Abstract
Glioma, especially high-grade glioma, is highly malignant with high rate of recurrence and poor prognosis. The mechanisms of glioma progression and recurrence have not been elucidated. Previous studies showed that long non-coding RNAs (lncRNAs) involved in the development and progression of glioma. However, the roles of lncRNAs in the recurrence of glioma remain unknown. We use high throughput microarray to screen the differentially expressed lncRNAs and mRNAs in recurrence gliomas compared with primary gliomas. We found a total of 1,111 lncRNAs were differentially expressed in recurrent group. Among these, 639 lncRNAs were up-regulated, while 472 lncRNAs were down-regulated (fold Change ≥2.0). GO (Gene ontology) and pathway analysis revealed that the potential functions of differentially expressed lncRNAs were closely connected with the processes of cancer progression and pathogenesis. LncRNA classification and subgroup analysis further identified three important clusters of differentially expressed lncRNA-mRNA pairs which have potential gene regulatory functions. This study for the first time showed abundant differentially expressed lncRNAs in recurrent gliomas. Some lncRNAs may play important roles in glioma recurrence, such as previously reported H19, CRNDE, HOTAIRM1 or unreported AC016745.3, XLOC_001711, RP11-128A17.1. Moreover, this study set a basis for future researches on specific lncRNA which may contribute to the recurrence of glioma. Further studies on these lncRNAs will help to elucidate the mechanism of glioma recurrence at genetic level and find therapeutic targets for glioma patients.
Keywords: lncRNA, microarray, glioma, recurrent
Introduction
Glioma is the most common malignant brain tumor in adult. Based on the pathological features, gliomas can be classified as World Health Organization (WHO) grade I-IV. Almost all high-grade (WHO grade III-IV) gliomas recur after tumor resection. The median survivals are 30-39 weeks for patients with high grade gliomas [1]. The high rates of recurrence and inefficient treatments contribute to the poor prognosis of patients with gliomas. Despite the development of multi-mode treatments that include surgical resection, radiotherapy, chemotherapy as well as target therapy in the past decades, the outcome of glioma, especially high grade glioma, is still unsatisfactory [2]. To improve treatment efficiency, a further understanding of molecular mechanism of glioma recurrence is urgently needed.
Genome-wide profiling studies have revealed that only about 2% of the human genome sequence encodes protein, while more than 90% of the genome is actively transcribed [3-5]. It suggests that most of the non-coding transcripts may have regulatory function. Among these, one kind of the non-coding RNAs (ncRNAs), microRNAs (miRNAs) are well documented to play important roles in regulating gene expression. NcRNAs can be grouped into two classes based on transcripts length: Short ncRNAs and lncRNAs. Short ncRNAs have a length under 200 nucleotides which include small interfering RNAs (siRNAs), miRNAs and so on. LncRNAs are mRNA-like transcripts lacking significant open reading frames and longer than 200 nucleotides in length. Most of them are transcribed by RNA polymerase II and polyadenylated [6,7]. Initially, lncRNAs are regarded to be transcriptional noise [8]. However, the discovery of imprinted H19 gene and X-inactive-specific transcript (XIST) gene makes lncRNAs fade in people’s sight. A increasing number of lncRNAs have been identified to be involved in some important biological processes, such as imprinting control, cell differentiation, tumorigenesis and so on [6]. Recent evidences showed that lncRNAs were not only differentially expressed between glioma and normal brain tissue, but also differentially expressed among different WHO grades. Some of these aberrant lncRNAs were regarded to regulate the tumorigenesis in glioma even reflect patient prognosis, although the precise mechanism is not well clarified [9-11]. However, there is no study of lncRNA expression profile between primary and recurrent gliomas. It remains unknown whether lncRNAs are associated with the process of glioma recurrence.
To explore the association between lncRNAs and the process of glioma recurrence, we identified the expression profiles of lncRNA and mRNA between recurrent gliomas and primary gliomas by microarray. The potential functions of the lncRNAs were analyzed by GO and pathway analysis based on the differentially expressed mRNAs. For further screening important lncRNAs, lncRNA classification and subgroup analysis were performed. This study was aimed to set a basis for searching specific lncRNAs which might be helpful to elucidate the mechanism of glioma recurrence at genetic level.
Materials and methods
Acquirement of patient samples
The three paired samples were obtained from 3 recurrent glioma patients (Table 1) of Changzheng Hospital, Shanghai, China. None of them received chemotherapy or radiation before the first surgery. Each patient underwent gross total resection of primary glioma under microscope. Two of them who were diagnosed as WHO III and IV glioma received treatment of temozolomide (TMZ) and radiation after the first surgery. The other one who was diagnosed as primary WHO II glioma received treatment of TMZ only after the first surgery. They all recurred and received the second surgery for recurrent gliomas. The diagnosis was confirmed by histopathology. All patients were provided written informed consent before samples were collected and frozen in liquid nitrogen. The study was in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards and approved by the Specialty Committee on Ethics of Biomedicine Research, Second Military Medical University of China.
