Abstract
Small regulatory RNAs, known as microRNAs, regulate gene expression at the post-transcriptional level; such as protein translation inhibition or mRNA degradation. Altered miRNA expressions have been implicated in various cancers. In present studies, it was demonstrated that microRNA-29a (miR-29a) expressions were significantly lower in prostate cancer (PCa) patient samples, but the role of microRNA-29s in PCa remains unclear. KDM5B was highly expressed in PCa cancer cells. Bioinformatics analysis revealed a conserved target site for miR-29a in the 3-untranslated region (UTR) of KDM5B. Gain-of-function studies using mature miR-29a were performed to investigate cell proliferation and apoptosis in two PCa cell lines (LNCaP and PC-3). We utilized gene expression analysis and in silico database analysis to identify miR-29a-mediated molecular pathways and targets. We showed that miR-29a significantly suppressed the activity of a lucifarice reporter containing KDM5B-3’UTR, which was not observed in cells transfected with mutated KDM5B-3’UTR. Gene expression data demonstrated that KDM5B expression was lower in noncancerous prostatic cell WPMY-1 than in the four PCa cell lines (LNCaP, 22RV1, PC-3 and DU145). Moreover, the enforced expression of miR-29a in PC-3 and LNCaP cells inhibited proliferation, and induced apoptosis by repressing the expression of KDM5B. This study revealed that the aberrant expression of miR-29a in PCa cells regulated KDM5B expression levels associated with tumor dissemination. These findings may be utilized in developing novel therapeutic tools for PCa.
Keywords: Prostate cancer, miR-29a, KDM5B, human, histone demethylase
Introduction
Prostate cancer is one of the most common cancer in men and accounts for 28% of the main causes of cancer related deaths from all newly diagnosed cancer cases in men, as indicated by prostate specific antigen (PSA) testing [1]. In recent years, the incidence of PCa in China has also sharply increased [2]. A tumor cell develops the ability of invading its surrounding tissue during the tumor cell development process; inducing angiogenesis and metastasis as well. It was proven that 30% of PCa in patients were clinical types, as diagnosed by clinical studies [3]. Unfortunately, after 18 to 24 months of androgen deprivation therapy, approximately 80% of androgen-dependent PCa patients eventually progressed to a castration-resistant or hormone independent form of cancer [4,5]. Hence, effective new therapies and accurate prognostic indicators are needed to improve clinical care of prostate cancer patients.
MicroRNAs, a group of small endogenously noncoding RNAs, are fundamental critical regulators of gene expressions [6,7]. Mature miRNAs negatively regulate their target genes through imperfect complementary sequence pairing to the 3’-untranslated region (UTR) of target genes, resulting to either mRNA degradation or translational repression [8]. Recent studies have shown that the global alteration of miRNA expression has become a hallmark of tumorigenesis, and serves as a diagnosis and prognosis signature [9]. So far, over 1,000 miRNAs have been identified in human cells, and their diverse functions in normal cell homeostasis and various different diseases have been thoroughly investigated during the past decade [10].
MiR-29s, currently one of the most interesting miRNA families in humans, consists of three mature members: miR-29a, miR-29b, miR-29c; encoded in two genetic clusters. Members of this family have been shown to be silenced or down-regulated in many different types of cancer, such as acute myeloid leukemia [11], chronic lymphocytic leukemia [12], and some solid tumors [13,14]; which has also been shown to mediate either tumor suppressive or oncogenic functions in distinct malignancies. Several potential oncogenes have been reported to be silenced by miR-29s, including antiapoptotic p53 up stream inhibitors p85a and CDC42 [15], DNA methyltransferase [16], and extracellular matrix proteins [17]. These may partake in abnormal migration and cell invasion or proliferation, and favor cancer development as well. Our recent studies revealed that microRNA-29s were significantly downregulated and was a putative tumor-suppressive miRNA family in PCa. However, the role of microRNA-29s in PCa remains unclear.
KDM5B was the specific demethylase of H3K4 and its upregulation could reduce H3K4 methylation levels. Studies by Charlie Degui Chen have showed high KDM5B expressions in PCa cells [18]. Bioinformatics analysis revealed a conserved target site for miR-29a in the 3’-untranslated region (UTR) of KDM5B. MiR-29a affected the methylation status of H3K4 through regulating the expression of KDM5B; which controlled PCa development. Thus, understanding the roles of miR-29a in PCa and identifying relevant mRNA targets that mediate its tumor suppressor or oncogenic activities are essential in developing miR-29a as a therapeutic target.
