Abstract
Infection in neonates, particular the neonatal sepsis continues to be a global problem with significant morbidity and mortality. The diagnosis of neonatal sepsis is complicated by nonspecific clinical symptomatology, a high-false negative rate, and a delay in obtaining blood culture results. MicroRNAs (miRNAs) have recently been used as finger prints for sepsis, and have been validated to be potential sepsis biomarker recently. In the present study, we investigated the level of several miRNAs, such as miR-15a, miR-16, miR-15b, and miR-223, which have been identified as a biomarker in adult sepsis, in neonatal sepsis patients, and then we analyzed the association of miR-15a/16 with the patient prognosis. Results demonstrated that the level of miR-15a/16 was up-regulated in neonatal sepsis patients than in normal neonatal subjects; however, no statistical difference was disclosed in the miR-15b and miR-223 level between two groups. And the ROC analysis indicated the miR-15a and miR-16 were potent fingerprints for diagnosing neonate sepsis. In order to explore the miR-15a/16 function on the lipopolysaccharide (LPS)-induced inflammatory pathway, the mice macrophage RAW264.7 cells were transiently transfected with miR-15a/16 mimics. And it was demonstrated that the miR-15a/16 transfection down-regulated the Toll-like receptor 4 (TLR4) and Interleukin-1 receptor-associated kinase 1 (IRAK-1) transcription level with a statistical difference in the LPS treated cells. And the suppression capability of miR-15a/16 on the expression of TLR-4 and IRAK-1 were evaluated by western blot. Thus, in present study, we identified miR-15a/16 as potential biomarker for the diagnosis and prognosis of neonatal sepsis, and the upregulated miR-15a/16 downregulated the LPS-induced inflammatory pathway.
Keywords: miR-15a/16, neonatal sepsis, TLR4, IRAK-1
Introduction
Neonatal sepsis is a blood infection mainly caused by bacteria that occurs in an infant within 28 days old. Neonatal sepsis is categorized as early-onset or late-onset. Early-onset sepsis takes place in the first 3 days of life, while late-onset usually occurs at 4-28 days of life. The pathogens causing the neonatal sepsis majorly include Group B Streptococcus (GBS), Escherichia coli, Coagulase-negative Staphylococcus, Haemophilus influenza, and Listeria monocytogenes etc [1]. The incidence of neonatal sepsis in the developed countries is 1-8 per 1000 newborns, yet it is approximately three times in developing countries [2]. In China, the incidence of neonatal sepsis varies considerably among different regions; based on the limited clinical statistical data of few developed areas, the incidence of neonatal sepsis is 3-6.5 per 1000 newborns [3]. For the immature of infant’s immune system, the neonatal sepsis has distinct clinical symptoms compared with that of children or adults sepsis, suggesting a unique immune response against infection in the infant [4]. Due to the non-specific clinical symptom, the diagnosis of neonatal sepsis often presents a highly-false negative rate and a delay in obtaining blood culture results. Thus, biomarkers for distinguishing the sepsis accurately at earlier stage are urgently needed.
MicroRNAs (miRNAs) are endogenous small RNAs of ~22 nt lengths that regulate gene expression via binding target mRNAs for cleavage or translational suppression [5]. Unique patterns of serum or plasma miRNAs levels can be used as biomarkers for diagnosis or prognosis of various diseases lately [6-8]. Among them, miR-223 and miR-146a had been identified with diagnostic value for sepsis, another one plasma miRNA, miR-150, was identified as a potential biomarker for sepsis prognosis [9]. Recently, one report described miR15a was associated with the sepsis progress [10]. As we knew, the neonatal sepsis displayed the different symptoms in comparison with that of children or adults sepsis because of the infant’s immature immune system. It’s not clear whether these miRNA biomarkers can be used as fingerprints to evaluate the prognosis of neonatal sepsis, and there have been few studies on the evaluation of miRNA level in infant sepsis, thus it is necessary to confirm whether these miRNAs can function as biomarkers in neonatal sepsis patients.
