Abstract
Eukaryotic cells fold and assemble membrane and secreted proteins in the endoplasmic reticulum (ER), before delivery to other cellular compartments or the extracellular environment. Correctly folded proteins are released from the ER, and poorly folded proteins are retained until they achieve stable conformations; irreparably misfolded proteins are targeted for degradation. Diverse pathological insults, such as amino acid mutations, hypoxia, or infection, can overwhelm ER protein quality control, leading to misfolded protein buildup, causing ER stress. To cope with ER stress, eukaryotic cells activate the unfolded protein response (UPR) by increasing levels of ER protein-folding enzymes and chaperones, enhancing the degradation of misfolded proteins, and reducing protein translation. In mammalian cells, three ER transmembrane proteins, inositol-requiring enzyme-1 (IRE1; official name ERN1), PKR-like ER kinase (PERK; official name EIF2AK3), and activating transcription factor-6, control the UPR. The UPR signaling triggers a set of prodeath programs when the cells fail to successfully adapt to ER stress or restore homeostasis. ER stress and UPR signaling are implicated in the pathogenesis of diverse diseases, including neurodegeneration, cancer, diabetes, and inflammation. This review discusses the current understanding in both adaptive and apoptotic responses as well as the molecular mechanisms instigating apoptosis via IRE1 and PERK signaling. We also examine how IRE1 and PERK signaling may be differentially used during neurodegeneration arising in retinitis pigmentosa and prion infection.
The endoplasmic reticulum (ER) is an essential organelle responsible for folding of secreted and membrane proteins and lipid and sterol biosynthesis, and it is a major site of free calcium storage within the cell. Cells have evolved a unique homeostatic mechanism, termed the unfolded protein response (UPR), to ensure that the ER can adapt to changing environmental and physiological demands of its functions. In mammalian cells, the UPR is controlled by the ER resident transmembrane proteins, inositol-requiring enzyme-1 (IRE1; official name ERN1), PKR-like ER kinase (PERK; official name EIF2AK3), and activating transcription factor-6 (ATF6).
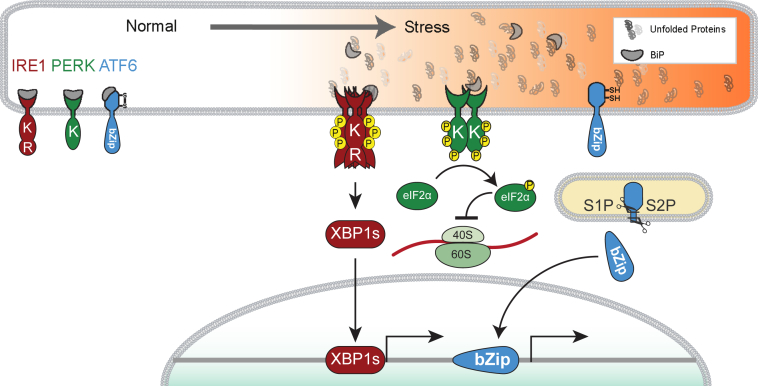
IRE1 is a transmembrane protein that controls a UPR signal transduction pathway conserved from yeast to mammals (Figure 1).1 IRE1 bears a luminal domain coupled across the ER membrane to cytosolic kinase and endoribonuclease (RNase) domains.1 In response to ER stress, IRE1 undergoes oligomeric assembly, transautophosphorylation by its kinase domain, and activation of its distal RNase activity.2,3 In metazoans, the RNase activity of activated IRE1 and the RtcB tRNA ligase splice out a small intron from the X-box binding protein-1 (Xbp1) mRNA to produce XBP1s transcription factor.4–8
Figure 1.
Endoplasmic reticulum (ER) stress activates inositol-requiring enzyme 1 (IRE1), PKR-like ER kinase (PERK), and activating transcription factor-6 (ATF6) intracellular signal transduction pathways of the unfolded protein response (UPR). In normal condition, the UPR transducers, IRE1, PERK, and ATF6, associate with BiP to prevent UPR. On accumulation of misfolded proteins in the ER lumen, BiP dissociates to activate UPR transducers. IRE1 bears a luminal domain coupled across the ER membrane to cytosolic kinase (K) and endoribonuclease (RNase) domains (R). In response to ER stress, IRE1 undergoes oligomeric assembly, transautophosphorylation by its kinase domain, activating its distal RNase activity. Activated IRE1 splices out small intron from the X-box binding protein-1 (Xbp1) mRNA to generate active transcription factor XBP1s. The PERK protein bears luminal domain coupled across the ER membrane to K. In response to ER stress, PERK dimerizes and subsequently activates its cytosolic kinase domain. PERK's kinase recognizes and phosphorylates eukaryotic translation initiation factor 2 subunit alpha (eIF2α), leading to attenuation of global protein translation. ATF6 bears an ER-tethered transcription factor. In response to ER stress, ATF6 migrates from the ER to the Golgi apparatus, where site 1 and 2 proteases (S1P and S2P, respectively) cleave its luminal and transmembrane domains, and release the cytosolic portion of ATF6 containing the bZIP transcriptional activator domain. Cleaved ATF6 fragment translocates to the nucleus to serve as a transcription factor.
The transcriptional targets of XBP1 are highly enriched for ER-associated protein degradation (ERAD) factors, ER chaperones, and enzymes required for lipid biosynthesis and protein glycosylation across diverse mammalian cell types.9–14 Up-regulation of these molecules by IRE1-to-XBP1s induction therefore enhances the ER's capacity to better fold new proteins as well as target irreparably damaged proteins for retrotranslocation out of the ER for degradation by proteasomes in the cytosol (Figure 1).
