Abstract
PTEN (10q23.3) is a negative regulator of the phosphatidylinositol 3-kinase (PIK3)/Akt survival pathway and a tumor suppressor frequently deleted in prostate cancer. PTEN genomic deletion is among the most common genetic aberrations in human prostate cancer. At present, the prognostic value of PTEN genomic deletion is unclear. We performed a systematic review and meta-analysis to clarify the association between PTEN genomic deletion and a higher Gleason score or a higher possibility of capsular penetration. A comprehensive, computerized literature search of PubMed was carried out until May 27, 2014. Studies were included according to specific inclusion criteria. Pooled hazard ratio was estimated using the fixed effects model or random effects model according to heterogeneity between studies. Seven eligible studies meeting the specific inclusion criteria were selected for further analysis; all were retrospective studies. Overall meta-analysis demonstrated that PTEN genomic deletion was associated with a higher Gleason score (OR 0.319; 95% confidence interval: 0.153-0.666; P = 0.000) and a higher possibility of capsular penetration (OR 0.393; 95% confidence interval: 0.185-0.837; P = 0.015). None of the studies materially altered the original results and no evidence of publication bias was found. Conclusion: PTEN genomic deletion in operable localized prostate cancer indicates a higher Gleason score and a higher probability of capsular penetration, indicating a worse prognosis. Further studies should be conducted in order to investigate the effect of PTEN genomic deletion on clinical outcomes in different histological types of prostate cancer or its function in castration-resistant prostate cancer.
Keywords: Prostate cancer, PTEN deletion, Gleason score, capsular penetration
Introduction
Prostate cancer (PCa) is the most commonly diagnosed cancer and a leading cause of cancer-related death in American men in addition to strongly affecting men all over the world [1]. With a more comprehensive understanding of PCa and new protocols for treatment, the outcome for PCa patients has improved in the past few decades. However, we are still not completely aware of the factors that affect the prognosis of patients with PCa. Identifying potential biomarkers that could serve as prognostic factors for PCa patients is crucial for individual treatment. Several biomarkers have been demonstrated to affect the survival of PCa patients so far, including androgen receptor variants [2], circulating tumor cells [3], and cysteine-rich secretory protein 3 [4]. PTEN genomic deletion has been detected in human tissues representing all stages of PCa development [5] and progression including high-grade prostatic intraepithelial neoplasia (HGPIN) [6], clinically localized PCa, metastatic PCa, and castration-resistant prostate cancer (CRPC) [7]. To clarify the association of PTEN deletion with the Gleason score and capsular penetration in patients with PCa, we conducted the first comprehensive meta-analysis of the existing published literature on this topic in patients with operable localized PCa.
Materials and methods
Literature search
A comprehensive, computerized literature search of PubMed was carried out until May 27, 2014. Potentially relevant studies were identified using “prostate cancer” (i.e., “prostate cancer”, “prostate carcinoma”, “prostate neoplasm”) and “PTEN”, “PI3K” and “pAkt” groups of search terms. The references from relevant papers, especially from review articles, were checked to identify studies overlooked in the original search. This systematic review and meta-analysis was planned, conducted, and reported in adherence to the standards of quality for reporting meta-analyses. Studies meeting all of the following inclusion criteria were deemed eligible and included in the analysis: (1) published in English, and (2) explored the relation between PTEN deletion and pathological outcome of operable localized PCa. All studies that did not satisfy the inclusion criteria as well as any data obtained from reviews, animal experiments, or cell line studies were excluded. Study quality was assessed using the Newcastle-Ottawa Scale. A flowchart of the literature search, study selection, and results of each step is presented in Figure 1.
Figure 1.