In this study, we investigated the miR-29a expression in PC-3 cell lines, and it identified miR-29a as an essential regulator for PCa by targeting KDM5B in LNCaP and PC-3 cells.
Materials and methods
Patients
Tumor samples were extracted from 75 prostate cancer patients, 10 cases of normal prostate tissues and 30 cases of benign prostatic hyperplasia tissues were included in this study. The normal prostate tissue was from bladder cancer patients after total cystectomy, BPH tissues obtained in BPH patients and prostate cancer tissue were from prostate cancer patients after RP in Tong Ji Hospital, subsidiary of Shanghai Tong Ji University. The prostate cancer patients undergoing RP and regional lymph node dissection were from Tong Ji Hospital, subsidiary of Shanghai Tong Ji University, between January 2001 and December 2013, who did not receive any pre-operation treatment. The Research Ethics Committee of Tong Ji Hospital approved this protocol and verbal consent was obtained from all patients.
Cell culture
PCa cell line LNCaP, 22Rv1, PC-3 and DU145, as well as WPMY-1 were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China); which were authenticated by mycoplasma detection, DNA-Fingerprinting, isozyme detection and cell vitality detection. Four PCa cell lines were maintained in an RPMI 1640 medium supplemented with 10% FBS, 1% penicillin/streptomycin, 1% nonessential amino acids and 1% (mg/ml) sodium pyruvate. The cell lines were cultured at 37°C with 5% CO2. WPMY-1 cells were cultured in a DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin. All reagents used in cell culture were obtained from Life Technologies (New York, USA).
MiRNA mimics
Synthetic miR-29a mimics and its scrambled control, control miRNA (miR-NC), were from GenePharma (Shanghai, China); and were used at a concentration of 50 nM. SiRNA targeted sequences were as follows: miR-29a: 5’-UAGCACCAUCUGAAAUCGGUUA-3’ and 3’-ACCGAUUUCAGAUGGUGCUAUU-5’; Negative Control siRNA: 5’-UUCUCCGAACGUGUCACGUTT-3’ and 3’-ACGUGACACGUUCGGAGAATT-5’.
Transient transfection
Transient transfection was performed as previously described [20]. Transfection was carried out with Lipofectamine 2000 Transfection Reagent in accordance with manufacturer’s instructions. The day before transfection, cells were seeded in six-well plates. A 100-pmol sample of siRNA in a 250 μl Opti-MEM medium was mixed with 5 μl of Lipofectamine 2000 dissolved in 250 μl of the same medium; which was then allowed to stand at room temperature for 20 minutes. The 500 μl transfection solution results were then added to each well, which already contained 1.5 ml of Opti-MEM. Four hours later, the cultures were replaced with 2 ml of fresh RPMI 1640 medium. Opti-MEM medium and Lipofectamine 2000 were both purchased from Life Technologies (Shanghai, China).
Real-time reverse transcription PCR (qRT-PCR)
Total RNA was extracted from 22RV1, DU145, PC-3 and LNCaP cells with Trizol (Invitrogen); and reverse transcription was performed according to the PrimeScript RT reagent kit manual (TaKaRa, Tokyo, Japan). Real-time quantitative PCR was performed using an ABI Prism 7900HT (Applied Biosystems, Foster City, California, USA). Primers for KDM5B were forward, 5’-AGCAGACTGGCATCTGTAAGG-3’ and reverse, 3’-GAAGTTTATCAACATCACATGCAA-5’. The primers for β-actin were forward, 5’-CCTCTCCCAAGTCCACACAG-3’ and reverse, 3’-GGGCACGAAGGCTCATCATT-5’ (synthesized by SBS Gentech, Shanghai, China). Primers for RNU6B were forward, 5’-CGCTTCGGCAGCACATATACTAA-3’ and reverse, 3’-TATGGAACGCTTCACGAATTTGC-5’. For miRNA quantification, cDNA was synthesized from total RNA with MiScript Reverse Transcription kit (Qiagen, California, USA). The specific mature miR-29a primers were obtained from Qiagen (Catalog No. MS00031430), forward, 5’-GTGGAGGGTCCGAGGT-3’ and reverse, 3’-CACCATCTGAAATCGGTTAGT-5’.
