Abstract
Arterial medial calcification (AMC) is a hallmark of aging, diabetes, and chronic kidney disease. Smooth muscle cell (SMC) transition to an osteogenic phenotype is a common feature of AMC, and is preceded by expression of runt-related transcription factor 2 (Runx2), a master regulator of bone development. Whether SMC-specific Runx2 expression is required for osteogenic phenotype change and AMC remains unknown. We therefore created an improved targeting construct to generate mice with floxed Runx2 alleles (Runx2f/f) that do not produce truncated Runx2 proteins after Cre recombination, thereby preventing potential off-target effects. SMC-specific deletion using SM22–recombinase transgenic allele mice (Runx2ΔSM) led to viable mice with normal bone and arterial morphology. After vitamin D overload, arterial SMCs in Runx2f/f mice expressed Runx2, underwent osteogenic phenotype change, and developed severe AMC. In contrast, vitamin D–treated Runx2ΔSM mice had no Runx2 in blood vessels, maintained SMC phenotype, and did not develop AMC. Runx2 deletion did not affect serum calcium, phosphate, fibroblast growth factor-23, or alkaline phosphatase levels. In vitro, Runx2f/f SMCs calcified to a much greater extent than those derived from Runx2ΔSM mice. These data indicate a critical role of Runx2 in SMC osteogenic phenotype change and mineral deposition in a mouse model of AMC, suggesting that Runx2 and downstream osteogenic pathways in SMCs may be useful therapeutic targets for treating or preventing AMC in high-risk patients.
Arterial medial calcification (AMC) is prevalent in aging, diabetic, and chronic kidney disease (CKD) patients.1–3 In contrast to arterial intimal calcification (AIC) associated with atherosclerosis, AMC occurs in the absence of inflammation, is the earliest type of vascular calcification found in children with CKD, and is considered a hallmark of CKD mineral and bone disorder in adults.4–6 Although AIC and AMC both are associated with increased cardiovascular mortality,7 AIC is associated with plaque rupture and myocardial infarction,8–10 whereas AMC leads to vessel stiffening, increased pulse-wave velocity, reduced cardiac perfusion, and, ultimately, left ventricular hypertrophy and heart failure.1,7,11,12 Importantly, heart failure is a predominant cardiovascular cause of death in aged, diabetic, and CKD patients.1–3,13 Furthermore, in randomized clinical trials, statins do not improve survival in CKD patients, suggesting that noninflammatory arteriosclerotic disease featuring AMC may underlie the high cardiovascular mortality risk in these patients.14–18 Thus, understanding the cellular and molecular mechanisms mediating AMC is critical for improved therapeutics for high-risk patients.
Analyses of arteries from children4 and experimental animals19,20 have indicated that cells expressing an osteogenic phenotype appear in the media before extracellular matrix mineralization in AMC.21,22 Furthermore, genetic fate mapping studies show that SMCs are the predominant progenitors of osteogenic cells in a mouse model of AMC,23 supporting a critical role for SMCs in this pathology. Mechanistically, osteogenic reprogramming of vascular SMCs is preceded by de novo expression of runt-related transcription factor 2 (Runx2).23,24 Runx2, also known as Cbfa1 or AML3, is a transcription factor that absolutely is required for skeletal formation and remodeling.25,26 Mutations in the Runx2 gene lead to cleidocranial dysplasia in humans,27 and targeted deletion in mice results in perinatal death caused by defective bone formation and respiratory failure.25,26 However, it is not yet known whether Runx2 expression specifically in SMCs is required for either osteogenic phenotype change or AMC under conditions of calcifying vascular disease. We therefore generated mice with SMC-specific Runx2 inactivation alleles (Runx2ΔSM) to determine whether selective ablation of Runx2 in vascular SMCs prevents SMC osteogenic differentiation and mineral deposition in a mouse model of AMC.
Materials and Methods
Generation of a Runx2 Conditional Allele that Targets the Runt Homology Domain
A Runx2 targeting vector was created such that exon 4, the second exon of the runt homology domain, was flanked by a single loxP sequence on the 5′ side and a FRT-flanked and a loxP-flanked neomycin-resistant (Neo) cassette oriented opposite of the Runx2 gene on the 3′ side. This targeting vector then was linearized and electroporated into C57BL/6 × 129SvEv hybrid embryonic stem cells. On G418 antibiotic selection and Southern blotting using a probe that recognizes sequences outside of the homologous recombination arms of the allele, embryonic stem cell clones with proper homologous recombination were identified (data not shown). Two positive clones subsequently were injected into C57BL/6 blastocysts and produced five chimeric floxed-Runx2 transgenic mice that showed a high percentage of agouti coat color. These chimeras then were bred onto a FLP recombinase background to remove the Neo cassette, and their genomic DNA was extracted from tail biopsy specimens.
Primers used to identify the hemizygous floxed-Runx2 mice (Runx2+/f) were as follows: LOX1 5′-GCTCAAGACCTGACTCGAGAC-3′ and SDL2 5′-GAAACACTTAAGGACAGAGAACACATGC-3′, which flank the single loxP site; A1 5′-AAACCAGCCAAAACTCAGAAAGCC-3′, which is downstream of the short homology arm; F3 5′-GCATAAGCTTGGATCCGTTCTTCGGAC-3′, which is inside of the Neo cassette; and NDEL1 5′-GTTAGGCTCTCTGGTGCAAG-3′ and NDEL2 5′-CTTGAAACCATCCACAGGTGAT-3′, which flank the Neo cassette. The Runx2+/f mice were back-crossed to C57BL/6 mice for three generations and then inbred to produce homozygotes (Runx2f/f). Mice hemizygous and homozygous for the conditional transgenic allele were viable, normal in size, fertile, and did not show gross physical or behavioral abnormalities.
The Runx2+/f mice were bred further with either Sox2-Cre transgenic mice (008454; The Jackson Laboratory, Bar Harbor, ME) to produce Runx2+/f:Sox2-Cre (Runx2+/Δ), or SM22α-Cre transgenic mice to produce Runx2f/f:SM22α-Cre (Runx2ΔSM) to deplete the Runx2 gene globally or specifically in SMCs, respectively. Genotyping of these mice was performed using PCR of DNA extracted from tail biopsy specimens. Primers used for genotyping were LOX1 and NDEL2 (see previous paragraph), which amplify regions from the 5′ side of the first loxP to the 3′ side of the second loxP, and generated amplicons of 1019 bp (wild-type allele), 1269 bp (floxed conditional allele), and 527 bp (Cre recombined allele).
Characterization of Runx2Δ/Δ Mice
Runx2+/+, Runx2+/Δ, and Runx2Δ/Δ embryos were collected at 18.5 days after coitum from timed inbreeding of Runx2+/Δ mice. The pregnant adult mice were euthanized with CO2 asphyxiation followed by exsanguination. Embryos were kept hypothermic followed by an immediate peritoneal injection of pentobarbital at a dosage of 450 mg/kg body weight. Sacrificed embryos were imaged for gross anatomy and tail biopsy specimens were taken for DNA extraction and genotyping. Subsets of Runx2+/+, Runx2+/Δ, and Runx2Δ/Δ embryos were either X-rayed, double-stained for cartilage and bone with Alcian blue and Alizarin red,28 or fixed in 10% buffered formalin for histology.
Vitamin D Induction of AMC in Mice
AMC was induced via vitamin D (cholecalciferol) overload as previously described.29 USP grade cholecalciferol (C1357; Sigma, St. Louis, MO) was dissolved in 100% ethanol and diluted freshly to 1.65 mg/mL containing 5% ethanol for each injection. Ten-week-old Runx2f/f and Runx2ΔSM mice received cholecalciferol subcutaneously at a dosage of 500,000 IU/kg/day for 4 consecutive days, and 5% ethanol in sterile water was used as a vehicle control. Animals were monitored daily and terminated for study 10 to 14 days after injection. Specifically, mice were sacrificed with 50 to 180 mg/kg pentobarbital intraperitoneally followed by exsanguination via cardiac puncture. Sera were collected for blood chemistry. Aortic arches and abdominal aortas plus common iliac arteries were collected for calcium quantification, and thoracic aortas, renal arteries, and kidneys were collected for histology.
All animals were maintained in a specific pathogen-free environment. Protocols were in compliance with the NIH Guideline for the Care and Use of Laboratory Animals and have been approved by the Institutional Animal Care and Use Committee at the University of Washington.