In this research, we investigated the level of miR-15a and miR16 in neonatal sepsis patients and analyzed the correlation of miR-15a/16 with the patient prognosis. Results confirmed the higher level of miR-15a/16 in neonatal sepsis patients than in normal neonatal subjects.
Materials and methods
Patients and blood samples collection
This study was mainly conducted in the department of pediatrics, the Yantai Yuhuangding Hospital of Qingdao University and was approved by the ethics committee of the hospital. We obtained 87 blood samples from neonates with sepsis and respiratory infection/pneumonia, including 46 neonates with sepsis and 41 neonates with respiratory infection/pneumonia. The blood samples from neonates with respiratory infection or pneumonia were used as controls. The characteristics of the subjects are shown in Table 1. The diagnosis of neonatal sepsis was performed as the criteria established at the 2003 Kunming Neonatal Sepsis Definitions Conference [3]. We collected the blood samples from the neonates with sepsis and the control subjects when they were diagnosed.
Table 1.
Clinical data for neonatal sepsis patients and control subjects
| Variant items | Neonatal sepsis (n=46) | Controlα (n=41) | p value |
|---|---|---|---|
| Gender (M/F) | 27/19 | 25/16 | 0.829A |
| Age (D) | 12.64±7.62 | 13.96±8.25 | 0.423B |
| Gestational age (W) | 38.24±2.16 | 38.86±1.96 | 0.346B |
| Weight (g) | 3463.25±525.67 | 3356.18±498.18 | 0.735B |
| PCT (ng/ml) | 4.26 (0.31, 13.26) | 1.74 (0.23, 5.39) | 0.006C |
| CRP (mg/L) | 13.71 (4.83, 22.36) | 10.16 (3.67, 20.56) | 0.134C |
| WBC (×109/L) | 12.78 (4.68, 18.37) | 11.03 (5.37, 16.42) | 0.087C |
Control subjects involved 17 (9/6) patients with supper respiratory infection and 24 (16/10) patients with pneumonia;
M, Male; F, Female; D, Days; W, Weeks;
Chi Square tests;
Student t test;
Mann-Whitney U tests.
Serum total RNA isolation
Briefly, blood samples were collected in special tubes containing separating gel and clot activator and centrifuged at 3000 rpm for 15 mins at room temperature. The supernatants were collected and centrifuged at 15,000 rpm for 30 mins to precipitate cell debris, and then the supernatants were stored at -80°C until RNA extraction. Serum total miRNAs was isolated using mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer‘s instructions; whereas the serum total mRNA or mRNA in RAW264.7 cells were extracted by using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA).
Evaluation of RNA transcription level using qRT-PCR
Reverse transcription (RT) was done in a reaction volume of 15 μl containing 1.5 μl of 10× reverse transcription buffer, 0.5 μl of RNase inhibitor, 1 μl of dNTPs, 0.5 μl of reverse transcriptase, 1.5 μl of miRNA-specific stem-loop RT primer (Applied Biosystems), 2.5 μl (5 ng) of RNA preparations and 7.7 μl RNase-free water. RT reaction was performed as 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. Then, 4.5 μl of 5:28 diluted RT product was combined with 5.0 μl of TaqMan gene expression Master Mix and 0.5 μl of Taqman miRNA assay. The qPCR was made using an ABI PRISM 7300 detection system at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All qPCR reactions were performed in triplicate and the raw Ct (threshold cycle) of each sample was the mean value of three independent Ct values. Housekeeping gene β-actin or U6 was simultaneously amplified to standardize the amount of sample RNA. Relative quantification of gene transcription level was performed by the -ΔΔct method [11] .
Cell lines and transfection
Mice macrophage RAW264.7 cells were maintained in the DMEM (GIBCO, Rockville, MD, USA) supplemented with 10% fetal bovine serum (Hyclone, Pittsburgh, PA, USA), 50 units/mL penicillin, and 50 mg/mL streptomycin in a humidified incubator under 5% CO2 at 37°C. The miRNA transfection was performed with the lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) as described in its protocol.
