Abstract
Objective: This study was designed to investigate the clinical microbiology of fungal ball (FB) rhinosinusitis by culturing fungal clumps collected under endoscopic surgery. Methods: From April to November of 2012, fungal clumps were sampled by endoscopic surgery from patients diagnosed with FB using clinical and histopathological methods. The specimens were subjected to smear microscopy, and cultured for bacteria and fungi analysis. Results: Out of the 81 specimens from 80 patients, 69 (69/81, 85.19%) specimens were detected a mixed infection of bacteria and fungi. However, only 25 (25/81, 30.86%) specimens resulted in fungal growth. There were 12 (12/81, 14.81%) specimens with fungal infections alone. The cultured fungi included 36 strains belonging to five genera, and most of them were Aspergillus spp. (30/36, 83.3%). The cultured bacteria included 94 strains belonging to 16 genera, and the most frequently seen was Staphylococcus spp. (23/94, 35.94%). When it was fungal and Pseudomonas aeruginosa mixed infection, the fungal growth was inhibited (P = 0.002). Conclusion: Patients with fungal ball usually have mixed fungal and bacterial infections. The fungi from these samples are sometimes difficult to culture, which may be the result of the inhibition by bacteria in vitro and in vivo.
Keywords: Fungal ball, fungal rhinosinusitis, clinical microbiology, microbial culture, mixed infections
Introduction
Fungal ball (FB) rhinosinusitis is one of the main types of noninvasive fungal rhinosinusitis [1]. Along with fungi, bacteria are often isolated from the fungal clumps of patients with FB [2]. Fungal rhinosinusitis (FRS) is an important clinical problem with diverse manifestations. The disease was classified as being noninvasive or invasive based on whether the fungi have invaded the sinonasal submucosal tissue, which could result in tissue necrosis and destruction [3]. FB and allergic fungal rhinosinusitis (AFRS) are considered as the types of noninvasive FRS. Invasive disease is characterized as either acute or chronic based on the length of time the symptoms exist before presentation [4]. FB is the major form of noninvasive FRS in China [5]. FB can be cured only by surgical intervention [6]; however, the role of microorganisms in FB remains unknown. Aspergillus spp. is the most commonly reported microorganism to cause fungal balls of the sinuses [7]. In previous studies, researchers focused more on fungal culture and ignored bacterial culture of the samples from FB patients. Mixed infections of fungi and bacteria are often detected in clinical samples. Microorganisms or pathogens were also determined as one of the responsible agents in the etiopathogenesis of FB. Therefore, it is necessary to analyze the microbiological characteristics of FB, including fungi and bacteria.
The aim of this study was to characterize the microbes found in the fungal clumps of patients diagnosed with FB using histopathology. Traditional culture methods for fungi and bacteria and smear microscopy were used in this study.
Materials and methods
Samples
A total of 81 samples were obtained from 80 patients histologically diagnosed with fungal balls. The fungal clumps were sampled under endoscopy from April to November of 2012.
Microorganism cultures
All fungal clump samples were cut into small pieces (the size of each one is about 1 mm3) under sterile conditions. Pieces were cultured in liquid Sabouraud medium, agar slant Sabouraud medium and Sabouraud dextrose agar medium [8]. The media were incubated at 26°C for 3 weeks for fungal cultures. Additionally, pieces were cultured in blood agar medium, chocolate agar medium and Makanke medium (Oxoid, Basingstoke, UK). These media were incubated at 37°C for 72 hr for the culturing of aerobic bacteria. Other pieces were cultured in anaerobic media at 37°C under anaerobic conditions for 5 days to culture anaerobic bacteria. For bacteria, depending on the morphology, we selected colonies and identified the species using MALDI-TOF MS (Bruker Daltonics, Bremen, Germany). As for fungi, we selected colonies and identified them by phenotype depending on the morphology.
Fungal smears
Fungal smears of each specimen were subjected to lactic acid phenol Medan staining and 10% KOH staining. A small piece of the fungal clump (the size of the small piece clump is about 1 mm3) was placed on a slide and pressed with a coverslip. The coverslip was removed, and a drop of lactic acid phenol Medan dye or 10% KOH was added, and then a new coverslip was placed on top.
Bacterial smears
A small piece of the fungal clump was smeared on a glass slide, dried and fixed, and Gram stained.
Statistical analysis
SPSS 17.0 statistical software was used for data analysis (SPSS, Chicago, IL, USA). Continuous values were expressed as mean ± standard deviation and analyzed with t-tests. Categorical variables are presented as absolute numbers and analyzed by Chi-square test. Two-sided p-values < 0.05 indicated statistical significance.