Histochemical and Immunohistochemical Staining
Specimens were fixed with modified Clark's fixative (75% methanol, 25% glacial acetic acid) and embedded in paraffin. Serial sections (5 μm thick) were collected and used for hematoxylin and eosin staining. Calcium-phosphate minerals were detected by Alizarin red and von Kossa staining.19,23 Immunohistochemistry was used to determine SMCs and osteochondrogenic precursors. In brief, sections were deparaffinized and rehydrated, blocked for endogenous peroxidase activity and nonspecific binding, and incubated with primary antibodies that recognize SM22α (ab10135; Abcam, Cambridge, MA), SMMHC (ab683; Abcam), ACTA2 (A2547; Sigma), Runx2 (MAB2006; R&D systems, Minneapolis, MN), or osteopontin (AF808; R&D systems). The presence of vascular SMCs or osteochondrogenic precursors then were visualized through biotin-conjugated secondary antibodies followed by avidin-biotin complex amplification (PK-6100; Vector Labs, Burlingame, CA) and 3, 3′-diaminobenzidine staining (D-0426; Sigma) or fluorescent secondary antibodies.23,24
Serum Analysis
Mice were fasted for 4 hours before blood collection into serum separator tubes. Sera were used to determine calcium and blood urea nitrogen levels colorimetrically using the o-cresolphthalein complexone kit (C503-480; Teco Diagnostics, Anaheim, CA) and the QuantiChrom Urea Assay kit (DIUR-500; BioAssay Systems, Hayward, CA), respectively. Fibroblast growth factor-23 was measured using an enzyme-linked immunosorbent assay kit from Immuotopics (#60-6300; San Clemente, CA), and phosphorus by a standard bioanalyzer.19
Isolation and Culture of Vascular SMCs
Vascular SMCs were isolated from aortas of 4-week-old wild-type, Runx2f/f, and Runx2ΔSM mice as previously described.30 Briefly, thoracic and the upper part of the abdominal aorta were incubated first with 1 mg/mL collagenase, 0.23 mg/mL elastase, 0.375 mg/mL soybean trypsin inhibitor, and 2 mg/mL bovine serum albuminA at 37°C for 5 minutes to separate aortic media from adventitia. Endothelial cells were removed with a cotton swab, and the medial layer then was stripped off carefully under a dissection microscope and cut into 1-mm pieces. After digestion in 1 mg/mL collagenase for 20 minutes to remove residual endothelial and adventitial cells, the aortic media pieces were rinsed with culture medium and dispersed in a mixture of 0.64 mg/mL collagenase and 0.44 mg/mL elastase in culture medium containing 20% fetal bovine serum. After incubation at 37°C for 40 minutes to 1 hour with occasional gentle agitation, medial cells were released. The cell suspension was centrifuged at 800 × g for 5 minutes, and the cell pellet was washed and resuspended in Dulbecco’s modified Eagle’s medium containing 100 U/mL penicillin, 100 μg/mL streptomycin, 0.25 μg/mL fungizone, and 20% fetal bovine serum. Aortic SMCs were seeded at a density of 1 × 105 cells/mL for primary culture, and split 1:2 at confluency. Cells used for the experiments were from the third to ninth passages. Subcultured SMCs were maintained in Dulbecco’s modified Eagle’s medium containing 100 U/mL penicillin, 100 μg/mL streptomycin, 0.25 μg/mL fungizone, and 10% fetal bovine serum.30 SMCs of more than 96% purity as determined by the presence of SM22α, SMMHC, and ACTA2 were used for the study (data not shown).
Western Blotting
Nuclear lysates were prepared from SMC monolayers using a nuclear extraction kit (78,833; Thermo Scientific, Waltham, MA). The protein content of the lysates was measured by the Micro BCA assay (Life Technologies, Grand Island, NY). Equal amounts of the protein from each sample were separated by SDS-PAGE followed by a transfer to a polyvinylidene difluoride membrane (Perkin Elmer, Waltham, MA). Runx2 protein then was detected using an antibody specific for mouse Runx2 (D130-3; MBL Int Corp, Woburn, MA; or sc-10758; Santa Cruz Biotechnology, Inc., Dallas, TX), a horseradish peroxidase–conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), and the SuperSignal WestDura chemiluminescence detection kit (Thermo Scientific). Probing of the same membrane with antibody recognizing lamin B (sc-6217; Santa Cruz) was used to monitor sample loading.
Reverse Transcription-PCR and Quantitative Real-Time PCR
Total RNA was extracted from blood vessels or SMC monolayers using the RNeasy Mini kit (Qiagen, Valencia, CA). The contaminating genomic DNA was digested by RNase-free DNase I (Qiagen). Total RNA (0.5 to 1 μg) was used to synthesize first-strand cDNA using Omniscript (Qiagen) at 37°C for 1 hour, and the cDNA produced was used to determine the expression of Runx2 and osteogenic genes, osteopontin (Opn, official name Spp1), alkaline phosphatase (Alpl), and osteocalcin (Ocn, official name Bglap) using TaqMan quantitative real-time PCR. Primer and probe sequences were as follows: Runx2 forward primer: 5′-CACCGACAGTCCCAACTTCCT-3′, Runx2 reverse primer: 5′-ACGGTAACCACAGTCCCATCTG-3′, and Runx2 probe: 5′-FAM-CCTTCAAGGTTGTAGCCCT-MGB-3′; Opn forward primer: 5′-TGAGGTCAAAGTCTAGGAGTTTCC-3′, Opn reverse primer: 5′-TTAGACTCACCGCTCTTCATGTG-3′, and Opn probe: 5′-FAM-TTCTGATGAACAGTATCCTG-MGB-3′; Alpl forward: 5′-CAAGGACATCGCATATCAGCTAA-3′, Alpl reverse: 5′-CAGTTCTGTTCTTCGGGTACATGT-3′, and Alpl probe: 5′-FAM-AGGATATCGACGTGATCAT-MGB-3′; Ocn forward: 5′-CTGGCTGCGCTCTGTCTCT-3′, Ocn reverse: 5′-GACATGAAGGCTTTGTCAGACTCA-3′, and Ocn probe: 5′-FAM-TGACCTCACAGATGCCAA-MGB-3′. Probe sequences were created to span an exon–exon junction of the desired genes to avoid amplification of residual genomic DNA. The 18s ribosomal RNA expression was determined using TaqMan Ribosomal RNA Control Reagents (Life Technologies) to control sample loading. Gene expression levels were normalized to 18s ribosomal RNA levels and expressed as a fold of control samples. The Runx2 primers also were used to amplify exon 4 to determine the deletion efficiency of the Runx2 gene in the Runx2ΔSM alleles. Thirty cycles of PCR were used to allow exponential amplification of the desired genes, and glyceraldehyde-3-phosphate dehydrogenase was used to monitor sample loading.
Calcium Quantification
Approximately 6 mm of the aortic arch and 4 mm of the abdominal aorta plus two common iliac arteries from each mouse were collected and lyophilized to a constant weight. Calcium was extracted from the lyophilized tissue with 0.6 N HCl at 37°C for 48 hours on a shaker, determined colorimetrically using the o-cresolphthalein complexone kit (C503-480; Teco Diagnostics), and normalized to the tissue dry weight.19,20 To determine calcification of SMC cultures, cells first were rinsed with ice-cold phosphate-buffered saline and decalcified with 0.6 N HCl at 4°C for 24 hours on a shaker. Calcium released from the cell cultures was determined colorimetrically as described earlier and normalized to cellular protein of the culture and expressed as micrograms of calcium per milligram of cellular protein.30
Statistics
Data, shown as means ± SEM, were analyzed with the Student's t-test or one-way analysis of variance with the Dunn post hoc test. Data were considered statistically significant at P < 0.05.