The ER transmembrane protein PERK regulates another UPR signaling pathway in metazoans15 (Figure 1). On ER stress, PERK oligomerizes and activates its cytosolic kinase domain.15 PERK's kinase phosphorylates eukaryotic translation initiation factor 2 subunit alpha (eIF2α) on Ser51, inhibiting the guanine nucleotide exchange factor eIF2B, which converts inactive GDP-bound eIF2 to its active GTP form.15,16 Active eIF2 complex is needed to form the GTP-tRNAMet ternary complex required for translation initiation. Therefore, eIF2α phosphorylation leads to translation inhibition that helps alleviate ER stress by reducing the load of new polypeptides that require assembly and folding in the ER compartment.17
ATF6 bears an ER-tethered bZip transcription factor that regulates a third UPR signal transduction pathway18 (Figure 1). In response to ER stress, ATF6 migrates from the ER to the Golgi apparatus, where site 1 protease and site 2 protease cleave its luminal and transmembrane domains to release the cytosolic portion of ATF6.18,19 The cytosolic portion of ATF6 contains the bZIP transcriptional activator domain, and after cleavage, this ATF6 fragment migrates to the nucleus to transcriptionally up-regulate ER chaperones and ERAD components, thereby enhancing ER protein-folding capacity and efficiency of ERAD.10,12,18,20 Interestingly, ATF6 also transcriptionally up-regulates Xbp1, thereby facilitating IRE1 signal transduction by increasing levels of IRE1's RNase substrate, Xbp1 mRNA.21
Put together, these initial transcriptional and translational effects of IRE1, PERK, and ATF6 signaling help cells adapt to ER stress by enhancing the fidelity of protein folding, increasing the degradation of damaged/misfolded proteins, and suppressing new protein synthesis. However, if these actions fail to restore ER homeostasis and ER stress persists, UPR signaling consequently triggers maladaptive proapoptotic programs, many of which are specifically activated through the IRE1 and PERK pathways.
IRE1 Signaling through RIDD, c-Jun N-Terminal Kinase, and BCL2
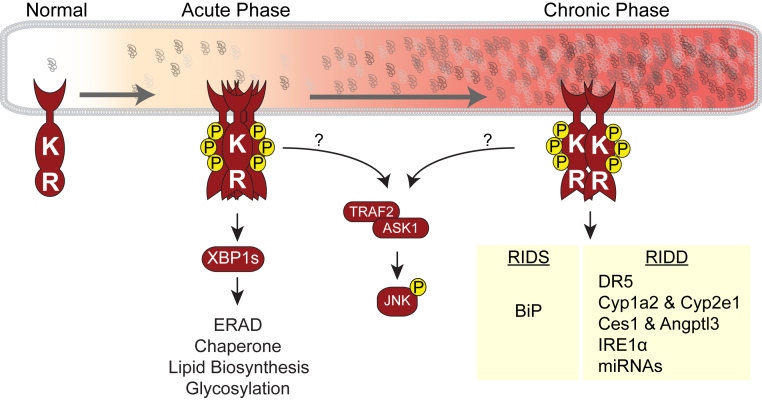
Seminal mechanistic studies from the laboratory of Walter and colleagues have revealed that IRE1 undergoes dynamic conformational and functional changes as a function of the duration of ER stress. In response to acute ER stress, IRE1 quickly forms oligomeric clusters in the ER plane, but IRE1 subsequently dissociates if ER stress persists (Figure 2).22,23 Interestingly, Xbp1 mRNA splicing only occurs during the acute phase.23–25 During the chronic phase of ER stress, IRE1's RNase substrate specificity is altered to cleave primarily ER-targeted mRNAs in a process termed regulated IRE1-dependent mRNA decay (RIDD)26–28 (Figure 2).
Figure 2.
Consequences of acute and chronic inositol-requiring enzyme 1 (IRE1) activation. In response to acute endoplasmic reticulum (ER) stress, IRE1 undergoes oligomeric assembly, undergoes transautophosphorylation by its kinase domain, and activates its distal RNase activity. Activated IRE1's RNase splices out a small intron from the X-box binding protein-1 (Xbp1) mRNA to produce the active transcription factor XBP1s. IRE1-to-XBP1s induction enhances the ER's capacity by up-regulation of gene sets involved in ER-associated protein degradation (ERAD), ER chaperones, lipid biosynthesis, and protein glycosylation. In the chronic phase of IRE1 activation, IRE1's RNase domain cleaves ER-targeted mRNAs in a phenomenon termed regulated IRE1-dependent mRNA decay (RIDD). Most RIDD-targeted mRNAs are disposed. In contrast, IRE1-dependent cleavage of the 3′ untranslated region of BiP mRNA in Schizosaccharomyces pombe stabilizes BiP mRNA, thereby increasing BiP protein levels to cope with ER stress. There is likely a regulated IRE1-dependent mRNA stabilization (RIDS) rather than RIDD, which may be a new mode by which IRE1's RNase positively regulates mRNAs. The TRAF2 adaptor protein binds to IRE1 and the MAPKKK ASK1 to activate downstream molecules such as c-Jun N-terminal kinase (JNK), p38, and extracellular signal–regulated kinase (ERK). However, it is unclear which phase of IRE1 activity can interact with TRAF2-ASK1. DR5, death receptor 5.
In contrast to Xbp1 mRNA, RIDD targets are not ligated after IRE1 cleavage, and most mRNA fragments cleaved through RIDD are degraded. RIDD's physiological significance varies widely and can confer protective or proapoptotic effects, depending on the cellular function of the mRNA being targeted. RIDD-mediated cleavage of death receptor 5 (DR5) mRNA enhances cell survival during ER stress by reducing production of proapoptotic DR5 protein.29 RIDD-mediated cleavage of cytochrome P450 enzyme mRNAs in liver confers resistance to liver damage after acetaminophen overdose by preventing P450-mediated generation of hepatotoxic by-products.30 The mRNA fragments produced by RIDD cleavage can trigger inflammation by engaging with the cytosolic RIG1 RNA virus innate immunity sensor.31 RIDD-mediated loss of lipid metabolism mRNAs can alter plasma lipid levels in mice.32 Ire1 mRNA itself is a RIDD substrate, and RIDD may act as an autoregulatory brake on IRE1 signaling by down-regulating Ire1 mRNA levels.33 miRNA precursors have also been identified as RIDD substrates in vitro.34 Disruption of miRNA maturation by RIDD cleavage may further affect multiple biological processes by modulating miRNA-mRNA interactions throughout the cell.
