Abstract
Two highly homologous microRNAs (miRNAs, miRs), miR-222 and miR-221, act as a cluster in cellular regulation. We have previously reported that miR-221 promoted the growth of human non-small cell lung cancer cell line H460. However, the role of miR-222 in regulating the growth of H460 is unclear. H460 cells were transfected with miR-222 mimics, inhibitors or their negative controls and their effects were confirmed by Real-time quantitative reverse transcription polymerase chain reactions (qRT-PCRs). Cell viability was assessed by Cell Counting Kit-8 (CCK-8) while cell proliferation was determined by 5-Ethynyl-2’-deoxyuridine (EdU) assay. P27 and P57, two putative targets of miR-222, were checked by qRT-PCRs. We found that miR-222 overexpression increased cell viability and proliferative rate in H460 cells while opposite effects were obtained by down-regulation of miR-222. P27 but not P57 was identified as a potential target of miR-222 in H460 cells as P27 was negatively regulated by miR-222 in the protein level. In summary, the present study indicates that miR-222 controls the growth of H460 likely by targeting P27. Inhibition of miR-222 might be a novel therapy for human non-small cell lung cancer.
Keywords: MicroRNA, human non-small cell lung cancer, cell viability, proliferation
Introduction
Lung cancer is one of the leading causes of unnatural death worldwide. Among all types of lung cancers, non-small cell lung cancer (NSCLC) is the most prevalent one, which accounts for about 80% of total cases, including squamous cell carcinoma, adenocarcinoma, adenosquamous cell carcinoma, large cell carcinoma, and several other histologic types [1-3]. Despite the advances in medical technology including surgical therapy, radiotherapy, chemotherapy and targeting therapy, the prognosis for NSCLC patients is very poor as evidenced by the fact that the overall 5 year survival rate of NSCLC patients remains as low as 15% [3,4]. Besides that, it is estimated that lung cancer, especially NSCLC, will continue to be a major medical issue worldwide of mankind for the next 40 to 50 years [5]. Therefore, development of effective therapies for NSCLC is urgently needed.
MicroRNAs (miRNAs, miRs) are a group of small, non-coding RNAs, which regulate gene expression in a relatively flexible sequence-specific manner [6-10]. To date, thousands of miRs have been identified in almost all metazoan genomes, including worms, flies, plants and mammals. MiRNAs have diverse expression patterns (eg. tissue-specific or non-tissue-specific/intracellular or extracellular) and regulate various developmental and physiological processes [11]. miRNAs have been demonstrated to be involved in the initiation and development of a variety of cancers [12-16].
MiR-222 belongs to the miR-221/222 family, and has been reported to play a potential oncogenic role in multiple cancer types including hepatocellular carcinoma, breast cancer, prostate cancer, bladder cancer, endometrial carcinoma and glioblastoma [17-23]. A recent study has indicated that miR-222 overexpression was related to clinicopathological factors and prognosis of NSCLC [24], indicating that miR-222 may play crucial roles in NSCLC. In addition, we also reported that miR-221 promoted the growth of human non-small cell lung cancer cell line H460 [25]. However, the role of miR-222 in regulating the growth of H460 is unclear. Here we intended to investigate the role of miR-222 in regulating the growth of human non-small cell lung cancer cell line H460.
Methods
Cell culture and transient transfection
Human NSCLC H460 cell line was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). H460 cells were maintained in RAMI supplemented with 10% FBS (Hyclone, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin, at 37°C in a humidified atmosphere of 5% CO2.
miR-222 mimics, inhibitors and their negative controls were purchased from RiboBio (Guangzhou, China). H460 cells were starved for 6 hours before transfection. miR-222 mimics (50 nM), miR-222 inhibitors (100 nM) and their negative controls were transfected into H460 cells with Lipofectamine 2000 for 48 hours according to the manufacturer’s instructions.