Results
Demographic data
The ages of the 80 patients ranged from 9 to 78 years old (mean age: 52.98 ± 11.72). The study group was made up of 24 (30%) males and 56 (70%) females. In total, 57 (71.25%) cases occurred in the maxillary sinus, 16 (20%) in the sphenoid sinus, 5 (6.25%) in the ethmoid sinus, 1 (1.25%) in the nasal sphenoethmoidal recess and 1 (1.25%) in the side of the maxillary sinus and contralateral ethmoid (Table 1).
Table 1.
Clinical summary of patients with fungal ball rhinosinusitis
| Age | n (%) | M:F | Site of infection | Average course of disease (months) | No. of mixed infection (n, %) | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Maxillarysinus | Sphenoid sinus | Eethmoid sinus | |||||
| ≤ 29 | 3 (3.8) | 1:2 | 2 | - | 1 | 34.7 | 1 (1.4) |
| 30-59 | 54 (67.5) | 15:39 | 40 | 10 | 4 | 32.7 | 47 (68.1) |
| ≥ 60 | 23 (28.2) | 7:16 | 16 | 6 | 1 | 43.7 | 20 (28.9) |
Footnote: M:F = Male : Female.
Microbial cultures
Of the 81 specimens from the 80 patients, smear microscopy detected mixed bacterial and fungal infections in 69 (69/81, 85.19%) samples; however, only 25 (25/81, 30.86%) specimens showed fungal culture growth. The other 44 (44/81, 54.32%) mixed infection samples did not display fungal colony growth. There were 12 (12/81, 14.81%) specimens with only fungal infections. The cultured fungi included 36 strains belonged to 6 genera, and the most often seen was Aspergillus spp. (30/36, 83.3%) (Table 2). The cultured bacteria included 94 strains belonged to 19 genera, and the most often seen were Staphylococcus spp. (23/94, 35.94%) (Table 2). There were no patients with a positive anaerobic bacteria culture. Patients with FB usually had mixed fungal and bacterial infections. The fungi from these samples were difficult to culture, which may be the result of the bacterial inhibition in vitro and in vivo (Figure 1). When the mixed infection contained P. aeruginosa, the ability to culture the fungi was lower than when a different bacteria was present (P = 0.002). Additionally, the phenomenon of sparse and even broken mycelium (shading poor) in specimens with mixed fungal and bacterial infections was found by smear microscopy (Figure 2).
Table 2.
Microorganisms summary of fungal ball rhinosinusitis
| Fungal culture-negative in mixed infection | Fungal culture-positive in mixed infection | Fungal infections alone | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Bacteria (Strain) | Bacteria (Strain) | Fungi (Strain) | Fungi (Strain) | ||||
| Staphylococcus sp | 23 | Proteus | 1 | Candida | 1 | Aspergillus flavus | 7 |
| Pseudomonas sp. | 10 | Staphylococcus sp. | 18 | A. flavus | 13 | Aspergillus fumigatus | 2 |
| Streptococcus sp | 8 | Streptococcussp | 4 | A. fumigatus | 7 | Aspergillus versicolor | 2 |
| Klebsiellasp | 7 | Klebsiellasp | 1 | Scedosporium apiospermum | 1 | Schizophyllum commune | 1 |
| Enterobacter sp. | 3 | Citrobactersp | 1 | A. versicolor | 1 | ||
| Acinetobacter sp. | 2 | Corynebacterium sp. | 1 | S. commune | 1 | ||
| Corynebacteriumsp | 2 | Brenham spp. | 1 | Alternaria alternate | 1 | ||
| Neisseria sp. | 1 | Haemophilussp | 1 | ||||
| Haemophilus sp. | 1 | Acinetobactersp | 1 | ||||
| Escherichia sp. | 1 | Streptomycessp | 1 | ||||
| Citrobacter sp. | 1 | ||||||
| Serratia sp. | 1 | ||||||
| Eikenella sp. | 1 | ||||||
| Leclercia sp. | 1 | ||||||
| Stenotrophomonas sp. | 1 | ||||||
| Fusobacteriumsp | 1 | ||||||
| 16 | 64 | 10 | 30 | 7 | 25 | 4 | 12 |
Figure 1.

Summary of bacteria in mixed infection and their role.
Figure 2.