Dual luciferase assay
LNCaP cells were co-transfected in 24-well plates with 20-pmol miR-29a or miR-NC, together with 0.8 μg of firefly luciferase report construct, containing wild-type or mutant KDM5B-3’-UTR and 8 ng control vector pRL-TK (Promega, Madison, WI). After 48 hours of transfection, firefly and renilla luciferase activities were measured with GloMax® 96 Microplate Luminometer (Promega, Madison, WI).
Western blot
Western blot was performed as previously described [19]. Forty-eight hours after siRNA transfection, cells were harvested for protein using a 1 x SDS lysis buffer (50 mM Tris-HCl, pH 6.8; 100 mM DTT; 2% SDS; 0.1% bromphenol blue; 10% glycerol). Protein concentration was determined using BCA Protein Assay reagent kit (Thermo Fisher Scientific, Waltham, MA, USA), according to manufacturer’s instructions. Then, proteins were separated by SDS–PAGE in 12% (w/v) polyacrylamide gels and transferred to membranes with antibodies, against KDM5B (1:1000, Sigma-Aldrich, MO, USA; 1:500, Proteintech, Chicago, USA) and β-Actin (1:4000, Sigma-Aldrich). Secondary antibodies were purchased from Sigma-Aldrich (St. Louis, MO, USA). Western blot signal intensity was quantified with Quantity One Software (Bio-Rad, California, USA).
Cell proliferation assay
Cell proliferation was performed as described [20]. Cells were briefly transfected with miR-29a and their negative control; then examined at 0, 24, 48 and 72 hours. At each time-point, a CCK-8 reagent was added into the cells and incubated for two hours. Absorbance was measured at 450 nm using a multi-mode microplate reader (BioTek, Winooski, VT). The absorbance at 630 nm was used as reference. Each experiment was performed in sextuplicate.
Cell cycle assay
Cell cycle was performed as described [21]. Cells were briefly incubated with propidium iodide (10 μg/ml) (Sigma-Aldrich) for 15 minutes in the dark. The fractions of viable cells in G1, S, and G2 cell cycle phases were measured with a FACStar flow cytometer (Becton-Dickinson, San Jose, CA) and analyzed with ModFit software (Verity Software House, Topsham, ME).
Apoptosis assays
MiR-29a and miR-NC were transfected as described above. 48 hours after transfection, cells were collected and assayed with FITC Annexin V Apoptosis Detection Kit (Becton-Dickinson) on a FACScalibur flow cytometer (Becton-Dickinson), following manufacturer’s instructions.
Statistical analysis
The differential expressions of miR-29a between groups were analyzed using χ2 analysis. To analyze the correlation between miR-29a and KDM5B expression, the T-test, χ2 analysis were employed according to the test condition, respectively. The statistical analysis was performed based on SPSS 17.0 software. Statistical significance was defined as P<0.05.
For experiments in cell lines, means and standard deviations of individual groups (n≥3) were calculated. P values were assessed by performing two-tailed Student’s t-test.
Result
Expression of miR-29a and KDM5B in PCa tissues and cell lines
We first detected the expressions of miR-29a and KDM5B in 75 cases of PCa tissues, 10 cases of normal prostate tissues and 30 cases of benign prostatic hyperplasia tissues. Compared to normal prostate tissues and BPH tissues, miR-29a expression was lower in the 75 samples of PCa tissues (Figure 1A). Conversely, the expression of KDM5B was significantly high in tumor tissues, based on the qRT-PCR analysis of microarray dataset GSE6919 (Figure 1B). In prostate cancer cell lines (DU145, PC-3 and LNCaP), miR-29a expression was generally low (Figure 1C). We further measured the expression of miR-29a and KDM5B in four PCa cell lines (LNCaP, 22RV1, PC-3 and DU145) and noncancerous prostatic cell WPMY-1. QRT-PCR showed that WPMY-1 had a relatively higher miR-29a expression (Figure 1D). Reversely, KDM5B expression was lower in WPMY-1, while 22RV1 had the highest KDM5B expression (Figure 1D).
Figure 1.