Recently, IRE1's RNase was found to cleave the 3′ untranslated region of BiP mRNA in Schizosaccharomyces pombe, but this truncation surprisingly stabilized, rather than promoted, the decay of the remaining BiP mRNA, thereby increasing BiP protein levels during ER stress.35 Regulated IRE1-dependent mRNA stabilization, rather than RIDD, may be a new mode by which IRE1's RNase positively regulates mRNAs (Figure 2). Recent biochemical studies have suggested that IRE1 uses RIDD at intense ER stress levels, whereas Xbp1 mRNA splicing is initiated at much lower levels of ER stress25 (Figure 2). IRE1's decision to trigger RIDD, regulated IRE1-dependent mRNA stabilization, or Xbp-1 mRNA splicing may be dependent on the intensity of the stress and the ability of various cell types to respond to that stress.
The IRE1 oligomeric clusters formed by ER stress also act as molecular scaffolds to recruit other proteins and nucleate the formation of stress signal transduction sites at the ER lipid bilayer22,23 (Figure 2). For example, TRAF2 adaptor protein binds to IRE1 as well as the ASK1 MAPKKK to activate cytosolic signaling kinases, such as C-Jun N-terminal kinase, p38, and extracellular signal–regulated kinase during ER stress36,37 (Figure 2). IRE1 also interacts with the RACK1 adaptor protein to recruit phosphatases to the ER membrane during ER stress.38 IRE1 forms protein-protein interactions with BCL2 family proteins, such as BAX and BAK, and the BI-1 BCL2 regulatory protein.39,40 IRE1 also binds cytoskeletal nonmuscle myosin II during ER stress.41 These IRE1-centered protein-protein interactions can influence the sensitivity of IRE1 activation in response to ER stress. IRE1-centered protein-protein interactions could also act conversely to influence cellular signaling processes and structures in other cellular compartments during ER stress.
Proapoptotic Consequences of PERK Signaling
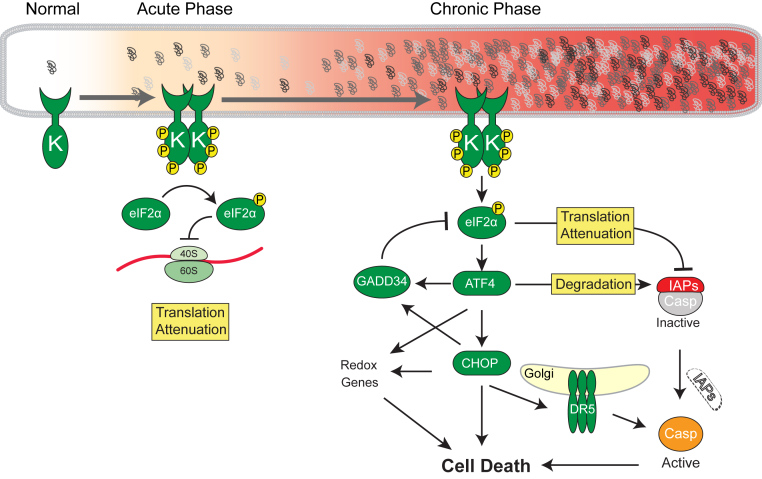
PERK signaling down-regulates translation from most mRNAs, thereby restricting de novo peptide loading onto the ER (Figure 1). This cytoprotective event by PERK to eIF2αP occurs rapidly (Figure 3). Rare mRNAs bearing 5′ upstream open reading frames, including the mRNAs encoding the ATF4, ATF5, and CHOP transcriptional activators, are paradoxically translated more efficiently during the phosphorylated state of eIF2α (Figure 3).42–44 ATF4's transcriptional targets include genes involved in amino acid metabolism, oxidoreductases required for disulfide bond formation in the ER, several ubiquitin ligases, GADD34 phosphatase, and CHOP transcription factor.45–47 ATF4-null mouse embryonic fibroblasts are sensitive to oxidative stress and require supplemental reducing compounds for survival and growth in cell culture.46 Interestingly, ATF4 overexpression in mouse embryonic fibroblasts and neurons also evokes oxidative stress and increases cell death.45,47 These findings point to ATF4 as an important determinant in regulating cell fate during ER stress, with too little and too much ATF4 both producing deleterious effects.
Figure 3.
Consequences of acute and chronic PKR-like endoplasmic reticulum (ER) kinase (PERK) activation. PERK has a kinase domain (K), and phosphorylates eukaryotic translation initiation factor 2 subunit alpha (eIF2α). In the acute phase, PERK-eIF2αP attenuates overloading the proteins into ER. On chronic activation of PERK signaling, expression of activating transcription factor-4 (ATF4) is transnationally up-regulated, which regulates cell fate. GADD34 dephosphorylates eIF2αP to eIF2α, and protein translation is reinitiated. Expression of ATF4 causes oxidative stress. Proapoptotic transcription factor CHOP is transcriptionally induced by ATF4, and its translation is also enhanced by ATF4. Death receptor 5 (DR5; official name TNFRSF10B) is a CHOP target gene, and abundant DR5 protein forms oligomer at the Golgi apparatus, which activates caspase (Casp)8 without requirement of any ligand. Inhibitors of apoptosis proteins (IAPs) are key cell death regulators in metazoans, through their suppression of caspases. Recent studies link PERK-eIF2αP-ATF4 signaling to IAP regulation during ER stress. In response to chronic ER stress, IAP levels decrease specifically through the actions of PERK, but not IRE1 or ATF6 branches of the unfolded protein response. The eIF2αP attenuates de novo IAP synthesis, particularly X-linked IAP, and ATF4's transcriptional activity destabilizes extant XIAP protein.