Cell viability
Cell viability was determined with Cell Counting Kit 8 (CCK-8, Dojindo, Japan). In brief, H460 cells were planted into 96-well plate (2 × 103/well), and then cells were allowed to adhere overnight. After transfection of miR-222 mimics, inhibitors or their negative controls, 10 μL CCK-8 agent was added into each well. Cells were incubated for 1 hour at 37°C. Absorbance at 450 nm was detected by Microplate Absorbance Reader (Bio-rad).
Cell proliferation
The proliferative rate of H460 cells was detected with 5-ethynyl-2’-deoxyuridine (EdU) assays. H460 cells was planted into 24-well plate at 2 × 104/ml, and then cells were allowed to adhere overnight. After transfection of miR-222 mimics, inhibitors or their negative controls, cells were incubated with EdU for 8 hour before fluorescent detection. Cells were fixed with 4% paraformaldehyde for 30 minutes, and stained with Cell-Light™ EdU Apollo®488 In Vitro Imaging Kit (RioBio, China) according to the manufacturer’s instructions.
Quantitative reverse transcription polymerase chain reactions (qRT-PCRs)
Total RNA was isolated from H460 cells using TRIZOL reagent (Invitrogen, USA). For p27 and p57 mRNA detection, reverse transcription reaction was performed using 400 ng of total RNA and reverse transcribed into cDNA using Bio-Rad iScripTM cDNA Synthesis Kit (Bio-Rad, USA). Then cDNA was subjected into 40 cycles of quantitative PCR with Takara SYBR Premix Ex TaqTM (Takara, Japan) in CFX96TM Real-Time PCR Detection System (Bio-Rad, USA), using the following cycling conditions: 95°C for 3 min; 95°C for 15 s, 60°C for 30 s, 72°C for 30 s, 40 cycles. GAPDH was used to normalize p27 and p57 genes. The sequence of p27 primer: FOR AACGTGCGAGTGTCTAACGG (5’-3’), REV CCCTCTAGGGGT TTGTGATTCT (5’-3’). The sequence of p57 primer: FOR GCGGCGATCAAGAA GCTGT (5’-3’), REV GCTTGGCGAAGAAATCGGAGA (5’-3’). The sequence of GAPDH primer: FOR TGTGGGCATCAATGGATTTGG (5’-3’), REV ACACCATG TATTCCGGGT CAAT (5’-3’).
For miR-222 detection, reverse transcription reaction was performed using Bulge-Loop™ miRNA qRT-PCR Primer Set (Ribobio, China) according to the manufacturer’s instructions. Then cDNA was subjected into 40 cycles of quantitative PCR with Takara SYBR Premix Ex TaqTM (Takara, Japan) in CFX96TM Real-Time PCR Detection System (Bio-Rad, USA), using the following cycling conditions: 95°C for 10 min; 95°C for 10 s, 60°C for 20 s, 72°C for 10 s, 40 cycles. U6 was used to normalize miR-222.
Western blot
H460 cells were plate in a 6-well plate at a density of at 2 × 104/ml and were allowed to adhere overnight. After transfection of miR-222 mimics, inhibitors or their negative controls, cells were harvested. The cells were scraped in cold PBS and lysed in in RIPA buffer (Keygen, China) containing 1 mM PMSF. Western blotting was performed using standard 12% SDS-PAGE gel, loading 40 µg of protein per lane, probed with anti-p27 Ab (Bioworld, USA) and were detected using HRP-linked secondary Ab and the ECL System (Bio-rad, USA). β-actin was used as loading control.
Statistical analysis
All data were represented as mean ± SEM. An independent Student T-test or one-way ANOVA was conducted with a Bonferroni’s post-hoc test. P value < 0.05 was considered as statistically significant. All analyses in the study were performed using IBM SPSS 19.0 for Windows.