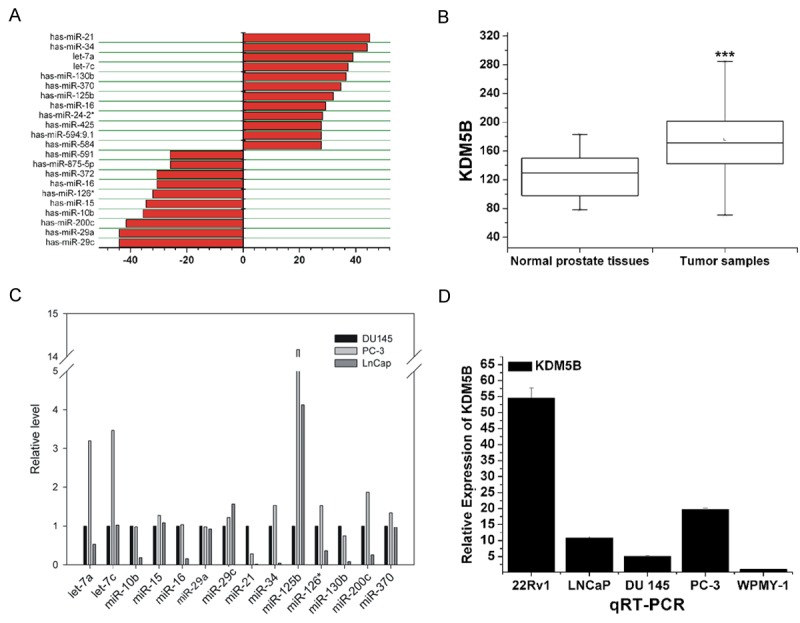
The expression levels of miR-29a and KDM5B in prostate cancer tissues and cell lines. Compared with normal prostate tissues and BPH tissues, miR-29a expression levels were lower in PCa tissues (A). The expression levels of KDM5B were significantly high in tumor tissues, based on microarray dataset GSE6919 analysis (B), (C, D) The qRT-PCR analysis results are illustrated. There were generally low miR-29a expression levels in PCa cell lines (DU145, PC-3 and LNCaP). Reversely, the expression levels of KDM5B were lowest in WPMY-1 cells and highest in 22Rv1 cells, as detected in the five prostate cell lines (22Rv1, LNCaP, DU145, PC-3 and WPMY-1). ***P<0.005.
MiR-29a influenced PCa cell proliferation and cell cycle status
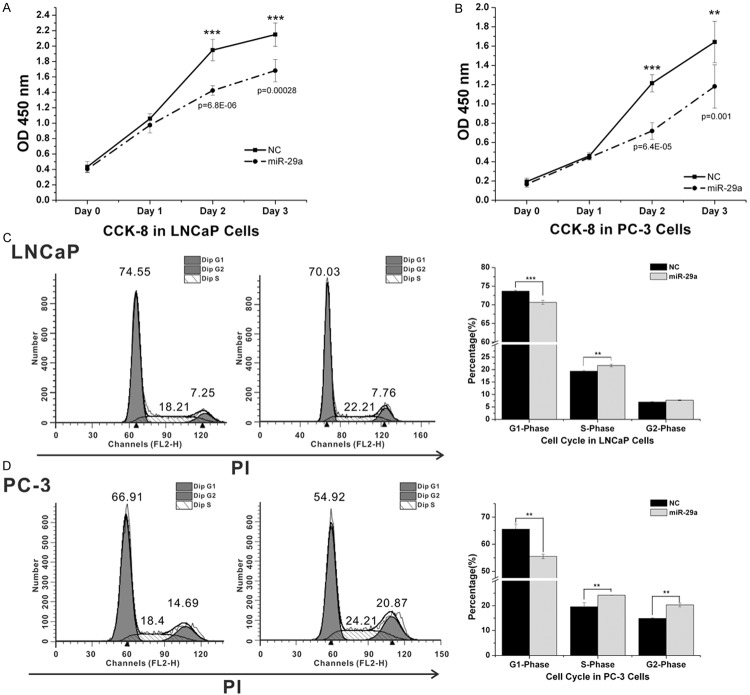
MiR-29a was transfected into two PCa cell lines (LNCaP and PC-3) to determine its effects in vitro. As shown in Figure 2, the enforced expression of miR-29a significantly decreased the growth rate of LNCaP and PC-3 cells. Therefore, these results suggest that the overexpression of miR-29a inhibits prostate cancer growth.