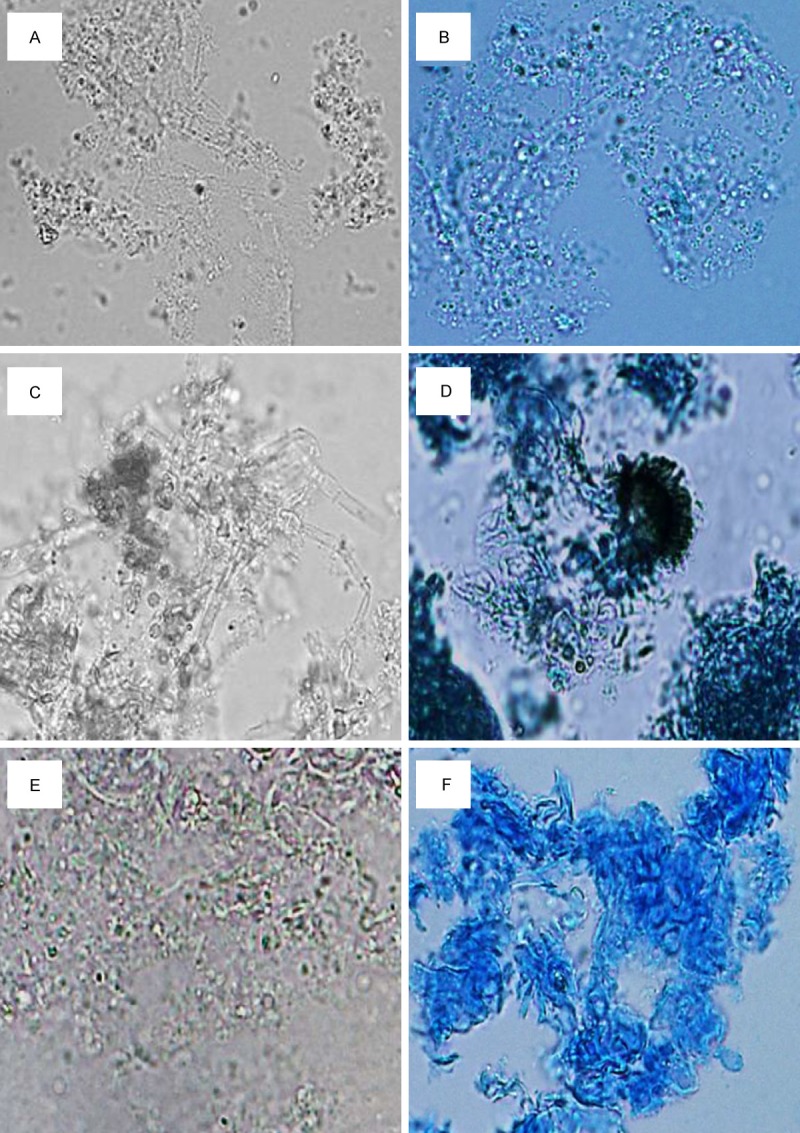
Fungal smears under light microscope. A-F: Fungal smears under light microscope with lactic acid phenol Medan staining and 10% KOH staining. A, B: In mixed fungal and P. aeruginosa infection, sparse fungal mycelium shading poor, fewer spores can be found. C, D: In mixed fungal and the other bacterial infection, thicker hyphae shading good, more spores and even conidia can be found, with active hyperplasia. E, F: In fungal infections alone, dense mycelium tightly wound, shading good, and dyed dark coloring by lactic acid phenol Medan.
Discussion
Fungal sinusitis is classified as invasive (acute invasive fungal sinusitis, chronic invasive fungal sinusitis, and granulomatous invasive fungal sinusitis) and noninvasive (allergic fungal sinusitis and sinonasal fungal ball). A fungal ball is described as the noninvasive accumulation of a dense conglomeration of fungal hyphae in one sinus cavity, usually the maxillary sinus. However, the disease may affect other sinuses and resulted in multiple sinuses [1,9]. The results of our study, in which 71.25% of cases involved the maxillary sinus, were consistent with previous studies. Fungal sinusitis is more common in middle-aged and elder female individuals in contrast to all forms of invasive and chronic aspergillosis, which are more common in male individuals [3,4,10]. Again, our results (female and male ratio of 2.3:1) are consistent with previous findings.