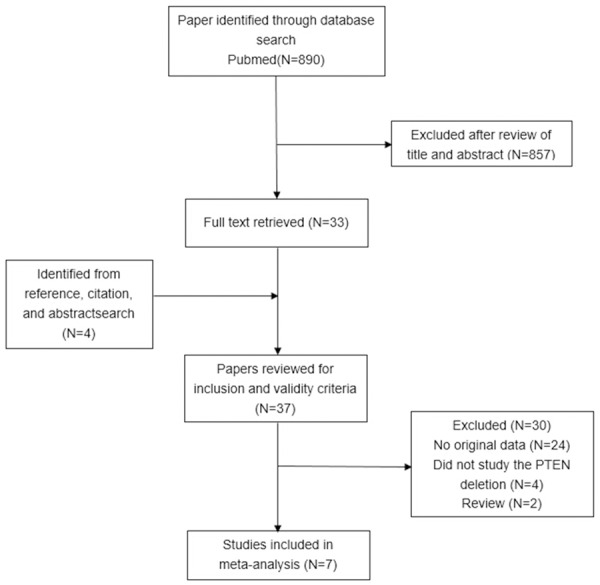
The literature search process. Notes: Eight hundred and ninety studies were identified in the primary literature search. 37 potentially relevant studies were further evaluated and seven studies were finally included in the analysis according to the inclusion criteria.
Data extraction and outcomes
In order to ensure homogeneity of the data gathering and to preclude subjectivity in the data collection and entry, two reviewers independently assessed studies for inclusion, and disagreements were resolved through open discussion. The following information about each study was recorded: first author names, journal and year of publication, patient nationality, total number of patients, median age of patients at diagnosis, the median stage, the median Gleason score, and the number of patients with PTEN deletion.
Statistical analysis
First, we assessed the heterogeneity between studies using the Q-test and I2 statistic to measure the proportion of total estimate variation that was attributable to study heterogeneity, and either a P-value < 0.10 or I2 > 50% was considered statistically significant. The pooled HR was estimated using the fixed effects model unless heterogeneity was found and was unexplainable, in which case, the random effects model was applied. We used the random effects model to analyze the relationship between PTEN deletion and Gleason score as the heterogeneity between studies was statistically significant (I2 = 82.6%; P = 0.000). And the random effects model was also applied to analyze the relationship between PTEN deletion and capsular penetration as the heterogeneity between studies was statistically significant (I2 = 69.7%; P= 0.019). We performed a sensitivity analysis by removing each individual study from the meta-analysis. Several methods were used to assess potential publication bias. Potential bias of publication was examined by using the Begg funnel plot and Egger linear regression test (All reported P values were two-sided, and P values < 0.05 were considered statistically significant). All statistical analyses performed in this study were carried out using Stata software (v 12.0; StataCorp LP, College Station, TX, USA).
Results
The literature search process and the result of each step are presented in Figure 1. Studies were identified in the primary literature, of which 37 potentially relevant studies were further evaluated after review of their titles and abstracts. A total of seven studies meeting the inclusion criteria were finally included in this study. The main characteristics of the eligible studies, all of which were retrospective cohort studies, are shown in Table 1. The analyzed studies were published between 1999 and 2013. All seven studies reported the relationship between PTEN deletion and a detailed cancer Gleason score (or sufficient data by which these could be calculated) [8-14], while four of them analyzed the relationship between PTEN deletion and capsular penetration [9,11-13]. One study defined the PTEN classification as three different types which include positive, negative and mixed [8]. So we excluded the patients who were divided into the group of “mixed”. Of all the studies analyzed, six studies presented a less PTEN deletion rate than positive rate while only one study reported the opposite. Data on the percentage of PTEN genomic deletion associated with PCa biochemical recurrence were also recorded. However, we were unable to obtain sufficient data to render any further analysis.
Table 1.
Studies included in this meta-analysis
| Study | Country | Number of patients | Median patient age, years | Treatment | ethnicity | PTEN positive rate | QUADAS score |
|---|---|---|---|---|---|---|---|
| Mcmenamin (1999) | USA | 39 | 66 | radical prostatectomy | Caucasian | 17/39 | 12 |
| Reid (2010) | UK | 322 | 69 | TURPT | Caucasian | 266/322 | 11 |
| Bismar (2010) | USA | 659 | 64 | radical prostatectomy | Caucasian | 454/659 | 12 |
| Yoshimoto (2007) | Canada | 107 | 63 | radical prostatectomy | Caucasian | 60/107 | 12 |
| Lotan (2011) | USA | 397 | * | radical prostatectomy | Caucasian | 254/397 | 10 |
| Nagle (2013) | USA | 90 | 63 | radical prostatectomy | Caucasian | 71/90 | 12 |
| Tina (2004) | Germany | 86 | 63 | radical prostatectomy | Caucasian | 48/86 | 12 |
Figure 2A presents a forest plot of meta-analysis for the Gleason score, including OR, 95% CIs, and the weight of each study in the analysis. As the heterogeneity between studies was statistically significant (I2 = 82.6%; P = 0.000), the random effects model was applied. The combined OR was 0.319 (95% CI: 0.153-0.666; P = 0.000). To further test the robustness of our study, we performed publication bias analysis by Begg’s test (Figure 3A) and found no evidence of publication bias (P = 0.133). The result indicates that loss of PTEN expression in PCa correlates with a higher Gleason score. Figure 2B presents the forest plot of meta-analysis for the possibility of capsular penetration, also including OR, 95% CIs, and the weight of each study in the analysis. As the heterogeneity between studies was not statistically significant (I2 = 69.7%; P = 0.019), the random effects model was applied. The combined OR was 0.393 (95% CI: 0.185-0.837; P = 0.015). We also performed a publication bias analysis by Begg’s test (Figure 3B), which showed no evidence of publication bias (P = 0.734). Thus, the loss of PTEN expression in PCa correlates with a higher possibility of capsular penetration and a more advanced pathological stage. To further test the robustness of our study, we also performed sensitivity analysis by omitting one study each time. We found that no single study altered the original results significantly.