Figure 2.
MiR-29a influenced cell proliferation and cell cycle status in PCa cells. A,B. Cell proliferation analysis graphs. Cells were seeded into a 96-well plate at 5,000 cells/well and examined at 0, 24, 48 and 72 hours after transfection. Each experiment was performed in octuplicate (n=8). MiR-29a overexpression inhibited cell proliferation in LNCaP (A) and PC-3 (B). C,D. Cell cycle analysis graphs. Cells were transfected with miR-29a and miR-NC for 48 hours, stained with Propidium Iodide (PI), and subjected to cell cycle analysis. MiR-29a overexpression caused a decrease in the G1 phase and an increase in the S phase in LNCaP (C) and PC-3 (D).
Cell cycle distribution is a parameter that reflects cell growth; we assessed the function of miR-29a on the cell cycle profile of LNCaP and PC-3 cells by flow cytometry. The overexpression of miR-29a in LNCaP and PC-3 cells induced an increase in the S phase, and a decrease in the G1 phase, compared with miR-NC (P<0.05).
MiR-29a promotes cell apoptosis
We further explored the role of miR-29a on PCa cell apoptosis. LNCaP and PC-3 cells were treated with miR-29a and negative control, respectively; and the cells were subsequently stained with Annexin V-FITC/PI, followed by flow cytometry analyses. As shown in Figure 3, the enforced expression of miR-29a significantly reduced the fraction of living cells and boosted apoptotic cells (P<0.05).
Figure 3.
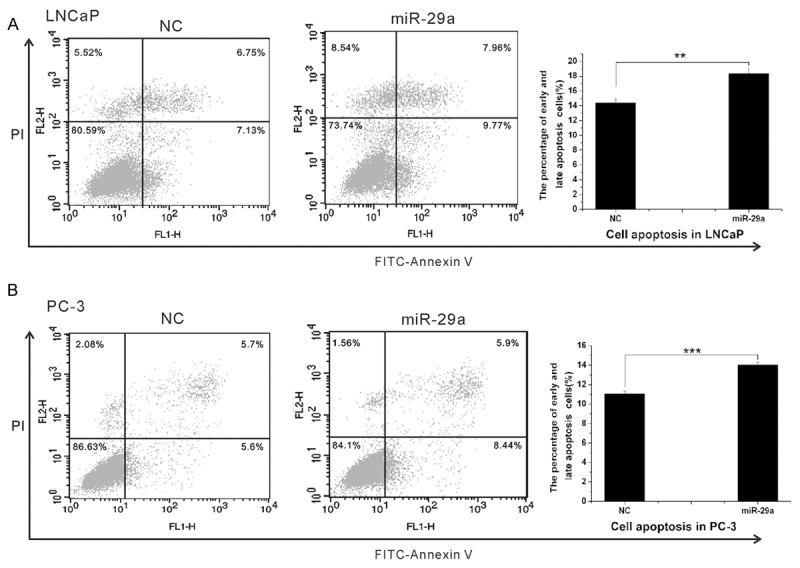
MiR-29a promoted PCa cell apoptosis. Cells were transfected with MiR-29a or miR-NC for 48 hours, and then, subjected to cell apoptosis (stained with PI and FITC-Annexin V). A. In LNCaP cells, MiR-29a overexpression caused an increase in early and late apoptotic cells, and a decrease in living cells. B. In PC-3 cells, miR-29a overexpression caused a decrease in living cells and an increase in early apoptotic cells. Each experiment was performed in triplicate.