Sinonasal fungal ball is known to be the most common form of fungal sinusitis, and it can be completely cured by proper surgical removal [3]. Currently, the definitive diagnosis of sinonasal fungal ball is made by histopathological finding of a high number of hyphae and mycotic colonization and is confirmed by culture of specimens. Culture of pathogens is critical for the correct diagnosis and appropriate clinical therapy of fungal balls [11]. In our study, Aspergillus was the most commonly found fungi in fungal balls, and this result is consistent with previous studies [12]. Dufour et al. reported that most fungal balls comprise mixed fungal and bacterial infections [13]. However, a majority of studies have focused on fungal cultures, and ignored bacterial cultures. In our study, 69 (85.19%) samples comprised mixed bacterial and fungal infections, and only 25 (30.86%) samples were fungal culture-positive. These results confirmed that the fungal culture-positive rate of fungal balls was low and that fungi were difficult to culture in vitro [7]. Molecular biological examination should be recommended for identification of the fungal balls that cannot be cultured. Previous studies confirmed that molecular technique of internal transcribed spacer sequencing used to identify pathogenic fungal species from fungal balls can improve the culture-positive rate to 80% [14]. In addition, Willinger et al. reported that PCR amplification and sequence analysis with universal fungal primers for 28 s ribosomal DNA was a more sensitive technique than hybridization and culture [7]. Thus, molecular biological methods can compensate for the lack of a positive fungal ball culture. Such methods are also important for an etiological diagnosis of fungal ball rhinosinusitis. Therefore, Metagenomics approach will be used to analyze the etiology of fungal balls in our next study.
Importantly, the present study revealed that in fungal balls formed by mixed fungal and P. aeruginosa infection, the positive fungal culture rate is significantly lower than that involved other bacteria. Smear microscopy revealed sparse and even broken mycelium within the fungal balls formed by mixed fungal and bacterial infections, especially those involved P. aeruginosa. The reason for this phenomenon may be due to that bacteria, specifically P. aeruginosa, can inhibit fungal growth both in vivo and in vitro [15]. Interactions between bacteria and fungi have dramatic effects on the survival, colonization, and pathogenesis of these organisms. Some bacteria could provide fungi with compounds that enhance the production of fungal virulence determinants. Other bacteria produce factors that are likely to inhibit pathogenesis by repressing fungal filamentation [16]. For example, the opportunistic pathogen P. aeruginosa appeared to inhibit the growth of both Candida albicans and dermatophyte fungi [17]. Such interactions could have evolved through competition with fungi in soil, in association with plants, or in the context of host-associated infections [18]. Our result was consistent with the results of previous studies. The 3-oxo-C12-acyl homoserine lactone-signaling molecule produced by P. aeruginosa was reported to inhibit C. albicans hyphal formation [19]. Another possible mechanism involved the fact that most bacteria can produce chitinase, which can break down fungal cell wall component to inhibit fungal growth [20]. Furthermore, some mixed bacterial-fungal biofilms have properties that are distinct from their single-species counterparts. Therefore, interactions between bacteria and fungi in fungal balls should be paid more attention. Clinical studies of the interactions between bacteria and fungi in fungal balls in combination with in vitro model systems are necessary to understand how bacterial-fungal interactions affect the development of sinonasal fungal balls in vivo. In-depth analysis of the etiology of fungal balls using metagenomics methods will help to understand the bacterial-fungal interactions within fungal balls.
Acknowledgements
This study was supported by National Natural Science Foundation of China (Grant No. 81070768) and Wu Jieping Clinical Research Special Assistance Fund (No. 32067501294).
Disclosure of conflict of interest
None.
