Abstract
Objective: Chemokine (C-C motif) receptor 6 gene (CCR6) polymorphism has been reported to be associated with rheumatoid arthritis (RA) in different ethnic populations. Moreover, its inhibition by monoclonal antibody in mouse model has suppressed arthritis. However, few replication studies have reported conflicting results about this association. Therefore, to establish that CCR6 indeed is a risk factor associated with RA among different ethnic populations, a comprehensive meta-analysis study was conducted. Methods: PubMed and MEDLINE databases were searched using the term ‘CCR6’ for all articles published before May 2014. All the replication studies examining the association between CCR6 and RA were reviewed for meta-analysis. Data were summarized using random-effects meta-analysis. The heterogeneity and publication bias among studies were examined by χ2 -based Q statistic test and Egger’s test, respectively. Results: A total of 24955 RA patients and 56129 controls from seven articles were included in the meta-analysis. While CCR6 was a risk factor in Asian (OR = 1.19, 95% CI: 1.14-1.24) and European (OR = 1.14, 95% CI: 1.08-1.21) populations, it was indicated as a protective factor in African Americans (OR = 0.79, 95% CI: 0.62-0.96). Conclusions: Our meta-analysis study concludes that there is a significant association between CCR6 and RA in all racial groups except African-American subgroup, which require a large sample size for concrete prediction.
Keywords: Rheumatoid arthritis, CCR6, population genetics, susceptibility, meta-analysis
Introduction
Rheumatoid arthritis (RA) is one of the most common chronic systemic inflammatory diseases that cause joint destruction. The clinical presentation of RA is usually symmetrical invasion in the small joints, especially in the hands and feet [1]. Although the definite etiology of RA is not clear, more and more research have been suggested that it likely involve an interaction between genetic and environmental factors.
Recently, three independent GWAS studies identified the chemokine (C-C motif) receptor 6 gene (CCR6) as a susceptibility locus for RA [2-4], and this finding was validated in several independent replication cohorts [5-8]. In Japanese population it was observed that there was a strong association between CCR6 and RA and the evidence supported it play an important pathogenic role in the disease. However, an inconsistent result was reported in later studies using African American populations [8,9]. Due to this discrepancy, the role of CCR6 polymorphisms in RA remains controversial and is necessary to be assessed.
To our knowledge there has been no published systematic review of the literature that has characterized the magnitude of these associations. Therefore, expecting to investigate these conflicting results and reveal the role of CCR6 in RA, we conducted the meta-analysis of all published studies on the association between CCR6 polymorphisms and RA risk.
Materials and methods
Identification of eligible studies
The first association study of CCR6 with RA was published in May 2010. We systematically searched PubMed and MEDLINE for all articles published between May 2010 and May 2014. The gene name ‘CCR6’ and disease name (‘rheumatoid arthritis’ or ‘RA’) was respectively used to retrieve the appropriate articles. Moreover, additional hand searching was performed to identify potentially relevant studies.
Selection
Manuscripts were selected if they met all of the following criteria: (1) The diagnosis of RA was determined according to the revised criteria of the American College of Rheumatology 1987 criteria for Rheumatoid Arthritis [11]; (2) the odds ratio (OR) with 95% confidence intervals (CIs) in patients and in controls were available or could be calculated; (3) the study was published as a full paper, not as a meeting abstract or review. The exclusion criteria included: (1) not a population study; (2) studied other SNPs; (3) the duplicate studies. A flow chart depicting the selection process for the studies and the reasons for exclusion is presented in Figure 1. The qualities of all included studies were assessed through a table (Table S1).
Figure 1.
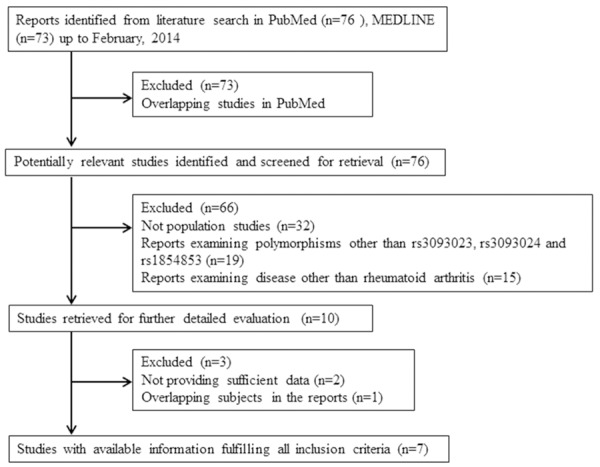
Flow chart for the selection of studies and specific reasons for exclusion of studies from the meta-analysis.