One mechanism by which ATF4 can promote cell death is via transcriptional up-regulation of the GADD34 phosphatase (Figure 3). GADD34 dephosphorylates eIF2αP to eIF2α.48 Dephosphorylation of eIF2αP removes the translational brake initially generated by PERK activation and leads to more protein synthesis and thereby protein folding demands on the ER. Indeed, Han et al45 found that ATF4 overexpression increased protein synthesis concomitant to increasing cell death. Furthermore, chemical inhibition of GADD34's dephosphorylation of eIF2αP by the salubrinal and guanabenz compounds protects cells from ER stress–induced cell death.49–51
A second mechanism by which ATF4 promotes cell death is via transcription of the proapoptotic Chop gene (official name DDIT3), whose translation is also enhanced by eIF2αP (Figure 3).16,43,44,52,53 By dual transcriptional and translational up-regulation, CHOP is highly enriched when PERK is strongly activated. CHOP's role as a proapoptotic transcription factor has been clearly shown in vitro where CHOP-null cells are resistant to cell death induced by the chemical ER toxins, tunicamycin, and thapsigargin.53 Several of CHOP's transcriptional targets are implicated in apoptosis, including the apoptotic Bim (official name BCL2L11) and Puma (official name BBC3) Bcl-2 family genes,54,55 Trb3 (official name TAS2R13), and Dr5 (official name TNFRSF10B).29,56,57 DR5 can signal cell death by activating caspase-8.29,57 During ER stress–induced cell death, DR5 protein accumulates in Golgi apparatus, where it oligomerizes, leading to activation of cytosolic caspase 8. Interestingly, DR5 mRNA is also down-regulated by IRE1-mediated RIDD, and the balance between the opposing effects of IRE1 and PERK on DR5 levels may tip whether UPR selects cell survival or cell death during ER stress.29
Some types of ER stress–induced damage and cell death are unlikely to be mediated via CHOP induction. Comprehensive RNA-sequencing and microarray studies saw minimal transcriptional induction of previously identified apoptotic genes after forced Chop expression.45 Furthermore, forced CHOP expression itself does not trigger cell death in vitro,45,58 indicating that other proapoptotic hits are necessary for ER stress–induced cell death.
Inhibitors of apoptosis proteins (IAPs) are key cell death regulators in metazoan organisms through their suppression of caspases.59 Recent studies link PERK-eIF2αP-ATF4 signaling to IAP regulation during ER stress. In response to chronic ER stress, IAP levels decrease significantly in many mammalian cell types, specifically through the actions of the PERK, but not IRE1 or ATF6, branches of the UPR.60–63 PERK signaling attenuates de novo X-linked IAP (XIAP) protein synthesis via eIF2αP and also promotes extant XIAP protein degradation by ATF4 transcriptional activity (Figure 3). In contrast, CHOP had no effect on XIAP levels. Loss of XIAP enhances sensitivity to ER stress–induced cell death, and overexpression of XIAP protects cells from ER stress, and interestingly, synergizes with the absence of CHOP to induce even greater resistance to ER stress–induced cell death than Chop−/− alone.58 These findings show that PERK-eIF2αP-ATF4 signaling promotes multiple proapoptotic hits within the cell, including the induction of CHOP and suppression of IAPs (Figure 3). These effects of chronic PERK signaling, therefore, generate a cellular milieu conducive for efficient caspase activation by removal of caspase inhibitors.
The ability of IRE1 and PERK signaling to activate multiple distinct proapoptotic circuits provides attractive mechanisms to link ER stress to disease pathogenesis and progression. Physiological ER stresses vary tremendously with respect to their intensity and their cause (eg, hypoxia versus genetic mutation). Important questions for defining the role of UPR as disease mechanism include the following: Which UPR signaling events are activated by a physiological ER stress? What is the consequence of UPR activation in the cellular and tissue context of a specific disease? In the subsequent sections, we examine how UPR activation and function contribute to the pathogenesis of two diseases associated with ER stress, retinitis pigmentosa and prion infection.
Divergent Mechanisms of ER Stress–Induced Neurodegeneration
ER-Associated Degradation in Retinitis Pigmentosa
Retinitis pigmentosa is a human blinding disease arising from photoreceptor cell death in the eye. Photoreceptors are specialized sensory neurons that detect light and activate retinal circuitry to transmit visual information to the brain. Photoreceptors accomplish this feat using the visual pigment, rhodopsin, a G-protein–coupled transmembrane receptor protein covalently linked to 11-cis-retinal.64 Rhodopsin is essential for photoreceptor function and survival, and rhodopsin knockout mice (Rho−/−) develop early retinal degeneration.65,66 More than 100 rhodopsin mutations have been identified in families with heritable types of retinitis pigmentosa.67 Many of these mutations generate misfolded rhodopsin proteins that are aggregation prone and retained in the ER.24,68–71 Recent studies in mouse models of retinitis pigmentosa have shed light into how the UPR in photoreceptors copes with mutant rhodopsins and influences the disease process.
