Abstract
Background: Previous studies of the association between COL1A1 polymorphisms and high myopia risk have yielded conflicting results. To help resolve the discrepancies, we performed a meta-analysis to estimate the relationship between COL1A1 polymorphisms and high myopia risk. Methods: We searched for case-control and cohort studies in MEDLINE, EMBASE, and OVID. Odds ratios (OR) with 95% confidence intervals (CI) were derived for single-nucleotide polymorphisms (SNPs). We also analyzed heterogeneity and publication bias. Results: This meta-analysis was based on five studies of rs2075555 (1,944 high myopia cases and 3,060 controls), and three studies of rs2269336 (1,454 high myopia cases and 1,512 controls). The combined results showed an association between rs2075555 and high myopia in the dominant (OR = 0.86, 95% CI = 0.71-0.99) and homozygote models (OR = 0.79, 95% CI = 0.64-0.97). In the recessive model for rs2269336, OR was 1.26 (95% CI = 1.05-1.50); in the heterozygote model, OR was 0.81 (95% CI = 0.69-0.96). Begg’s and Egger’s tests for rs2075555 showed no evidence of publication bias. Conclusions: This meta-analysis suggests COL1A1 rs2075555 is a potential low risk factor for high myopia.
Keywords: COL1A1, polymorphisms, high myopia, meta-analysis
Introduction
Myopia is caused by lengthening of the ocular axis and focusing of light rays on the front of the retina; people with this condition can see close objects more clearly than distant ones [1]. It is a major global cause of visual impairment, has adverse social, educational, and economic consequences, and affects quality of life [2]. Epidemiological evidence suggests the prevalence of myopia has increased 28.1% and 19.4% in white and black populations [3,4], whereas in Asians, the incidence has increased from 40% to 80% [5] and is still growing. As an extreme form of myopia, high myopia is usually defined as a refractive error of at least -6.00 diopter (D) or an axial eye length greater than 26 mm. This serious form of myopia is now considered the fourth most common cause of irreversible blindness [6]. Individuals with high myopia are predisposed to many pathologic ocular abnormalities such as cataracts, retinal detachment, glaucoma, chorioretinal degeneration, myopic foveoschisis, or choroidal neovascularization [7,8], which may also lead to irreversible vision impairment or blindness. Epidemiological, experimental, and clinical studies provide convincing evidence that environmental and genetic factors each play a role in the occurrence of myopia. Less outdoor activity and more near-work activity are known risk factors for myopia [9]. High heritability in family-based and twin studies supports the theory that genetic factors are also responsible for high myopia [10-12]. Whole genome linkage analysis and genome-wide association studies have mapped more than 20 known chromosomal loci. [7,13,14] and candidate genes associated with high myopia have been reported, including collagen type I (COL1A1).
Published studies of the association between COL1A1 and high myopia risk are inconclusive, and no meta-analyses have been conducted in this area. We performed a meta-analysis by using strict criteria to include or exclude potentially relevant studies in order to examine the association between COL1A1 variants and high myopia risk.
Methods
We performed a systematic review of the published literature. The study was performed according to the MOOSE guidelines and the PRISMA statement [15,16] for meta-analysis of observational studies.
Search strategy
We performed systematic literature searches of MEDLINE (1966 to June 1, 2014), EMBASE (1980 to June 1, 2014), and OVID (1950 to June 1, 2014) with no language limitation and using medical subject headings (MeSH) or free text, with the following terms and keywords: “COL1A1”, “polymorphism (s)”, “variant (s)”, “mutation (s)”, and outcomes (“myopia”, “refraction”, “refraction error”, “refractive error”). We also checked the reference lists of all relevant publications for other potentially relevant studies.