Western blotting analysis
The treated RAW264.7 cells were lysed in RIPA lysis buffer (Pierce, Rockford, IL, USA) with 1 mM phenylmethanesulfonyl fluoride (PMSF). The cell lysates were subjected to SDS-PAGE and blotted onto PVDF membrane. Blocking was done with 5% non-fat milk. The membrane was incubated with rabbit polyclone antibodies to TLR-4 (Abcam, Cambridge, UK) or IRAK-1 (Abcam, Cambridge, UK) at 1:1000 dilutions for 1 hour and subsequently reacted with second IgG at 1:5000 dilutions for 30 minutes. Membranes were washed with PBST buffer for 4 times after each step. Western-light substrate was then applied to the membrane for 5 minutes. Once the membrane was dried, X-ray films were exposed to the membrane and developed by a Kodak processor.
Statistical methods
The ROC curves of selected miRNAs were mapped as previously described by the soft SPSS16.0 [12]. The U-test was used for the comparison of miRNAs levels in the sera between the neonatal groups and control groups. The student’s t-test was applied in comparison of the transcription and expression level between two groups after miRNA transfection.
Results
Clinical characterizations and miRNA profiles in the neonate sepsis patients and the control patients
46 neonate sepsis patients and 41 control patients (17 patients with supper respiratory infection and 24 patients with pneumonia) were enrolled in the study. There were no significant differences in the gender, age, gestational age, weight, and CRP and WBC values between the two groups. However, PCT value was significantly different between sepsis patients and control subjects (Table 1). To explore the miRNA profiles in two groups, a panel of miRNAs associated with the inflammation response was quantified using real-time RT-PCR, the fold changes of serum miRNA in neonatal sepsis patients compared with control subjects were listed in Table 2, miR15a and miR16 were up-regulated significantly with statistical differences.
Table 2.
Fold-changes of serum miRNAs in patients with neonatal sepsis (S), compared to control subjects (C)
| miRNAs | Fold-change (S/C)Ψ | p value# |
|---|---|---|
| miR-15a | 3.36 | <0.001 |
| miR-15b | 1.98 | 0.057 |
| miR-16 | 2.97 | <0.001 |
| miR-206 | -1.78 | 0.064 |
| miR-223 | 1.72 | 0.102 |
| miR-378 | -2.56 | 0.042 |
| miR-451 | 2.76 | 0.036 |
Fold change formula: Fold-change = log2 (D/S).
Student t test.
miR-15a and miR-16 levels increased in patients of neonate sepsis
The transcription levels of miR-15a, miR-16, miR-15b, and miR-223 of neonate sepsis patients and control group were measured by Real-time RT-PCR respectively. As shown in the Figure 1C and 1D, no statistical difference was disclosed in the miR-15b and miR-223 level between two groups, however the miR-15a and miR-16 levels were both greatly improved compared with the control group (Figure 1A and 1B) (P<0.0001). In detail, the mean relative miR-15a and miR-16 level increased 2.5 and 3 times in comparison with control, separately. The results indicated the miR-15a and miR-16 might be the appropriate biomarker for diagnosing the Neonate Sepsis.
Figure 1.