Figure 2.
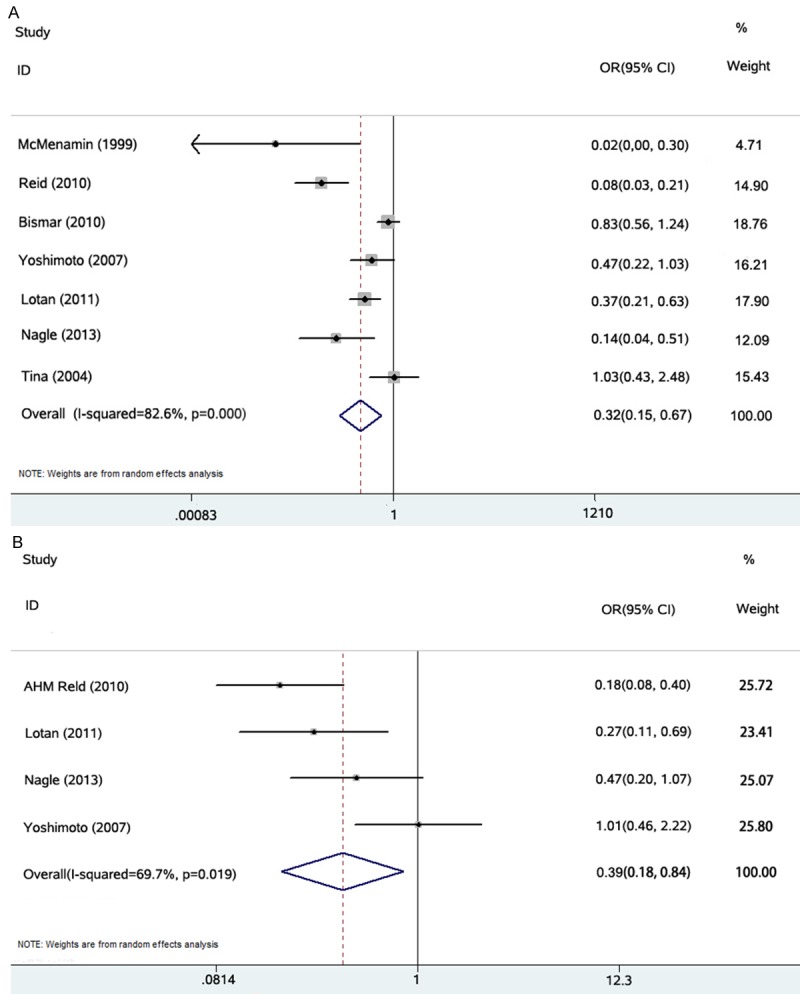
A. Individual study and overall ORs of relationships between PTEN deletion and higher Gleason score (≥ 7). B. Individual study and overall ORs of relationships between PTEN deletion and capsular penetration.
Figure 3.