Data extraction
The following information was extracted from each study: first author, year of publication, region, and study population, the type of study design, the numbers and sex ratio of the patients and controls, estimated OR with 95% confidence interval. Data were extracted by two authors (P.C. and Y.Z.) independently and in duplicate. All disagreements and uncertainties were discussed and resolved by consensus, with the involvement of another author (F.G.) if necessary.
Data analysis
Based on the 1000 genomes project, the single nucleotide polymorphism (SNP) rs3093024 was in strong linkage disequilibrium (LD) with rs3093023 and rs1854853 (D’ = 1.0, r2 > 0.9). Therefore, the SNP rs3093024, which tags rs3093023 and rs1854853, is most likely the best proxy to evaluate the effect of this gene.
The population-wide impact of CCR6 polymorphism on susceptibility to RA was assessed by pooling together the per-allele ORs data weighted by their inverse variance from each independent study. The random-effects model was used to calculate the pooled OR. Heterogeneity was evaluated by χ2-based Q statistic and I 2 statistic [12,13]. To evaluate the reliability and stability of our results, publication bias was evaluated with Egger’s linear regression and Begger’s funnel plot [14-16], and the influence of each study on the pooled-OR was investigated in a sensitivity test by excluding one study each time. All analyses were carried out using Stata SE 12.0 data analysis and statistical software.
Results
Search results and study characteristics
Our search strategy resulted in 76 articles in PubMed and 73 articles in MEDLINE. Of these, 73 were excluded for overlapping. In these 76 articles, 32 were excluded because they were not population studies, 19 because of obvious irrelevant polymorphisms and 15 because of not involving RA. The full texts of the 10 remaining articles were obtained for detailed review. After the exclusion of 2 articles for insufficient data and 1 article for overlapping subjects in the reports, a total of 7 independent articles with 11 studies were finally included in the current meta-analysis, which contained information for 56129 healthy subjects and 24955 RA patients. Of the included studies, 8 focused on Asian-descent populations, 2 focused on Caucasian- or European-descent populations, and 1 study reported data concerning populations of African-American descent. Of these studies, three are GWAS (Genome wide association study) studies (2 on Asian and 1 on European population) [2,4] that are part of our meta-analysis. The detailed characteristics of the included studies are listed in Table 1. Additionally, after scoring for the studies, we found all included studies had a high level quality.
Table 1.
Information of all the studies included in the meta-analysis
| Study | Country/region | Population | Study design | GWAS | Cases | Controls | COHORT name | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| First Author | Year | (YES/NO) | Total | FEMALE% | Total | FEMALE% | |||||
| 1a | Kochi et al. 1 [3] | 2010 | Japan | Japanese | Population based case-control | YES | 2301 | 81.40% | 3368 | 44.40% | NA |
| 1b | Kochi et al. 2 [3] | 2010 | Japan | Japanese | Population based case-control | NO | 3662 | 81.50% | 15873 | 34.10% | NA |
| 1c | Kochi et al. 3 [3] | 2010 | Japan | Japanese | Population based case-control | NO | 1106 | 79.20% | 1486 | 60.60% | NA |
| 2a | Stahl et al. 1 [4] | 2010 | USA/Canada/UK/North American/The Netherlands | European | Population based case-control | YES | 5539 | NA | 20169 | NA | NA |
| 2b | Stahl et al. 2 [4] | 2010 | USA/Canada/UK/North American/The Netherlands | European | Population based case-control | NO | 6768 | NA | 8806 | NA | NA |
| 3 | Hughes et al. [8] | 2010 | USA | African Americans | Population based case-control | NO | 556 | 83.88% | 804 | 83.88% | CLEAR |
| 4 | Teng et al. [6] | 2012 | Japan | Asian | Hospital based case-control | NO | 556 | 82% | 440 | 45% | TTSH |
| 5 | Prasad et al. [7] | 2012 | India | North Indians | Population based case-control | NO | 983 | NA | 1007 | NA | AIIMS, R & R |
| 6 | Chang et al. [5] | 2012 | Taiwan | Han Chinese | Hospital based case-control | NO | 400 | 81.79% | 680 | 49.93% | NA |
| 7a | Jiang et al. 1 [2] | 2014 | China | Han Chinese | Hospital based case-control | YES | 952 | NA | 943 | NA | NA |
| 7b | Jiang et al. 2 [2] | 2014 | China | Han Chinese | Population based case-control | NO | 2132 | NA | 2553 | NA | NA |
Case definition: Diagnosis was determined according to the revised criteria of the American College of Rheumatology 1987 criteria for Rheumatoid Arthritis.