Table 1.
Characteristics of three cases
| Case | Sex | Age | Sample | Pathological Diagnosis | WHO Grade | Extent of resection for primary glioma | Chemotherapy (temozolomide) | Radiation | Recurrence interval (month) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Female | 43 | P1 | Diffuse Astrocytoma | II | Gross total resection under microscope | Yes | No | 33 |
| R1 | Anaplastic Astrocytoma | III | |||||||
| 2 | Male | 59 | P2 | Glioblastoma | IV | Gross total resection under microscope | Yes | Yes | 14 |
| R2 | Glioblastoma | IV | |||||||
| 3 | Male | 58 | P3 | Anaplastic Astrocytoma | III | Gross total resection under microscope | Yes | Yes | 9 |
| R3 | Anaplastic Astrocytoma | III |
RNA extraction
The RNAs were isolated from the primary and recurrent glioma tissue using TRIzol reagent (Invitrogen, CA) according to the manufacturer’s protocol. The NanoDrop ND-1000 spectrophotometer was used to measure RNA quantity and quality. The RNA integrity was assessed by standard denaturing agarose gel electrophoresis.
Microarray and data analysis
We performed sample labeling and array hybridization in accordance with the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology) with minor modifications. Concisely, mRNA was purified from total RNA by removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre). Then, a random priming method (Arraystar Flash RNA Labeling Kit, Arraystar) was applied to amplify and tanscribe each sample into fluorescent cRNA along the entire length of the transcripts without 3’ bias. The Human LncRNA Array v3.0 (8 x 60 K, Arraystar) was employed to hybridize the labeled cRNAs. About 30,586 lncRNAs and 26,109 coding transcripts could be detected by this third-generation lncRNA microarray. We carefully constructed the lncRNAs according to the most authoritative public transcriptome databases (Refseq, UCSC knowngenes, Gencode, etc), as well as landmark publications. The specific exon or splice junction probe, which could identify individual transcript accurately, was used to represent each transcript. Positive probes for housekeeping genes as well as negative probes were printed onto the array for hybridization quality control. The hybridized arrays were washed, fixed and scanned with the Agilent DNA Microarray Scanner (part number G2505C).
The array images were analyzed by Agilent Feature Extraction software (version 11.0.1.1). The GeneSpring GX v12.1 software package (Agilent Technologies) was used to perform quantile normalization and data processing. When the raw data had been quantile normalized, lncRNAs and mRNAs that at least 3 out of 6 samples had flags in Present or Marginal (“All Targets Value”) were chosen for further data analysis. Volcano Plot filtering was used to identify differentially expressed lncRNAs and mRNAs with statistical significance between the two groups. Fold Change filtering was used to identify differentially expressed lncRNAs and mRNAs (fold change ≥2.0, P<0.05).
GO and pathway analysis
GO analysis was performed to analyze the function of differentially expressed mRNAs by associating them with GO categories. Derived from Gene Ontology (www.geneontology.org), the GO categories include three integrated networks of defined terms which describe gene property. Pathway analysis was used to determine the significant biological pathways of these differentially expressed mRNAs. We performed the pathway analysis based on the latest KEGG (Kyoto Encyclopedia of Genes and Genomes) database. The P value of Fisher’s exact test denoted the significance of the pathway. The less the P value was, the more significant of the pathway was (P <0.05 was considered statistically significant).
LncRNA classification and subgroup analysis
Enhancer lncRNAs, LincRNAs (large intergenic noncoding RNAs) and antisense lncRNAs were three clusters of lncRNAs with gene regulatory functions. They were classified based on Orom UA’s study [12], Guttman M’s papers [13,14] and data base annotation, respectively. By integrating these lncRNAs with the differentially expressed mRNAs, we performed lncRNA-mRNA co-expression analysis. Differentially expressed enhancer lncRNA and nearby coding gene (distance <300 kb) pairs were screened by the criterion that both enhancer lncRNA and its nearby coding genes were changed more than 2.0 fold. The lincRNA and associated coding gene pairs were screened in the same way. Antisense lncRNA and corresponding sense gene pairs were also screened when they both were changed more than 2.0 fold.
Statistical methods
The differentially expressed lncRNAs and mRNAs between the primary glioma group and recurrent glioma group were compared using a paired t-test (P <0.05 was considered statistically significant). Fisher’s exact test was used for pathway analysis to select significant pathways, respectively (P <0.05 was considered statistically significant). False discovery rate (FDR) was calculated from Benjamini Hochberg FDR to correct the P value.