Results
Global Deletion of Runx2 Inhibits Fetal Bone and Cartilage Development
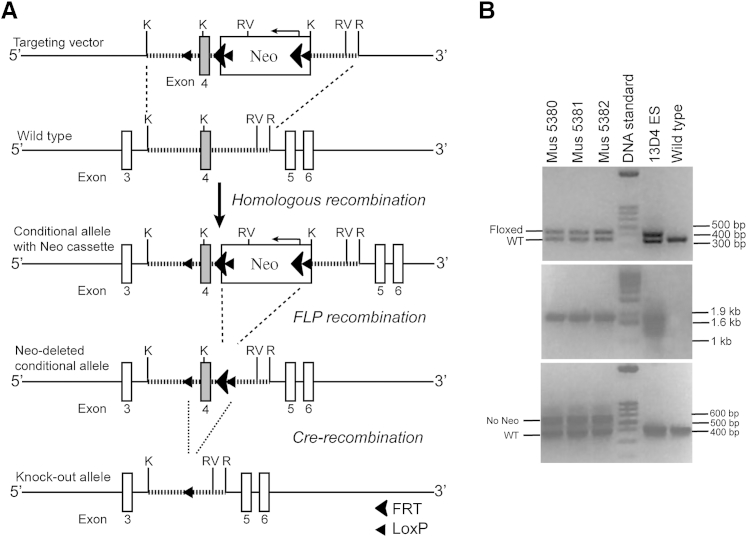
Mice carrying a Runx2 conditional allele that targets exon 4, the first exon of the runt homology domain of the Runx2 gene, were generated (Figure 1A). On Cre recombination, exon 4 is removed, resulting in a reading frame shift and early stop codon (Figure 1A). This strategy was chosen to avoid the generation of runt homology domain-containing truncated proteins that previously were reported to retain DNA binding ability and act as a dominant-negative competitor.31 In this way, we were able to avoid off-target effects on related runt homology domain-containing family members (Runx1 and Runx3) that potentially could confound our studies. By using Runx2 targeting vector–containing embryonic stem cell clone (13D4) as positive control, three mice were identified to have appropriate loxP sites (Figure 1B) after homologous recombination at the Runx2 locus (Figure 1B) and in the absence of the Neo cassette (Figure 1B). We also verified sequences of the floxed exon 4 region through DNA sequencing (data not shown).
Figure 1.
Design and characterization of a conditional runt-related transcription factor 2 (Runx2) allele. A: Conditional targeting strategy for the mouse Runx2 gene. Runx2 genomic sequence (striped line) containing exon 4 (grey box) was cloned into targeting vector. Homologous recombination (dashed line) between targeting vector and wild-type locus resulted in the targeted allele. Neo cassette was used to select positive embryonic stem cell clones and was removed from the conditional allele of the chimeric mice through breeding to a FLP transgenic background. On Cre recombination (dotted line), exon 4 was removed, leading to a shift in reading frame that resulted in a stop codon. Restriction enzyme site K, Kpn I; R, EcoR I; RV, EcoR V. B: Characterization of the neo cassette–deleted conditional allele by PCR. Genomic DNA was extracted from the tail biopsy specimens of mice 5380 to 5382. Specific primers were used to identify the presence of a single LoxP site (upper, 428 bp), the homologous recombination of the targeting vector, and the wild-type Runx2 locus (middle, 1.8 kb), and the absence of the neo cassette in the conditional allele (lower, 592 bp). Fragments amplified from wild-type (WT) Runx2 allele were 366 bp (top panel) and 414 bp (bottom panel). No fragment was amplified from the wild-type Runx2 allele (middle panel). Genomic DNA isolated from 13D4 embryonic stem cells was used as a positive control for the conditional allele.
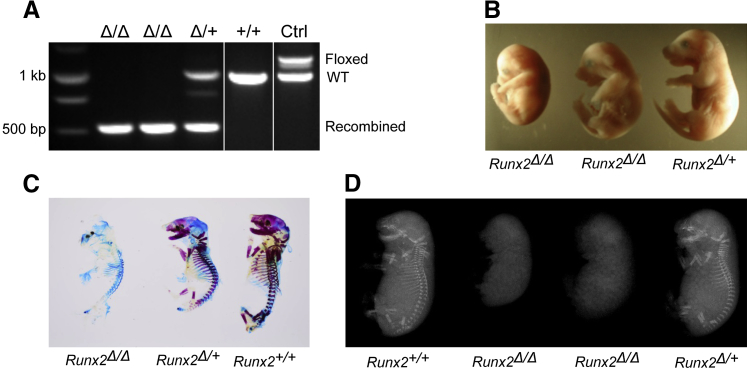
To confirm that the conditional allele led to appropriate Runx2 deletion, male Runx2f/f mice were bred with female Sox2-Cre recombinase transgenic mice to produce Runx2+/Δ mice. Inbreeding of the Runx2+/Δ mice produced Runx2+/+, Runx2+/Δ, and Runx2Δ/Δ embryos at a rate similar to that reported previously for traditionally generated Runx2 knockout embryos.25,26 Figure 2 shows representative embryos of each genotype. Genotypes were identified using PCR primers described in Materials and Methods. Three different sizes of amplicon indicate wild-type (1019 bp), floxed (1219 bp), and Cre-recombined (527 bp) alleles (Figure 2A). The global Runx2Δ/Δ embryos generated by Cre recombination of the conditional Runx2 alleles were smaller in size than Runx2+/Δ and Runx2+/+ mice, and had foreshortened noses (Figure 2B and data not shown). Importantly, Alizarin red S staining of skeletons of the Runx2+/+ embryo at 18.5 days after coitum had ossification in the skull, vertebral arches, ribs, clavicles, scapulae, and long bones, whereas the Runx2Δ/Δ embryonic littermates showed neither calcified cartilage nor mineralized bone (Figure 2C). Furthermore, there was no skeleton element detected in the skull, mandibles, upper and lower extremities, ribs, and vertebrate of the Runx2Δ/Δ embryos by soft X-ray emission spectroscopy, a technique often used for determining skeleton structure of experimental animals (Figure 2D). Runx2+/+ and Runx2+/Δ embryos were normal in size and showed no difference in skeletal development in radiography. The defects in skeletal development observed in the Runx2Δ/Δ embryos were similar to those observed in conventional targeted Runx2 knockout mice,25,26 confirming that appropriate targeting of the conditional Runx2 allele had been achieved.
Figure 2.
Global deletion of the runt-related transcription factor 2 (Runx2) gene leads to bone and cartilage abnormality and perinatal lethality in mice. A: Global deletion of the Runx2 gene. A Runx2+/flox male mouse was bred to a female Sox2-Cre recombinase transgenic mouse followed by interbreeding of the Runx2+/Δ mice to generate Runx2 nulls (Runx2Δ/Δ). Embryos at 18.5 days after coitum were genotyped (A), and then imaged by a dissection scope (B). Skeletons of mice with representative genotypes were determined by Alcian blue and Alizarin red stain (C) and by soft X-ray (D). Ctrl, control; Runx2+/+, wild-type; Runx2Δ/Δ, knockout; Runx2+/Δ, heterozygote; WT, wild type.
Generation and Characterization of SMC-Specific Runx2 Knockout Mice
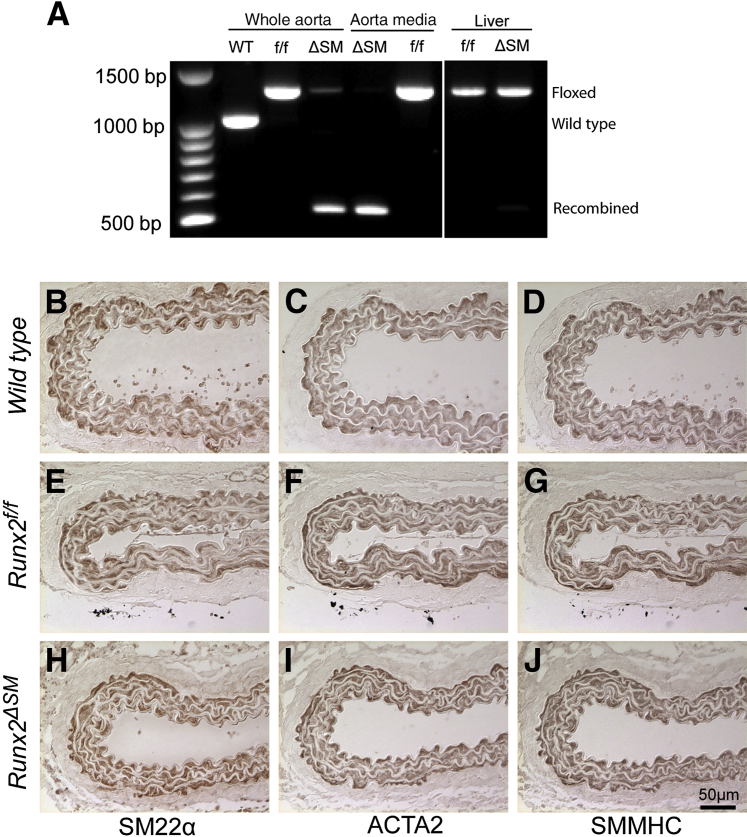
To generate mice with Runx2 deletion specifically in SMCs, we introduced the SM22α-Cre recombinase transgenic allele into the Runx2f/f mice (Runx2ΔSM). The 2.8-kb SM22α promoter used to drive Cre recombinase expression in this transgenic allele contains regulatory sequences that direct SMC-restricted expression32 and has been used successfully for in vivo loss-of-function studies in vascular SMCs33,34 and genetic fate mapping studies of vascular SMCs.23,24 By using primers that amplify the sequence containing the floxed exon 4 region, genomic DNA rearrangement of the conditional Runx2 alleles directed by the SM22α-Cre recombinase was determined. Exon 4 was deleted efficiently from the aortas of the Runx2ΔSM mice and was absent in the aortic media, compared with wild-type and Runx2f/f counterparts (Figure 3A). In contrast, no SM22α-Cre–mediated DNA rearrangement of the Runx2 gene was observed in liver, similar to previous observations by us and other investigators.23,32,33 Runx2f/f and Runx2ΔSM mice were viable, normal in size, and fertile (data not shown). No developmental defects were observed in the arteries of the Runx2f/f and Runx2ΔSM mice as determined by hematoxylin and eosin staining (data not shown), and immunohistochemical staining for SMC lineage markers SM22, ACTA2, and SMMHC (Figure 3, B–J).