Quantitative assessment of all current evidence
The overall estimate suggested a significant association between the CCR6 rs3093024-A allele and RA risk (OR, 1.15; 95% CI, 1.10-1.21; P < 0.001) (Figure 2), but with a high level of heterogeneity (Q = 35.79; I 2 = 72.1%; P < 0.001). Interestingly, when considered separately by ethnicity, a contrasting effect of this variant on RA was observed (subgroup difference x2 = 21.19; P < 0.001). The results from Asian studies indicated that the A-allele may be associated with an increased risk of RA (OR, 1.19; 95% CI, 1.14-1.24; P < 0.001), with moderate heterogeneity observed (Q = 11.22; I 2 = 37.6%; P = 0.129). The European studies also show similar trend (OR, 1.14; 95% CI, 1.08-1.21; P < 0.001), but have high level of significant heterogeneity (Q = 3. 83; I 2 = 70.4%; P = 0.066). Conversely, in African-Americans, the A-allele was associated with a decreased risk of RA (OR, 0.79; 95% CI, 0.62-0.96; P = 0.015). Given the limited sample size of African American studies, the current meta-analysis may be under-powered to draw conclusive insight into this discrepancy.
Figure 2.
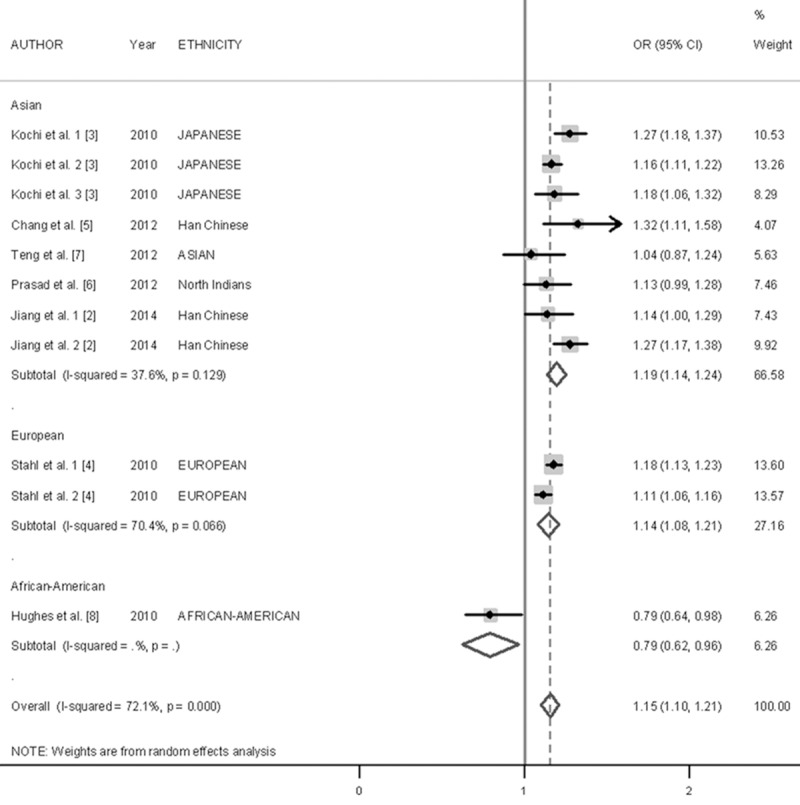
Forest plot describing the association between the CCP6 rs3093024 variant and RA risk. Squares indicate the OR in each study, with square sizes inversely proportional to the standard error of the OR. Horizontal lines represent 95% CI. Analysis was stratified according to ethnicity.