Selection criteria
Studies were included in the meta-analysis if they met the following criteria: (1) original case-control or cohort studies evaluating at least one COL1A1 polymorphism and high myopia risk, and (2) providing sufficient data on each genotype and/or allele in both case and control groups. When the same patient population was included in several publications, we enrolled the most recent or complete study in our meta-analysis. Reviewers independently evaluated published quantitative estimates of the association between COL1A1 and high myopia for inclusion in the meta-analysis. Studies that did not meet the above inclusion criteria were excluded during initial review. Any disagreement in extracted data was resolved by discussion.
Data extraction
The reviewers independently extracted essential data using a standardized data collection form. Discrepancies in data interpretation were resolved by arbitration. The following data were extracted from each study: first author, publication year, country, participant ethnicity, gender, age, study size, specific COL1A1 SNPs, genotyping method, extent of refractive degree and axial length for cases and controls, and number of eligible and genotyped cases and controls.
Statistical analyses
We evaluated the Hardy-Weinberg equilibrium (HWE) of genetic frequency distributions for the controls by using the χ2 test, with P < 0.05 regarded as evidence of unequal genetic distributions. As for individual studies or non-HWE studies, we performed a none-way sensitivity analysis by sequential omission to test the robustness of the association. “A” was used to denote a major allele, and “a” to denote a minor allele. We selected the dominant model (aa + Aa vs. AA), recessive model (aa vs. AA + Aa), homozygote comparison model (aa vs. AA), and heterozygote comparison model (Aa vs. AA) to dissect the association patterns. The odds ratios (ORs) and 95% confidence intervals (95% CIs) were used as the common measure across studies in fixed-effect (using the Mantel-Haenszel method) and random-effects models (using the DerSimonian and Larid method) [17]. The model was chosen according to the heterogeneity assumption by the Q test; P > 0.10 indicated a lack of heterogeneity. If P < 0.10, heterogeneity was considered significant and the random-effects model was used to calculate the pooled OR; otherwise, the fixed-effects model was employed. Cochran I 2 statistics quantifying the proportion of total variation attributable to between-study heterogeneity were also calculated [18]. I 2 represents the percentage of total variation across studies which were attributable to heterogeneity rather than chance. As suggested by Higgins et al., I 2 values of 25%, 50%, and 75% were considered as low, moderate, and high heterogeneity, respectively [19]. We generated a funnel plot of the overall OR, produced a standard error (ER) to detect publication bias, and used Egger’s and Begg’s regression tests. We conducted stratified analyses to identify associations between COL1A1 and high myopia, and relevant study characteristics such as ethnicity were analyzed. All analyses described above were conducted using Stata 12 (StataCorp, College Station, TX). Statistical significance was defined as a P-value < 0.05.
Results
Five studies met the inclusion criteria [20-24], and our meta-analysis included 1,944 cases and 3,060 controls. The included cases consisted of 1,057 males and 887 females. The study selection process is presented in Figure 1 as a flow chart.
Figure 1.
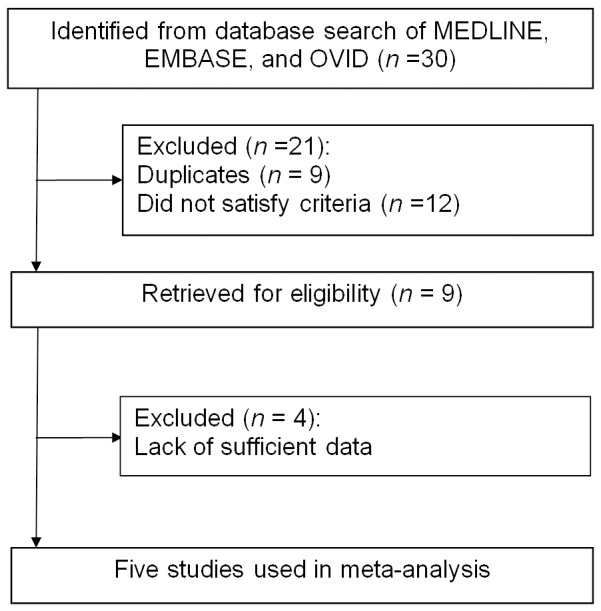
Flowchart of the study selection process.