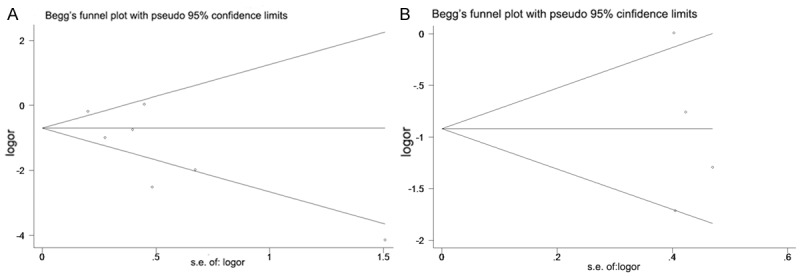
A. Test of publication bias of the analysis of PTEN deletion and higher Gleason score (≥ 7). B. Test of publication bias of the analysis of PTEN deletion and capsular penetration.
Discussion
PTEN loss is proposed to be a critically important and frequently occurring molecular event in prostate carcinogenesis. PTEN targets proteins in signaling pathways that regulate cell growth, survival, and genome stability [15]. PTEN is a phosphoinositide 3-phosphatase located on chromosome 10 and acts as a tumor suppressor gene by negatively regulating the PI3K signaling pathway [16]. It is a negative regulator of the phosphatidylinositol 3-kinase (PIK3)/Akt survival pathway and a tumor suppressor frequently deleted in PCa [17]. The deletion of PTEN has been detected in human tissues representing all stages of PCa development and progression [18], and initiates numerous signaling events involved in oncogenesis including cell proliferation, cell invasion, metastasis, and survival [19-23]. Numerous aberrations of the PI3K-Akt pathway, which contains PTEN deletion has been observed in several human malignancies including PCa, breast cancer, gastric cancer, and colorectal cancer [24-28]. Previous studies using loss of heterozygosity analyses of 10q deletions showed that PTEN loss is present in nearly 50% cases of advanced PCas [8,29], and in approximately 40% of localized PCas [30]. In advanced disease, fluorescence in situ hybridization has identified hemizygous and homozygous PTEN deletions, with the incidence of PTEN deletion approaching 70-80% in CRPC [31]. In particular, recent publications have confirmed the relationship between PTEN deletion and tumor growth in mouse models. Moreover, even though the function and mechanism of PTEN in the human body has not yet been fully elucidated, we can still affirm the value of PTEN from previous studies. For example, some studies have examined the prognostic significance of PTEN deletions by fluorescence in situ hybridization with small patient cohorts and biochemical recurrence as the outcome [32,33]. Although there is an abundance of information on the association of these genomic changes and clinical outcomes in PCa [34], data on their distribution in individual Gleason grades and capsular penetration are limited. Some studies found that in PCa, decreased expression of PTEN is associated with high Gleason score and more advanced stage tumors, suggesting that PTEN gene alterations may be associated with tumor progression [10,11,35-38]. However, other studies have shown that patients’ prognosis cannot be predicted using PTEN loss alone by analysis of the relationship between Gleason score, TNM stage, and PTEN deletion [14]. Since the results from previous studies are inconclusive, we performed a systematic review and meta-analysis to clarify the relationship between PTEN deletion and PCa grade and stage. In our analysis, we formulated a unified standard where only the patients with operable localized PCa were included. Seven eligible studies with PTEN deletion detected by immunohistochemistry were included in this study. Finally, we concluded that PTEN genomic deletion was associated with a higher Gleason score and a higher possibility of capsular penetration. PTEN is also related to the differentiation and invasion of PCa cells; its deletion indicates a poor prognosis. PTEN deletion can be a good biomarker to judge the prognosis of patients. Moreover, we also analyzed the relationship between PTEN deletion and seminal vesicle invasion, using three studies, which published relevant data and determined that there is no statistical significance between PTEN deletion and seminal vesicle invasion (Figure 4).
Figure 4.
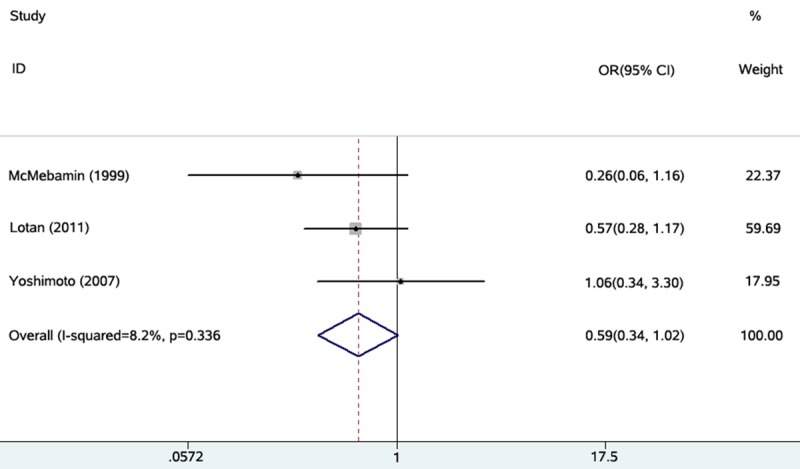
Individual study and overall ORs of relationships between PTEN deletion and seminal vesicle invasion.
