Abstract
Microbial decomposition of organic matter is an essential process in the global carbon cycle. The soil bacteria Pseudopedobacter saltans and Flavobacterium johnsoniae are both able to degrade complex organic molecules, but it is not fully known how their membrane structures are adapted to their environmental niche. The membrane lipids of these species were extracted and analyzed using high performance liquid chromatography-electrospray ionization/ion trap/mass spectrometry (HPLC-ESI/IT/MS) and high resolution accurate mass/mass spectrometry (HRAM/MS). Abundant unknown intact polar lipids (IPLs) from P. saltans were isolated and further characterized using amino acid analysis and two dimensional nuclear magnetic resonance (NMR) spectroscopy. Ornithine IPLs (OLs) with variable (hydroxy) fatty acid composition were observed in both bacterial species. Lysine-containing IPLs (LLs) were also detected in both species and were characterized here for the first time using HPLC-MS. Novel LLs containing hydroxy fatty acids and novel hydroxylysine lipids with variable (hydroxy) fatty acid composition were identified in P. saltans. The confirmation of OL and LL formation in F. johnsoniae and P. saltans and the presence of OlsF putative homologs in P. saltans suggest the OlsF gene coding protein is possibly involved in OL and LL biosynthesis in both species, however, potential pathways of OL and LL hydroxylation in P. saltans are still undetermined. Triplicate cultures of P. saltans were grown at three temperature/pH combinations: 30°C/pH 7, 15°C/pH 7, and 15°C/pH 9. The fractional abundance of total amino acid containing IPLs containing hydroxylated fatty acids was significantly higher at higher temperature, and the fractional abundance of lysine-containing IPLs was significantly higher at lower temperature and higher pH. These results suggest that these amino acid-containing IPLs, including the novel hydroxylysine lipids, could be involved in temperature and pH stress response of soil bacteria.
Keywords: lysine lipid, hydroxylysine lipid, Pseudopedobacter saltans, Flavobacterium johnsoniae, stress response, soil bacteria
Introduction
Intact polar lipids (IPLs) are useful biomarker molecules because their structures can be specific to microbial taxa or environmental conditions (Sturt et al., 2004; Schubotz et al., 2009). For example, marine phytoplankton from multiple regions of the ocean produce non-phosphorus containing lipids in response to phosphorus scarcity (Van Mooy et al., 2009), which demonstrates the importance of microbial membrane modification. IPLs containing the amino acid ornithine as the polar head group (ornithine lipids, OLs) are common phosphorus-free membrane IPLs among bacteria (Figure 1A). Approximately 50% of bacterial species whose genomes have been sequenced are predicted to have the capacity to form OLs, but they have not been predicted in eukaryotes or archaea (Lopez-Lara et al., 2003; Geiger et al., 2010; Vences-Guzmán et al., 2012, 2014). In various bacteria, OL production increases under phosphorus limitation (Weissenmayer et al., 2002; Gao et al., 2004), and in other microbes the fatty acids of OLs are hydroxylated under thermal or acid stress (Taylor et al., 1998; Rojas-Jimenez et al., 2005; Vences-Guzman et al., 2011). In eight Desulfovibrio strains isolated from intertidal sediments of the North Sea, relative OL content was found to increase at higher growth temperature (Seidel et al., 2013). It has also been suggested that OLs are important for Gram-negative bacterial outer membrane stability due to their zwitterionic nature (Freer et al., 1996) and essential to maintain a constant level of extracytoplasmic cytochromes in Rhodobacter capsulatus (Aygun-Sunar et al., 2006).
Figure 1.