Sensitivity analysis and publication bias
Sensitivity analysis (exclusion of 1 study at a time) indicated that no single study changed the pooled ORs qualitatively (Figure 3), which suggested that the results of the meta-analysis were reliable. Egger’s test suggested no publication bias in the current meta-analysis (P = 0.662), and the shape of the funnel plots appeared symmetrical (Figure 4). Thus, publication bias likely does not have a significant influence on the result of this meta-analysis.
Figure 3.
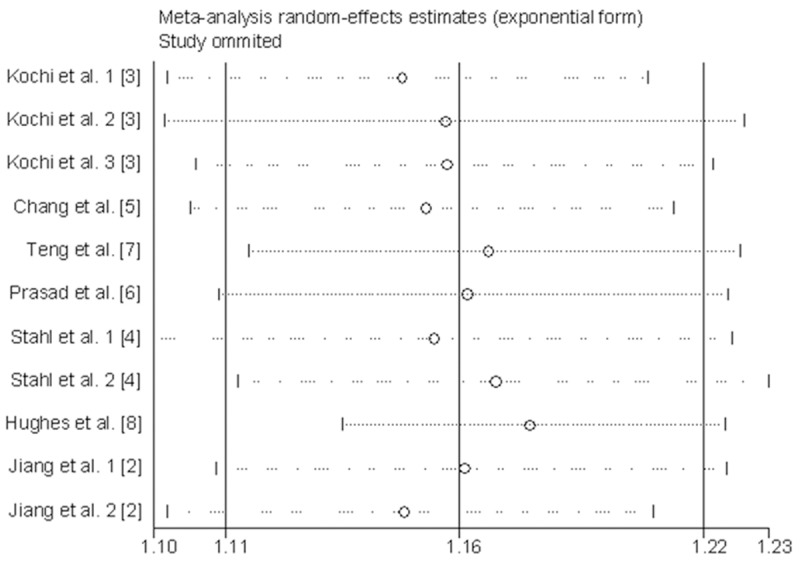
Sensitivity analyses of the CCP6 rs3093024 variant in an additive model by omitting one study at a time. The summary OR (95% CI) was indicated by each horizontal line when the labeled study was omitted, and the remaining studies were then reanalyzed.
Figure 4.
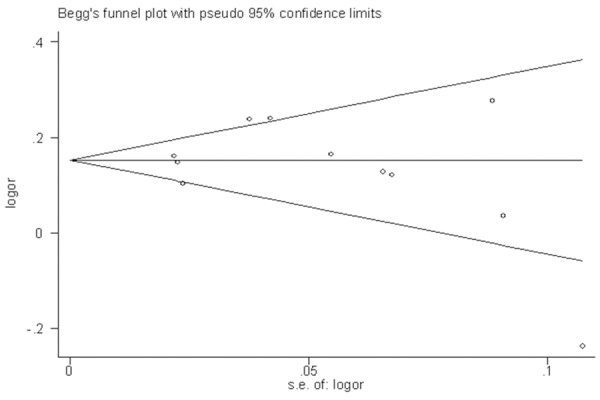
Begg’s funnel plot of studies on the association between CCP6 rs3093024 variant and RA risk. Each point represents a separate study for the indicated association.
Discussion
The present meta-analysis included 7 articles representing 11 studies on single-nucleotide polymorphisms of CCR6 (rs3093024, rs3099023 and rs1854853) among 81084 subjects. The overall data analysis demonstrated that CCR6 is a susceptibility gene for RA across populations, with an overall OR for the risk allele of 1.15. However, high heterogeneity (I 2 = 72.1%, P < 0.001) leads to an underpowered result. To make these conclusions more complete and reliable, subgroup analyses were conducted based on ethnic populations. While CCR6 was a risk factor in Asians (OR = 1.19, 95% CI: 1.14-1.24) and Europeans (OR = 1.14, 95% CI: 1.08-1.21), it was indicated as a protective factor in African Americans (OR = 0.79, 95% CI: 0.62-0.96). Therefore, population groups may be the main cause of the overall high heterogeneity.