Figure 3.
Mice with smooth muscle–specific deletion of the runt-related transcription factor 2 (Runx2) gene are viable and show normal vascular morphology. Runx2flox/flox mice were bred with SM22-Cre transgenic mice to generate smooth muscle cell (SMC)-specific Runx2 knockout mice (Runx2ΔSM). A: Tissue-specific deletion of Runx2 was examined by PCR using primers that amplify the loxP-flanked targeting sequence. Genomic DNA isolated from aorta and liver of Runx2f/f and Runx2ΔSM mice was used as a template. In addition, aortic media were isolated carefully with enzymatic digestion and showed complete removal of Runx2 exon 4. (B–J) Upper abdominal aortas dissected from wild-type (B–D), Runx2f/f (E–G), and Runx2ΔSM (H–J) mice were stained for SMC markers, SM22α (B, E, and H), ACTA2 (C, F, and I), and SMMHC (D, G, and J).
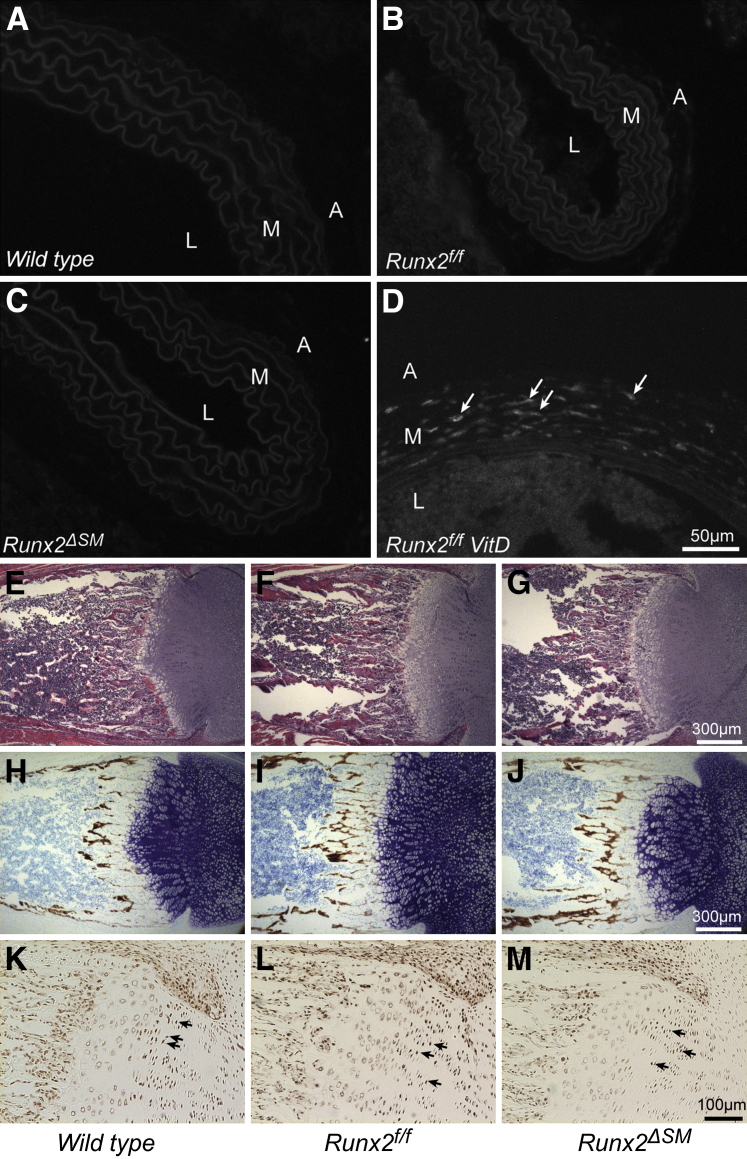
Consistent with previous studies in mice and humans,23,24,35 Runx2 was not detected in untreated blood vessels of Runx2+/+, Runx2f/f, and Runx2ΔSM mice (Figure 4, A–C). On the other hand, Runx2 expression was detected easily after vitamin D (VitD) treatment of Runx2f/f mice (Figure 4D). Hematoxylin and eosin and von Kossa staining of mouse femurs showed no differences in morphology and mineralization in the Runx2f/f and Runx2ΔSM mice in comparison with their wild-type counterparts (Figure 4, E–J), and the Runx2 expression pattern was similar among various genotypes as well (Figure 4, K–M).
Figure 4.
Smooth muscle cell (SMC)-specific removal of runt-related transcription factor 2 (Runx2) does not alter normal bone development. Thoracic aortas dissected from Runx2+/+ (A), Runx2f/f (B), and Runx2ΔSM (C) mice were negative for Runx2 by immunofluorescent staining. Runx2 was up-regulated in vitamin D (VitD)-treated Runx2f/f mice (D). Arrows designate Runx2+ cells in aortic media. Femurs of 7-day-old wild-type (E, H, and K), Runx2f/f (F, I, and L), and Runx2ΔSM (G, J, and M) mice were stained with hematoxylin and eosin (H&E) for gross morphology (E–G), von Kossa, and toluidine blue counterstain for bone development (H–J), and immunohistochemically for Runx2 (K–M). Similar morphology and Runx2 expression patterns were observed among sections of different genotypes. A, adventitia; M, media; L, lumen.
Runx2 Expression in SMC Is Required for VitD-Induced AMC in Mice
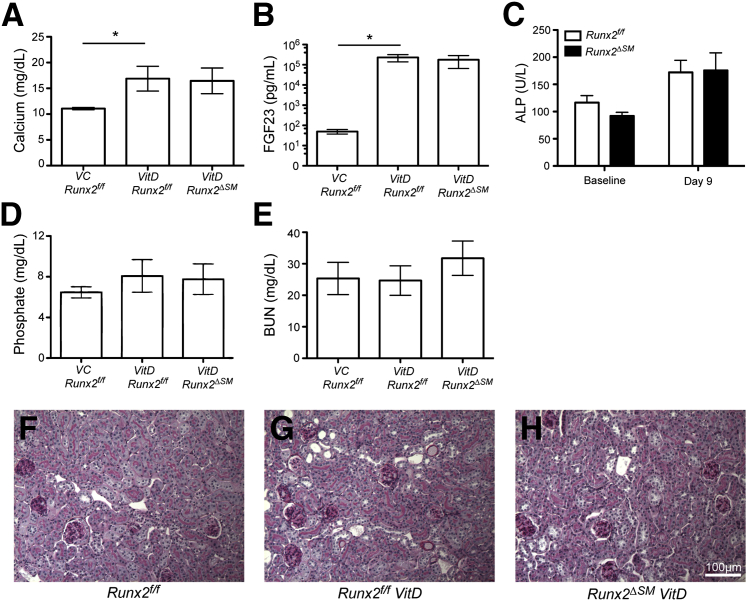
To determine whether Runx2 expression in SMCs was required for AMC, we induced disordered mineral homeostasis in Runx2f/f and Runx2ΔSM mice via subcutaneous injection of VitD at a dosage of 500,000 IU/kg/day for 4 consecutive days. VitD treatment of both Runx2f/f and Runx2ΔSM mice led to significantly increased serum calcium, fibroblast growth factor-23, and alkaline phosphatase levels compared with vehicle controls (Figure 5, A–C). Serum phosphate level was increased in VitD-treated mice compared with vehicle controls, but did not achieve statistical significance, likely owing to the highly increased serum fibroblast growth factor-23 levels that occurred in response to VitD treatment (Figure 5D). Blood urea nitrogen levels were similar in VitD- and vehicle-treated mice (Figure 5E). Furthermore, no significant differences in these serum parameters were observed between VitD-treated Runx2f/f and Runx2ΔSM animals (Figure 5, A–E). Consistent with blood urea nitrogen findings, kidneys from VitD-treated Runx2f/f and Runx2ΔSM mice showed normal renal histology compared with vehicle-treated Runxf/f mice, and the absence of renal calcification. (Figure 5, F–H).