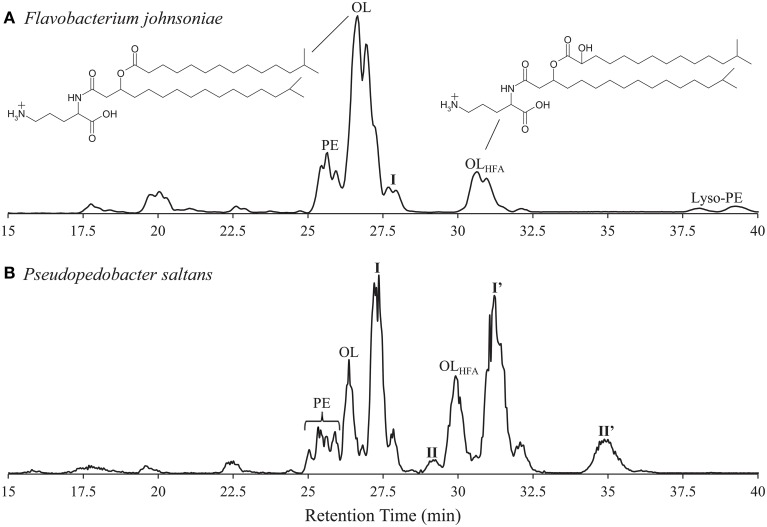
High pressure liquid chromatography-electrospray/ion trap/mass spectrometry (HPLC-ESI/IT/MS) base peak chromatograms of (A) Flavobacterium johnsoniae (low abundance glycine lipids at retention time 6.5 min not shown; ornithine lipid structure included with variable fatty acid hydroxy group), and (B) Pseudopedobacter saltans lipid extracts. PE, phosphatidylethanolamine; OL, ornithine lipid; OLHFA, Ornithine lipid with hydroxylated fatty acid; I, I', II, II”, unknown intact polar lipids. Chromatographic separation was performed on a Lichrosphere diol column (250 mm by 2.1 mm; 5-μm particles; Grace Alltech Associates Inc.). Elution was achieved with hexane–2-propanol–formic acid–14.8 M aqueous NH3 (79:20:0.12:0.04, v/v/v/v) (A) and 2-propanol–water–formic acid–14.8 M aqueous NH3 (88:10:0.12:0.04, v/v/v/v) (B) starting at 10% B, followed by a linear increase to 30% B in 10 min, followed by a 20-min hold and a further increase to 65% B at 45 min. The flow rate was 0.2 ml min−1, and the total run time was 60 min, followed by a 20 min re-equilibration period.
The combination of bacterial specificity, OL production and modification in response to environmental conditions, along with a suggested role in membrane stability makes this class of lipids useful potential biomarker molecules for microbial populations. Besides OLs, other amino acid-containing IPLs have also been identified including glycine lipids (Kawazoe et al., 1991; Batrakov et al., 1999), lysine lipids (Tahara et al., 1976a), glutamine lipid (Zhang et al., 2009), and an ornithine-taurine linked lipid in Gluconobacter cerinus (Tahara et al., 1976b) using various combinations of thin layer chromatography, infrared spectrometry, gas chromatography-mass spectrometry, electrospray ionization-mass spectrometry, and 1H nuclear magnetic resonance (NMR) spectroscopy. None of the lipids from these studies were identified using high performance liquid chromatography/mass spectrometry (HPLC/MS) methods, which are far less laborious than the above mentioned methods, and extremely effective for identifying IPLs in complex bacterial cultures and environmental samples (Sturt et al., 2004; Schubotz et al., 2009). The LC/MS chromatographic and fragmentation behavior of OLs is well known (Hilker et al., 1978, 1979; Tomer et al., 1983; Cerny et al., 1986 Linscheid et al., 1997; Geiger et al., 2010), but not described for various other amino acid containing IPLs. The characteristic multi-stage MS fragmentation of OLs includes the sequential loss of an H2O molecule from the head group, a fatty acid moiety, and the βOH-fatty acid moiety, resulting in a diagnostic m/z 115 cyclic fragment (Hilker et al., 1978; Cerny et al., 1986; Geiger et al., 2010). This fragmentation pattern can be used as an example for identifying other amino acid-containing IPLs via LC/MS multistage fragmentation in bacteria which are important in organic matter recycling.