References
- 1.Montone KT, Livolsi VA, Feldman MD, Palmer J, Chiu AG, Lanza DC. Fungal rhinosinusitis: a retrospective microbiologic and pathologic review of 400 patients at a single university medical center. Int J Otolaryngol. 2012;2012:684835. doi: 10.1155/2012/684835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang JH, Lee BJ, Jang YJ. Bacterial coinfection and antimicrobial resistance in patients with paranasal sinus fungus balls. Ann Otol Rhinol Laryngol. 2010;119:406–411. doi: 10.1177/000348941011900608. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti A, Denning DW, Ferguson BJ, Ponikau J, Buzina W, Kita H. Fungal rhinosinusitis: a categorization and definitional schema addressing current controversies. Laryngoscope. 2009;119:1809–1818. doi: 10.1002/lary.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driemel O, Wagner C, Hurrass S, Muller-Richter U, Kuhnel T, Reichert TE, Reichert TE, Kosmehl H. [Allergic fungal sinusitis, fungus ball and invasive sinonasal mycosis - three fungal-related diseases] . Mund Kiefer Gesichtschir. 2007;11:153–159. doi: 10.1007/s10006-007-0058-4. [DOI] [PubMed] [Google Scholar]
- 5.Yang X, Lei K. The clinical characters and acoustic rhinometry analyses of 98 cases fugal ball sinusitis. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2011;25:783–785. [PubMed] [Google Scholar]
- 6.Dufour X, Kauffmann-Lacroix C, Ferrie JC, Goujon JM, Rodier MH, Karkas A. Paranasal sinus fungus ball and surgery: a review of 175 cases. Rhinology. 2005;43:34–39. [PubMed] [Google Scholar]
- 7.Willinger B, Obradovic A, Selitsch B, Beck-Mannagetta J, Buzina W, Braun H. Detection and identification of fungi from fungus balls of the maxillary sinus by molecular techniques. J Clin Microbiol. 2003;41:581–585. doi: 10.1128/JCM.41.2.581-585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pine L, Peacock CL. Studies on the growth of histoplasma capsulatum. IV. Factors influencing conversion of the mycelial phase to the yeast phase. J Bacteriol. 1958;75:167–174. doi: 10.1128/jb.75.2.167-174.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Shazo RD, Chapin K, Swain RE. Fungal sinusitis. N Engl J Med. 1997;337:254–259. doi: 10.1056/NEJM199707243370407. [DOI] [PubMed] [Google Scholar]
- 10.Chakrabarti A, Das A, Panda NK. Controversies surrounding the categorization of fungal sinusitis. Med Mycol. 2009;47(Suppl 1):S299–308. doi: 10.1080/13693780802213357. [DOI] [PubMed] [Google Scholar]
- 11.Babinski D, Narozny W, Skorek A, Rzepko R, Stankiewicz C. Noninvasive fungal sinusitis (fungus ball)--diagnostic difficulties. Otolaryngol Pol. 2007;61:694–697. doi: 10.1016/S0030-6657(07)70508-5. [DOI] [PubMed] [Google Scholar]
- 12.Howard SJ, Cerar D, Anderson MJ, Albarrag A, Fisher MC, Pasqualotto AC, Laverdiere M, Arendrup MC, Perlin DS, Denning DW. Frequency and evolution of Azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis. 2009;15:1068–1076. doi: 10.3201/eid1507.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dufour X, Kauffmann-Lacroix C, Ferrie JC, Goujon JM, Rodier MH, Klossek JM. Paranasal sinus fungus ball: epidemiology, clinical features and diagnosis. A retrospective analysis of 173 cases from a single medical center in France, 1989-2002. Med Mycol. 2006;44:61–67. doi: 10.1080/13693780500235728. [DOI] [PubMed] [Google Scholar]
- 14.Li LL, Zhao ZT, Wan Z, Li RY, Liu HG. Application of PCR combined with reverse line blot assay in detection and identification of common pathogenic Aspergillus in fungal sinusitis. Zhonghua Bing Li Xue Za Zhi. 2012;41:6–10. [PubMed] [Google Scholar]
- 15.Kerr JR, Taylor GW, Rutman A, Hoiby N, Cole PJ, Wilson R. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol. 1999;52:385–387. doi: 10.1136/jcp.52.5.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wargo MJ, Hogan DA. Fungal--bacterial interactions: a mixed bag of mingling microbes. Curr Opin Microbiol. 2006;9:359–364. doi: 10.1016/j.mib.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Z, Wang Q, Wang K, Brian K, Liu C, Gu Y. Study of the antifungal activity of Bacillus vallismortis ZZ185 in vitro and identification of its antifungal components. Bioresour Technol. 2010;101:292–297. doi: 10.1016/j.biortech.2009.07.071. [DOI] [PubMed] [Google Scholar]
- 19.Gould TA, Schweizer HP, Churchill ME. Structure of the Pseudomonas aeruginosa acylhomoserinelactone synthase Lasi. Mol Microbiol. 2004;53:1135–1146. doi: 10.1111/j.1365-2958.2004.04211.x. [DOI] [PubMed] [Google Scholar]
- 20.Guan Y, Ramalingam S, Nagegowda D, Taylor PW, Chye ML. Brassica juncea chitinase Bjchi1 inhibits growth of fungal phytopathogensand agglutinates Gram-negative bacteria. J Exp Bot. 2008;59:3475–3484. doi: 10.1093/jxb/ern197. [DOI] [PMC free article] [PubMed] [Google Scholar]