Study characteristics and quality
Main study characteristics are listed in Table 1. The most frequently studied genetic variants were rs2075555 and rs2269336, and our meta-analysis focused largely on these SNPs. All five included studies addressed the association between rs2075555 and high myopia risk (1,944 high myopia cases and 3,060 controls); three of these studies also addressed rs2269336 (1,454 high myopia cases and 1,512 controls). None of the studies of rs2075555 showed a deviation from HWE in the controls; for polymorphism rs2269336, however, one study had a deviation from HWE at a P value of 0.016.
Table 1.
Study characteristics
| Study (year) | Country | Ethnicity | Genotyping method | SNP ID | Gender (M/F) | Age (mean ± SD, a) | Sample size | Refractive degree (diopter) | Axial length (mm) | HWE | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | ||||||
| Zhang et al. (2011) | China | Asian | Dye terminator-based SNaPshot | rs2075555 | 317/380 | 352/411 | 34.3 ± 11.9 | 54.5 ± 9.2 | 697 | 762 | ≤ -6.00 | > -1.0 | ≥ 26 | NA | rs2075555: 0.09 |
| rs2269336 | rs2269336: 0.05 | ||||||||||||||
| Vatavuk et al. (2009) | Croatia | Caucasian | HumanHap Genotyping BeadChip | rs2075555 | 4/15 | NA | NA | NA | 19 | 925 | ≤ -6.00 | NA | NA | NA | rs2075555: 0.922 |
| Nakanisbi et al. (2009) | Japan | Asian | TaqMan | rs2075555 | 134/293 | 194/226 | 57.6 ± 14.1 | 44.3 ± 12.1 | 427 | 420 | ≤ -5.00 | NA | ≥ 26.5 | NA | rs2075555: 0.234 |
| rs2269336 | rs2269336: 0.655 | ||||||||||||||
| Liang et al. (2007) | Taiwan | Asian | TaqMan | rs2075555 | 471/0 | 623/0 | 18-25 | NA | 471 | 623 | ≤ -6.0 D in one eye and ≤ –4.0 D in the other eye | ≥ -1.5 | NA | NA | rs2075555: 0.459 |
| Inamori et al. (2007) | Japan | Asian | PCR | rs2075555 | 131/199 | 131/199 | 37.82 ± 11.97 | 37.82 ± 11.97 | 330 | 330 | ≤ -9.25 | NA | 27.78 ± 1.30 | NA | rs2075555: 0.663 |
| rs2269336 | rs2269336: 0.016 | ||||||||||||||
PCR: Polymerase chain reaction; RFLP: Restriction fragment length polymorphism; NA: Not applicable; HWE: Hardy-Weinberg equilibrium.
Main analysis
Our meta-analysis of rs2075555 and rs2269336 in COL1A1 and high myopia risk is presented in Table 2. For rs2075555 a reduced risk for high myopia was apparent in the dominant model (pooled OR = 0.86, 95% CI = 0.74-0.99; P Heterogeneity = 0.376, I 2 = 5.3%) or in homozygotes (pooled OR = 0.79, 95% CI = 0.64–0.97; P Heterogeneity = 0.261, I 2 = 24%); no association was observed between rs2075555 and high myopia risk in the other models (OR 0.90, 95% CI = 0.77-1.06; P Heterogeneity = 0.422, I 2 = 0% for the recessive model; OR 0.87, 95% CI = 0.75-1.02; P Heterogeneity = 0.593, I 2 = 0% for heterozygotes). rs2269336 studies revealed an increased risk in the recessive model (OR = 1.26, 95% CI = 1.05-1.50; P Heterogeneity = 0.123, I 2 = 52.2%) and a reduced risk in heterozygotes with a pooled OR of 0.81 (95% CI = 0.69-0.96; P Heterogeneity = 0.351, I 2 = 4.6%). No significant association was observed in other models (OR = 0.88, 95% CI = 0.75-1.03; P Heterogeneity = 0.164, I 2 = 44.6% and OR = 1.10, 95% CI = 0.89-1.36; P Heterogeneity = 0.091, I 2 = 58.2% for the dominant model and homozygotes, respectively).