Microbial organic matter decomposition is an important part of the global carbon cycle with contributions to climate change through carbon cycle feedbacks (Bardgett et al., 2008; Singh et al., 2010). The bacterial genus Pedobacter is of great interest to understand organic matter decomposition in soils because many species within the genus are able to degrade heparin, the biomolecule with the greatest known negative charge density (Steyn et al., 1998; Liolios et al., 2011). Pseudopedobacter saltans (Cao et al., 2014), originally classified in the Pedobacter genus (Steyn et al., 1998), has a distinct taxonomic position and its genome has the highest number of heparinase coding genes (Liolios et al., 2011). As P. saltans degrades highly negatively charged molecules like heparin and chondroitin, this species may be involved in the degradation of complex organic molecules in the soil such as other mucopolysaccharides or humic acids. Other microbes with the ability to degrade recalcitrant biomacromolecules are important in organic matter decomposition as well. For example, Flavobacterium johnsoniae, formerly Cytophaga johnsonae, another bacterium found in soil, rapidly digests chitin and many other macromolecules (Stanier, 1947; Larkin, 1989) and has been studied extensively for its gliding motility (McBride, 2001; McBride et al., 2003; Nelson and McBride, 2006). Both P. saltans and F. johnsoniae belong to the Cytophaga-Flavobacterium-Bacteroides (CFB) group (Stanier, 1947; Larkin, 1989; Steyn et al., 1998; Liolios et al., 2011).
Despite their important role in soil organic matter decomposition, it is not known how the membrane structural components of P. saltans and F. johnsoniae adapt to their different environmental niches. OLs and glycine lipids have both been identified in F. johnsoniae (Pitta et al., 1989; Kawazoe et al., 1991, 1992; Okuyama and Monde, 1996), and amino lipids have been identified in P. saltans (Cao et al., 2014), but the full complement of IPLs have not been described for both species. Amino acid-containing IPLs may be involved in environmental stress response by these consequential species. Here, we study the IPLs, and specifically the amino acid-containing lipids, of P. saltans and F. johnsoniae by HPLC/MS with further structural characterization by NMR spectroscopy. We report the IPL content of P. saltans, and F. johnsoniae including the structural elucidation of novel amino acid-containing lipids and discuss the potential genes and environmental factors that influence their production in P. saltans.
Materials and methods
Strains and culture conditions
F. johnsoniae (DSM 2064T) and P. saltans (DSM 12145T) were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ) in Braunschweig, Germany. F. johnsoniae was grown in liquid CYE medium (g per liter of distilled water): Casitone, 10.0; yeast extract, 5.0; MgSO4, 0.1; Tris buffer, 1.2; pH 7.2; 25°C. P. saltans was grown in DSMZ Medium 948 (g per liter of distilled water): Lab-Lemco powder (Oxoid), 1.0; yeast extract, 2.0; peptone, 5.0; NaCl, 5.0; pH 7.2; 25°C. Additional P. saltans cultures were grown in triplicate at 30°C/pH 7, 15°C/pH 7, and 15°C/pH 9 in temperature controlled rooms to determine changes in IPL fractional abundance due to different environmental conditions. The temperatures and pH levels were chosen based on temperature growth range reported by Liolios et al. (2011) and initial growth experiments, in which P. saltans cultures did not readily grow at temperatures below 15°C or below pH 7. The pH was adjusted by addition of HCl or NaOH solutions to achieve the desired values. Biomass from 250 ml cultures was collected by centrifugation at the stationary growth phase and freeze dried for lipid extraction and further analysis.
Lipid extraction
Lipids were extracted from freeze dried biomass of each culture by a modified Bligh & Dyer method (Rutters et al., 2002). Biomass was fully submerged and extracted three times in methanol/dichloromethane/phosphate-buffer (MeOH/DCM/P-buffer, 2/1/0.8, v/v/v) extraction solvent for 10 min in an ultrasonic bath (P-buffer: 8.7 g K2HPO4 L−1 bi-distilled water adjusted to pH 7-8 with 1 N HCl). Extracts were centrifuged for 2 min at 1400 g to separate DCM phase from the MeOH/P-buffer phase, and the lower DCM layer was pipetted into a separate vial. The MeOH/P-buffer layer was washed twice more with DCM, centrifuged, and the resulting DCM layers were combined with the original DCM layer. DCM was removed under a stream of nitrogen, the residue was dissolved in injection solvent (hexane:2-propanol:H2O, 718:271:10, v/v/v), and filtered through a 0.45 μm, 4 mm diameter True™ Regenerated Cellulose syringe filter (Grace Davison) prior to injection. Extracts were dried down and stored at −80°C until analysis.
