Abstract
Objective: We aimed to investigate the relation between adiponectin gene (ADIPOQ) polymorphisms and type 2 diabetes mellitus (T2DM) in a Chinese population. Methods: The present study included 510 subjects with normal glucose tolerant (NGT) and 510 patients with type 2 diabetic. Five SNPs (rs2241767, rs3821799, rs182052, rs1501299 and rs7627128) were genotyped by TaqMan methods. Results: Of these 5 SNPs, three SNPs (rs1501299, rs182052, and rs7627128) were found to be significantly associated with T2DM. The haplotypes AAT (Construction of rs1501299, rs182052, and rs7627128) was frequent in T2DM patients (OR=2.051, 95% CI: 1.439~2.923, P<0.001), but GAT (Construction of rs1501299, rs182052, and rs7627128) was frequent in the control group (OR=0.65, 95% CI: 0.540~0.805, P<0.001). Conclusion: The ADIPOQ gene variants and haplotype were associated with the risk for development of type 2 diabetes.
Keywords: ADIPOQ, diabetes, genetic polymorphism, case-control study
Introduction
Type 2 diabetes mellitus (T2DM) is a common chronic disease worldwide [1]. Although the exact pathogenesis of T2DM is unclear, it is general accepted that T2DM is a multifactor disorders resulting from genetic polymorphisms and several environmental factors [2]. The previous studies indicated that genetic polymorphisms such as SAA gene [3], CYP17A1 gene [4], and CCR5 gene [5] were associated with the risk for T2DM. However, these results can only explain a small fraction of the susceptibility for T2DM.
Previous studies suggested the role of adipose tissue is of great significance where it contributes to the pathogenesis of diabetes as well as obesity by secreting a variety of secretory proteins [6]. Among them, adiponectin is the major adipocyte secretory protein most abundantly found in human plasma with potent roles in insulin sensitivity in muscle and liver, regulating energy homeostasis and glucose tolerance [7]. Adiponectin is a product of the ADIPOQ gene which spans approximately 16 kb with three exons on chromosome 3q27 and has been linked to metabolic syndrome, type 2 diabetes and cardiovascular disease [8,9]. However, the relation between ADIPOQ genetic polymorphisms and T2DM in Chinese population is unclear. Therefore, in this study we investigate the relation between genetic polymorphism in ADIPOQ and T2DM in a Chinese population.
Subjects and methods
Ethnics
The present study has been performed with the approval of the ethics committee of Third Military Medical University and is in compliance with the Helsinki Declaration. The informed consents of the study were collected from all the candidate subjects.
Subjects
We recruited 1,020 unrelated study subjects, including 510 controls subjects with normal glucose tolerant (NGT) and 510 patients with T2DM from July 2010 to June 2013. Oral glucose tolerance test (OGTT) was utilized to confirm diabetes or NGT. According to World Health Organization (WHO) criteria, the subjects who have 2-h plasma glucose value ≥11.1 mmol/l were identified as diabetes and those with 2-h plasma glucose value <7.8 mmol/l were confirmed as NGT [10]. The characteristics of participants were shown in Table 1.
Table 1.
Characteristics of participants
| Groups | N | Sex ratio (Male/Female) | Age (years) | Family history (n, %) | BMI (Kg/m2) | GLU (mmol/L) | TG (mmol/L) | TC (mmol/L) | HDL-C (mmol/L) | LDL-C (mmol/L) |
|---|---|---|---|---|---|---|---|---|---|---|
| T2DM group | 510 | 365/145 | 53.3±10.2 | 147 (28.82) | 24.8±2.7 | 5.52±0.49 | 1.61±0.27 | 2.85±0.68 | 0.89±0.22 | 1.45±0.49 |
| Control group | 510 | 37/139 | 53.9±10.1 | 15 (2.94) | 23.2±1.5 | 4.32±0.43 | 1.52±0.17 | 2.64±0.56 | 0.90±0.21 | 1.27±0.34 |
| P | 0.987 | 0.321 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.893 | <0.001 |
Phenotype measurements
We collected the characteristics of the participants including height, weight, and waist using standardized methods. Fasting plasma glucose, serum cholesterol, serum triglycerides, high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), glycated hemoglobin (HbA1c), and serum insulin concentration were measured according to previous literatures. Total serum adiponectin was measured by radioimmunoassay.