Table 2.
Meta-analysis of COL1A1 rs2075555 and rs2269336 and high myopia risk
| SNPs | Models tested | Number of studies | Pooled OR (95% CL) | P | Heterogeneity | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Q | PQ | I 2, % | |||||
| rs2075555 | Dominant model | 5 | 0.86 (0.74, 0.99) | 0.037 | 4.23 | 0.376 | 5.3 |
| Recessive model | 5 | 0.90 (0.77, 1.06) | 0.197 | 3.88 | 0.422 | 0 | |
| Homozygote | 5 | 0.79 (0.64, 0.97) | 0.024 | 5.26 | 0.261 | 24.0 | |
| Heterozygote | 5 | 0.87 (0.75, 1.02) | 0.076 | 2.79 | 0.593 | 0 | |
| rs2269336 | Dominant model | 3 | 0.88 (0.75, 1.03) | 0.115 | 3.61 | 0.164 | 44.6 |
| Recessive model | 3 | 1.26 (1.05, 1.50) | 0.012 | 4.19 | 0.123 | 52.2 | |
| Homozygote | 3 | 1.13 (0.81, 1.59) | 0.477 | 4.79 | 0.091 | 58.2 | |
| Heterozygote | 3 | 0.81 (0.69, 0.96) | 0.016 | 2.10 | 0.351 | 4.6 | |
“A”, major allele; “a”, minor allele; dominant model, aa + Aa vs. AA; recessive model, aa vs. AA + Aa; homozygote comparison, aa vs. AA; heterozygote, Aa vs. AA.
Stratified analysis
Stratified analysis of the association between rs2075555 and high myopia risk showed a risk association of Asian ethnicity and rs2075555 in the dominant model and homozygotes (OR = 0.86, 95% CI = 0.74-0.99, P Heterogeneity = 0.238; OR = 0.79, 95% CI = 0.64-0.97, P Heterogeneity = 0.154, respectively). No statistical association was observed for the recessive model and heterozygotes (OR = 0.90, 95% CI = 0.77-1.06, P Heterogeneity = 0.276; OR = 0.87, 95% CI = 0.75-1.02, P Heterogeneity = 0.426, respectively).
Publication bias
Begg’s and Egger’s tests were performed to assess publication bias for COL1A1 rs2075555, and both tests showed consistent results, indicating no publication biases in the dominant and recessive models (P = 0.462 for Begg’s test and P = 0.838 for Egger’s test in the dominant model; P = 1.000 for Begg’s test and P = 0.887 for Egger’s test in the recessive model). The shape of the funnel plot of rs2075555 in Figure 2 revealed no evidence of asymmetry in the dominant and recessive models.
Figure 2.
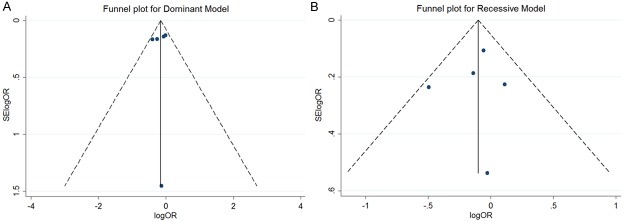
Publication bias analysis for rs2075555. A and B show the Begg’s funnel plot of studies in the dominant and recessive models, respectively. The horizontal axis indicates the logOR and the vertical axis indicates the standard error of logOR (SElogOR). The vertical and sloping lines in the funnel plot demonstrate the fixed-effects summary OR, and the expected 95% CI for a given standard error, respectively.