Figure 5.
Vitamin D (VitD) overload increases serum calcium, fibroblast growth factor (FGF)-23, and alkaline phosphatase (ALP) levels without affecting phosphate levels, blood urea nitrogen (BUN) levels, and kidney histology. Vitamin D and vehicle (VC) were prepared and delivered as described in Materials and Methods. Fasting serum and kidney were collected from vehicle-injected Runx2f/f and vitamin D–injected Runx2f/f or Runx2ΔSM mice. Serum calcium (A), FGF-23 (B), and ALP (C) levels were increased by vitamin D overload, but no differences were observed between genotypes. Serum phosphate level (D), BUN level (E), and gross kidney anatomy (periodic acid-Schiff stain) (F–H) were not affected significantly by vitamin D overload. Data are presented as means ± SD (n = 3 to 5). ∗P < 0.05 as determined by one-way with ANOVA the Dunn post hoc test.
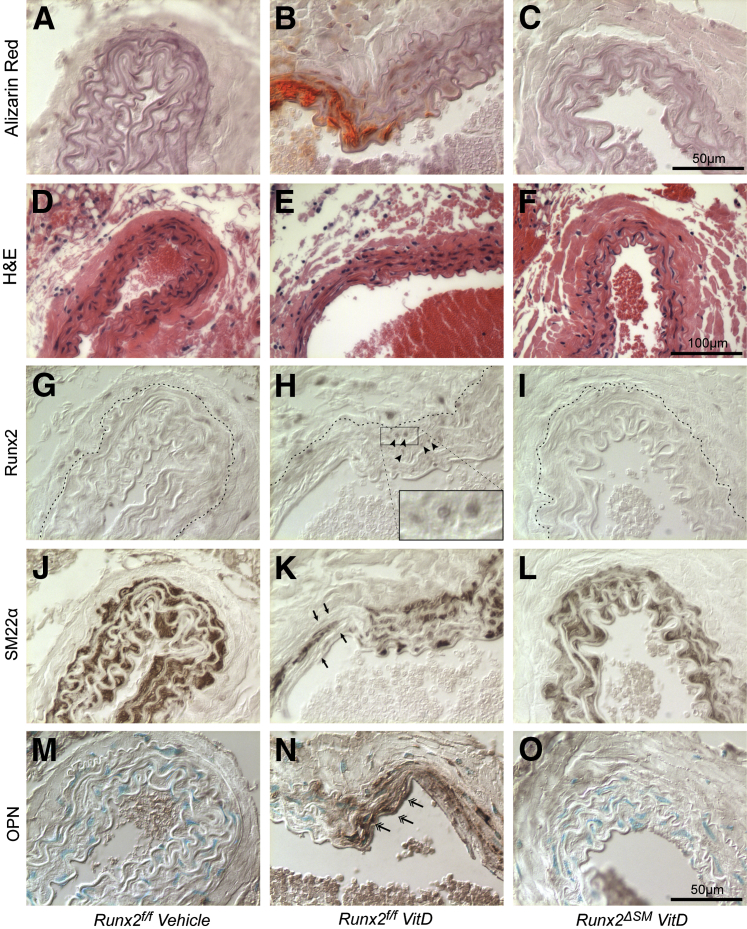
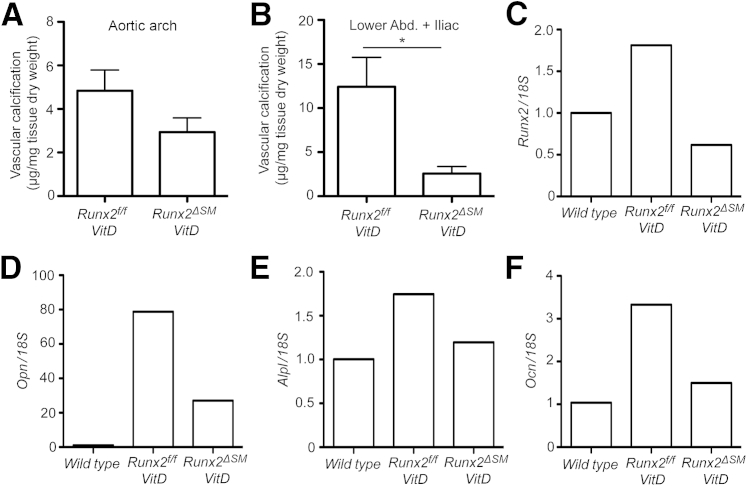
In association with altered mineral homeostasis, VitD treatment rapidly induced calcification in the arteries of Runx2f/f mice. Calcification was restricted to arterial media (Figure 6B), was highly associated with the deformed and fragmented elastic lamina (Figure 6E), and was undetectable in VitD-treated Runx2ΔSM vessels (Figure 6C) and vehicle control vessels (Figure 6A). Calcium content was increased greatly in aortic arches and abdominal aortas plus common iliac arteries on VitD treatment, ranging from 4 to 12 μg/mg dry weight on average (Figure 7, A and B). Importantly, inactivation of the Runx2 gene in SMCs significantly reduced AMC by 79% in abdominal aortas plus common iliac arteries of Runx2ΔSM mice (Figure 7B) in comparison with Runx2f/f mice. The same trend (approximately 39% reduction in AMC; P = 0.26) also was observed in aortic arches, but most likely did not attain statistical significance because aortic arches calcified to a much lower extent than the lower abdominal aorta and iliac arteries in this model (Figure 7A). Finally, no inflammatory lesions were observed in the vasculature of VitD-treated mice (Figure 6, D–F).
Figure 6.
Smooth muscle cell (SMC)-specific deletion of Runx2 prevents SMC osteochondrogenic phenotype change. Runx2f/f (A, B, D, E, G, H, J, K, M, and N) and Runx2ΔSM (C, F, I, L, and O) mice were treated with either vitamin D (VitD) (B, C, E, F, H, I, K, L, N, and O) or vehicle control (A, D, G, J, and M) as described in Materials and Methods. Abdominal aortas were collected to visualize calcium deposition through Alizarin Red S stain (A–C), as well as hematoxylin and eosin (H&E) (D–F), Runx2 (brown nuclear stain) (G–I), SMC marker protein, SM22α (brown stain) (J–L), and osteopontin (OPN) (brown stain) (M–O, double arrows). Dashed lines in G–I designate external elastic lamina. Note the bright red calcium stain in the aortic media of Runx2f/f mice challenged with vitamin D (B) and the presence of Runx2-positive cells in the media layer around the calcification site (H, arrowheads), but not in vehicle-injected (A) or Runx2ΔSM vessels (C). Note also the concomitant loss of SM22α at the calcification site (K, arrows).
Figure 7.
Smooth muscle cell (SMC)-specific deletion of Runx2 prevents vascular calcification. Total calcium deposition in blood vessels [aortic arches (A) or abdominal + iliac aorta (B)] of Runx2f/f and Runx2ΔSM mice with [vitamin D (VitD)] or without (vehicle) vitamin D treatment was quantified and normalized to tissue dry weight. Total RNA was extracted from pooled thoracic aortas (n = 4) and used to determine the expression of Runx2, Ocn, Opn, and Alpl by quantitative real-time PCR with 18S rRNA as a normalizing control and untreated wild-type samples as a calibrator (C–F). Data for vascular calcification are presented as means SEM (n = 3 to 5) (A and B). Data are presented as the fold-change compared with vehicle control (C–F). ∗P < 0.05 as determined by the nonparametric t-test.
Runx2 Expression in SMC Is Required for the VitD-Induced Osteogenic Phenotype Change in Mice
To determine whether Runx2 expression in SMC was required for osteogenic phenotype transition in AMC, immunostaining for Runx2, SM22α, and OPN was performed. As expected, Runx2 was not detected in vehicle-treated Runx2f/f mice (Figure 6G), but was induced in medial cells after treatment with VitD in Runx2f/f (Figure 6H) and absent when Runx2 was depleted in Runx2ΔSM mice (Figure 6I). The SMC lineage marker SM22α was decreased dramatically in the medial cells within calcified areas of VitD-treated Runx2f/f vessels compared with vehicle-treated controls (Figure 6, J and K). In contrast, VitD-treated Runx2ΔSM showed SM22α levels equivalent to that observed in vehicle-treated Runx2f/f mice (Figure 6L). Furthermore, the osteogenic marker OPN was increased dramatically in the medial cells of VitD-treated Runx2f/f mice compared with vehicle-treated controls (Figure 6, M and N), whereas no OPN was detectable in medial cells in VitD-treated Runx2ΔSM mice (Figure 6O). We also quantified expression levels of Runx2, Opn, Alpl, and Ocn in these vessels via quantitative real-time PCR. The mRNA levels of Runx2 were increased in the VitD-treated Runx2f/f vessels but no increase was observed in VitD-treated Runx2ΔSM vessels, suggesting that SMC is the major source of Runx2 signaling during VitD-induced vascular calcification (Figure 7C). Finally, deletion of the Runx2 gene in vascular SMCs greatly reduced the expression of Opn and completely blocked the up-regulation of the osteoblast marker genes, Alpl and Ocn, in response to VitD (Figure 7, D–F).