Genetic analysis
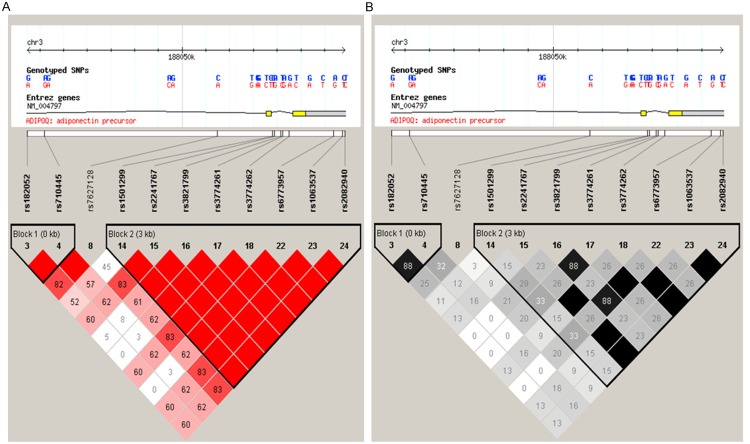
As shown in Figure 1, we obtained five tagging SNPs (rs2241767, rs3821799, rs182052, rs1501299, and rs7627128) for Chinese Han, using the Haploview 4.2 software and the HapMap phrase II database. Genomic DNA was extracted from the whole blood by the phenol chloroform method. Genotyping was confirmed by TaqMan method as described previously [11].
Figure 1.
Genetic variation at human ADIPOQ gene. Using the Haploview 4.2 software and the HapMap phrase II database, we scanned 11 genotyped single-nucleotide polymorphisms (SNPs) in Chinese Han. Linkage disequilibrium (LD) blocks across the locus in Chinese Han. LD blocks derived by solid spline method in Haploview 4.2. A: LD value shown: |D’|×100; |D’| colour scheme: |D’|=0: white; 0<r2<1: shades of Pink; |D’|=1: red; B: LD value shown: r2×100; r2 color scheme: r2=0: white; 0<r2<1: shades of grey; r2=1: black.
Statistical analysis
We utilized SPSS 17.0 software (SPSS, Chicago, IL) to perform the statistical analyses. Gene counting methods were utilized to estimate the allele frequencies. And a χ2 test was used to test the Hardy-Weinberg expectations (HWE). Student’s t test was utilized to compare of the difference of the means between the two groups. We utilized the χ2 test to compare the proportions of genotypes or alleles and utilized the one way analysis of variance to compare groups for continuous variables. We utilized the SHEsis software (http://analysis2.bio-x.cn/myAnalysis.php) [12,13] to perform linkage disequilibrium (LD) analysis and haplotype analysis. Statistical significance was established at P<0.0125.
Results
The genotype distribution of each SNP was in line with the Hardy-Weinberg equilibrium (data not shown). For total participants, the genotype (P=0.008, P=0.006, P<0.001, respectively) and the allele (P=0.006, P=0.005, P<0.001, respectively) distribution of rs182052, rs1501299, and rs7627128 was significant different between the T2DM patients and the control subjects. The genotype and the allele distributions of other two SNPs (rs3821799, and rs2241767) were not different between the T2DM patients and the control participants (Table 2).
Table 2.
Distributions of ADIPOQ genotype
| Groups | N | SNP | Genotypes (n, %) | P value | Allele | OR (95% CI) | P value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| rs2241767 | AA | AG | GG | A | G | |||||
| T2DM group | 510 | 265 (0.520) | 180 (0.353) | 65 (0.127) | 0.325 | 710 (0.696) | 310 (0.304) | 0.90 (0.74-1.09) | 0.307 | |
| Control group | 510 | 271 (0.531) | 189 (0.371) | 50 (0.098) | 731 (0.717) | 289 (0.283) | ||||
| rs3821799 | CC | CT | TT | |||||||
| T2DM group | 510 | 95 (0.186) | 220 (0.431) | 195 (0.382) | 0.231 | 410 (0.402) | 610 (0.598) | 1.05 (0.87-1.25) | 0.543 | |
| Control group | 510 | 83 (0.163) | 232 (0.455) | 195 (0.382) | 398 (0.390) | 622 (0.610) | ||||
| rs182052 | AA | AG | GG | |||||||
| T2DM group | 510 | 101 (0.198) | 257 (0.504) | 152 (0.298) | 0.008 | 459 (0.450) | 561 (0.550) | 1.27 (1.07-1.52) | 0.006 | |
| Control group | 510 | 87 (0.171) | 224 (0.439) | 199 (0.390) | 398 (0.390) | 622 (0.610) | ||||
| rs1501299 | AA | AG | GG | |||||||
| T2DM group | 510 | 87 (0.171) | 301 (0.590) | 122 (0.239) | 0.0006 | 475 (0.466) | 545 (0.534) | 1.28 (1.07~1.53) | 0.005 | |
| Control group | 510 | 80 (0.157) | 253 (0.496) | 177 (0.347) | 413 (0.405) | 607 (0.595) | ||||
| rs7627128 | AA | AC | CC | |||||||
| T2DM group | 510 | 61 (0.120) | 293 (0.575) | 156 (0.306) | <0.0001 | 415 (0.407) | 605 (0.593) | 1.40 (1.16-1.6) | 0.0002 | |
| Control group | 510 | 55 (0.108) | 226 (0.443) | 229 (0.449) | 336 (0.329) | 684 (0.671) | ||||
According to the |D’| and r2 values of these five SNPs, there are three SNPs (rs1501299, rs182052, and rs7627128) are located in one haplotype block. In the haplotype-based case-control analysis, haplotypes were established through the use of all these three SNPs. As shown in Table 3, the haplotypes AAT (Construction of rs1501299, rs182052, and rs7627128) was frequent in T2DM patients (OR=2.051, 95% CI: 1.439~2.923, P<0.001), but GAT (Construction of rs1501299, rs182052, and rs7627128) was frequent in the control group than that in the T2DM group (OR=0.65 95% CI: 0.540~0.805, P<0.001).