Results
MiR-222 controls H460 cells’ viability
To know if miR-222 could regulate human NSCLC cell line H460 cells’ viability, we firstly transfected miR-222 mimics, inhibitors or their negative controls to H460 cells. The transfection rate of mimics and inhibitors has been previously shown [25]. As determined by qRT-PCR, we confirmed that miR-222 mimics or inhibitors used in the present study successfully took effects in increasing or decreasing miR-222 levels in H460 cells, which is evidenced by the fact that 48 h after transfection of miR-222 mimics, miR-222 levels were significantly upregulated in H460 cells, while miR-222 inhibitors downregulated miR-222 levels (Figure 1). Based on that, using CCK-8 assays, we showed that miR-222 mimics increased cell viability of H460 cells while miR-222 inhibitors decreased that (Figure 2). These data confirm that miR-222 might be responsible for the tumor properties of H460 cells by regulating cell viability.
Figure 1.
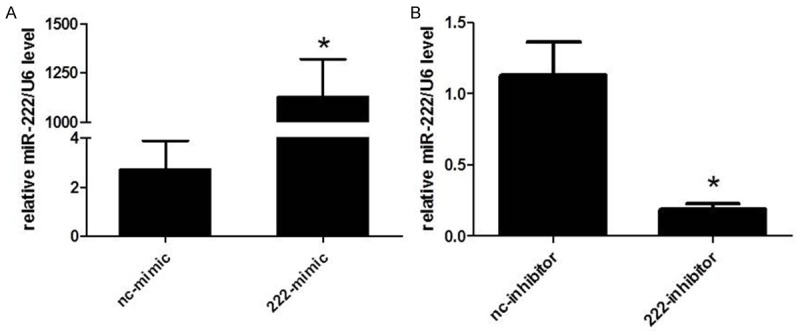
Quantitative reverse transcription polymerase chain reactions (qRT-PCRs) prove that miR-222 mimics and inhibitors successfully take effects in H460 cells. A. miR-222 mimics upregulate miR-222 levels in H460 cells. *P < 0.05. B. miR-222 inhibitors downregulate miR-222 levels in H460 cells. *P < 0.05.
Figure 2.
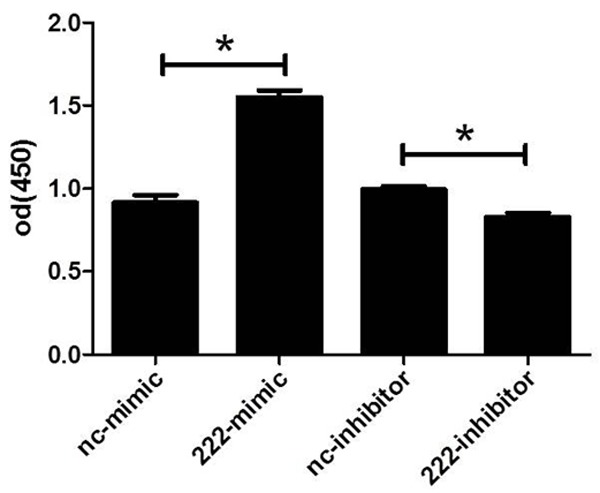
miR-222 regulates cell viability of H460 cells. Cell Counting Kit-8 assays indicate that miR-222 mimics increase cell viability of H460 (A) while miR-222 inhibitors decrease cell viability of H460 (B). *P < 0.05.
MiR-222 induces H460 cells’ proliferation
To check the effects of miR-222 in regulating H460 cell’ proliferation, in this study we used EdU assays. We showed that up-regulation of miR-222 with miR-222 mimics increased the percentage of EdU positive cells, indicating that miR-222 induces H460 cells’ proliferation. In addition, down-regulation of miR-222 with miR-222 inhibitors deceased the percentage of EdU positive cells (Figure 3). These data indicate that miR-222 may be responsible for the tumor properties of H460 cells by promoting cell proliferation.
Figure 3.