Discussion
Five studies of COL1A1 and high myopia risk were included in this meta-analysis, including 1,944 high myopia cases and 3,060 controls. The studies addressed the association between two COL1A1 polymorphisms and high myopia risk (rs2075555 and rs2269336). SNP rs2075555 is likely a low risk factor for high myopia, as the OR was 0.86 in the dominant model (95% CI = 0.74-P Heterogeneity = 0.376, I 2 = 5.3%; P Heterogeneity = 0.261, I 2 = 24.0% respectively). Further analysis of rs2269336 revealed an OR of 1.26 in the recessive model (95% CI = 1.05-1.50), and 0.81 in heterozygotes (95% CI = 0.69-0.96). Begg’s and Egger’s tests showed no evidence of publication bias for either polymorphism.
Previous studies have shown a significant genotypic association between COL1A1 variants and high myopia in a Japanese population [20], although this finding could not be replicated by another Japanese population study [22]. No significant association between COL1A1 polymorphisms and high myopia risk was observed in another Asian population [21]. Contradictory conclusions have also been drawn in other ethnicities [23,25].
Dysfunction of type I collagen genes has been associated with disorders such as osteogenesis imperfecta [26] and ocular disorder [27-29]. COL1A1, which is located on chromosome 17q21 near MYP5 [30], has been demonstrated in experimental myopia models to play an important role in pathogenesis. The development of high myopia can be explained by altered scleral morphology associated with changes in collagen fibril ultrastructure and increased numbers of small-diameter collagen fibrils [27]. In selected collagen subtypes screened by RT-PCR and sequencing, COL1A1 was present in the sclera, thus validating the possible relationship between COL1A1 polymorphisms and risk of high myopia. Gentle et al. showed reduced type I collagen mRNA expression and scleral collagen accumulation in the sclera of myopic tree shrews [29], suggesting COL1A1 variations control the development of myopia through scleral thinning, tissue loss, and altered tissue morphology. The findings of our meta-analysis showed that COL1A1 rs2075555 was inversely associated with high myopia.
We found a converse interaction between rs2269336 and high myopia, with an OR of 1.26 in the recessive model (95% CI = 1.05-1.50), and 0.81 in heterozygotes (95% CI = 0.69-0.96). For the three enrolled studies of rs2269336, all of which were performed in Asian populations, one suggested a significant association with high myopia in Japanese [20]; the other two suggested no association [22,24]. One of these studies had a deviation from HWE at a P value of 0.016 [20]. It is possible that this ambivalence is due to bias, as our analysis included a limited number of studies. Studies in other ethnic populations are required to clarify the role of these variations in high myopia risk.
The results of this meta-analysis should be treated with cautionary attention to its limitations. Among the five enrolled studies, one study conducted by Vatavuk et al [23]. Only included 19 cases of high myopia, and only one COL1A1 polymorphism (rs2075555) was available in their scan, which failed to find an association for this polymorphism. Additional, larger studies should be evaluated, and a haplotype-based approach is needed for a more objective evaluation. Multiple hypotheses complicate the interpretation of this positive result, as it might be attributed to the small number of studies enrolled in the stratified analysis.
In conclusion, we found COL1A1 rs2075555 is a potential low risk factor for high myopia, and identified a contradictory risk association for rs2269336. Larger, more comprehensive genetic and molecular biological studies are needed to determine the contribution of COL1A1 to the incidence of high myopia.
Disclosure of conflict of interest
None.