Table 3.
Haplotype analyses results
| Variables | Case (n, frequency) | Control (n, frequency) | P value | OR (95% CI) |
|---|---|---|---|---|
| A A C | 378.36 (0.371) | 363.57 (0.356) | 0.519032 | 1.061 [0.886~1.272] |
| A A T | 96.62 (0.095) | 49.43 (0.048) | <0.001 | 2.051 [1.439~2.923] |
| G A T | 226.36 (0.222) | 307.10 (0.301) | <0.001 | 0.659 [0.540~0.805] |
| G G C | 22.98 (0.023) | 23.52 (0.023) | 0.929743 | 0.974 [0.545~1.743] |
| G G T | 287.00 (0.281) | 265.47 (0.260) | 0.296449 | 1.110 [0.913~1.350] |
Discussion
T2DM is common geriatric diseases and their incidences increase over age. Its pathogeneses are complex and are usually influenced by genetic and environmental factors. Multiple genes have been found to be associated with T2DM. The present study found that the ADIPOQ gene rs1501299, rs182052, and rs7627128 genotype distribution frequencies were significant between T2DM patients and healthy control subjects, which indicated that ADIPOQ gene was associated with T2DM in Chinese population.
Previously, Gu and the colleagues found that ADIPOQ gene polymorphisms were associated with T2DM in Swedish Caucasians [12]. There were several studies related to the association of ADIPOQ gene polymorphisms with T2DM have been carried out. However, the results from each study were found to be controversial. Ye and colleagues found there was significant association ADIPOQ gene polymorphism and the serum adiponectin level and the T2DM hereditary risk [13]. Sun et al. [14] also found T2DM risk was related to the ADIPOQ gene variant. However, Shi et al. [15] and Li et al [16] did not verify this association mentioned above.
Previous study suggested that analysis based on haplotypes has advantages over analysis based on individual SNPs for genes with multiple susceptibilities [17]. In the present study, we carried out a haplotype-based case-control study to investigate the association of ADIPOQ polymorphisms with T2DM, and we found rs182052, rs1501299, and rs7627128 polymorphisms were significantly associated with T2DM. In addition, the haplotypes AAT (Construction of rs1501299, rs182052, and rs7627128) was frequent in T2DM patients (OR=2.051, 95% CI: 1.439~2.923, P<0.001), but GAT (Construction of rs1501299, rs182052, and rs7627128) was frequent in the control group than that in the T2DM group (OR=0.65, 95% CI: 0.540~0.805, P<0.001).
In conclusion, we found ADIPOQ gene polymorphisms was associated with T2DM. The AAT haplotype appear to be a useful genetic marker and the GAT haplotype might be protective factors of T2DM in Chinese people.
Disclosure of conflict of interest
None.