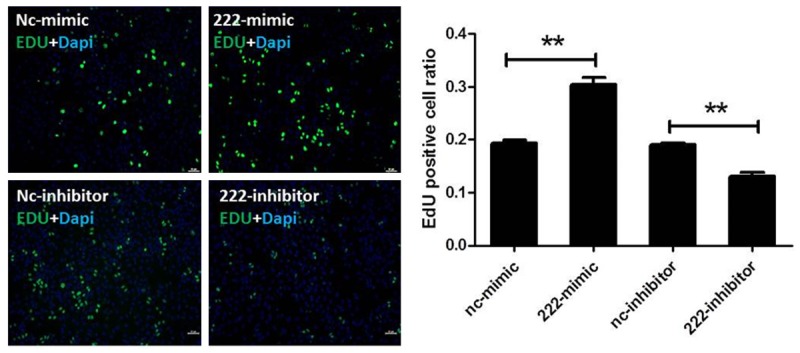
miR-222 controls cell proliferation of H460 cells. 5-Ethynyl-2’-deoxyuridine (EdU) stainings show that miR-222 mimics increase the proliferation of H460 cells (A) while miR-222 inhibitors decrease the proliferation of H460 cells (B). **P < 0.01.
P27 is a potential target gene of miR-222 in H460 cells
P27 and p57, also respectively known as cyclin-dependent kinase inhibitor 1B and cyclin-dependent kinase inhibitor 1C, are members of the Cip/Kip family of cyclin-dependent kinase inhibitors and function to negatively control cell proliferation [26-30]. In addition, they are well-established target genes of miR-222 in multiple cell types [31-34]. To determine if P27 and/or P57 are putative target genes of miR-222 in H460 cells, we detected the mRNA levels of p27 and p57 in H460 cells firstly. As demonstrated with qPCRs, mRNA levels of p27 were down-regulated by overexpression of miR-222 while remained unchanged by miR-222 inhibition (Figure 4A). However, mRNA levels of P57 were not affected by either overexpression or down-regulation of miR-222 (Figure 4A). To further confirm that p27 is a potential target gene of miR-222 in H460 cells, we next detected the protein levels of p27. As shown in Figure 4B, the protein levels of p27 were decreased by miR-222 overexpression while increased by miR-222 downregulation in H460 cells, indicating that P27 might be a target gene of miR-222 in H460 cells.
Figure 4.
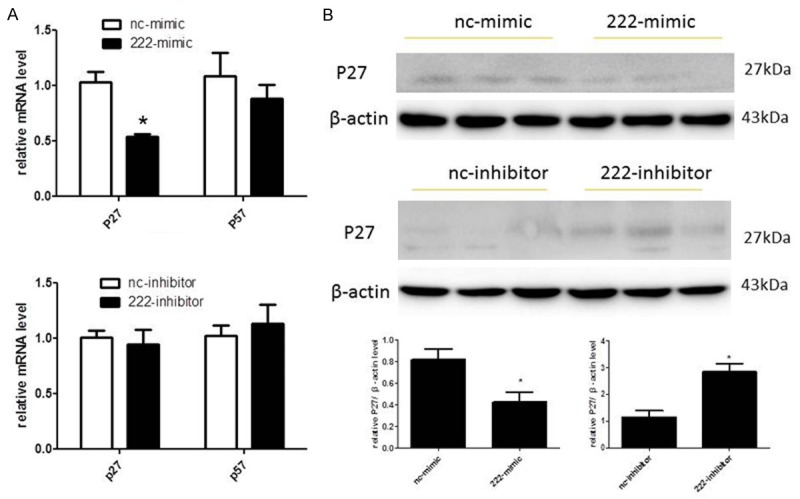
P27 is a potential target gene of miR-222 in H460 cells. A. miR-222 negatively regulates p27 but not P57 expression levels at the mRNA level. *P < 0.05. B. miRNA-221 negatively regulates p27 expression levels at the protein levels. *P < 0.05.