KDM5B is a potential target of miR-29a
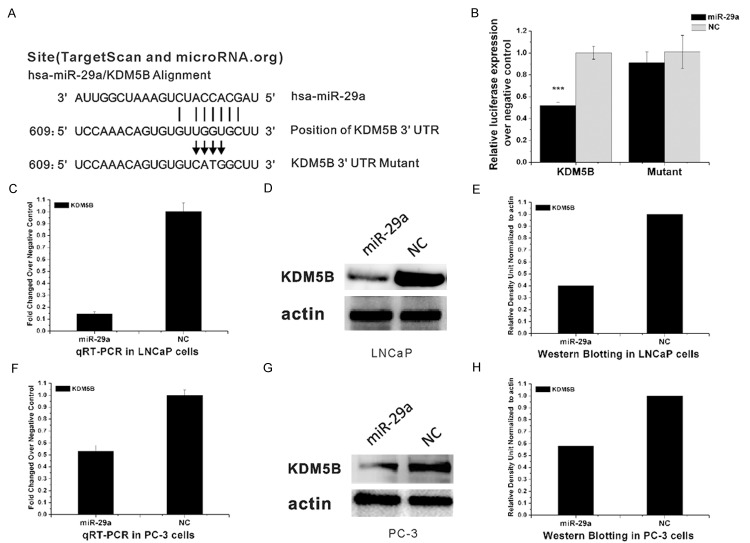
To identify possible miR-29a target genes, we performed a computational screen for genes with complementary sites of miR-29a in their 3’UTR using open-access software. The software included TargetScan (www.targetscan.org), PicTar (http://pictar.bio.nyu.edu), Sanger microRNA target (http://microrna.sanger.ac.uk), and Miranda (www.microrna.org). We focused our attention on KDM5B, a histone lysine demethylase of Jumonji family.
To investigate interactions between miR-29a and KDM5B, we cloned the miR-29a binding sites from the 3’-UTR of KDM5B into a luciferase reporter plasmid containing a constitutively-active promoter and subsequently transfected HEK293T cells (Figure 4B). Co-transfection of miR-29a with luciferase reporter plasmid resulted in less luciferase activities than in transfecting the reporter plasmid alone. Additionally, miR-29a transfection did not reduce the luciferase activity of the reporter construct transfected with mutant 3’UTR of KDM5B. Negative control (NC) miRNA did not affect the luciferase activity of reporters containing either the 3’UTR of KDM5B or the mutant KDM5B construct (Figure 4A, 4B). These results indicated that miR-29a directly interacts with KDM5B.
Figure 4.
MiR-29a directly targeted KDM5B. A. The binding site of MiR-29a to the 3’-UTR of KDM5B. Vertical arrows represent the mutated bases in the KDM5B-3’UTR mutant reporter constructs. B. HEK293T cells were seeded into a 24-well plate, KDM5B reporter construct (wild-type or mutant) or the empty reporter vector was co-transfected with miR-29a and pRL-TK, or co-transfected with miR-NC and pRL-TK. MiR-29a overexpression decreased the luciferase activity of KDM5B-3’UTR wild-type (KDM5B), but not the empty reporter vector (empty). C. Expression of KDM5B mRNA after transfection with miR-29a in LNCaP cells. D, E. Western blot analysis of KDM5B protein levels in LNCaP cells 24 h after transfection. F. Expression of KDM5B mRNA after transfection with miR-29a in PC-3 cells. G, H. Western blot analysis of KDM5B protein levels in PC-3 cells 24 h after transfection. E, H show the relative gray values of each band (normalized to β-actin). Protein bands were quantified using Quantity One software (Bio-Rad, USA).
We also examined the effects of the overexpression of miR-29a on the KDM5B protein expressions in LNCaP and PC-3 cells. Compared with cells transfected with miRNA control, cells transfected with miR-29a showed a significant increase in miR-29a mRNA expression (P<0.01) (Figure 4C, 4F), accompanied by a significant reduction in KDM5B protein expression (P<0.001) (Figure 4D, 4G) Collectively, our results demonstrated that KDM5B is a direct target of miR-29a.
Discussion
MiRNA expression profiles of numerous solid malignancies have been reported [22]. Compared to traditional mRNA and protein markers, miRNA expression patterns are more reliable and sensitive to changes in cell biology. mRNAs and its translated protein levels are not often proportional; one important cause is the regulatory influence of epigenetic mechanisms, including those mediated by miRNAs [23].
Specifically, miRNAs played critical regulatory roles in diverse biological processes, including metabolism [24,25] and tumorigenesis [26,27]; and some miRNAs have been shown to repress well-known oncogenes or tumor suppressors [28]. Recently, more and more evidence indicate that abnormal miRNA expressions are involved in tumorigenesis or metastasis pathways [29,30]. MicroRNAs may regulate the progress of tumors through epigenetic mechanisms. Epigenetic mechanisms usually refer to: histone modification, DNA methylation [31]. According to the new miRbase, humans have about 1,000 mature miRNAs, but it was reported that only approximately 50 miRNAs have abnormal expressions in prostate cancer. However, only a few were involved in the occurrence and development of prostate cancer [32].