Result
Quality assessment of lncRNAs and mRNAs data
We used Box-Plot to visualize the distributions of the intensities from all samples. The distributions of the log2-ratios among the six samples were almost the same after quantile normalization (Figure 1). We used Scatter-Plot to visualize the lncRNAs and mRNAs expression variation between two groups. The Scatter-Plots showed that abundant lncRNAs and mRNAs were changed more than 2.0 fold between two compared groups (Figure 2).
Figure 1.
Box plots of lncRNAs (A) and mRNAs (B). “P” represents primary glioma, “R” represents recurrent glioma.
Figure 2.
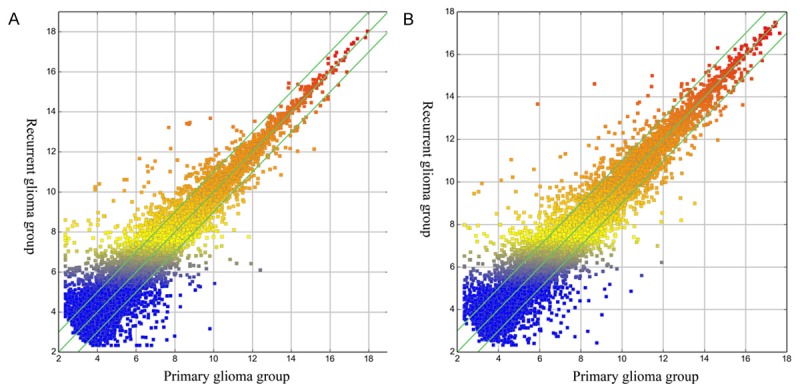
Scatter plots of lncRNAs (A) and mRNAs (B). They are used to visualize the expression variation between two groups. The values of X and Y axes in the Scatter-Plot were the averaged normalized log2 scaled signal values of groups. The green lines indicate where fold change value is 2.0. The lncRNAs and mRNAs above the top green line and below the bottom green line indicated more than 2.0 fold change between two compared groups.
Hierarchical clustering
Hierarchical Clustering was used for analysis of lncRNAs expression data of six samples. Based on their expression levels, cluster analysis arranged samples into groups. Therefore, we could hypothesize the relationship among samples. The lncRNAs expression patterns of six samples were showed by dendrogram (Figure 3).
Figure 3.
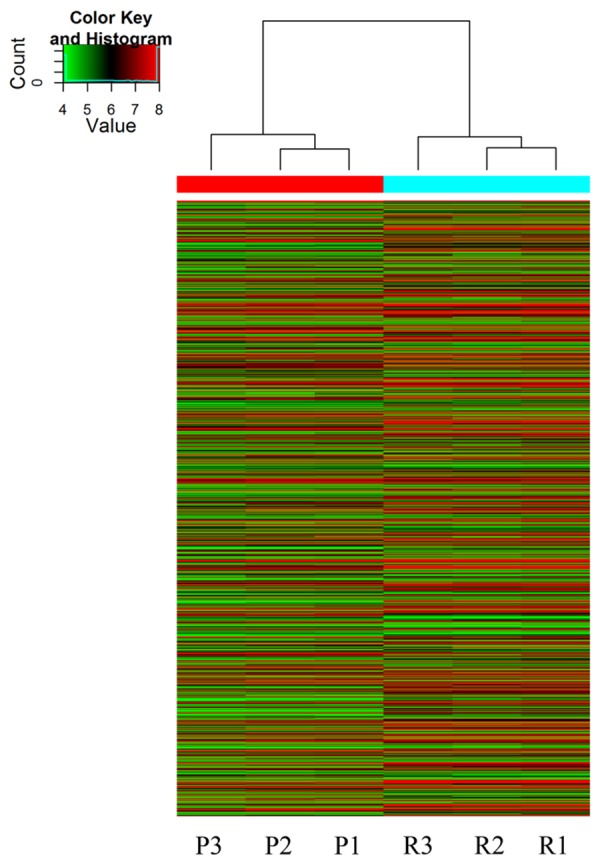
Hierarchical Clustering of lncRNAs. The relationships among the expression levels of six samples are showed clearly by this dendrogram. “Red” indicates high relative expression and “green” indicates low relative expression. “P” indicates primary glioma and “R” indicates recurrent glioma.
Expression profile of lncRNAs and mRNAs
Volcano Plot filtering was applied to screen the differentially expressed lncRNAs and mRNAs with statistical significance (fold change ≥2.0, P <0.05) between the two groups (Figure 4). A total of 1,111 lncRNAs and 1,233 mRNAs were differentially expressed between the recurrent and the primary glioma group. Among these, 639 lncRNAs and 627 mRNAs were up-regulated in the recurrent group compared with the primary group, while 472 lncRNAs and 606 mRNAs were down-regulated (fold change ≥2.0, P <0.05). It was noteworthy that lncRNAs H19, HOTAIRM1 (HOX antisense intergenic RNA myeloid 1) and CRNDE (Colorectal neoplasia differentially expressed) was up-regulated for 3.0 fold, 4.1 fold and 11.2 fold, respectively. The three lncRNAs were previously demonstrated to play important roles in glioma. The top 10 up-regulated (Table 2) and down-regulated lncRNAs (Table 3) are listed.