References
- 1.Pandya N, Wei W, Meyers JL, Kilpatrick BS, Davis KL. Burden of sliding scale insulin use in elderly long-term care residents with type 2 diabetes mellitus. J Am Geriatr Soc. 2013;61:2103–10. doi: 10.1111/jgs.12547. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Salem A, Ajina M, Suissi M, Daher HS, Almawi WY, Mahjoub T. Polymorphisms of transcription factor-7-like 2 (TCF7L2) gene in Tunisian women with polycystic ovary syndrome (PCOS) Gene. 2014;533:554–7. doi: 10.1016/j.gene.2013.09.104. [DOI] [PubMed] [Google Scholar]
- 3.Xie X, Ma YT, Yang YN, Li XM, Zheng YY, Fu ZY, Ma X, Liu F, Huang Y, Chen BD. SAA1 genetic polymorphisms are associated with plasma glucose concentration in non-diabetic subjects. Clin Chem Lab Med. 2013;51:2331–4. doi: 10.1515/cclm-2013-0097. [DOI] [PubMed] [Google Scholar]
- 4.Yang SJ, Lee ST, Kim WJ, Park SE, Park SW, Kim JW, Park CY. Genetic variation in CYP17A1 is associated with arterial stiffness in diabetic subjects. Exp Diabetes Res. 2012;2012:827172. doi: 10.1155/2012/827172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buraczynska M, Zukowski P, Wacinski P, Berger-Smyka B, Dragan M, Mozul S. Chemotactic cytokine receptor 5 gene polymorphism: relevance to microvascular complications in type 2 diabetes. Cytokine. 2012;58:213–7. doi: 10.1016/j.cyto.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Yang FS, Yun CH, Wu TH, Hsieh YC, Bezerra HG, Liu CC, Wu YJ, Kuo JY, Hung CL, Hou CJ, Yeh HI, Lee JJ, Bulwer BE, Cury RC. High pericardial and peri-aortic adipose tissue burden in pre-diabetic and diabetic subjects. BMC Cardiovasc Disord. 2013;13:98. doi: 10.1186/1471-2261-13-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamauchi T, Waki H, Kamon J, Murakami K, Motojima K, Komeda K, Miki H, Kubota N, Terauchi Y, Tsuchida A, Tsuboyama-Kasaoka N, Yamauchi N, Ide T, Hori W, Kato S, Fukayama M, Akanuma Y, Ezaki O, Itai A, Nagai R, Kimura S, Tobe K, Kagechika H, Shudo K, Kadowaki T. Inhibition of RXR and PPARgamma ameliorates diet-induced obesity and type 2 diabetes. J Clin Invest. 2001;108:1001–13. doi: 10.1172/JCI12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francke S, Manraj M, Lacquemant C, Lecoeur C, Leprêtre F, Passa P, Hebe A, Corset L, Yan SL, Lahmidi S, Jankee S, Gunness TK, Ramjuttun US, Balgobin V, Dina C, Froguel P. A genome-wide scan for coronary heart disease suggests in Indo-Mauritians a susceptibility locus on chromosome 16p13 and replicates linkage with the metabolic syndrome on 3q27. Hum Mol Genet. 2001;10:2751–65. doi: 10.1093/hmg/10.24.2751. [DOI] [PubMed] [Google Scholar]
- 9.Vionnet N, Hani EH, Dupont S, Gallina S, Francke S, Dotte S, De Matos F, Durand E, Leprêtre F, Lecoeur C, Gallina P, Zekiri L, Dina C, Froguel P. Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21-q24. Am J Hum Genet. 2000;67:1470–80. doi: 10.1086/316887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15:539–53. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 11.Xiang X, Ma YT, Fu ZY, Yang YN, Xiang M, Chen BD, Wang YH, Fen Liu. Haplotype analysis of the CYP8A1 gene associated with myocardial infarction. Clin Appl Thromb Hemost. 2009;15:574–80. doi: 10.1177/1076029608329581. [DOI] [PubMed] [Google Scholar]
- 12.Gu HF, Abulaiti A, Ostenson CG, Humphreys K, Wahlestedt C, Brookes AJ, Efendic S. Single nucleotide polymorphisms in the proximal promoter region of the adiponectin (APM1) gene are associated with type 2 diabetes in Swedish caucasians. Diabetes. 2004;53:S31–S35. doi: 10.2337/diabetes.53.2007.s31. [DOI] [PubMed] [Google Scholar]
- 13.Ye F, He L, Li JN, Dong CP, Yu J. Association of SNPs in the proximal promoter region of the APM1 gene with obesity and type 2 diabetes. Journal of Xian Jiaotong University (Medical Sciences) 2008;29:74–76. [Google Scholar]
- 14.Sun H, Wang SM, Zhuang J, Liu HL, Zhou HH, Wu J, Liu ZQ. The association of adiponectin allele 45T/G and -11377C/G polymorphisms with Type 2 diabetes and rosiglitazone response in Chinese patients. Br J Clin Pharmacol. 2008;65:917–26. doi: 10.1111/j.1365-2125.2008.03145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi XH, Jin F, Wang L, Yang Z. Association of SNPs-11377 C/G in proximal promoter region of adiponectin gene with type 2 diabetes in northern Chinese Han population. Journal of Clinical Rehabilitative Tissue Engineering Research. 2007;11:6815–6818. [Google Scholar]
- 16.Li YP, Zhang Y, Song DP, Yang Y, Yao YF. Single nucleotide polymorphism haplotypes of adiponectin gene are associated with type 2 diabetes in Han population. Chin J Diabetes. 2011;19:101–104. [Google Scholar]
- 17.Morris RW, Kaplan NL. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet Epidemiol. 2002;23:221–33. doi: 10.1002/gepi.10200. [DOI] [PubMed] [Google Scholar]