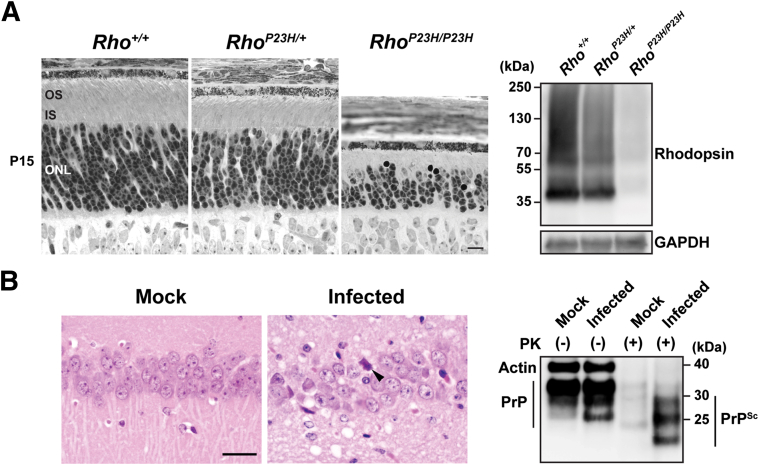
The P23H rhodopsin mutation is the most common cause of heritable retinitis pigmentosa in North America, and photoreceptors carrying the mutation generate misfolded rhodopsin proteins that do not traffic normally to the rod photoreceptor outer segment. A P23H rhodopsin knock-in mouse closely mirrors the spatial distribution and temporal progression of photoreceptor cell death and vision loss found in patients with this mutation.72 Analysis of these mice indicates that these photoreceptors use an unusual, customized UPR tailored to cope with P23H rhodopsin.73 IRE1's induction of XBP1s, and the transcriptional up-regulation of ERAD by XBP1s, was seen in photoreceptors expressing P23H rhodopsin.73 Concomitant with ERAD up-regulation, P23H rhodopsin protein is found to be robustly ubiquitinated and almost entirely degraded in these photoreceptors73 (Figure 4A). In contrast, other IRE1-mediated signaling events, including c-Jun N-terminal kinase activation or RIDD, are not observed in these photoreceptors.73 Furthermore, minimal activation of the PERK signaling pathway is seen, with no changes in ATF4, CHOP, or IAP levels in P23H rhodopsin-expressing photoreceptors.73 These findings reveal that the dominant effect of UPR in photoreceptors of P23H rhodopsin knock-in mice is the induction of ERAD to degrade and clear mutant rhodopsin, which is accomplished through a preferential use of the parts of the UPR regulating ERAD, such as IRE1's induction of XBP1. PERK's proapoptotic functions through modulation of ATF4, CHOP, and IAPs are not used in these photoreceptors. Consistently, Chop−/− fails to delay retinal degeneration in P23H rhodopsin knock-in photoreceptors, and in other genetically modified mouse lines expressing other rhodopsin mutations seen in retinitis pigmentosa.73–75
Figure 4.
A: Rhodopsin (Rho) is robustly degraded during retinal degeneration. Light micrographs of wild-type and P23H knock-in mouse retinas at postnatal (P) day 15. At P15, both rod outer segments (OSs) and rod inner segments (ISs) are shorter in RhoP23H/+ mice compared with those of the Rho+/+ mice, and significantly shortened in RhoP23H/P23H mice. The outer nuclear layer (ONL) is also significantly thinner in RhoP23H/P23H mice. Rhodopsin protein levels are significantly diminished in RhoP23H/P23H mice. Retinal protein lysates were collected from Rho+/+, RhoP23H/+, and RhoP23H/P23H mice at P15. Rhodopsin is detected by immunoblotting. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a loading control. B: High prion protein (PrP) levels are maintained during prion infection. Hematoxylin and eosin–stained hippocampal sections from mock- or prion-inoculated transgenic mice expressing human PrP reveal neuronal necrosis (arrowhead) and spongiform degeneration only in prion-infected mice. Total PrP levels (PrPC + PrPSc) in brain are similar in mock- and prion-infected mice by immunoblotting (20 μg protein per well). Samples treated with 100 μg/mL proteinase K (PK) reveal PK-resistant PrPSc only in prion-infected brain (100 μg protein per well). Actin served as a loading control (the actin bands are above the PrP in the undigested lanes). This blot was developed using monoclonal antibodies 3F4 against PrP (Millipore, Billerica, MA) and GT5412 against actin (Genetex, Irvine, CA). PrP contains two potential n-glycosylation sites and, thus, migrates as three bands corresponding to diglycosylated, monoglycosylated, or unglycosylated PrP. Scale bars: 10 μm (A); 50 μm (B).
What drives photoreceptor cell death if PERK's proapoptotic signals are not activated? Rhodopsin is essential for photoreceptor function, structure, development, and survival, and Rho−/− mice develop early retinal degeneration.65,66 In P23H rhodopsin knock-in animals, rhodopsin protein degradation occurs as soon as photoreceptors are born, and loss of rhodopsin precedes any photoreceptor cell death (Figure 4A).73 Disruption of rhodopsin protein homeostasis by ERAD is likely to be a key trigger for photoreceptor cell death.
PERK Signaling in Prion Diseases
Prion diseases are fatal neurodegenerative disorders arising from conversion of the normal cellular prion protein, PrPC, into a misfolded and self-templating conformer, PrPSc. PrPC is highly conserved among mammals and ubiquitously expressed, yet the functions attributed to PrPC are diverse and include maintenance of myelin, nomal synaptic function, and neuroprotection.76–79 PrPC is a glycosylphosphatidylinositol-anchored glycoprotein that undergoes folding and post-translational modifications within the ER and secretory pathway. In cell culture, PrPSc replication triggers ER stress, resulting in the aberrant accumulation of PrP in the cytosol and further enhancing the formation of PrPSc.80–82 ER stress and UPR activation have also been observed in prion infection in vivo83–85 as well as transgenic mice expressing mutant PrPC.86
Genetic and chemical modulation of different UPR pathways in prion-infected models has revealed intriguing and surprising differences from mutant rhodopsin models in the role of UPR signaling pathways in cellular degeneration. PrPC-to-PrPSc conversion and neurodegeneration are unchanged in mice deficient in neuronal Xbp1−/− compared with controls.85 In contrast, genetic or chemical inhibition of PERK pathway signaling using GADD34 overexpression or the PERK inhibitor glycogen synthetase kinase 2606414 ameliorates neurodegeneration in prion-infected mice, whereas activating the PERK pathway using salubrinal worsens prion-associated neurotoxicity.83,84 These studies reveal a direct role for the PERK pathway in prion disease pathogenesis, and suggest that IRE1 signaling, at least through XBP1s generation, is dispensable in this process.