Figure 4.
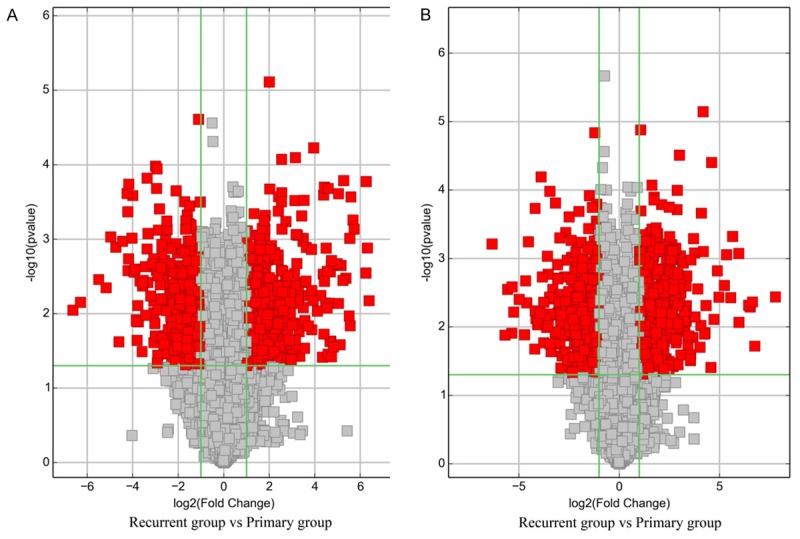
Volcano plots of lncRNAs (A) and mRNAs (B). Volcano plot is used for visualizing differential expression between two groups. The vertical lines correspond to 2.0-fold up and down, respectively, and the horizontal line represents a p-value of 0.05. So the red point in the plot represents the differentially expressed lncRNAs and mRNAs with statistical significance.
Table 2.
Top 10 up-regulated lncRNAs in the recurrent group compared with the primary group
| Seqname | GeneSymbol | Fold Change | P-value | Regulation |
|---|---|---|---|---|
| ENST00000415338 | RP5-998N21.4 | 83.4596327 | 0.007 | up |
| ENST00000428289 | RP11-196G18.3 | 79.1648301 | 0.001 | up |
| NR_051996 | MGC32805 | 76.3391949 | 0.000 | up |
| ENST00000457996 | RP11-439A17.9 | 75.7762829 | 0.003 | up |
| ENST00000471990 | ADAMTS9-AS1 | 52.7837886 | 0.001 | up |
| ENST00000449386 | RP4-792G4.2 | 51.0472876 | 0.001 | up |
| ENST00000493124 | ADAMTS9-AS1 | 47.8919752 | 0.000 | up |
| ENST00000437267 | BX571672.2 | 46.8798484 | 0.014 | up |
| ENST00000454263 | TRIM43B | 46.2437653 | 0.001 | up |
| uc004abd.1 | DQ573539 | 45.5054089 | 0.011 | up |
Table 3.
Top 10 down-regulated lncRNAs in the recurrent group compared with the primary group
| Seqname | GeneSymbol | Fold Change | P-value | Regulation |
|---|---|---|---|---|
| TCONS_00001368 | XLOC_000669 | 99.0401229 | 0.009 | down |
| ENST00000490341 | TUBA4B | 78.7176547 | 0.007 | down |
| NR_038200 | M1 | 45.2325513 | 0.003 | down |
| ENST00000439198 | RP11-324I22.2 | 36.1382223 | 0.004 | down |
| ENST00000511064 | RP11-679C8.2 | 31.3017411 | 0.001 | down |
| NR_038199 | M1 | 26.7071748 | 0.001 | down |
| TCONS_00011943 | XLOC_005452 | 24.3911391 | 0.024 | down |
| TCONS_00023300 | XLOC_011169 | 21.8388805 | 0.001 | down |
| NR_024539 | POPDC3 | 19.0093338 | 0.000 | down |
| NR_038220 | LOC170425 | 18.6276796 | 0.000 | down |
GO and pathway analysis
In order to reveal the potential roles of differentially expressed lncRNAs, GO analysis and KEGG pathway annotation were applied to the differentially expressed mRNAs which was the target gene pool of lncRNAs. The Gene ontology (GO) covers three domains: biological process, cellular component and molecular function. Using GO analysis, we found that most up-regulated mRNAs were involved in biological regulation (biological process), membrane (cellular component) and binding (molecular function). The most down-regulated mRNAs were involved in response to stimulus (biological process), cytoplasm (cellular component) and protein binding (molecular function). The classifications of GO molecular function for up-regulated and down-regulated mRNAs were listed (Figure 5).