The inconsistent OR of African Americans might involve several factors. First, this may be due to weak correlations between the causal allele and actual risk of RA, according to the International Haplotype Mapping Project (Hap-Map) [17]. What’s more, Dickson.S.P et al [18] use simple computer simulations to show us that uncommon or rare genetic variants can easily create synthetic associations that are credited to common variants in some conditions, which needs to be considered seriously in GWAS. Second, genetic heterogeneity may account for the difference; the Asian and European risk allele can function differently in the effect on RA risk for African-Americans. For instance, Johanna Hadler et al [19] suggest that the association of the same SNP with the specific disease varies across ancestor groups, but association was seen in different groups with different SNPs at the same loci. The different associations may also be caused by genetically distinct subsets through some unknown ways, which are still not explored by researchers [19,20]. Third, the inconsistency may simply be accidental. There is only one study included in the assay with limited cases and controls, so the weak association in African-Americans population needs to be further validated by including some additional studies. To sum up, the subgroup analysis conclusions are reliable to an extent, and further experiments with more individuals in different genetic background are necessary for a better understanding of possible explanations.
In recent studies, similar to the Th17 cell differentiation transcription factor RORγt, CCR6 is considered as a marker for Th17 cells, which is a novo subset of CD4+ T cell [21,22]. Upregulation of RORγt induces both IL-17 and CCR6 in naive T cells, blocking CCR6 with monoclonal antibody mainly suppressed arthritis in mouse model [22]. Haas, J. D and his colleagues also proved that only CCR6+ δγ T cells produced IL-17A and CCR6+ δγT cells are more responsive to TCR stimulation [23]. In innate immune responses to microbial stimulation, the CCR6/CCL20 chemokine loop increase B cells rapidly through a TNF-α dependent pathway [24]. Except for T cells and B cells, the chemoattraction of another important player in RA, dendritic cells, was also involved CCR6. All of these suggest CCR6 as a susceptibility gene for the aberrant immune environment in RA [24-26].
The relationship between CCR6 and RA has been discussed certainly, which would lead us to enrich the treatment for RA targeting CCR6. Despite the efficacy of specific blockade of CCR6 on RA has only been tested in an animal model so far [22,27], the development of chronic IBD was found to be inhibited through regulating CCR6 biological function, and the alteration of CCR6 uses by viruses may influence the susceptibility of CD4+ CCR6+ T-cells and dendritic cell subsets in vivo [27,28]. These findings all indicate a promising role for CCR6 in ameliorating RA. In humans, the majority of circulating Th17 cells expresses CCR6 [21,22], and its ligand, CCL20, is also detected in inflamed synovial tissues [29]. Therefore, the efficacy and safety of long-term blockade of CCR6 in treating RA are promising and warrant further investigation.
Several limitations of the current meta-analysis should be noted. First, although we included all articles on the susceptibility of CCR6 (rs3093024, rs3099023 and rs1854853) on RA, this was a total of only 11 studies. Second, the overall study was underpowered because of high heterogeneity, but the subgroup results show us a positive association. Future studies of large, well-characterized cohorts of different racial groups are necessary to better understand this association. We believe that detailed genetic studies of CCR6 with RA will lead to important insights into the pathogenesis of this disease. Additionally, the difference in associations between females and males is worthy of further discussion and analysis (data not shown).
In conclusion, pooled results for 81804 subjects demonstrated a significant association between CCR6 and RA. However, the subgroup analysis confirmed different effects of CCR6 on RA within different racial groups, and the meta-analysis results of associations of these SNPs with RA are required for further evaluation in larger samples from African American populations.