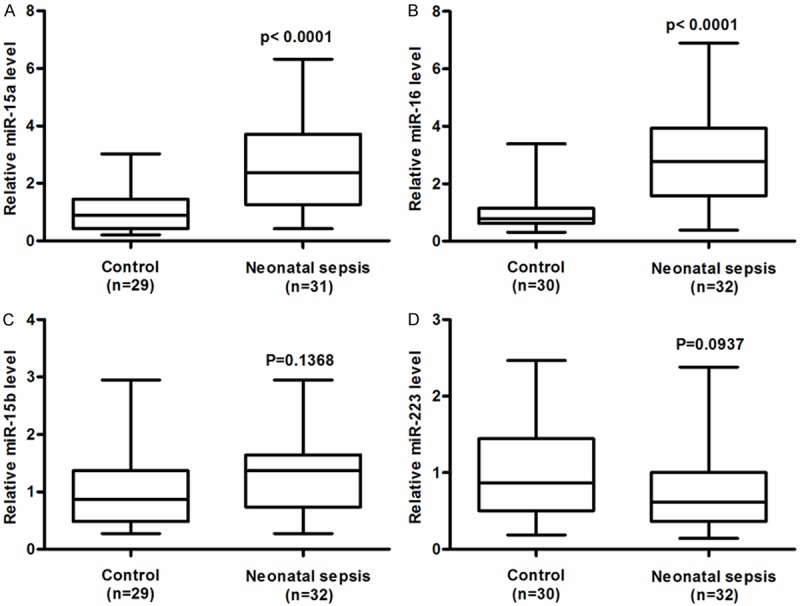
Relative serum levels of miRNA-15a, miRNA-16, miRNA-15b, miRNA-223 in neonatal sepsis patients. The relative amounts of miR-15a (A), miR-16 (B), miR-15b (C), and miR-223 (D), were examined by RT-qPCR in the serum of neonatal sepsis patients, the normal neonatal subjects were taken as control. The level of each miRNA in control neonatal subjects were designated as “1”. All other samples were expressed as a relative value to the control sample. The sample numbers and the statistical significance were shown respectively.
ROC analysis indicated the miR-15a and miR-16 were potent fingerprints for diagnosing neonate sepsis
To investigate the predictive mortality value of the four selected miRNAs, receiver operating characteristic (ROC) curves were generated and the areas under the curves (AUCs) were calculated. The AUCs for the four miRNAs ranged from 0.62850 to 0.790. The AUCs for miR-15a, miR-16, miR-15b and miR-223 were, 0.8543, 0.8688, 0.6285, and 0.6323, respectively (Figure 2). The AUC for miR-16 had the highest value, followed by that for miR-15a, and both displayed dramatic statistical differences (Figure 2A and 2B).
Figure 2.
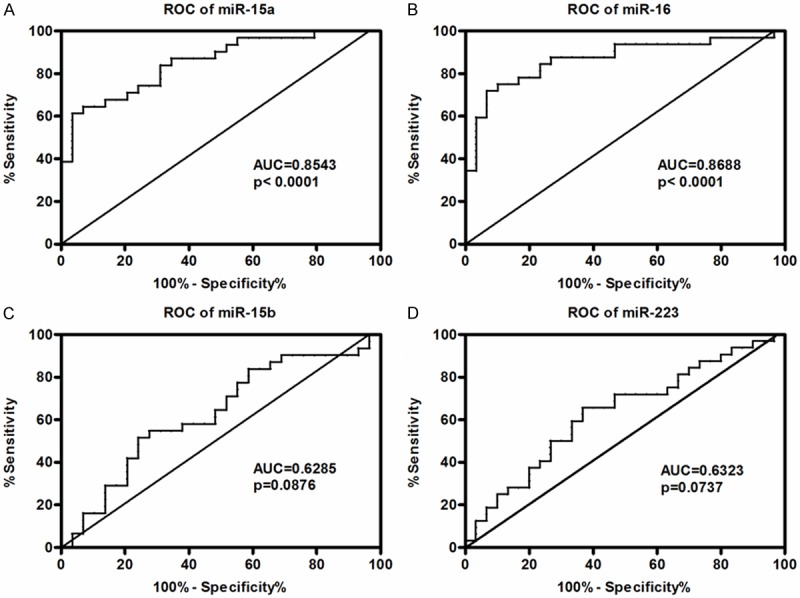
ROC analyses of the serum levels of miRNA-15a, miRNA-16, miRNA-15b, miRNA-223 in neonatal sepsis patients. The area under curve (AUC) and the statistical significance were shown in each figure.