How is the PERK signaling critical in the pathogenesis of prion disease, yet unnecessary for mutant rhodopsin-induced cell death? One fundamental difference between these diseases is that prion conversion likely occurs on the cell membrane or in an endolysosomal compartment; thus, PrPSc escapes the misfolded protein clearance and degradation mechanisms triggered during UPR activation86 (Figure 4B). Indeed, levels of the PrPSc isoform increase substantially by the terminal stage of disease (Figure 4B). The accumulation of PrP in the brain likely causes chronic ER stress, leading to strong PERK signaling and its subsequent proapoptotic signaling cascade (Figure 2). In contrast, misfolded rhodopsin is degraded so quickly and efficiently (Figure 4A) that the strength and duration of ER stress do not increase to a threshold necessary for strong PERK activation. Thus, prion infection may represent a class of ER stress–associated diseases in which PERK's proapoptotic signaling output prevails when UPR protein quality control mechanisms fail to remove the misfolded proteins. Misfolded rhodopsins may represent another class of ER stress–associated diseases in which UPR protein quality control mechanisms remove the misfolded proteins but, in doing so, disrupt vital cellular structures or processes required for cell viability.
Tailoring the UPR to Fit a Specific Disease
The UPR signaling pathways are found in all cell types and activate broad transcriptional, translational, and post-translational programs to help cells cope with ER stress. The PERK and IRE1 UPR pathways can promote cell death through multiple downstream effectors. Comparison of two ER stress–associated diseases (retinitis pigmentosa and prion disease) reveals that different UPR signaling events are activated during pathogenesis of these diseases. Customization and tailoring of the UPR to fit a physiological ER stress and specific cell types are likely to occur in other ER stress–associated disease processes.
Acknowledgment
We thank Dr. Luke Wiseman (The Scripps Research Institute, La Jolla, CA) for his helpful comments.
Footnotes
Supported by NIH grants R01EY020846, R01NS069566, R01NS076896, and R01NS088485 and VA grant BX002284.
Disclosures: None declared.
The American Society for Investigative Pathology Cotran Early Career Investigator Award recognizes early career investigators with demonstrated excellence as an investigator with recently established or emerging independence and with a research focus leading to an improved understanding of the conceptual basis of disease. Jonathan H. Lin, recipient of the ASIP 2013 Cotran Early Career Investigator Award, delivered a lecture entitled Endoplasmic Reticulum Stress in Disease Pathogenesis on April 22, 2013, at the annual meeting of the American Society for Investigative Pathology in Boston, MA.
References
- 1.Hetz C., Glimcher L.H. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox J.S., Shamu C.E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 3.Shamu C.E., Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 4.Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y., Liang F.X., Wang X. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol Cell. 2014;55:758–770. doi: 10.1016/j.molcel.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jurkin J., Henkel T., Nielsen A.F., Minnich M., Popow J., Kaufmann T., Heindl K., Hoffmann T., Busslinger M., Martinez J. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J. 2014;33:2922–2936. doi: 10.15252/embj.201490332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosmaczewski S.G., Edwards T.J., Han S.M., Eckwahl M.J., Meyer B.I., Peach S., Hesselberth J.R., Wolin S.L., Hammarlund M. The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep. 2014;15:1278–1285. doi: 10.15252/embr.201439531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee A.H., Iwakoshi N.N., Glimcher L.H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shoulders M.D., Ryno L.M., Genereux J.C., Moresco J.J., Tu P.G., Wu C., Yates J.R., 3rd, Su A.I., Kelly J.W., Wiseman R.L. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 2013;3:1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto K., Yoshida H., Kokame K., Kaufman R.J., Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem. 2004;136:343–350. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 13.Shaffer A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A., Hurt E.M., Petroulakis E., Sonenberg N., Yewdell J.W., Calame K., Glimcher L.H., Staudt L.M. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z.V., Deng Y., Gao N., Pedrozo Z., Li D.L., Morales C.R., Criollo A., Luo X., Tan W., Jiang N., Lehrman M.A., Rothermel B.A., Lee A.H., Lavandero S., Mammen P.P., Ferdous A., Gillette T.G., Scherer P.E., Hill J.A. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell. 2014;156:1179–1192. doi: 10.1016/j.cell.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding H.P., Zhang Y., Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 16.Harding H.P., Calfon M., Urano F., Novoa I., Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- 17.Ron D., Harding H.P. Protein-folding homeostasis in the endoplasmic reticulum and nutritional regulation. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a013177. pii: a013177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye J., Rawson R.B., Komuro R., Chen X., Dave U.P., Prywes R., Brown M.S., Goldstein J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 20.Wu J., Rutkowski D.T., Dubois M., Swathirajan J., Saunders T., Wang J., Song B., Yau G.D., Kaufman R.J. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida H., Matsui T., Hosokawa N., Kaufman R.J., Nagata K., Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 22.Aragon T., van Anken E., Pincus D., Serafimova I.M., Korennykh A.V., Rubio C.A., Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Korennykh A.V., Behrman S.L., Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci U S A. 2010;107:16113–16118. doi: 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., Shokat K.M., Lavail M.M., Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam A.B., Koong A.C., Niwa M. Ire1 has distinct catalytic mechanisms for XBP1/HAC1 splicing and RIDD. Cell Rep. 2014;9:850–858. doi: 10.1016/j.celrep.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollien J., Weissman J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 27.Hollien J., Lin J.H., Li H., Stevens N., Walter P., Weissman J.S. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X.T., McCullough K.D., Wang X.J., Carpenter G., Holbrook N.J. Oxidative stress-induced phospholipase C-gamma 1 activation enhances cell survival. J Biol Chem. 2001;276:28364–28371. doi: 10.1074/jbc.M102693200. [DOI] [PubMed] [Google Scholar]