Acknowledgements
We want to especially acknowledge all of the participants in this study. This work was supported by grants from the National Natural Science Foundation of China (No. 81171696 and No. 81300828).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 2.Jiang L, Yin J, Ye L, Yang J, Hemani G, Liu AJ, Zou H, He D, Sun L, Zeng X, Li Z, Zheng Y, Lin Y, Liu Y, Fang Y, Xu J, Li Y, Dai SM, Guan J, Jiang L, Wei Q, Wang Y, Li Y, Huang C, Zuo X, Liu Y, Wu X, Zhang L, Zhou L, Zhang Q, Li T, Chen L, Xu Z, Yang X, Qian F, Xie W, Liu W, Guo Q, Huang S, Zhao J, Li M, Jin Y, Gao J, Lv Y, Wang Y, Lin L, Guo A, Danoy P, Willner D, Cremin C, Hadler J, Zhang F, Zhao Y, Li M, Yue T, Fan X, Guo J, Mu R, Li J, Wu C, Zeng M, Wang J, Li S, Jin L, Wang B, Wang J, Ma X, Sun L, Zhang X, Brown MA, Visscher PM, Su DF, Xu H. Novel risk loci for rheumatoid arthritis in han chinese and congruence with risk variants in europeans. Arthritis Rheumatol. 2014;66:1121–1132. doi: 10.1002/art.38353. [DOI] [PubMed] [Google Scholar]
- 3.Kochi Y, Okada Y, Suzuki A, Ikari K, Terao C, Takahashi A, Yamazaki K, Hosono N, Myouzen K, Tsunoda T, Kamatani N, Furuichi T, Ikegawa S, Ohmura K, Mimori T, Matsuda F, Iwamoto T, Momohara S, Yamanaka H, Yamada R, Kubo M, Nakamura Y, Yamamoto K. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat Genet. 2010;42:515–519. doi: 10.1038/ng.583. [DOI] [PubMed] [Google Scholar]
- 4.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, Guiducci C, Chen R, Alfredsson L, Amos CI, Ardlie KG, Barton A, Bowes J, Brouwer E, Burtt NP, Catanese JJ, Coblyn J, Coenen MJ, Costenbader KH, Criswell LA, Crusius JB, Cui J, de Bakker PI, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Lee AT, Liu X, Martin P, Morgan AW, Padyukov L, Posthumus MD, Radstake TR, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van MA, van der Horst-Bruinsma IE, van der Schoot CE, van Riel PL, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth BP, Wijmenga C, Karlson EW, Toes RE, de Vries N, Begovich AB, Worthington J, Siminovitch KA, Gregersen PK, Klareskog L, Plenge RM. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang WC, Woon PY, Wei JC, Chang CM, Hsu YW, Guo YC, Hwang DY, Kochi Y, Yen JH. A single-nucleotide polymorphism of CCR6 (rs3093024) is associated with susceptibility to rheumatoid arthritis but not ankylosing spondylitis, in a Taiwanese population. J Rheumatol. 2012;39:1765–1766. doi: 10.3899/jrheum.120040. [DOI] [PubMed] [Google Scholar]
- 6.Teng E, Leong KP, Li HH, Thong B, Koh ET, Loi PL, Zhao Y, Tan EK. Analysis of a genome-wide association study-linked locus (CCR6) in Asian rheumatoid arthritis. Dna Cell Biol. 2012;31:607–610. doi: 10.1089/dna.2011.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad P, Kumar A, Gupta R, Juyal RC, Thelma BK. Caucasian and Asian specific rheumatoid arthritis risk loci reveal limited replication and apparent allelic heterogeneity in north Indians. PLoS One. 2012;7:e31584. doi: 10.1371/journal.pone.0031584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes LB, Reynolds RJ, Brown EE, Kelley JM, Thomson B, Conn DL, Jonas BL, Westfall AO, Padilla MA, Callahan LF, Smith EA, Brasington RD, Edberg JC, Kimberly RP, Moreland LW, Plenge RM, Bridges SJ. Most common single-nucleotide polymorphisms associated with rheumatoid arthritis in persons of European ancestry confer risk of rheumatoid arthritis in African Americans. Arthritis Rheum. 2010;62:3547–3553. doi: 10.1002/art.27732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkins EA, Landis D, Causey ZL, Edberg Y, Reynolds RJ, Hughes LB, Gregersen PK, Kimberly RP, Edberg JC, Bridges SJ. Association of single-nucleotide polymorphisms in CCR6, TAGAP, and TNFAIP3 with rheumatoid arthritis in African Americans. Arthritis Rheum. 2012;64:1355–1358. doi: 10.1002/art.33464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 11.Asimit J, Day-Williams A, Zgaga L, Rudan I, Boraska V, Zeggini E. An evaluation of different meta-analysis approaches in the presence of allelic heterogeneity. Eur J Hum Genet. 2012;20:709–712. doi: 10.1038/ejhg.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petitti DB. Approaches to heterogeneity in meta-analysis. Stat Med. 2001;20:3625–3633. doi: 10.1002/sim.1091. [DOI] [PubMed] [Google Scholar]