References
- 1.Waddell K. Spherical refraction for general eye workers. Community Eye Health. 2000;13:6–7. [PMC free article] [PubMed] [Google Scholar]
- 2.Saw SM, Katz J, Schein OD, Chew SJ, Chan TK. Epidemiology of myopia. Epidemiol Rev. 1996;18:175–187. doi: 10.1093/oxfordjournals.epirev.a017924. [DOI] [PubMed] [Google Scholar]
- 3.Pan CW, Ramamurthy D, Saw SM. Worldwide prevalence and risk factors for myopia. Ophthalmic Physiol Opt. 2012;32:3–16. doi: 10.1111/j.1475-1313.2011.00884.x. [DOI] [PubMed] [Google Scholar]
- 4.Vitale S, Ellwein L, Cotch MF, Ferris FL 3rd, Sperduto R. Prevalence of refractive error in the United States, 1999-2004. Arch Ophthalmol. 2008;126:1111–1119. doi: 10.1001/archopht.126.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun J, Zhou J, Zhao P, Lian J, Zhu H, Zhou Y, Sun Y, Wang Y, Zhao L, Wei Y, Wang L, Cun B, Ge S, Fan X. High prevalence of myopia and high myopia in 5060 Chinese university students in Shanghai. Invest Ophthalmol Vis Sci. 2012;53:7504–7509. doi: 10.1167/iovs.11-8343. [DOI] [PubMed] [Google Scholar]
- 6.Dandona R, Dandona L. Refractive error blindness. Bull World Health Organ. 2001;79:237–243. [PMC free article] [PubMed] [Google Scholar]
- 7.Hornbeak DM, Young TL. Myopia genetics: a review of current research and emerging trends. Curr Opin Ophthalmol. 2009;20:356–362. doi: 10.1097/ICU.0b013e32832f8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcus MW, de Vries MM, Junoy Montolio FG, Jansonius NM. Myopia as a risk factor for open-angle glaucoma: a systematic review and meta-analysis. Ophthalmology. 2011;118:1989–1994. e1982. doi: 10.1016/j.ophtha.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Lin Z, Vasudevan B, Jhanji V, Mao GY, Gao TY, Wang FH, Rong SS, Ciuffreda KJ, Liang YB. Near Work, Outdoor Activity, and their Association with Refractive Error. Optom Vis Sci. 2014;91:376–382. doi: 10.1097/OPX.0000000000000219. [DOI] [PubMed] [Google Scholar]
- 10.Siegwart JT Jr, Norton TT. Perspective: how might emmetropization and genetic factors produce myopia in normal eyes? Optom Vis Sci. 2011;88:E365–372. doi: 10.1097/OPX.0b013e31820b053d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitry D, Williams L, Charteris DG, Fleck BW, Wright AF, Campbell H. Population-based estimate of the sibling recurrence risk ratio for rhegmatogenous retinal detachment. Invest Ophthalmol Vis Sci. 2011;52:2551–2555. doi: 10.1167/iovs.10-6375. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Hur YM, Huang W, Ding X, Feng K, He M. Shared genetic determinants of axial length and height in children: the Guangzhou twin eye study. Arch Ophthalmol. 2011;129:63–68. doi: 10.1001/archophthalmol.2010.323. [DOI] [PubMed] [Google Scholar]
- 13.Ciner E, Wojciechowski R, Ibay G, Bailey-Wilson JE, Stambolian D. Genomewide scan of ocular refraction in African-American families shows significant linkage to chromosome 7p15. Genet Epidemiol. 2008;32:454–463. doi: 10.1002/gepi.20318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawthorne F, Feng S, Metlapally R, Li YJ, Tran-Viet KN, Guggenheim JA, Malecaze F, Calvas P, Rosenberg T, Mackey DA, Venturini C, Hysi PG, Hammond CJ, Young TL. Association mapping of the high-grade myopia MYP3 locus reveals novel candidates UHRF1BP1L, PTPRR, and PPFIA2. Invest Ophthalmol Vis Sci. 2013;54:2076–2086. doi: 10.1167/iovs.12-11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 16.