Runx2 Function in Vascular Calcification Is SMC Autonomous
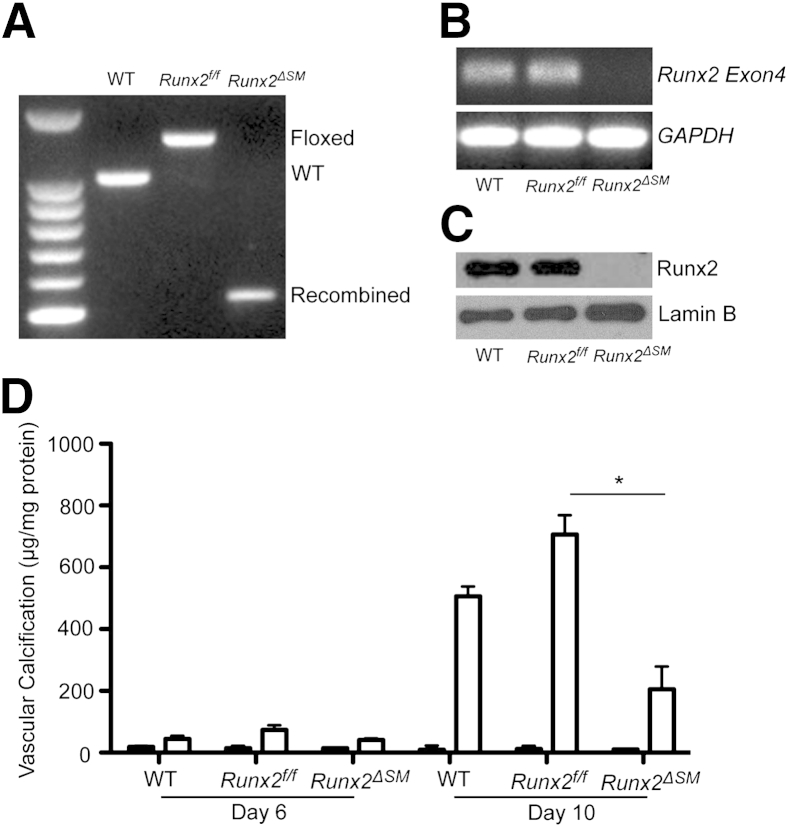
To determine whether the function of Runx2 in AMC was SMC autonomous, we isolated vascular SMCs from aortas of wild-type, Runx2f/f, and Runx2ΔSM mice and determined their calcification susceptibility in response to the calcification stimulus, increased phosphate. Genotypes of the SMCs were determined by genomic DNA rearrangement at exon 4 of the Runx2 gene (Figure 8A) and the expression of Runx2 at mRNA and protein levels (Figure 8, B and C). SMCs isolated from Runx2ΔSM mice were found to have a complete loss of Runx2 even when cells were treated with increased phosphate, whereas the Runx2f/f SMCs retained appropriate Runx2 level in response to phosphate induction (Figure 8, B and C). Increased phosphate levels induced matrix calcification in SMCs of wild-type and Runx2f/f genotypes in a time-dependent manner (Figure 8D). In contrast, SMCs isolated from the Runx2ΔSM mice showed significantly attenuated phosphate-induced matrix calcification compared with Runx2f/f (Figure 8D).
Figure 8.
Runx2-deficient smooth muscle cells (SMCs) have decreased susceptibility to calcification in vitro. A: SMCs were isolated from wild-type (WT), Runx2f/f, and Runx2ΔSM mice. Genomic DNA was extracted and the presence of WT, floxed, and knockout alleles was determined as described in Materials and Methods. B and C: WT, Runx2f/f, and Runx2ΔSM SMCs were treated with 2.4 mmol/L inorganic phosphate for 4 days, and total RNA and nuclear protein were extracted and used to determine the presence of exon 4 transcripts of Runx2 by reverse transcriptase-PCR (B), and Runx2 protein by Western blotting (C). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and lamin B were used to monitor sample loading. D: SMCs of various genotypes were cultured in the normal (0.9 mmol/L; black bars) or increased (2.4 mmol/L; white bars) inorganic phosphate (Pi) for 6 and 10 days. SMC calcification was determined and normalized to total cellular protein. Data are presented as means ± SD (n = 3). ∗P < 0.05 as determined by one-way analysis of variance with the Dunn post hoc test.
Discussion
AMC is highly prevalent in aging, diabetic, and CKD patients, and is a major risk factor for cardiovascular morbidity and mortality.1–3 Runx2, a master regulator of bone formation, has been proposed as a key transcription factor controlling SMC osteogenic phenotype change and calcification, but definitive in vivo evidence for this is lacking. Here, we developed and characterized mice carrying an improved Runx2 conditional targeting allele for SMC-specific deletion. By using these mice, we showed that SMC-specific Runx2 expression was required for pathologic AMC development induced by VitD overload. In addition, Runx2 knockout in SMC prevented osteogenic phenotype change. Finally, SMCs isolated from Runx2ΔSM mice were completely devoid of Runx2 protein and unable to respond to high-phosphate–induced calcification in vitro.
Although previous studies have implicated Runx2 as an important mediator of vascular SMC calcification in vitro,30,36 the current studies are the first to show that Runx2 expression specifically in SMCs is required for AMC in vivo. Our studies also confirm and extend findings by Han et al37 that used Runx2 haploinsufficient mice to examine functional cooperation between VitD-receptor and Runx2 in VitD-treated mice. Although they observed less AMC in Runx2+/− mice treated with VitD, bone remodeling activity, as measured by serum alkaline phosphatase levels, also was reduced. Because alkaline phosphatase degrades pyrophosphate, a major serum inhibitor of vascular calcification,38 decreases in alkaline phosphate may have protected vessels against AMC, thereby confounding interpretation of the studies. The conditional targeting strategy used here allowed us to delete Runx2 selectively from SMC without changes in serum alkaline phosphatase levels, osteoblast expression, or bone morphology, thereby allowing us to distinguish between the skeletal and vascular effects of Runx2.
The appearance of vascular medial cells with an osteogenic phenotype is a common finding in AMC in people and experimental animals, and these cells are known major mediators of vascular calcification.19,20,23,39–41 Lineage tracing studies have confirmed that vascular SMCs could undergo lineage reprogramming and give rise to Runx2-expressing osteochondrogenic progenitors in AMC23 as well as AIC.24,42 The present findings show for the first time that Runx2 expression in SMCs is required for osteogenic lineage reprogramming in vessels undergoing AMC. AMC in VitD-treated mice was associated with increased Runx2, Opn, Alpl, and Ocn, and loss of SM22α expression in the tunica media in Runx2f/f but not Runx2ΔSM mice. Furthermore, SMCs isolated from Runx2ΔSM mice were much less susceptible to phosphate-induced calcification than those from Runx2f/f and Runx2+/+ mice. Together, these findings suggest an SMC autonomous role of Runx2 in AMC via control of osteogenic lineage reprogramming.
AMC occurs within the tunica media typically starting along the elastic lamina, and proceeding in the absence of inflammation.19,43 Indeed, in our hands, no signs of vascular inflammation were detected in VitD-treated mice, regardless of genotype. In contrast, AIC is characterized by mineral deposition in highly inflamed and necrotic atherosclerotic lesions of the tunica intima. Interestingly, Sun et al44 reported that Runx2 deficiency in SMCs prevented AIC in ApoE null mice fed a high-fat diet. In that study, mice with conditional Runx2 targeting had decreased expression of receptor activator of NF-κB ligand in SMCs, and greatly reduced macrophage infiltration in atherosclerotic lesions, suggesting a role for Runx2 in regulating proinflammatory effects of SMCs. Thus, the mechanism(s) by which Runx2 promotes vascular calcification are likely to be different in AMC versus AIC, and highly dependent on disease context. However, it should be noted that Sun et al44 used a Runx2 exon 8 targeting construct that resulted in truncated Runx2 proteins containing the Runt homology domain to be synthesized in SMCs. Such truncated Runx2 proteins have been shown to act as dominant-negative inhibitors and potentially can interfere with other runt homology domain family members: Runx1 and Runx3.31 In addition, recent evidence has shown that the truncated Runx2 still retains various degrees of transcriptional activity depending on the promoter.45 Thus, possible dominant-negative inhibition of Runx1 and Runx3 as an alternative explanation for the effects observed by Sun et al44 cannot be ruled out. Clearly, future studies using a Runx2 targeting vector that leads to complete deletion of Runx2 in SMCs, as was developed in the present study, are warranted to distinguish between these mechanisms in AIC.