HPLC-MS analysis
Intact polar lipids were analyzed by high performance liquid chromatography-electrospray ionization/ion trap mass spectrometry (HPLC-ESI/IT/MS) and HPLC-high resolution accurate mass/mass spectrometry (HRAM/MS) according to Moore et al. (2013). The HPLC-ESI/IT/MS methods closely followed Sturt et al. (2004) with some modifications (Sinninghe Damsté et al., 2011). An Agilent 1200 series HPLC, with thermostatted auto-injector, was coupled to a Thermo Scientific™ LTQ XL™ linear ion trap mass spectrometer with Ion Max source and ESI probe (Thermo Fisher Scientific, Waltham, MA). Chromatographic separation was performed on a Lichrosphere diol column (250 mm by 2.1 mm; 5-μm particles; Grace Alltech Associates Inc.). Elution was achieved with hexane–2-propanol–formic acid–14.8 M aqueous NH3 (79:20:0.12:0.04, v/v/v/v) (A) and 2-propanol–water–formic acid–14.8 M aqueous NH3 (88:10:0.12:0.04, v/v/v/v) (B) starting at 10% B, followed by a linear increase to 30% B in 10 min, followed by a 20-min hold and a further increase to 65% B at 45 min. The flow rate was 0.2 ml min−1, and the total run time was 60 min, followed by a 20 min re-equilibration period. Lipid extracts were analyzed by scanning the mass range of m/z 400–2000 in positive-ion mode, followed by data-dependent, dual-stage tandem MS (MS2), in which the four most abundant masses in the mass spectrum were fragmented successively (normalized collision energy, 25; isolation width, 5.0; activation Q, 0.175). Each MS2 was followed by data-dependent, triple-stage tandem MS (MS3), where the base peak of the MS2 spectrum was fragmented under identical fragmentation conditions to those described for MS2. The performance of HPLC-ESI/IT/MS was monitored by regular injections of platelet-activating factor (PAF) standard (1-O-hexadecyl-2-acetyl-snglycero-3-phosphocholine).
HPLC-HRAM/MS analysis was accomplished on a Thermo Scientific™ Dionex™ UltiMate™ 3000 series LC with thermostatted auto-injector coupled to a Thermo Scientific™ Q Exactive™ Orbitrap™ mass spectrometer. Higher-energy Collisional Dissociation (HCD) and product ion scan were used for mass fragmentation of sample masses. The chromatographic conditions and column were the same as those described above for HPLC-ESI/IT/MS. The positive-ion ESI settings were as follows: capillary temperature, 275°C; sheath gas (N2) pressure, 35 arbitrary units (AU); auxiliary gas (N2) pressure, 10 AU; spray voltage, 4.0 kV; probe heater temperature, 300°C; S-lens, 50 V. Target lipids were analyzed with a mass range of m/z 400–1000 (resolution, 70,000), followed by data dependent MS2 (resolution, 17,500), in which the five most abundant masses in the mass spectrum were fragmented successively (normalized collision energy, 35; isolation width, 1.0). IPL fractional abundances were calculated based on HPLC-ESI/IT/MS chromatogram base peak area of each lipid group. Student's t-tests were performed using the GraphPad t-test Calculator (GraphPad Software, Inc. La Jolla, CA) in order to identify statistically significant differences in the fractional abundances of IPLs under different growth conditions; p < 0.05 were considered statistically significant.
Fatty acid analysis
Aliquots of Bligh and Dyer extracts of F. johnsoniae and P. saltans were hydrolyzed with 1.5 N HCl in MeOH by refluxing for 3 h. The hydrolysate was adjusted to pH 4 with 2 N KOH-MeOH (1:1, v/v). Water was added to give a final ratio of 1:1 H2O-MeOH and this mixture was extracted three times with DCM. The DCM fractions were collected and dried over sodium sulfate. The extract was methylated with diazomethane (Sinninghe Damsté et al., 2004), followed by silylation in pyridine with N,O bis(trimethylsilyl)trifluoroacetamide (BSTFA) at 60°C for 20 min. The methylated-silylated extracts were dissolved in ethyl acetate for gas chromatography (GC)-MS analysis (Sinninghe Damsté et al., 2011). GC was performed with a Hewlett-Packard gas chromatograph (HP6890) equipped with an on-column injector and a flame ionization detector. GC-MS was performed on a Finnigan Trace Ultra gas chromatograph interfaced with a Finnigan Trace DSQ mass spectrometer operated at 70 eV with a mass range of m/z 40–800 and a cycle time of 1.7 s (resolution, 1000).