miR-15a/16 suppressed the LPS-promoted TLR-4/IRAK signaling
In order to explore the miR-15a /16 function on the LPS-induced inflammatory pathway, the mice macrophage RAW264.7 cells were transiently transfected with miR-15a/16 mimic or miR control, respectively. Then the copies of miR-15a/16 and house-keeping gene U6 were subsequently measured by RT-qPCR. As shown in Figure 3A, the relative miR-15a/16 level to U6 increase significantly at 25 nM post-transfection, and the level rose to a higher level at the concentration of 50 nM. In the next step, we checked the effect of miR-15a/16 on TLR-4 expression in the LPS treated RAW264.7 cells. In the cells without LPS treatment, no statistical difference was observed between the miR Control-transfected group and the miR-15a/16-transfected group, nevertheless the miR-15a/16 transfection down-regulated the TLR4 transcription level with a statistical difference in the LPS treated cells (Figure 3B). The suppression capability of miR-15a/16 on the expression of TLR-4 and IRAK-1 were evaluated by Western Blot. Obviously, the miR15a/16 did not influence the expression of TLR-4 or IRAK-1 without LPS treatment, however the TLR-4 and IRAK-1 expressions were significantly decreased by miR15a/16 under the LPS treatment (Figure 3C). Furthermore, statistical difference was clearly disclosed between the LPS-treated miR-con groups and the LPS-treated miR15a/16 groups (Figure 3D).
Figure 3.
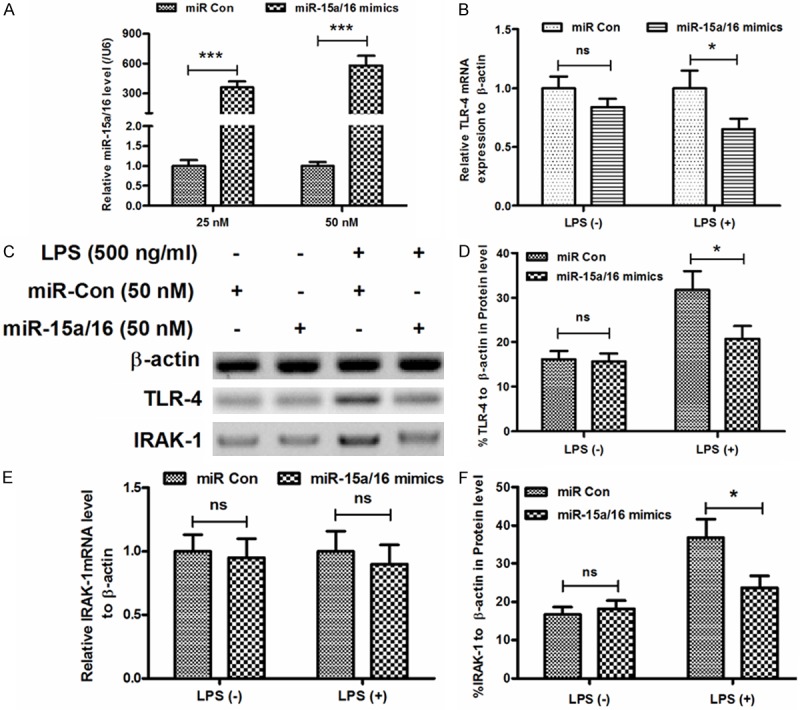
Manipulation of miR-15a/16 level in mice macrophage RAW264.7 cells downregulated the LPS-promoted TLR-4/IRAK signaling. A: Up-regulation of miR-15a/16 level in RAW264.7 cells by the transfection with miR-15a/16; B: Downregulation of the LPS-promoted TLR-4 mRNA level in RAW264.7 cells by the transfection with miR-15a/16; C, D, F: Western blot analysis of TLR-4 and IRAK-1 in the LPS-treated RAW264.7 cells with the transfection with miR-15a/16; E: Downregulation of the LPS-promoted IRAK-1 mRNA level in RAW264.7 cells by the transfection with miR-15a/16. All experiments were performed in triplicate. Statistical significance was considered with P<0.05 (*), P<0.001 (***) or no significance (ns).