In conclusion, with an improved Runx2 conditional inactivation model, we have shown the importance of SMC-specific Runx2 in AMC induced by disordered mineral metabolism. Mechanistically, this effect likely was owing to the inhibition of SMC reprogramming into osteochondrogenic progenitors. Furthermore, Runx2 function was SMC autonomous because systemic mineral homeostasis was not altered and decreased susceptibility of Runx2-deficient SMCs to calcification was recapitulated in vitro. Taken together, our study showed a critical role of SMC-specific Runx2 in SMC osteochondrogenic phenotype change and AMC development, and points to pathways controlling Runx2 expression as potential therapeutic targets.
Acknowledgments
We thank Dr. Veena Naik for assistance in animal breeding and data collection from Runx2 global deletion mice and Ngoc B. Nguyen for assistance in animal breeding and specimen collection from Runx2 SMC-specific deletion mice.
Footnotes
Supported by NIH grants R01 HL081785 (C.M.G.), R01 HL62329 (C.M.G.), and K01 DK075665 (M.Y.S.).
M.-E.L. and T.C. contributed equally to this work.
Disclosures: None declared.
Contributor Information
Mei Y. Speer, Email: yfwang@u.washington.edu.
Cecilia M. Giachelli, Email: ceci@u.washington.edu.
References
- 1.Guerin A.P., Pannier B., Metivier F., Marchais S.J., London G.M. Assessment and significance of arterial stiffness in patients with chronic kidney disease. Curr Opin Nephrol Hypertens. 2008;17:635–641. doi: 10.1097/mnh.0b013e32830dcd5c. [DOI] [PubMed] [Google Scholar]
- 2.Gu K., Cowie C.C., Harris M.I. Mortality in adults with and without diabetes in a national cohort of the U.S. population, 1971-1993. Diabetes Care. 1998;21:1138–1145. doi: 10.2337/diacare.21.7.1138. [DOI] [PubMed] [Google Scholar]
- 3.Dickstein K., Cohen-Solal A., Filippatos G., McMurray J.J., Ponikowski P., Poole-Wilson P.A., Strömberg A., van Veldhuisen D.J., Atar D., Hoes A.W., Keren A., Mebazaa A., Nieminen M., Priori S.G., Swedberg K., Committee for Practice Guidelines (CPG) ESC guidelines for the diagnosis and treatment of acute and chronic heart failure 2008: the Task Force for the diagnosis and treatment of acute and chronic heart failure 2008 of the European Society of Cardiology. Developed in collaboration with the Heart Failure Association of the ESC (HFA) and endorsed by the European Society of Intensive Care Medicine (ESICM) Eur J Heart Fail. 2008;10:933–989. doi: 10.1016/j.ejheart.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Shroff R.C., McNair R., Figg N., Skepper J.N., Schurgers L., Gupta A., Hiorns M., Donald A.E., Deanfield J., Rees L., Shanahan C.M. Dialysis accelerates medial vascular calcification in part by triggering smooth muscle cell apoptosis. Circulation. 2008;118:1748–1757. doi: 10.1161/CIRCULATIONAHA.108.783738. [DOI] [PubMed] [Google Scholar]
- 5.Goodman W.G., London G., Amann K., Block G.A., Giachelli C., Hruska K.A., Ketteler M., Levin A., Massy Z., McCarron D.A., Raggi P., Shanahan C.M., Yorioka N. Vascular calcification in chronic kidney disease. Am J Kidney Dis. 2004;43:572–579. doi: 10.1053/j.ajkd.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Moorthi R.N., Moe S.M. CKD-mineral and bone disorder: core curriculum 2011. Am J Kidney Dis. 2011;58:1022–1036. doi: 10.1053/j.ajkd.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.London G.M., Guerin A.P., Marchais S.J., Metivier F., Pannier B., Adda H. Arterial media calcification in end-stage renal disease: impact on all-cause and cardiovascular mortality. Nephrol Dial Transplant. 2003;18:1731–1740. doi: 10.1093/ndt/gfg414. [DOI] [PubMed] [Google Scholar]
- 8.Beadenkopf W.G., Daoud A.S., Love B.M. Calcification in the coronary arteries and its relationship to arteriosclerosis and myocardial infarction. Am J Roentgenol Radium Ther Nucl Med. 1964;92:865–871. [PubMed] [Google Scholar]
- 9.Kelly-Arnold A., Maldonado N., Laudier D., Aikawa E., Cardoso L., Weinbaum S. Revised microcalcification hypothesis for fibrous cap rupture in human coronary arteries. Proc Natl Acad Sci U S A. 2013;110:10741–10746. doi: 10.1073/pnas.1308814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehara S., Kobayashi Y., Yoshiyama M., Shimada K., Shimada Y., Fukuda D., Nakamura Y., Yamashita H., Yamagishi H., Takeuchi K., Naruko T., Haze K., Becker A.E., Yoshikawa J., Ueda M. Spotty calcification typifies the culprit plaque in patients with acute myocardial infarction: an intravascular ultrasound study. Circulation. 2004;110:3424–3429. doi: 10.1161/01.CIR.0000148131.41425.E9. [DOI] [PubMed] [Google Scholar]
- 11.Kelly R.P., Tunin R., Kass D.A. Effect of reduced aortic compliance on cardiac efficiency and contractile function of in situ canine left ventricle. Circ Res. 1992;71:490–502. doi: 10.1161/01.res.71.3.490. [DOI] [PubMed] [Google Scholar]
- 12.Niederhoffer N., Lartaud-Idjouadiene I., Giummelly P., Duvivier C., Peslin R., Atkinson J. Calcification of medial elastic fibers and aortic elasticity. Hypertension. 1997;29:999–1006. doi: 10.1161/01.hyp.29.4.999. [DOI] [PubMed] [Google Scholar]
- 13.Raggi P., Bellasi A., Ferramosca E., Islam T., Muntner P., Block G.A. Association of pulse wave velocity with vascular and valvular calcification in hemodialysis patients. Kidney Int. 2007;71:802–807. doi: 10.1038/sj.ki.5002164. [DOI] [PubMed] [Google Scholar]
- 14.Tonelli M., Moyé L., Sacks F.M., Kiberd B., Curhan G., Cholesterol and Recurrent Events (CARE) Trial Investigators Pravastatin for secondary prevention of cardiovascular events in persons with mild chronic renal insufficiency. Ann Intern Med. 2003;138:98–104. doi: 10.7326/0003-4819-138-2-200301210-00010. [DOI] [PubMed] [Google Scholar]
- 15.Palmer S.C., Craig J.C., Navaneethan S.D., Tonelli M., Pellegrini F., Strippoli G.F. Benefits and harms of statin therapy for persons with chronic kidney disease: a systematic review and meta-analysis. Ann Intern Med. 2012;157:263–275. doi: 10.7326/0003-4819-157-4-201208210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fellström B.C., Jardine A.G., Schmieder R.E., Holdaas H., Bannister K., Beutler J., Chae D.W., Chevaile A., Cobbe S.M., Grönhagen-Riska C., De Lima J.J., Lins R., Mayer G., McMahon A.W., Parving H.H., Remuzzi G., Samuelsson O., Sonkodi S., Sci D., Süleymanlar G., Tsakiris D., Tesar V., Todorov V., Wiecek A., Wüthrich R.P., Gottlow M., Johnsson E., Zannad F., Aurora Study Group Rosuvastatin and cardiovascular events in patients undergoing hemodialysis. N Engl J Med. 2009;360:1395–1407. doi: 10.1056/NEJMoa0810177. [DOI] [PubMed] [Google Scholar]
- 17.Wanner C., Krane V., März W., Olschewski M., Mann J.F., Ruf G., Ritz E., German Diabetes and Dialysis Study Investigators Atorvastatin in patients with type 2 diabetes mellitus undergoing hemodialysis. N Engl J Med. 2005;353:238–248. doi: 10.1056/NEJMoa043545. [DOI] [PubMed] [Google Scholar]
- 18.Cowell S.J., Newby D.E., Prescott R.J., Bloomfield P., Reid J., Northridge D.B., Boon N.A., Scottish Aortic Stenosis and Lipid Lowering Trial, Impact on Regeression (SALTIRE) Investigators A randomized trial of intensive lipid-lowering therapy in calcific aortic stenosis. N Engl J Med. 2005;352:2389–2397. doi: 10.1056/NEJMoa043876. [DOI] [PubMed] [Google Scholar]