Target lipid isolation
Isolation of target lipids from P. saltans and known OL from F. johnsoniae lipid extracts was accomplished using an Agilent Technologies (Santa Clara, CA) 1100 series LC equipped with an auto-injector, and a fraction collector (Foxy Jr., Isco, Inc., Lincoln, NE). A first isolation was achieved on a semi-preparative LiChrospher diol column (10 × 250 mm, 5 μm; Grace Alltech Associates Inc.) according to Boumann et al. (2009) with the same gradient program as described above for HPLC-ESI/IT/MS, but at a flow rate of 3 ml min−1. Typical injection volume was 200 μL containing up to 2 mg material per injection. Column effluent was collected in 1 min fractions, which were screened for the presence of target lipids by flow injection analysis cf. Smittenberg et al. (2002) using the ESI/IT/MS (5 μl injection of each fraction, ESI source settings same as described above for HPLC-ESI/IT/MS with a scan range of m/z 400–2000). Fractions containing target lipids were pooled and further purified on a second LiChrospher diol column (4.6 × 250 mm, 5 μm; Grace Alltech Associates Inc.). Typical injection volumes were 65 μl containing up to 0.65 mg of material. Lipids were eluted using the identical gradient program and conditions as described above for HPLC-ESI/IT/MS at a flow rate of 1 ml min−1, however mobile phases A and B did not contain NH3 or formic acid. Column effluent was collected in 15 s fractions and screened as described above. Fractions containing target lipids were again combined and elution solvent was removed under a stream of nitrogen. Purity was assessed by HPLC-ESI/IT/MS as described above for IPL analysis. Isolated lipids were stored at −80°C prior to further analysis.
Nuclear magnetic resonance
The purified lipids were dissolved in 99.9% CDCl3 at concentrations of 1.88 and 1.83 mmol ml −1, and NMR experiments were performed at 298 K on a Bruker 600-MHz Avance spectrometer equipped with 5 mm TCI cryoprobe and running under TOPSPIN 2.1. One-dimensional (1D) 1H and 1H-decoupled 1D 13C and DEPT-135 experiments were recorded with spectral widths/offsets of 24 ppm/6 ppm, 200 ppm/100 ppm, and 200 ppm/100 ppm and with 16,384 (16k), 4096 (4k), and 4096 (4k) complex points, respectively. The 12- by 12-ppm 2D correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), and nuclear Overhauser effect spectroscopy (NOESY) experiments were performed with 512 by 200 complex points (2k by 256 for the COSY) and an offset frequency of 5 ppm. Mixing times were 60 ms and 250 ms for TOCSY and NOESY, respectively. The (1H, 13C)-heteronuclear single-quantum correlation spectroscopy (HSQC) and (1H, 13C)-heteronuclear multiple-bond correlation spectroscopy (HMBC) experiments were recorded with spectral widths/offsets of 12/5 ppm for protons, 200/100 ppm for 13C HSQC (512 × 60 complex points) and 150/75 ppm for the HMBC (1k × 80 complex points). All spectra were calibrated with respect to internal residually protonated CHCl3 at 7.24 ppm (1H) and 77.0 ppm (13C).