Figure 5.
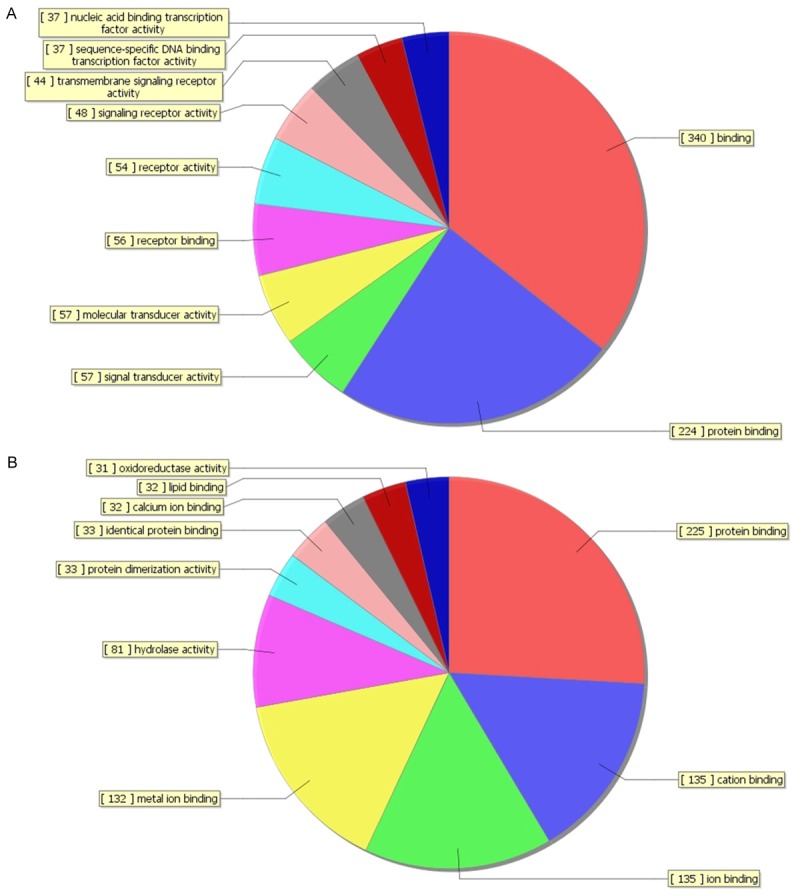
The classification of GO molecular function for up-regulated mRNAs (A) and down-regulated mRNAs (B).
Pathway analysis indicated that 41 pathways corresponded to the up-regulated mRNAs and 27 pathways corresponded to down-regulated mRNAs. The up-regulated mRNAs were involved in systemic lupus erythematosus, antigen processing and presentation, etc. On the other hand, the down-regulated mRNAs were involved in FoxO signaling pathway, GnRH signaling pathway, ErbB signaling pathway, MAPK signaling pathway, Pathways in cancer, etc. Many of them were cancer related pathways which might contribute to the recurrence of glioma. Top 10 enriched pathway for up-regulated and down-regulated mRNAs were listed (Figure 6).
Figure 6.
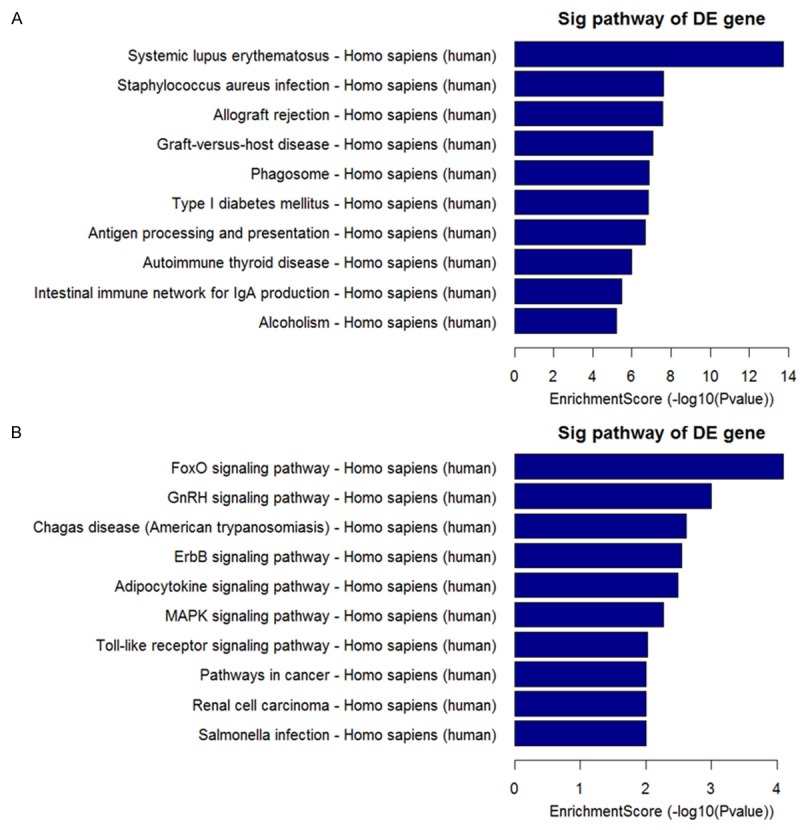
Top 10 enriched pathways for up-regulated mRNAs (A) and down-regulated mRNAs (B).