In our meta-analysis, even though all studies used IHC staining to assess PTEN expression, seven eligible studies that included the Gleason score analysis showed heterogeneity, and four studies that included the capsular penetration associate analysis were without heterogeneity. Although IHC analysis is simple and cost-effective, tremendous variation exists in the experimental procedures, which may influence the results and may in part be responsible for the observed heterogeneity. For example, when restricted to studies using IHC staining with the same antibody, patients with reduced PTEN expression were related to a higher Gleason score and capsular penetration, and the heterogeneity between studies may reduce. The clinical significance of this study includes the following: First, this analysis solved the contradictory results that exist in previous research, and confirmed that PTEN plays an important role during the development of PCa. PTEN deletion indicates a worse prognosis and results in faster PCa progression. Second, for patients with clinically localized prostate cancer which requires radical surgery, examining the PTEN deletion status should be recommended for providing more information to determine patient prognosis. Third, PTEN and its downstream Akt signaling pathways may become a treatment target in the future.
Our study is not devoid of limitations. First, the number of studies included in our analysis was small, and all of the included studies were retrospective, indicating low levels of evidence in evidence-based medicine. Second, our meta-analysis was based on data only from studies meeting our inclusion criteria, and there were many other published studies that did not meet these criteria. In addition, we could not obtain updated data on individual patients. The use of individual patient data could further enhance the accuracy and reduce the uncertainty of our estimates. Third, all the tissues in our analysis were from patients with clinically localized PCa who underwent radical prostatectomy; patients with a more advanced stage of PCa and patients with CRPC were not included in our analysis. Finally, publication bias may also be a concern. It was unavoidable that some data would remain unobtainable even after we tried to identify all relevant information. However, after examining the Begg funnel plots and performing the Egger linear regression test, we found that the association between PIK3CA mutation and clinical outcome remained unchanged.
In conclusion, the findings of our meta-analysis support PTEN as a tumor suppressor gene in PCa progression. Indeed, loss of PTEN expression can be an important negative prognostic indicator. Furthermore, the loss of PTEN induced by a majority of the mechanisms through which gene products are inactivated can be detected using immunohistochemistry.
Acknowledgements
This study was supported in part by the Grants for International Cooperation and Exchange of Science and Technology Commission of Shanghai Municipality (No. 12410709300), grants from Guide Project of Science and Technology Commission of Shanghai Municipality (No. 124119a7300), and Grants from Outstanding young talent training plan of Shanghai Municipal Commission of Health and Family Planning (No. XYQ2013102).
Disclosure of conflict of interest
None.