Decreased MiR-29s expressions have been reported in many cancers, including rhabdomyosarcoma, cholangiocarcinoma, acute myelogenous leukemia (AML), lung cancer, and nasopharyngeal carcinoma [18,19]. Further, miR-29s have subsequently been predominantly attributed with tumor-suppressing properties; albeit, exceptions have been described in its tumor-promoting functions. Restoration of miR-29 sensitized cholangiocarcinoma and AML cells to apoptotic stimuli, and inhibited rhabdomyosarcoma growth in lung cancer [33,34]. MiR-29s targets diverse protein expressions, such as collagens, transcription factors, and methyltransferases; these may partake in abnormal cell migration, invasion or proliferation; and may also favor cancer development. In our previous study, microRNAs chip screening results of the pathological specimens, including 75 PCa patients, revealed 22 differentially expressed miRNAs; the low miR-29a expression in PCa has been confirmed by RT-PCR. In this study, we also investigated the functional role of miR-29a in PCa. It has been found that miR-29a inhibited the proliferation of LNCaP and PC-3 cells, but stimulated the apoptosis of LNCaP and PC-3 cells. Therefore, miR-29a plays a tumor suppressor role in LNCaP and PC-3 cells. Consistent with this result, we observed that miR-29a inhibited cell proliferation and cell cycle progression, but promoted LNCaP and PC-3 cell apoptosis.
In this study, many oncogenic miR-29a targets have been identified on PCa cells; and the defects of histone methylation may function during the initiation and progression of cancer [35]. Seligson found that Histone 3 lysine 4 dimethylation (H3K4triMe) is associated with the prognosis of low-grade PCa patients [36]. KDM5B is the specific demethylase of H3K4; and its upegulation can reduce H3K4 methylation levels, without affecting other histone lysine methylation status. Histone lysine methylation plays a key role in the epigenetic regulation of eukaryotic genes; histone methylation disorders can lead to cancer [35]. High expressions of KDM5B in PCa have been confirmed [18], but its exact mechanism is unclear.
Recently, some scholars called some microRNAs as epi-miRNAs, which mutually influences with epigenetics [37,38]. MiR-1 and miR-140 can directly regulate the expression level of Histone acetyl transferase 4 (HDAC4). MiR-.1
KDM5B as a miR-29a target in two different PCa cell lines, indicating that miR-29a is a potent inhibitor of KDM5B protein production. Our data showed that KDM5B was upregulated in PCa cell lines. The main regulation occurring at translational level could be one explanation to the fact that the expression levels of miR-29a and KDM5B mRNA in prostate cell lines have negative correlations. In the present study, no positive data was obtained between PCa and KDM5B expression levels due to the small number of PCa samples. Further examination is necessary to elucidate the KDM5B levels in the PCa samples and its relationship with PCa patient survival.
To summarize, we found an increased expression of miR-29a in PCa cells and that miR-29a affected the expression of KDM5B; which suppressed PCa cell proliferation, induced PCa cell apoptosis and controlled PCa development. The results indicated that miR-29a functions as a tumor suppressor by targeting KDM5B, and these findings could potentially be beneficial as a novel therapeutic strategy in PCa.
