol. 266, 123-137. 10.1016/j.ydbio.2003.10.011 [DOI] [PubMed] [Google Scholar]
- Wang Z., Oron E., Nelson B., Razis S. and Ivanova N. (2012). Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10, 440-454. 10.1016/j.stem.2012.02.016 [DOI] [PubMed] [Google Scholar]
- Wardle F. C. and Papaioannou V. E. (2008). Teasing out T-box targets in early mesoderm. Curr. Opin. Genet. Dev. 18, 418-425. 10.1016/j.gde.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein D. C., Ruiz i Altaba A., Chen W. S., Hoodless P., Prezioso V. R., Jessell T. M. and Darnell J. E. Jr (1994). The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell 78, 575-588. 10.1016/0092-8674(94)90523-1 [DOI] [PubMed] [Google Scholar]
- Wilkinson D. G., Bhatt S. and Herrmann B. G. (1990). Expression pattern of the mouse T gene and its role in mesoderm formation. Nature 343, 657-659. 10.1038/343657a0 [DOI] [PubMed] [Google Scholar]
- Wilson V. and Beddington R. (1997). Expression of T protein in the primitive streak is necessary and sufficient for posterior mesoderm movement and somite differentiation. Dev. Biol. 192, 45-58. 10.1006/dbio.1997.8701 [DOI] [PubMed] [Google Scholar]
- Wilson V., Olivera-Martinez I. and Storey K. G. (2009). Stem cells, signals and vertebrate body axis extension. Development 136, 1591-1604. 10.1242/dev.021246 [DOI] [PubMed] [Google Scholar]
- Winzi M. K., Hyttel P., Dale J. K. and Serup P. (2011). Isolation and characterization of node/notochord-like cells from mouse embryonic stem cells. Stem Cells Dev. 20, 1817-1827. 10.1089/scd.2011.0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa K. O., Fujimoto H. and Urushihara H. (1981). Effects of the brachyury (T) mutation on morphogenetic movement in the mouse embryo. Dev. Biol. 87, 242-248. 10.1016/0012-1606(81)90147-0 [DOI] [PubMed] [Google Scholar]
- Yoon S.-J., Wills A. E., Chuong E., Gupta R. and Baker J. C. (2011). HEB and E2A function as SMAD/FOXH1 cofactors. Genes Dev. 25, 1654-1661. 10.1101/gad.16800511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W. et al. (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.