References
- 1.Nwosu V, Carpten J, Trent JM, Sheridan R. Heterogeneity of genetic alterations in prostate cancer: evidence of the complex nature of the disease. Hum Mol Genet. 2001;10:2313–2318. doi: 10.1093/hmg/10.20.2313. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, Morrissey C, Sun S, Ketchandji M, Nelson PS, True LD, Vakar-Lopez F, Vessella RL, Plymate SR. Androgen receptor variants occur frequently in castration resistant prostate cancer metastases. PLoS One. 2011;6:e27970. doi: 10.1371/journal.pone.0027970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reeves F, Sapre N, Corcoran N, Hovens C. Tumor vascularity in prostate cancer: an update on circulating endothelial cells and platelets as noninvasive biomarkers. Biomark Med. 2013;7:879–891. doi: 10.2217/bmm.13.100. [DOI] [PubMed] [Google Scholar]
- 4.Hoogland AM, Dahlman A, Vissers KJ, Wolters T, Schroder FH, Roobol MJ, Bjartell AS, van Leenders GJ. Cysteine-rich secretory protein 3 and beta-microseminoprotein on prostate cancer needle biopsies do not have predictive value for subsequent prostatectomy outcome. BJU Int. 2011;108:1356–1362. doi: 10.1111/j.1464-410X.2010.10059.x. [DOI] [PubMed] [Google Scholar]
- 5.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman JG, Isaacs WB, Bova GS, Sidransky D. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 6.Yoshimoto M, Cutz JC, Nuin PA, Joshua AM, Bayani J, Evans AJ, Zielenska M, Squire JA. Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet Cytogenet. 2006;169:128–137. doi: 10.1016/j.cancergencyto.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H, Freije D, Nusskern DR, Okami K, Cairns P, Sidransky D, Isaacs WB, Bova GS. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res. 1998;58:204–209. [PubMed] [Google Scholar]
- 8.McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high gleason score and advanced stage. Cancer Res. 1999;59:4291–4296. [PubMed] [Google Scholar]
- 9.Reid AH, Attard G, Ambroisine L, Fisher G, Kovacs G, Brewer D, Clark J, Flohr P, Edwards S, Berney DM, Foster CS, Fletcher A, Gerald WL, Moller H, Reuter VE, Scardino PT, Cuzick J, de Bono JS, Cooper CS, Grp TP. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer. 2010;102:678–684. doi: 10.1038/sj.bjc.6605554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bismar TA, Yoshimoto M, Vollmer RT, Duan QL, Firszt M, Corcos J, Squire JA. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int. 2011;107:477–485. doi: 10.1111/j.1464-410X.2010.09470.x. [DOI] [PubMed] [Google Scholar]
- 11.Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, Squire JA. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97:678–685. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu WN, Xu JF, Hicks JL, Park BH, Humphreys E, Partin AW, Han M, Netto GJ, Isaacs WB, De Marzo AM. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17:6563–6573. doi: 10.1158/1078-0432.CCR-11-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagle RB, Algotar AM, Cortez CC, Smith K, Jones C, Sathyanarayana UG, Yun S, Riley J, Nagy D, Dittamore R, Dalkin B, Brosh L, Pestano G. ERG Overexpression and PTEN Status Predict Capsular Penetration in Prostate Carcinoma. Prostate. 2013;73:1233–1240. doi: 10.1002/pros.22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreher T, Zentgraf H, Abel U, Kappeler A, Michel MS, Bleyl U, Grobholz R. Reduction of PTEN and p27kip1 expression correlates with tumor grade in prostate cancer. Analysis in radical prostatectomy specimens and needle biopsies. Virchows Arch. 2004;444:509–517. doi: 10.1007/s00428-004-1004-6. [DOI] [PubMed] [Google Scholar]
- 15.Verhagen PC, van Duijn PW, Hermans KG, Looljenga LH, van Gurp RJ, Stoop H, van der Kwast TH, Trapman J. The PTEN gene in locally progressive prostate cancer is preferentially inactivated by bi-allelic gene deletion. J Pathol. 2006;208:699–707. doi: 10.1002/path.1929. [DOI] [PubMed] [Google Scholar]
- 16.Feilotter HE, Nagai MA, Boag AH, Eng C, Mulligan LM. Analysis of PTEN and the 10q23 region in primary prostate carcinomas. Oncogene. 1998;16:1743–1748. doi: 10.1038/sj.onc.1200205. [DOI] [PubMed] [Google Scholar]
- 17.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 18.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J. Clin. Oncol. 2010;28:1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Ittmann MM, Ayala G, Tsai MJ, Amato RJ, Wheeler TM, Miles BJ, Kadmon D, Thompson TC. The emerging role of the PI3-K-Akt pathway in prostate cancer progression. Prostate Cancer Prostatic Dis. 2005;8:108–118. doi: 10.1038/sj.pcan.4500776. [DOI] [PubMed] [Google Scholar]