Similar results were also observed in the IRAK-1 expression, the miR-15a/16 transfection did not improve the IRAK-1 transcription level whether or not under the LPS treatment (Figure 3E), yet the miR15a/16 decreased the relative IRAK-1 protein level to β-actin compared with the miR control with LPS (Figure 3F).
Discussions
In this research, we compared the miRNA profiles between the neonatal sepsis patients and control subjects, and found the miR15a and miR16 expressions were improved in the serum of neonatal sepsis patients; the molecular mechanism of miR15a and miR16 were also evaluated in RAW264.7 cells by the transfection with miR-15a/16 mimics, miR15a/16 mimics transfection could suppress the expressions of sepsis related molecules, TLR4 and IRAK-1 in vitro. The miR-15/16 could be considered as potential biomarkers for the diagnosis and prognosis of neonatal sepsis.
Previous researches on miRNAs in sepsis patients mainly focused on the miRNA expression profiles of WBCs [13]. The expression level of miR-146b, miR-150, miR-342, and miR-let-7 g were found differentially in WBCs of healthy donors post E. Coli lipopolysaccharide (LPS) treatment, and miR-150, miR-182, miR-342-5p, and miR-486 were also identified in sepsis patients’ WBCs [14]. Recent researches reported miR-15a, miR-16, miR-193b, and miR-483-5p in patients’ sera were associated with death caused by sepsis [10,15]. However, these experiments were performed based on the adults’ sepsis, whether these biomarkers are fit for diagnosing neonatal sepsis patients were still unknown. One neonatal sepsis-based research even indicated miR16 was down regulated in G- infected neonatal sepsis [3]. Obviously, there is no common consensus regarding which miRNAs are qualified as biomarkers; our research clearly confirmed that miR15a and miR16 are potential fingerprints in diagnosing the neonatal sepsis.
Numerous researches had been performed on the identification of novel miRNAs as biomarkers for adults sepsis or neonatal sepsis using different methods [16-19], yet few studies were carried out to testify the mechanism of identified miRNA in inflammation response [20], although it is the pivotal step to explore the function of miRNAs in the LPS-induced chain reactions and whether the identified miRNAs were appropriate for the diagnostic criteria. Here, we checked the inhibition effectors of miR15a/16 on the key molecules (TLR4 and IRAK1) in the LPS-induced inflammatory pathway, we found the miR15a/16 could down-regulate the expression level of TLR4 and IRAK1, suggesting the miR15a/16 play an important role in the inflammatory responses of neonatal sepsis, thus, miR15a/16 are considered as ideal markers for neonatal sepsis.
This study also had some limitations. Firstly, the pathogenic bacteria causing neonatal sepsis were usually classified as Gram-positive bacteria and Gram-negative bacteria, these two kinds of bacteria have different pathogenesis that causes the differentially expressed miRNA profiles. In our research, we didn’t identify which kind of bacteria is responsible for the neonatal sepsis via blood incubation, and the sepsis patients were not divided into two groups: Gram-positive bacteria infected group and Gram-negative bacteria infected group, thus some characteristics miRNAs for these two groups were neglected, related research on the miRNAs profiles of these two groups will be performed in the future. Secondly, we only test the anti-TLR4/IRAK1 function of in vitro, thus a pilot study in animal models is necessary, and detailed research into the molecular mechanism on miR15a/16 should be carried out in the future.
Regardless of above limitations, the two miRNAs (miR15a/16) were found to be valuable predictors of neonatal sepsis, studying the kinetics of the expression levels of these two miRNAs during sepsis treatment would be even more valuable. Additionally, identifying the target genes of miR15a/16 and disclosing the function of target genes may provide new targets for the treatment of neonatal sepsis. In conclusion, the two serum miRNAs (miR15a/16) identified in our study can be used to diagnose the neonatal sepsis.