Discussion
Lung cancer is one of the most prevalent malignancies worldwide and is also the leading cause of cancer-related death. The NSCLC accounts for 80% of the total lung cancer cases, which shows very poor prognosis: the median overall survival of patients with advanced stage undergoing current standard chemotherapy is as short as approximately 10 months. Despite extensive efforts have been devoted into NSCLC studies, few novel therapeutic strategies have been proved to exhibit remarkable clinical effects.
Extensive evidence has suggested that miRNAs are crucially important in cancer gene regulation [12,15,35]. Particularly, a lot of miRNAs have been proved to promote tumorigenesis and cancer progression [36-41]. For these onco-miRNAs, proliferation promotion effect on tumor cells is a key property related to tumor development, of which a profound understanding is quite important for the potential use of miRNAs for therapeutic purposes.
MiR-222 belongs to the miR-221/222 family and is a well-established onco-miRNA. However, if miR-222 is involved in the progression of NSCLC remains unclear despite a study has indicated that the expression levels of miR-222 were elevated in the NSCLC tissue compared with that in adjacent normal tissue [24]. In this study, we investigated the effects of miR-222 on human NSCLC cell line H460, finding that overexpression of miR-222 enhances the cell viability and cell proliferation, as demonstrated by CCK-8 and EdU assay, whereas down-regulating miR-222 levels in H460 cells deceased the cell viability and cell proliferation. P27 is a target gene of miR-221 that we previously identified in H40 cells. Here we also indicates that p27 is regulated by miR-222 in H40 cells, indicating it might mediate the effects of miR-222 in H40 cells, though this needs to be further confirmed with reverse evidence in the future.
In conclusion, our investigation suggests that miR-222 controls H40 cells’ viability and proliferation, and might functions by targeting p27. This study may provide new evidence for understanding the role of miR-222 on NSCLC cells, as well as help develop novel therapeutic strategy for NSCLC.
Acknowledgements
This work was supported by the grants from Nantong University (to CJ Zhong).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson BC. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350:2008–2010. doi: 10.1056/NEJM200405063501921. [DOI] [PubMed] [Google Scholar]
- 3.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 4.Alevizakos M, Kaltsas S, Syrigos KN. The VEGF pathway in lung cancer. Cancer Chemother Pharmacol. 2013;72:1169–1181. doi: 10.1007/s00280-013-2298-3. [DOI] [PubMed] [Google Scholar]
- 5.Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin. 2011;61:91–112. doi: 10.3322/caac.20102. [DOI] [PubMed] [Google Scholar]
- 6.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Xu J, Cheng Y, Wang F, Song Y, Xiao J. Circulating microRNAs as mirrors of acute coronary syndromes: MiRacle or quagMire. J Cell Mol Med. 2013;17:1363–1370. doi: 10.1111/jcmm.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 11.Xiao J, Shen B, Li J, Lv D, Zhao Y, Wang F, Xu J. Serum microRNA-499 and microRNA-208a as biomarkers of acute myocardial infarction. Int J Clin Exp Med. 2014;7:136–141. [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Cheng S, Zhang Y, Li H, Huang J, Zhang P. Association between polymorphisms in the integrin gene predicted microRNA binding sites and bladder cancer risk. Int J Clin Exp Med. 2014;7:4398–4405. [PMC free article] [PubMed] [Google Scholar]
- 14.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bandopadhyay M, Banerjee A, Sarkar N, Panigrahi R, Datta S, Pal A, Singh SP, Biswas A, Chakrabarti S, Chakravarty R. Tumor suppressor micro RNA miR-145 and onco micro RNAs miR-21 and miR-222 expressions are differentially modulated by hepatitis B virus X protein in malignant hepatocytes. BMC Cancer. 2014;14:721. doi: 10.1186/1471-2407-14-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan R, Yang Y, Yang X, Zhao L, Lu J, Meng QH. Downregulation of miR-221/222 enhances sensitivity of breast cancer cells to tamoxifen through upregulation of TIMP3. Cancer Gene Ther. 2014;21:290–296. doi: 10.1038/cgt.2014.29. [DOI] [PubMed] [Google Scholar]