LncRNA classification and subgroup analysis
Enhancer lncRNAs are a cluster of lncRNAs with enhancer-like function. They can mediate positive regulation in their nearby gene [12]. LincRNAs (large intergenic noncoding RNAs) are a group of lncRNAs which are highly conserved and involved in diverse biological processes, including embryonic stem cell pluripotency, cell proliferation and so on [13,14]. Antisense lncRNAs are transcribed from the opposite DNA strands. They are proposed to regulate the sense mRNA with a variety of mechanisms including transcription-related modulation, RNA–DNA interactions, nuclear RNA duplex formation and cytoplasmic RNA duplex formation [15]. Here, we found 36 differentially expressed enhancer lncRNA-nearby gene pairs, 58 differentially expressed lncRNA-nearby gene pairs and 17 antisense lncRNA-corresponding sense mRNA pairs, respectively. Some of the differentially expressed enhancer lncRNA-nearby gene pairs (Table 4), lncRNA-nearby gene pairs (Table 5) and antisense lncRNA-corresponding sense mRNA pairs (Table 6) were listed.
Table 4.
Some differentially expressed enhancer lncRNAs and their nearby coding genes
| Seqname | GeneSymbol | Fold change - LncRNAs | Regulation - LncRNAs | Genome Relationship | NearbyGene | NearbyGeneSymbol | Fold change - mRNAs | Regulation - mRNAs |
|---|---|---|---|---|---|---|---|---|
| ENST00000422178 | AC016745.3 | 3.901481 | up | downstream | NM_012184 | FOXD4L1 | 2.6691191 | up |
| ENST00000453213 | RP11-492E3.1 | 2.0042102 | up | downstream | NM_016307 | PRRX2 | 2.6813676 | down |
| ENST00000440633 | LINC00545 | 2.2315919 | down | upstream | NM_001629 | ALOX5AP | 2.2498186 | up |
| ENST00000456100 | AL163953.3 | 2.5419899 | down | upstream | NM_006832 | FERMT2 | 2.0666781 | down |
| ENST00000454625 | GS1-421I3.2 | 2.1695855 | down | downstream | NM_001008535 | AKAP14 | 2.1786642 | down |
Table 5.
Some differentially expressed lincRNAs and their nearby coding genes
| Seqname | GeneSymbol | Fold change - LncRNAs | Regulation - LncRNAs | Genome Relationship | NearbyGene | NearbyGeneSymbol | Fold change - mRNAs | Regulation - mRNAs |
|---|---|---|---|---|---|---|---|---|
| TCONS_00003906 | XLOC_001711 | 2.1538904 | down | downstream | ENST00000325926 | RPRM | 11.962624 | down |
| TCONS_00029753 | XLOC_014147 | 2.1586285 | up | upstream | NM_001193414 | TUBA8 | 3.3856396 | up |
| TCONS_00029754 | XLOC_014147 | 2.060898 | up | upstream | NM_001193414 | TUBA8 | 3.3856396 | up |
| TCONS_00001281 | XLOC_000567 | 4.3178215 | down | upstream | NM_138794 | LYPLAL1 | 5.2807164 | down |
| TCONS_00002232 | XLOC_000566 | 3.6056213 | down | upstream | NM_138794 | LYPLAL1 | 5.2807164 | down |
Table 6.