Disclosure of conflict of interest
None.
References
- 1.Klinger G, Levy I, Sirota L, Boyko V, Reichman B, Lerner-Geva L. Epidemiology and risk factors for early onset sepsis among very-low-birthweight infants. Am J Obstet Gynecol. 2009;201:31–38. doi: 10.1016/j.ajog.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Thaver D, Zaidi AK. Burden of neonatal infections in developing countries: a review of evidence from community-based studies. Pediatr Infect Dis J. 2009;28:S3–S9. doi: 10.1097/INF.0b013e3181958755. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Jiang S, Cao Y, Yang Y. Altered miRNAs expression profiles and modulation of immune response genes and proteins during neonatal sepsis. J Clin Immunol. 2014;34:340–348. doi: 10.1007/s10875-014-0004-9. [DOI] [PubMed] [Google Scholar]
- 4.Futata EA, Fusaro AE, de Brito CA, Sato MN. The neonatal immune system: immunomodulation of infections in early life. Expert Rev Anti Infect Ther. 2012;10:289–298. doi: 10.1586/eri.12.9. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 9.Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, Ferracin M, Nicoloso MS, Barbarotto E, Popa M, Stanciulea O, Fernandez MH, Tulbure D, Bueso-Ramos CE, Negrini M, Calin GA. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One. 2009;4:e7405. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie L. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients’ mortality: a prospective observational study. PLoS One. 2012;7:e38885. doi: 10.1371/journal.pone.0038885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Feilotter HE, Pare GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R, Deng T, Liu H, Yin J, Wang S, Zen K, Ba Y, Zhang CY. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784–791. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt WM, Spiel AO, Jilma B, Wolzt M, Muller M. In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem Biophys Res Commun. 2009;380:437–441. doi: 10.1016/j.bbrc.2008.12.190. [DOI] [PubMed] [Google Scholar]
- 14.Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, Ferracin M, Nicoloso MS, Barbarotto E, Popa M, Stanciulea O, Fernandez MH, Tulbure D, Bueso-Ramos CE, Negrini M, Calin GA. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One. 2009;4:e7405. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie LX. Evidence for serum miR-15a and miR-16 levels as biomarkers that distinguish sepsis from systemic inflammatory response syndrome in human subjects. Clin Chem Lab Med. 2012;50:1423–1428. doi: 10.1515/cclm-2011-0826. [DOI] [PubMed] [Google Scholar]
- 16.Wang HJ, Zhang PJ, Chen WJ, Jie D, Dan F, Jia YH, Xie LX. Characterization and Identification of novel serum microRNAs in sepsis patients with different outcomes. Shock. 2013;39:480–487. doi: 10.1097/SHK.0b013e3182940cb8. [DOI] [PubMed] [Google Scholar]
- 17.Wang HJ, Zhang PJ, Chen WJ, Feng D, Jia YH, Xie LX. Four serum microRNAs identified as diagnostic biomarkers of sepsis. J Trauma Acute Care Surg. 2012;73:850–854. doi: 10.1097/TA.0b013e31825a7560. [DOI] [PubMed] [Google Scholar]
- 18.Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, Zhu KM. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun. 2010;394:184–188. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 19.Aziz M, Jacob A, Yang WL, Matsuda A, Wang P. Current trends in inflammatory and immunomodulatory mediators in sepsis. J Leukoc Biol. 2013;93:329–342. doi: 10.1189/jlb.0912437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wynn JL, Neu J, Moldawer LL, Levy O. Potential of immunomodulatory agents for prevention and treatment of neonatal sepsis. J Perinatol. 2009;29:79–88. doi: 10.1038/jp.2008.132. [DOI] [PMC free article] [PubMed] [Google Scholar]