Amino acid analysis
Approximately 8.4 nmol of each of the purified P. saltans lipids, and 40 nmol of a known ornithine containing lipid purified from F. johnsoniae used as a reference compound, were hydrolyzed in 0.5 ml of 6 M HCl at 110°C for 18 h to liberate the amino acid head groups from the fatty acids for amino acid analysis. Standardized methods for the analysis of the amino acid lipid hydrolysates follow Spackman et al. (1958), Commission Regulation (EC) no. 152 (2009)1, and Eur. Ph. Chapter 2.2.56 (2005)2 where they are described in detail. Added to each hydrolysate vial was 240 μl of Li-loading buffer (Li-citrate pH 2.2) and 80 μl of internal standard (IS) solution (0.5 mM norleucine in 5% 5-sulfosalicylic acid). After mixing and re-dissolving the mix was filtered using a PALL 0.2 μm, 13 mm GHP Acrodisk. Amino acid analysis (IS-method, one step calibration vs. standard solution A9906, Sigma-Aldrich) was performed using a Biochrom 30 amino acid analyzer by Ansynth Service B.V. (Roosendaal, Netherlands). Separation was performed on a weak acidic cation exchange resin stationary phase (200 × 4.6 mm column) with a mobile phase consisting of number of weak acidic Li-citrate buffers. Stepwise pH, temperature and salt concentration gradients were applied. Spectrophotometer detection occurred after post column derivatization with ninhydrin (135°C) at 570 or 440 nm.
Results and discussion
IPLs of F. johnsoniae
The IPLs of F. johnsoniae identified using HPLC-ESI/IT/MS analysis included abundant OLs, moderately abundant hydroxylated fatty acid OLs (OLHFA) and phosphatidylethanolamines (PEs), and low abundance lyso-PE (PE with one fatty acid), and glycine lipids, which make up 47, 11, 12, 2, and 1%, respectively of the total HPLC-ESI/IT/MS chromatogram base peak area (Figure 1A). OLHFA were hydroxylated at the α-position of the ester linked fatty acid (Figure 1A). In addition, two low abundance IPLs with OL-like fragmentation were observed at retention times 27.7 and 28.0 min with apparent protonated molecules ([M+H]+) at m/z values 639 and 625, respectively, making up 4.6% of HPLC-ESI/IT/MS chromatogram base peak area, which we will refer to as group I. The group I IPLs displayed sequential fragmentation loss of an H2O molecule, a fatty acid moiety, and a βOH-fatty acid, reminiscent of the fragmentation of OLs, but resulting in MS3 fragment ions at m/z 129, 130, and 147, instead of the characteristic m/z 115 OL MS3 fragment.
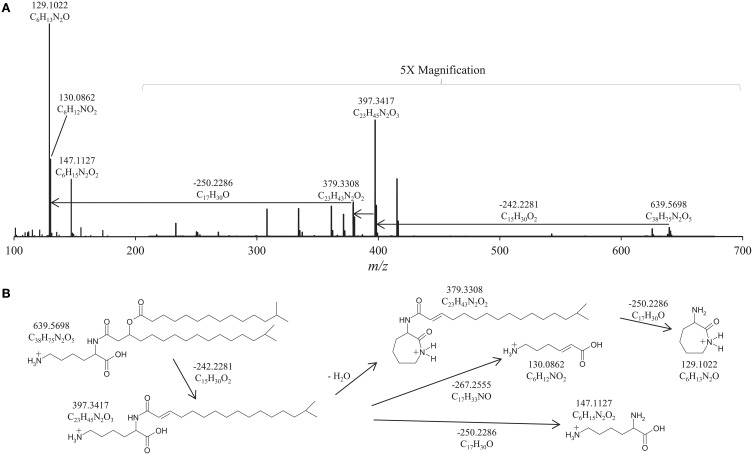
To further elucidate the identity of the group I lipids we analyzed the F. johnsoniae lipid extract by HPLC-HRAM/MS, which revealed the elemental composition of the m/z 129, 130, and 147 fragmentation products of group I (Figure 2, Table 1). During fragmentation the loss of fatty acids and H2O from the head group likely resulted in the formation of a ring structure as described for the fragmentation of OL (Zhang et al., 2009), giving the most abundant fragmentation product of m/z 129.1022 with elemental composition C6H13N2O. The loss of the fatty acid moieties without the loss of H2O results in the m/z 147.1127 fragmentation product, which has the elemental composition (C6H15N2O2) of protonated lysine, and further loss of NH3 from this fragment results in the m/z 130.0862 fragment ion (C6H12NO2). The elemental composition of the m/z 129.1022 fragment also corresponds to protonated lysine minus H2O. Elemental compositions of fatty acid losses corresponded to the expected molecular formulas of regular and βOH-fatty acids. Based on this information, we propose that group I IPLs in F. johnsoniae are lysine-containing lipids (LLs) with different fatty acid compositions (Table 1, lysine lipid, with 15C:0 and βOH-17C:0 fatty acids shown in Figure 2). GC/MS analysis showed that the most abundant 15C:0 and βOH-17C:0 fatty acids were iso branched (Figure 1A) as previously reported (Pitta et al., 1989; Okuyama and Monde, 1996).