- 29.Lu M., Lawrence D.A., Marsters S., Acosta-Alvear D., Kimmig P., Mendez A.S., Paton A.W., Paton J.C., Walter P., Ashkenazi A. Cell death: opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hur K.Y., So J.S., Ruda V., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Iwawaki T., Glimcher L.H., Lee A.H. IRE1alpha activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med. 2012;209:307–318. doi: 10.1084/jem.20111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho J.A., Lee A.H., Platzer B., Cross B.C., Gardner B.M., De Luca H., Luong P., Harding H.P., Glimcher L.H., Walter P., Fiebiger E., Ron D., Kagan J.C., Lencer W.I. The unfolded protein response element IRE1alpha senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling. Cell Host Microbe. 2013;13:558–569. doi: 10.1016/j.chom.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.So J.S., Hur K.Y., Tarrio M., Ruda V., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Lichtman A.H., Iwawaki T., Glimcher L.H., Lee A.H. Silencing of lipid metabolism genes through IRE1alpha-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 2012;16:487–499. doi: 10.1016/j.cmet.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tirasophon W., Lee K., Callaghan B., Welihinda A., Kaufman R.J. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Upton J.P., Wang L., Han D., Wang E.S., Huskey N.E., Lim L., Truitt M., McManus M.T., Ruggero D., Goga A., Papa F.R., Oakes S.A. IRE1α cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimmig P., Diaz M., Zheng J., Williams C.C., Lang A., Aragon T., Li H., Walter P. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. Elife. 2012;1:e00048. doi: 10.7554/eLife.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H.P., Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 37.Nishitoh H., Matsuzawa A., Tobiume K., Saegusa K., Takeda K., Inoue K., Hori S., Kakizuka A., Ichijo H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu Y., Mao T., Zhang Y., Shao M., You J., Ding Q., Chen Y., Wu D., Xie D., Lin X., Gao X., Kaufman R.J., Li W., Liu Y. A crucial role for RACK1 in the regulation of glucose-stimulated IRE1alpha activation in pancreatic beta cells. Sci Signal. 2010;3:ra7. doi: 10.1126/scisignal.2000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hetz C., Bernasconi P., Fisher J., Lee A.H., Bassik M.C., Antonsson B., Brandt G.S., Iwakoshi N.N., Schinzel A., Glimcher L.H., Korsmeyer S.J. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–576. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- 40.Lisbona F., Rojas-Rivera D., Thielen P., Zamorano S., Todd D., Martinon F., Glavic A., Kress C., Lin J.H., Walter P., Reed J.C., Glimcher L.H., Hetz C. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol Cell. 2009;33:679–691. doi: 10.1016/j.molcel.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y., Beatty A., Han X., Ji Y., Ma X., Adelstein R.S., Yates J.R., 3rd, Kemphues K., Qi L. Nonmuscle myosin IIB links cytoskeleton to IRE1alpha signaling during ER stress. Dev Cell. 2012;23:1141–1152. doi: 10.1016/j.devcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou D., Palam L.R., Jiang L., Narasimhan J., Staschke K.A., Wek R.C. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 43.Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 44.Palam L.R., Baird T.D., Wek R.C. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem. 2011;286:10939–10949. doi: 10.1074/jbc.M110.216093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han J., Back S.H., Hur J., Lin Y.H., Gildersleeve R., Shan J., Yuan C.L., Krokowski D., Wang S., Hatzoglou M., Kilberg M.S., Sartor M.A., Kaufman R.J. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D.F., Bell J.C., Hettmann T., Leiden J.M., Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 47.Lange P.S., Chavez J.C., Pinto J.T., Coppola G., Sun C.W., Townes T.M., Geschwind D.H., Ratan R.R. ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J Exp Med. 2008;205:1227–1242. doi: 10.1084/jem.20071460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novoa I., Zeng H., Harding H.P., Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papandreou I., Denko N.C., Olson M., Van Melckebeke H., Lust S., Tam A., Solow-Cordero D.E., Bouley D.M., Offner F., Niwa M., Koong A.C. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsaytler P., Harding H.P., Ron D., Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 2011;332:91–94. doi: 10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- 51.Boyce M., Bryant K.F., Jousse C., Long K., Harding H.P., Scheuner D., Kaufman R.J., Ma D., Coen D.M., Ron D., Yuan J. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 52.Lee H.C., Chen Y.J., Liu Y.W., Lin K.Y., Chen S.W., Lin C.Y., Lu Y.C., Hsu P.C., Lee S.C., Tsai H.J. Transgenic zebrafish model to study translational control mediated by upstream open reading frame of human chop gene. Nucleic Acids Res. 2011;39:e139. doi: 10.1093/nar/gkr645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H., Stevens J.L., Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puthalakath H., O'Reilly L.A., Gunn P., Lee L., Kelly P.N., Huntington N.D., Hughes P.D., Michalak E.M., McKimm-Breschkin J., Motoyama N., Gotoh T., Akira S., Bouillet P., Strasser A. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 55.Cazanave S.C., Elmi N.A., Akazawa Y., Bronk S.F., Mott J.L., Gores G.J. CHOP and AP-1 cooperatively mediate PUMA expression during lipoapoptosis. Am J Physiol Gastrointest Liver Physiol. 2010;299:G236–G243. doi: 10.1152/ajpgi.00091.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaguchi H., Wang H.G. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J Biol Chem. 2004;279:45495–45502. doi: 10.1074/jbc.M406933200. [DOI] [PubMed] [Google Scholar]