- 19.Liu M, Liu J, Wang L, Wu H, Zhou C, Zhu H, Xu N, Xie Y. Association of serum microRNA expression in hepatocellular carcinomas treated with transarterial chemoembolization and patient survival. PLoS One. 2014;9:e109347. doi: 10.1371/journal.pone.0109347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang Z, Li L, Qin J, Chen Y, Cho WC, Pestell RG, Liang L, Yu Z. miR-221/222 promotes S-phase entry and cellular migration in control of basal-like breast cancer. Molecules. 2014;19:7122–7137. doi: 10.3390/molecules19067122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B, Che Q, Qiu H, Bao W, Chen X, Lu W, Li B, Wan X. Elevated MiR-222-3p promotes proliferation and invasion of endometrial carcinoma via targeting ERalpha. PLoS One. 2014;9:e87563. doi: 10.1371/journal.pone.0087563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Yang Y, Gan R, Zhao L, Li W, Zhou H, Wang X, Lu J, Meng QH. Down-regulation of mir-221 and mir-222 restrain prostate cancer cell proliferation and migration that is partly mediated by activation of SIRT1. PLoS One. 2014;9:e98833. doi: 10.1371/journal.pone.0098833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang YF, Wang F, Xiao JJ, Song Y, Zhao YY, Cao Y, Bei YH, Yang CQ. MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HePG2 cells by downregulating p27. Int J Clin Exp Med. 2014;7:893–902. [PMC free article] [PubMed] [Google Scholar]
- 24.Mao KP, Zhang WN, Liang XM, Ma YR. MicroRNA-222 expression and its prognostic potential in non-small cell lung cancer. ScientificWorldJournal. 2014;2014:908326. doi: 10.1155/2014/908326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu YM, Zhong CJ, Ding SG, Huang HT, Shen ZY. MicroRNA-221 promotes human non-small cell lung cancer cell H460 growth. Int J Clin Exp Med. 2015;8:2024–2030. [PMC free article] [PubMed] [Google Scholar]
- 26.Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 27.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G (1)-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 28.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 29.Lee MH, Reynisdottir I, Massague J. Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 1995;9:639–649. doi: 10.1101/gad.9.6.639. [DOI] [PubMed] [Google Scholar]
- 30.Zhang PM, Liegeois NJ, Wong C, Finegold M, Hou H, Thompson JC, Silverman A, Harper JW, Depinho RA, Elledge SJ. Altered cell differentiation and proliferation in mice lacking p57 (KIP2) indicates a role in Beckwith-Wiedemann syndrome. Nature. 1997;387:151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Dai W, Chu X, Yang B, Zhao M, Sun Ye. Metformin inhibits lung cancer cells proliferation through repressing microRNA-222. Biotechnol Lett. 2013;35:2013–2019. doi: 10.1007/s10529-013-1309-0. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Li X, Kong X, Luo Q, Zhang J, Fang L. MiRNA-26b inhibits cellular proliferation by targeting CDK8 in breast ceancer. Int J Clin Exp Med. 2014;7:558–565. [PMC free article] [PubMed] [Google Scholar]
- 33.Gillies JK, Lorimer IA. Regulation of p27 (Kip1) by miRNA 221/222 in Glioblastoma. Cell Cycle. 2007;6:2005–2009. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- 34.Sun T, Wang Q, Balk S, Brown M, Lee GS, Kantoff P. The role of microrna-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69:3356–3363. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He L, He X, Lim LP, De Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–U1116. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM. miR-221 & 222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A, Falcone A, Danesi R, Campani D, Verheul HM, Boggi U. MicroRNA-21 in pancreatic cancer: correlation with clinical Outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 38.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD Are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takakura S, Mitsutake N, Nakashima M, Namba H, Saenko VA, Rogounovitch TI, Nakazawa Y, Hayashi T, Ohtsuru A, Yamashita S. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008;99:1147–1154. doi: 10.1111/j.1349-7006.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]