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronksley PE, Brien SE, Turner BJ, Mukamal KJ, Ghali WA. Association of alcohol consumption with selected cardiovascular disease outcomes: a systematic review and meta-analysis. BMJ. 2011;342:d671. doi: 10.1136/bmj.d671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins J, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins J, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 20.Inamori Y, Ota M, Inoko H, Okada E, Nishizaki R, Shiota T, Mok J, Oka A, Ohno S, Mizuki N. The COL1A1 gene and high myopia susceptibility in Japanese. Hum Genet. 2007;122:151–157. doi: 10.1007/s00439-007-0388-1. [DOI] [PubMed] [Google Scholar]
- 21.Liang CL, Hung KS, Tsai YY, Chang W, Wang HS, Juo SH. Systematic assessment of the tagging polymorphisms of the COL1A1 gene for high myopia. J Hum Genet. 2007;52:374–377. doi: 10.1007/s10038-007-0117-6. [DOI] [PubMed] [Google Scholar]
- 22.Nakanishi H, Yamada R, Gotoh N, Hayashi H, Otani A, Tsujikawa A, Yamashiro K, Shimada N, Ohno-Matsui K, Mochizuki M, Saito M, Saito K, Iida T, Matsuda F, Yoshimura N. Absence of association between COL1A1 polymorphisms and high myopia in the Japanese population. Invest Ophthalmol Vis Sci. 2009;50:544–550. doi: 10.1167/iovs.08-2425. [DOI] [PubMed] [Google Scholar]
- 23.Vatavuk Z, Skunca Herman J, Bencic G, Andrijevic Derk B, Lacmanovic Loncar V, Petric Vickovic I, Bucan K, Mandic K, Mandic A, Skegro I, Pavicic Astalos J, Merc I, Martinovic M, Kralj P, Knezevic T, Barac-Juretic K, Zgaga L. Common variant in myocilin gene is associated with high myopia in isolated population of Korcula Island, Croatia. Croat Med J. 2009;50:17–22. doi: 10.3325/cmj.2009.50.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D, Shi Y, Gong B, He F, Lu F, Lin H, Wu Z, Cheng J, Chen B, Liao S, Ma S, Hu J, Yang Z. An association study of the COL1A1 gene and high myopia in a Han Chinese population. Mol Vis. 2011;17:3379–3383. [PMC free article] [PubMed] [Google Scholar]
- 25.Metlapally R, Li YJ, Tran-Viet KN, Abbott D, Czaja GR, Malecaze F, Calvas P, Mackey D, Rosenberg T, Paget S, Zayats T, Owen MJ, Guggenheim JA, Young TL. COL1A1 and COL2A1 genes and myopia susceptibility: evidence of association and suggestive linkage to the COL2A1 locus. Invest Ophthalmol Vis Sci. 2009;50:4080–4086. doi: 10.1167/iovs.08-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marini JC, Blissett AR. New genes in bone development: what’s new in osteogenesis imperfecta. J Clin Endocrinol Metab. 2013;98:3095–3103. doi: 10.1210/jc.2013-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBrien NA, Jobling AI, Gentle A. Biomechanics of the sclera in myopia: extracellular and cellular factors. Optom Vis Sci. 2009;86:E23–30. doi: 10.1097/OPX.0b013e3181940669. [DOI] [PubMed] [Google Scholar]
- 28.McBrien NA, Metlapally R, Jobling AI, Gentle A. Expression of collagen-binding integrin receptors in the mammalian sclera and their regulation during the development of myopia. Invest Ophthalmol Vis Sci. 2006;47:4674–4682. doi: 10.1167/iovs.05-1150. [DOI] [PubMed] [Google Scholar]
- 29.Gentle A, Liu Y, Martin JE, Conti GL, McBrien NA. Collagen gene expression and the altered accumulation of scleral collagen during the development of high myopia. J Biol Chem. 2003;278:16587–16594. doi: 10.1074/jbc.M300970200. [DOI] [PubMed] [Google Scholar]
- 30.Paluru P, Ronan SM, Heon E, Devoto M, Wildenberg SC, Scavello G, Holleschau A, Makitie O, Cole WG, King RA, Young TL. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci. 2003;44:1830–1836. doi: 10.1167/iovs.02-0697. [DOI] [PubMed] [Google Scholar]