Some differentially expressed antisense lncRNAs and their corresponding sense mRNAs
| Seqname | GeneSymbol | Fold change - LncRNAs | Regulation - LncRNAs | Genome Relationship | NearbyGene | NearbyGeneSymbol | Fold change - mRNAs | Regulation - mRNAs |
|---|---|---|---|---|---|---|---|---|
| ENST00000559509 | RP11-128A17.1 | 2.2520785 | up | natural antisense | NM_170674 | MEIS2 | 3.5500179 | up |
| ENST00000559509 | RP11-128A17.1 | 2.2520785 | up | natural antisense | NM_170677 | MEIS2 | 2.0640461 | up |
| ENST00000433474 | RP11-109P14.9 | 3.3556245 | up | natural antisense | NM_001031740 | MANEAL | 4.2485947 | up |
| ENST00000515569 | CTC-575N7.1 | 4.2262584 | down | natural antisense | NM_175856 | CHSY3 | 2.5092305 | down |
| ENST00000554678 | RP11-203M5.8 | 2.0347683 | up | natural antisense | NM_000270 | PNP | 2.7215837 | up |
Discussion
There are increasing evidences that lncRNAs play important roles in cancer [16]. High throughput microarray analysis is a useful method to screen differentially expressed lncRNAs in cancer. Using this method, researchers have identified many important lncRNAs which play critical roles in glioma and help to clarify the pathogenesis at genetic level [17,18]. Certain lncRNAs not only regulate the process of proliferation and apoptosis in glioma, but also reflect the prognosis of glioma [19-21]. For example, Han et al. found 1,308 lncRNAs and peroxisome proliferator-activated receptor (PPAR) signaling pathway were aberrantly expressed in a GBM tissue compared with a normal control. Among these lncRNAs, ASLNC22381 and ASLNC2081 may target insulin like growth factor 1 (IGF-1) signaling and contribute to glioma progression and recurrence [18]. However, all existing studies have only analyzed the expression profile of lncRNA between gliomas and normal brain tissue or among different grade gliomas. It remains unknown whether lncRNAs are aberrant expressed between recurrent gliomas and primary gliomas, let alone the functions of lncRANs in glioma recurrence.
In this study, we gave an overview of lncRNAs profile between recurrent glioma and primary glioma. A total of 1,111 lncRNAs (639 up-regulated and 472 down-regulated) were differentially expressed in recurrent gliomas in comparison with primary gliomas. It is noteworthy that the three obviously up-regulated lncRNAs H19, HOTAIRM1 and CRNDE were previously reported to be up-regulated in glioma and other cancers. H19, which had been demonstrated as an oncogenic lncRNA in breast and colon cancer, was also identified to be higher expressed in high grade glioma than low grade glioma. The functional study revealed that H19 could induce invasion of glioma cell by driving miR-675 [25]. Since invasion is an important factor for glioma recurrence. The property of inducing invasion of glioma cell suggests H19 is an important lncRNA that related to glioma recurrence. Another lncRNA HOTAIRM1, which was found to be highly expressed in the fetal brain, was also found to be increased with ascending glioma grade [11]. The fetal lncRNA reactivated in high grade glioma suggested a critical role in malignant progression. The other lncRNA CRNDE was found to be up-regulated in glioma cell line. The siRNA-mediated knock down of it could promote cell apoptosis and inhibit cell proliferation. It suggested that CRNDE contributed to cell apoptosis and cell proliferation of glioma. All of these results revealed the potential roles of lncRNAs in glioma and their relationship to glioma malignant properties. These differentially expressed lncRNAs, such as H19, HOTAIRM1, CRNDE, in this study may relate to the recurrence of glioma.
For the reason that the functions of most lncRNAs have not been well studied, there is no data base can be applied to identify the functional annotations of lncRNAs. In order to find the functional roles of the differentially expressed lncRNAs, we performed GO and pathway analysis based on the differentially expressed mRNAs which are the potential targets of lncRNAs. Using this method, we showed the possible functions of the differentially expressed lncRNAs indirectly. The GO results gave us an overview of the biological process, cellular component and molecular function of the differentially expressed mRNAs. Most of the items were involved in critical processes in cancer pathogenesis such as protein binding, signal transducer activity, protein binding, etc. The pathway analysis showed that the potential targets were involved in many cancer related pathways including MAPK signaling pathway, pathway in cancer, etc. These results indicated the general functional roles of the differentially expressed lncRNAs in glioma recurrence.
In addition to the GO and pathway analysis, we performed lncRNA classification to screen three clusters of lncRNAs with potential gene regulatory function. Since lncRNAs can regulate gene in cis pattern [22], identification of differentially expressed lncRNA and nearby gene pairs may help to find some important lncRNA-mRNA pairs that contribute to the recurrence of glioma. Thus, we performed lncRNA-mRNA co-expression analysis and found many differentially expressed lncRNA-mRNA pairs in the recurrent group compared with the primary group. The mRNA in lncRNA-mRNA pair may be the target of lncRNA. Some of these targets are closely related to cancer or neural differentiation. For example, the up-regulated mRNA FOXD4L1 (forkhead box protein D4-like 1) corresponding to up-regulated enhancer lncRNA AC016745.3 could up-regulate genes to maintain proliferative neural precursors in an immature state [23]. The down-regulated mRNA RPRM (reprimo) corresponding to down-regulated lncRNA XLOC_001711 was demonstrate to be a tumor suppressor in pituitary tumors [24]. The up-regulated sense mRNA MEIS2 (myeloid ecotropic insertion site 2) corresponding to up-regulated antisense lncRNA RP11-128A17.1 was reported to highly expressed in human neuroblastoma cell lines and promote cell proliferation [25]. It suggests that these differentially expressed lncRNA-mRNA pairs may also play critical roles in glioma recurrence.