- 13.Song F, Gilbody S. Bias in meta-analysis detected by a simple, graphical test. Increase in studies of publication bias coincided with increasing use of meta-analysis. BMJ. 1998;316:471. [PMC free article] [PubMed] [Google Scholar]
- 14.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 16.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 17.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, Leo P, Cremin K, Pryce K, Harris J, Lee S, Joo KB, Shim SC, Weisman M, Ward M, Zhou X, Garchon HJ, Chiocchia G, Nossent J, Lie BA, Forre O, Tuomilehto J, Laiho K, Jiang L, Liu Y, Wu X, Bradbury LA, Elewaut D, Burgos-Vargas R, Stebbings S, Appleton L, Farrah C, Lau J, Kenna TJ, Haroon N, Ferreira MA, Yang J, Mulero J, Fernandez-Sueiro JL, Gonzalez-Gay MA, Lopez-Larrea C, Deloukas P, Donnelly P, Bowness P, Gafney K, Gaston H, Gladman DD, Rahman P, Maksymowych WP, Xu H, Crusius JB, van der Horst-Bruinsma IE, Chou CT, Valle-Onate R, Romero-Sanchez C, Hansen IM, Pimentel-Santos FM, Inman RD, Videm V, Martin J, Breban M, Reveille JD, Evans DM, Kim TH, Wordsworth BP, Brown MA. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45:730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terao C, Ohmura K, Ikari K, Kochi Y, Maruya E, Katayama M, Yurugi K, Shimada K, Murasawa A, Honjo S, Takasugi K, Matsuo K, Tajima K, Suzuki A, Yamamoto K, Momohara S, Yamanaka H, Yamada R, Saji H, Matsuda F, Mimori T. ACPA-negative RA consists of two genetically distinct subsets based on RF positivity in Japanese. PLoS One. 2012;7:e40067. doi: 10.1371/journal.pone.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, Parente E, Fili L, Ferri S, Frosali F, Giudici F, Romagnani P, Parronchi P, Tonelli F, Maggi E, Romagnani S. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirota K, Yoshitomi H, Hashimoto M, Maeda S, Teradaira S, Sugimoto N, Yamaguchi T, Nomura T, Ito H, Nakamura T, Sakaguchi N, Sakaguchi S. Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J Exp Med. 2007;204:2803–2812. doi: 10.1084/jem.20071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas JD, Gonzalez FH, Schmitz S, Chennupati V, Fohse L, Kremmer E, Forster R, Prinz I. CCR6 and NK1.1 distinguish between IL-17A and IFN-gamma-producing gammadelta effector T cells. Eur J Immunol. 2009;39:3488–3497. doi: 10.1002/eji.200939922. [DOI] [PubMed] [Google Scholar]
- 23.Paradis M, Mindt BC, Duerr CU, Rojas OL, Ng D, Boulianne B, McCarthy DD, Yu MD, Summers DL, Ward LA, Waldron JB, Philpott DJ, Gommerman JL, Fritz JH. A TNF-alpha-CCL20-CCR6 Axis Regulates Nod1-Induced B Cell Responses. J Immunol. 2014;192:2787–2799. doi: 10.4049/jimmunol.1203310. [DOI] [PubMed] [Google Scholar]
- 24.Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schutyser E, Struyf S, Van Damme J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 2003;14:409–426. doi: 10.1016/s1359-6101(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 26.Lee AY, Eri R, Lyons AB, Grimm MC, Korner H. CC Chemokine Ligand 20 and Its Cognate Receptor CCR6 in Mucosal T Cell Immunology and Inflammatory Bowel Disease: Odd Couple or Axis of Evil? Front Immunol. 2013;4:194. doi: 10.3389/fimmu.2013.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Islam S, Shimizu N, Hoque SA, Jinno-Oue A, Tanaka A, Hoshino H. CCR6 functions as a new coreceptor for limited primary human and simian immunodeficiency viruses. PLoS One. 2013;8:e73116. doi: 10.1371/journal.pone.0073116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee AY, Korner H. CCR6 and CCL20: emerging players in the pathogenesis of rheumatoid arthritis. Immunol Cell Biol. 2014;92:354–358. doi: 10.1038/icb.2013.97. [DOI] [PubMed] [Google Scholar]
- 29.Matsui T, Akahoshi T, Namai R, Hashimoto A, Kurihara Y, Rana M, Nishimura A, Endo H, Kitasato H, Kawai S, Takagishi K, Kondo H. Selective recruitment of CCR6-expressing cells by increased production of MIP-3 alpha in rheumatoid arthritis. Clin Exp Immunol. 2001;125:155–161. doi: 10.1046/j.1365-2249.2001.01542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