- 58.Hiramatsu N., Messah C., Han J., LaVail M.M., Kaufman R.J., Lin J.H. Translational and post-translational regulation of XIAP by eIF2α and ATF4 promotes ER stress-induced cell death during the unfolded protein response. Mol Biol Cell. 2014;25:1411–1420. doi: 10.1091/mbc.E13-11-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salvesen G.S., Duckett C.S. IAP proteins: blocking the road to death's door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 60.Chiang W.C., Hiramatsu N., Messah C., Kroeger H., Lin J.H. Selective activation of ATF6 and PERK endoplasmic reticulum stress signaling pathways prevent mutant rhodopsin accumulation. Invest Ophthalmol Vis Sci. 2012;53:7159–7166. doi: 10.1167/iovs.12-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hiramatsu N., Joseph V.T., Lin J.H. Monitoring and manipulating mammalian unfolded protein response. Methods Enzymol. 2011;491:183–198. doi: 10.1016/B978-0-12-385928-0.00011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin A.M., Fang S.F., Chao P.L., Yang C.H. Melatonin attenuates arsenite-induced apoptosis in rat brain: involvement of mitochondrial and endoplasmic reticulum pathways and aggregation of alpha-synuclein. J Pineal Res. 2007;43:163–171. doi: 10.1111/j.1600-079X.2007.00456.x. [DOI] [PubMed] [Google Scholar]
- 63.Lin J.H., Li H., Zhang Y., Ron D., Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lem J., Krasnoperova N.V., Calvert P.D., Kosaras B., Cameron D.A., Nicolo M., Makino C.L., Sidman R.L. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc Natl Acad Sci U S A. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Humphries M.M., Rancourt D., Farrar G.J., Kenna P., Hazel M., Bush R.A., Sieving P.A., Sheils D.M., McNally N., Creighton P., Erven A., Boros A., Gulya K., Capecchi M.R., Humphries P. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat Genet. 1997;15:216–219. doi: 10.1038/ng0297-216. [DOI] [PubMed] [Google Scholar]
- 67.Rattner A., Sun H., Nathans J. Molecular genetics of human retinal disease. Annu Rev Genet. 1999;33:89–131. doi: 10.1146/annurev.genet.33.1.89. [DOI] [PubMed] [Google Scholar]
- 68.Illing M.E., Rajan R.S., Bence N.F., Kopito R.R. A rhodopsin mutant linked to autosomal dominant retinitis pigmentosa is prone to aggregate and interacts with the ubiquitin proteasome system. J Biol Chem. 2002;277:34150–34160. doi: 10.1074/jbc.M204955200. [DOI] [PubMed] [Google Scholar]
- 69.Sung C.H., Schneider B.G., Agarwal N., Papermaster D.S., Nathans J. Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci U S A. 1991;88:8840–8844. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaushal S., Khorana H.G. Structure and function in rhodopsin, 7: point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994;33:6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- 71.Chiang W.C., Messah C., Lin J.H. IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol Biol Cell. 2012;23:758–770. doi: 10.1091/mbc.E11-08-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakami S., Maeda T., Bereta G., Okano K., Golczak M., Sumaroka A., Roman A.J., Cideciyan A.V., Jacobson S.G., Palczewski K. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J Biol Chem. 2011;286:10551–10567. doi: 10.1074/jbc.M110.209759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chiang W.C., Kroeger H., Sakami S., Messah C., Yasumura D., Matthes M.T., Coppinger J.A., Palczewski K., LaVail M.M., Lin J.H. Robust endoplasmic reticulum-associated degradation of rhodopsin precedes retinal degeneration. Mol Neurobiol. 2014 doi: 10.1007/s12035-014-8881-8. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nashine S., Bhootada Y., Lewin A.S., Gorbatyuk M. Ablation of C/EBP homologous protein does not protect T17M RHO mice from retinal degeneration. PLoS One. 2013;8:e63205. doi: 10.1371/journal.pone.0063205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adekeye A., Haeri M., Solessio E., Knox B.E. Ablation of the proapoptotic genes chop or ask1 does not prevent or delay loss of visual function in a P23H transgenic mouse model of retinitis pigmentosa. PLoS One. 2014;9:e83871. doi: 10.1371/journal.pone.0083871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bremer J., Baumann F., Tiberi C., Wessig C., Fischer H., Schwarz P., Steele A.D., Toyka K.V., Nave K.A., Weis J., Aguzzi A. Axonal prion protein is required for peripheral myelin maintenance. Nat Neurosci. 2010;13:310–318. doi: 10.1038/nn.2483. [DOI] [PubMed] [Google Scholar]
- 77.Collinge J., Whittington M.A., Sidle K.C., Smith C.J., Palmer M.S., Clarke A.R., Jefferys J.G. Prion protein is necessary for normal synaptic function. Nature. 1994;370:295–297. doi: 10.1038/370295a0. [DOI] [PubMed] [Google Scholar]
- 78.Steele A.D., Lindquist S., Aguzzi A. The prion protein knockout mouse: a phenotype under challenge. Prion. 2007;1:83–93. doi: 10.4161/pri.1.2.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Linden R., Martins V.R., Prado M.A., Cammarota M., Izquierdo I., Brentani R.R. Physiology of the prion protein. Physiol Rev. 2008;88:673–728. doi: 10.1152/physrev.00007.2007. [DOI] [PubMed] [Google Scholar]
- 80.Hetz C., Castilla J., Soto C. Perturbation of endoplasmic reticulum homeostasis facilitates prion replication. J Biol Chem. 2007;282:12725–12733. doi: 10.1074/jbc.M611909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orsi A., Fioriti L., Chiesa R., Sitia R. Conditions of endoplasmic reticulum stress favor the accumulation of cytosolic prion protein. J Biol Chem. 2006;281:30431–30438. doi: 10.1074/jbc.M605320200. [DOI] [PubMed] [Google Scholar]
- 82.Nunziante M., Ackermann K., Dietrich K., Wolf H., Gädtke L., Gilch S., Vorberg I., Groschup M., Schätzl H.M. Proteasomal dysfunction and endoplasmic reticulum stress enhance trafficking of prion protein aggregates through the secretory pathway and increase accumulation of pathologic prion protein. J Biol Chem. 2011;286:33942–33953. doi: 10.1074/jbc.M111.272617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moreno J.A. Sustained translational repression by eIF2[alpha]-P mediates prion neurodegeneration. Nature. 2012;485:507–511. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moreno J.A. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci Transl Med. 2013;5:206ra138. doi: 10.1126/scitranslmed.3006767. [DOI] [PubMed] [Google Scholar]
- 85.Hetz C. Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc Natl Acad Sci U S A. 2008;105:757–762. doi: 10.1073/pnas.0711094105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dametto P., Lakkaraju A.K., Bridel C., Villiger L., O'Connor T., Herrmann U.S., Pelczar P., Rülicke T., McHugh D., Adili A., Aguzzi A. Neurodegeneration and unfolded-protein response in mice expressing a membrane-tethered flexible tail of PrP. PLoS One. 2015;10:e0117412. doi: 10.1371/journal.pone.0117412. [DOI] [PMC free article] [PubMed] [Google Scholar]