- 20.Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 21.Yoeli-Lerner M, Toker A. Akt/PKB signaling in cancer-a function in cell motility and invasion. Cell Cycle. 2006;5:603–605. doi: 10.4161/cc.5.6.2561. [DOI] [PubMed] [Google Scholar]
- 22.Denning G, Jean-Joseph B, Prince C, Durden DL, Vogt PK. A short N-terminal sequence of PTEN controls cytoplasmic localization and is required for suppression of cell growth. Oncogene. 2007;26:3930–3940. doi: 10.1038/sj.onc.1210175. [DOI] [PubMed] [Google Scholar]
- 23.Bader AG, Kang SY, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 24.Dan S, Okamura M, Seki M, Yamazaki K, Sugita H, Okui M, Mukai Y, Nishimura H, Asaka R, Nomura K, Ishikawa Y, Yamori T. Correlating phosphatidylinositol 3-kinase inhibitor efficacy with signaling pathway status: in silico and biological evaluations. Cancer Res. 2010;70:4982–4994. doi: 10.1158/0008-5472.CAN-09-4172. [DOI] [PubMed] [Google Scholar]
- 25.Kang SY, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai J, Xu LJ, Tang HJ, Yang Q, Yi XQ, Fang Y, Zhu Y, Wang ZH. The role of the pten/pi3k/akt pathway on prognosis in epithelial ovarian cancer: ameta-analysis. Oncologist. 2014;19:528–535. doi: 10.1634/theoncologist.2013-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong ML, Loh M, Thakkar B, Pang B, Iacopetta B, Soong R. Phosphatidylinositol-3-kinase pathway aberrations in gastric and colorectal cancer: meta-analysis, co-occurrence and ethnic variation. Int J Cancer. 2014;134:1232–1238. doi: 10.1002/ijc.28444. [DOI] [PubMed] [Google Scholar]
- 28.Zhu XH, Qin X, Fei MG, Hou WM, Greshock J, Bachman KE, Wooster R, Kang JH, Qin CY. Combined phosphatase and tensin homolog (pten) loss and fatty acid synthase (fas) overexpression worsens the prognosis of chinese patients with hepatocellular carcinoma. Int J Mol Sci. 2012;13:9980–9991. doi: 10.3390/ijms13089980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whang YE, Wu X, Suzuki H, Reiter RE, Tran C, Vessella RL, Said JW, Isaacs WB, Sawyers CL. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci U S A. 1998;95:5246–5250. doi: 10.1073/pnas.95.9.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshimoto M, Ludkovski O, DeGrace D, Williams JL, Evans A, Sircar K, Bismar TA, Nuin P, Squire JA. PTEN genomic deletions that characterize aggressive prostate cancer originate close to segmental duplications. Genes Chromosomes Cancer. 2012;51:149–160. doi: 10.1002/gcc.20939. [DOI] [PubMed] [Google Scholar]
- 31.Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, Chen L, Ewing CM, Eisenberger MA, Carducci MA, Nelson WG, Yegnasubramanian S, Luo J, Wang Y, Xu J, Isaacs WB, Visakorpi T, Bova GS. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bedolla R, Prihoda TJ, Kreisberg JI, Malik SN, Krishnegowda NK, Troyer DA, Ghosh PM. Determining risk of biochemical recurrence in prostate cancer by immunohistochemical detection of PTEN expression and Akt activation. Clin Cancer Res. 2007;13:3860–3867. doi: 10.1158/1078-0432.CCR-07-0091. [DOI] [PubMed] [Google Scholar]
- 33.Choucair K, Ejdelman J, Brimo F, Aprikian A, Chevalier S, Lapointe J. PTEN genomic deletion predicts prostate cancer recurrence and is associated with low AR expression and transcriptional activity. BMC Cancer. 2012;12:543. doi: 10.1186/1471-2407-12-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCall P, Witton CJ, Grimsley S, Nielsen KV, Edwards J. Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br J Cancer. 2008;99:1296–1301. doi: 10.1038/sj.bjc.6604680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sircar K, Yoshimoto M, Monzon FA, Koumakpayi IH, Katz RL, Khanna A, Alvarez K, Chen GY, Darnel AD, Aprikian AG, Saad F, Bismar TA, Squire JA. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol. 2009;218:505–513. doi: 10.1002/path.2559. [DOI] [PubMed] [Google Scholar]
- 36.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, Palanisamy N, Tomlins SA, Chinnaiyan AM, Shah RB. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22:1083–1093. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al Bashir S, Alshalalfa M, Hegazy SA, Dolph M, Donnelly B, Bismar TA. Cysteine-rich secretory protein 3 (CRISP3), ERG and PTEN define a molecular subtype of prostate cancer with implication to patients’ prognosis. J Hematol Oncol. 2014;7:21. doi: 10.1186/1756-8722-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]