Acknowledgements
This work is supported by grant 81172426 of the National Natural Science Foundation of China and the Shanghai Edu-cation Commission Research and Innovation projects (No. 12ZZ034).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Can J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Zhang YF, Guan YB, Yang B, Wu HY, Dai YT, Zhang SJ, Wang JP, Anoopkumar-Dukie S, Davey AK, Sun ZY. Prognostic value of Her-2/neu and clinicopathologic factors for evaluating progression and disease-specific death in Chinese men with prostate cancer. Chin Med J (Engl) 2011;124:4345–4349. [PubMed] [Google Scholar]
- 3.Iversen P. Current topics in the treatment of hormone refractory prostate cancer. Eur Urol. 2003;2:3. [Google Scholar]
- 4.Small EJ, Reese DM, Vogelzang NJ. Hormone-refractory prostate cancer: an evolving standard of care. Semin Oncol. 1999;26:61–67. [PubMed] [Google Scholar]
- 5.Locke JA, Emma S, Guns ES, Lubik AA, Adomat HH, Hendy SC, Wood CA, Ettinger SL, Gleave ME, Nelson CC. Androgen Levels Increase by Intratumoral De novo Steroidogenesis during Progression of Castration-Resistant Prostate Cancer. Can Res. 2008;68:6407–6415. doi: 10.1158/0008-5472.CAN-07-5997. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Can. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Can. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Can Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 11.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, Havelange V, Volinia S, Blum W, Rush LJ, Perrotti D, Andreeff M, Bloomfield CD, Byrd JC, Chan K, Wu LC, Croce CM, Marcucci G. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloidleukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santanam U, Zanesi N, Efanov A, Costinean S, Palamarchuk A, Hagan JP, Volinia S, Alder H, Rassenti L, Kipps T, Croce CM, Pekarsky Y. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci U S A. 2010;107:12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sengupta S, den Boon JA, Chen IH, Newton MA, Stanhope SA, Cheng YJ, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci U S A. 2008;105:5874–5878. doi: 10.1073/pnas.0801130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Can Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 15.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncog. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 17.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltrans- ferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang Y, Zhu ZQ, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. PNAS. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mo W, Zhang J, Li X, Meng D, Gao Y, Yang S, Wan X, Zhou C, Guo F, Huang Y, Amente S, Avvedimento EV, Xie Y, Li Y. Identification of novel AR-targeted microRNAs mediating androgen signaling through critical pathways to regulate cell viability in prostate cancer. PLoS One. 2013;8:e56592. doi: 10.1371/journal.pone.0056592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Zhang D, Chen C, Ruan Z, Huang Y. MicroRNA-212 displays tumor-promoting properties in non-small cell lung cancer cells and targets the hedgehog pathway receptor PTCH1. Mol Biol Cell. 2012;23:1423–1434. doi: 10.1091/mbc.E11-09-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang D, Chen C, Li Y, Fu X, Xie Y, Huang Y. Cx31.1 acts as a tumour suppressor in non-small cell lung cancer (NSCLC) cell lines through inhibition of cell proliferation and metastasis. J Cell Mol Med. 2012;16:1047–1059. doi: 10.1111/j.1582-4934.2011.01389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Li RS, Li YH, Zhong S, Chen YY, Zhang CM, Hu MM, Shen ZJ. miR-21 as an independent biochemical recurrence predictor and potential therapeutic target for prostate cancer. J Urol. 2012;187:1466–1472. doi: 10.1016/j.juro.2011.11.082. [DOI] [PubMed] [Google Scholar]
- 23.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 24.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nat. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rayner KJ, Sua′rez Y, Da′valos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Ferna′ndez-Hernando C. miR-33 contributes to the regulation of cholesterol homeostasis. Sci. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nat. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self-renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 28.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Can Meta Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 29.Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Bioc Biop Res Comm. 2009;383:280–285. doi: 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 30.Greither T, Grochola L, Udelnow A, Lautenschläger C, Würl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumours associates with poorer survival. Int J Can. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 31.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Pang YX, Young CYF, Yuan HQ. MicroRNA and Prostate cancer. Acta Bioc Biop Sin. 2010;42:363–369. doi: 10.1093/abbs/gmq038. [DOI] [PubMed] [Google Scholar]
- 33.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncog. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, Andreeff M, Croce CM. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Bioc Biop Acta. 2011;1815:75–89. doi: 10.1016/j.bbcan.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nat. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 37.Valeri N, Vannini I, Fanini F, Calore F, Adair B, Fabbri M. Epigenetics MiRNAs, and human cancer: a new chapter in human gene regulation. Mamm Gen. 2009;20:573–580. doi: 10.1007/s00335-009-9206-5. [DOI] [PubMed] [Google Scholar]
- 38.Fabbri M, Calin GA. Epigenetics and miRNAs in human cancer. Adv Genet. 2010;70:87–99. doi: 10.1016/B978-0-12-380866-0.60004-6. [DOI] [PubMed] [Google Scholar]
- 39.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 41.Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. MiR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncog. 2009;28:1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 42.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, Brenner JC, Yu J, Kim JH, Han B, Tan P, Kumar-Sinha C, Lonigro RJ, Palanisamy N, Maher CA, Chinnaiyan AM. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Sci. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]