Since TMZ is the first-line chemotherapeutic agent for the treatment of glioma, it is hard to collect the recurrent glioma sample that acquired from no TMZ treated patient. All of the three patients had received TMZ treatment after the first surgery. Thus, the confounding of TMZ treatment effect on expression profile should be considered. However, it remains to be assessed whether TMZ treatment influences the expression profile of lncRNA. Besides, the small sample size and lack of RT-PCR to validate the results may also affect the precision and reliability of this study. The results of this study need to be further validated in future.
In conclusion, this study for the first time revealed a set of lncRNAs that were differentially expressed in the recurrent gliomas compared with the primary gliomas. Some of these lncRNAs may play important roles in glioma recurrence, such as previously reported H19, CRNDE, HOTAIRM1 or unreported AC016745.3, XLOC_001711, RP11-128A17.1. This study set a basis for future researches on specific lncRNA which may contribute to the recurrence of glioma. Further studies on these lncRNAs will help to elucidate the mechanism of glioma recurrence at genetic level and find therapeutic targets for glioma patients.
Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (No. 81172398, No. 81270038 and No. 81201565). The authors want to thank KangCheng Bio-tech Inc, Shanghai, China for the assistance in microarray and bioinformatic analysis.
Disclosure of conflict of interest
None.
References
- 1.Lamborn KR, Yung WK, Chang SM, Wen PY, Cloughesy TF, DeAngelis LM, Robins HI, Lieberman FS, Fine HA, Fink KL, Junck L, Abrey L, Gilbert MR, Mehta M, Kuhn JG, Aldape KD, Hibberts J, Peterson PM, Prados MD. Progression-free survival: an important end point in evaluating therapy for recurrent high-grade gliomas. Neuro Oncol. 2008;10:162–170. doi: 10.1215/15228517-2007-062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 4.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 5.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 9.Bian EB, Li J, Xie YS, Zong G, Zhao B. LncRNAs: New players in gliomas, with special emphasis on the interaction of lncrnas with EZH2. J Cell Physiol. 2015;230:496–503. doi: 10.1002/jcp.24549. [DOI] [PubMed] [Google Scholar]
- 10.Sun Y, Wang Z, Zhou D. Long non-coding RNAs as potential biomarkers and therapeutic targets for gliomas. Med Hypotheses. 2013;81:319–321. doi: 10.1016/j.mehy.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan Y, Zhang L, Jiang Y, Xu T, Mei Q, Wang H, Qin R, Zou Y, Hu G, Chen J, Lu Y. LncRNA and mRNA interaction study based on transcriptome profiles reveals potential core genes in the pathogenesis of human glioblastoma multiforme. J Cancer Res Clin Oncol. 2014;141:827–38. doi: 10.1007/s00432-014-1861-6. [DOI] [PubMed] [Google Scholar]
- 18.Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu S, Zhang A, Jia Z, Wang G, Yu S, Pu P, Dong L, Kang C. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40:2004–2012. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 19.Qin X, Yao J, Geng P, Fu X, Xue J, Zhang Z. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int J Clin Exp Pathol. 2014;7:3065–3072. [PMC free article] [PubMed] [Google Scholar]
- 20.Yao J, Zhou B, Zhang J, Geng P, Liu K, Zhu Y, Zhu W. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biol. 2014;35:7935–7944. doi: 10.1007/s13277-014-1949-2. [DOI] [PubMed] [Google Scholar]
- 21.Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J, Xue YX. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal. 2015;27:275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Orom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Curr Opin Genet Dev. 2011;21:194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein SL, Neilson KM, Orban J, Yaklichkin S, Hoffbauer J, Mood K, Daar IO, Moody SA. Conserved structural domains in FoxD4L1, a neural forkhead box transcription factor, are required to repress or activate target genes. PLoS One. 2013;8:e61845. doi: 10.1371/journal.pone.0061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu M, Knox AJ, Michaelis KA, Kiseljak-Vassiliades K, Kleinschmidt-DeMasters BK, Lillehei KO, Wierman ME. Reprimo (RPRM) is a novel tumor suppressor in pituitary tumors and regulates survival, proliferation, and tumorigenicity. Endocrinology. 2012;153:2963–2973. doi: 10.1210/en.2011-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zha Y, Xia Y, Ding J, Choi JH, Yang L, Dong Z, Yan C, Huang S, Ding HF. MEIS2 is essential for neuroblastoma cell survival and proliferation by transcriptional control of M-phase progression. Cell Death Dis. 2014;5:e1417. doi: 10.1038/cddis.2014.370. [DOI] [PMC free article] [PubMed] [Google Scholar]