- 19.Pai A., Leaf E.M., El Abbadi M., Giachelli C.M. Elastin degradation and vascular smooth muscle cell phenotype change precede cell loss and arterial medial calcification in a uremic mouse model of chronic kidney disease. Am J Pathol. 2011;178:764–773. doi: 10.1016/j.ajpath.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Abbadi M.M., Pai A.S., Leaf E.M., Yang H.Y., Bartley B.A., Quan K.K., Ingalls C.M., Liao H.W., Giachelli C.M. Phosphate feeding induces arterial medial calcification in uremic mice: role of serum phosphorus, fibroblast growth factor-23, and osteopontin. Kidney Int. 2009;75:1297–1307. doi: 10.1038/ki.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen N.X., Moe S.M. Arterial calcification in diabetes. Curr Diab Rep. 2003;3:28–32. doi: 10.1007/s11892-003-0049-2. [DOI] [PubMed] [Google Scholar]
- 22.Vattikuti R., Towler D.A. Osteogenic regulation of vascular calcification: an early perspective. Am J Physiol Endocrinol Metab. 2004;286:E686–E696. doi: 10.1152/ajpendo.00552.2003. [DOI] [PubMed] [Google Scholar]
- 23.Speer M.Y., Yang H.Y., Brabb T., Leaf E., Look A., Lin W.L., Frutkin A.D., Dichek D.A., Giachelli C.M. Smooth muscle cells give rise to osteochondrogenic precursors and chondrocytes in calcifying arteries. Circ Res. 2009;104:733–741. doi: 10.1161/CIRCRESAHA.108.183053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naik V., Leaf E.M., Hu J.H., Yang H.Y., Nguyen N.B., Giachelli C.M., Speer M.Y. Sources of cells that contribute to atherosclerotic intimal calcification: an in vivo genetic fate mapping study. Cardiovasc Res. 2012;94:545–554. doi: 10.1093/cvr/cvs126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 26.Otto F., Thornell A.P., Crompton T., Denzel A., Gilmour K.C., Rosewell I.R., Stamp G.W., Beddington R.S., Mundlos S., Olsen B.R., Selby P.B., Owen M.J. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 27.Otto F., Kanegane H., Mundlos S. Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Hum Mutat. 2002;19:209–216. doi: 10.1002/humu.10043. [DOI] [PubMed] [Google Scholar]
- 28.Kimmel C.A., Trammell C. A rapid procedure for routine double staining of cartilage and bone in fetal and adult animals. Stain Technol. 1981;56:271–273. doi: 10.3109/10520298109067325. [DOI] [PubMed] [Google Scholar]
- 29.Kang Y.H., Jin J.S., Yi D.W., Son S.M. Bone morphogenetic protein-7 inhibits vascular calcification induced by high vitamin D in mice. Tohoku J Exp Med. 2010;221:299–307. doi: 10.1620/tjem.221.299. [DOI] [PubMed] [Google Scholar]
- 30.Speer M.Y., Li X., Hiremath P.G., Giachelli C.M. Runx2/Cbfa1, but not loss of myocardin, is required for smooth muscle cell lineage reprogramming toward osteochondrogenesis. J Cell Biochem. 2010;110:935–947. doi: 10.1002/jcb.22607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ducy P., Starbuck M., Priemel M., Shen J., Pinero G., Geoffroy V., Amling M., Karsenty G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13:1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L., Miano J.M., Mercer B., Olson E.N. Expression of the SM22alpha promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J Cell Biol. 1996;132:849–859. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frutkin A.D., Shi H., Otsuka G., Leveen P., Karlsson S., Dichek D.A. A critical developmental role for tgfbr2 in myogenic cell lineages is revealed in mice expressing SM22-Cre, not SMMHC-Cre. J Mol Cell Cardiol. 2006;41:724–731. doi: 10.1016/j.yjmcc.2006.06.067. [DOI] [PubMed] [Google Scholar]
- 34.Holtwick R., Gotthardt M., Skryabin B., Steinmetz M., Potthast R., Zetsche B., Hammer R.E., Herz J., Kuhn M. Smooth muscle-selective deletion of guanylyl cyclase-A prevents the acute but not chronic effects of ANP on blood pressure. Proc Natl Acad Sci U S A. 2002;99:7142–7147. doi: 10.1073/pnas.102650499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyson K.L., Reynolds J.L., McNair R., Zhang Q., Weissberg P.L., Shanahan C.M. Osteo/chondrocytic transcription factors and their target genes exhibit distinct patterns of expression in human arterial calcification. Arterioscler Thromb Vasc Biol. 2003;23:489–494. doi: 10.1161/01.ATV.0000059406.92165.31. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka T., Sato H., Doi H., Yoshida C.A., Shimizu T., Matsui H., Yamazaki M., Akiyama H., Kawai-Kowase K., Iso T., Komori T., Arai M., Kurabayashi M. Runx2 represses myocardin-mediated differentiation and facilitates osteogenic conversion of vascular smooth muscle cells. Mol Cell Biol. 2008;28:1147–1160. doi: 10.1128/MCB.01771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han M.S., Che X., Cho G.H., Park H.R., Lim K.E., Park N.R., Jin J.S., Jung Y.K., Jeong J.H., Lee I.K., Kato S., Choi J.Y. Functional cooperation between vitamin D receptor and Runx2 in vitamin D-induced vascular calcification. PLoS One. 2013;8:e83584. doi: 10.1371/journal.pone.0083584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lomashvili K.A., Cobbs S., Hennigar R.A., Hardcastle K.I., O'Neill W.C. Phosphate-induced vascular calcification: role of pyrophosphate and osteopontin. J Am Soc Nephrol. 2004;15:1392–1401. doi: 10.1097/01.asn.0000128955.83129.9c. [DOI] [PubMed] [Google Scholar]
- 39.Zebger-Gong H., Müller D., Diercke M., Haffner D., Hocher B., Verberckmoes S., Schmidt S., D'Haese P.C., Querfeld U. 1,25-Dihydroxyvitamin D3-induced aortic calcifications in experimental uremia: up-regulation of osteoblast markers, calcium-transporting proteins and osterix. J Hypertens. 2011;29:339–348. doi: 10.1097/HJH.0b013e328340aa30. [DOI] [PubMed] [Google Scholar]
- 40.Moe S.M., Duan D., Doehle B.P., O'Neill K.D., Chen N.X. Uremia induces the osteoblast differentiation factor Cbfa1 in human blood vessels. Kidney Int. 2003;63:1003–1011. doi: 10.1046/j.1523-1755.2003.00820.x. [DOI] [PubMed] [Google Scholar]
- 41.Shroff R.C., McNair R., Skepper J.N., Figg N., Schurgers L.J., Deanfield J., Rees L., Shanahan C.M. Chronic mineral dysregulation promotes vascular smooth muscle cell adaptation and extracellular matrix calcification. J Am Soc Nephrol. 2010;21:103–112. doi: 10.1681/ASN.2009060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen N.B., Naik V., Speer M.Y. Diabetes mellitus accelerates cartilaginous metaplasia and calcification in atherosclerotic vessels of LDLr mutant mice. Cardiovasc Pathol. 2013;22:167–175. doi: 10.1016/j.carpath.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shanahan C.M., Cary N.R., Salisbury J.R., Proudfoot D., Weissberg P.L., Edmonds M.E. Medial localization of mineralization-regulating proteins in association with Monckeberg's sclerosis: evidence for smooth muscle cell-mediated vascular calcification. Circulation. 1999;100:2168–2176. doi: 10.1161/01.cir.100.21.2168. [DOI] [PubMed] [Google Scholar]
- 44.Sun Y., Byon C.H., Yuan K., Chen J., Mao X., Heath J.M., Javed A., Zhang K., Anderson P.G., Chen Y. Smooth muscle cell-specific runx2 deficiency inhibits vascular calcification. Circ Res. 2012;111:543–552. doi: 10.1161/CIRCRESAHA.112.267237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H., Ghori-Javed F.Y., Rashid H., Adhami M.D., Serra R., Gutierrez S.E., Javed A. Runx2 regulates endochondral ossification through control of chondrocyte proliferation and differentiation. J Bone Miner Res. 2014;29:2653–2665. doi: 10.1002/jbmr.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]