Figure 2.
High Resolution Accurate Mass/Mass Spectrometer (HRAM/MS) fragmentation of (A) Flavobacterium johnsoniae group I lipid at retention time 27.70 min; (B) proposed MS fragmentation of lysine lipids (LL) observed in F. johnsoniae. The relative abundance of the m/z 200-700 range is magnified 5×. The positive-ion ESI settings were as follows: capillary temperature, 275°C; sheath gas (N2) pressure, 35 arbitrary units (AU); auxiliary gas (N2) pressure, 10 AU; spray voltage, 4.0 kV; probe heater temperature, 300°C; S-lens, 50 V. Target lipids were analyzed with a mass range of m/z 400–1000 (resolution, 70,000), followed by data dependent MS2 (resolution, 17,500), in which the five most abundant masses in the mass spectrum were fragmented successively (normalized collision energy, 35; isolation width, 1.0).
Table 1.
Elemental composition of amino acid containing lipid fragmentation products observed by high performance liquid chromatography-high resolution accurate mass/mass spectrometry (HPLC-HRAM/MS) in the lipid extracts of Flavobacterium johnsoniae and Pseudopedobacter saltans.
| Species/Lipid | m/z | RT (min) | m/z, Product | Δmmu | m/z, Product | Δmmu | m/z, Product | Δmmu |
|---|---|---|---|---|---|---|---|---|
| Flavobacter johnsoniae | ||||||||
| I - 15C:0, βOH-17C:0 | 639 | 27.70 | 147, C6H15N2O2 | 0.074 | 130, C6H12NO2 | 0.055 | 129, C6H13N2O | 0.010 |
| I - 15C:0, βOH-16C:0 | 625 | 27.95 | 147, C6H15N2O2 | 0.114 | 130, C6H12NO2 | 0.075 | 129, C6H13N2O | 0.040 |
| Pseudopedobacter saltans | ||||||||
| I—15C:0, βOH-17C:0 | 639 | 27.53 | 147, C6H15N2O2 | 0.006 | 130, C6H12NO2 | 0.095 | 129, C6H13N2O | 0.100 |
| I—15C:0, βOH-16C:0 | 625 | 27.93 | 147, C6H15N2O2 | 0.006 | 130, C6H12NO2 | 0.095 | 129, C6H13N2O | 0.100 |
| I—15C:0, βOH-15C:0 | 611 | 28.05 | 147, C6H15N2O2 | 0.024 | 130, C6H12NO2 | 0.075 | 129, C6H13N2O | 0.080 |
| I'—OH-15C:0, βOH-17C:0 | 655 | 31.73 | 147, C6H15N2O2 | 0.026 | 130, C6H12NO2 | 0.105 | 129, C6H13N2O | 0.110 |
| I'—OH-15C:0, βOH-16C:0 | 641 | 32.44 | 147, C6H15N2O2 | 0.006 | 130, C6H12NO2 | 0.095 | 129, C6H13N2O | 0.100 |
| I'—OH-15C:0, βOH-15C:0 | 627 | 32.76 | 147, C6H15N2O2 | 0.016 | 130, C6H12NO2 | 0.105 | 129, C6H13N2O | 0.100 |
| II—15C:0, βOH-17C:0 | 655 | 29.59 | 163, C6H15N2O3 | 0.319 | 145, C6H13N2O2 | 0.126 | 128, C6H10NO2 | 0.145 |
| II'—OH-15C:0, βOH-17C:0 | 671 | 35.97 | 163, C6H15N2O3 | 0.189 | 145, C6H13N2O2 | 0.064 | 128, C6H10NO2 | 0.055 |
| II'—OH-15C:0, βOH-16C:0 | 657 | 36.91 | 163, C6H15N2O3 | 0.219 | 145, C6H13N2O2 | 0.156 | 128, C6H10NO2 | 0.155 |
| II'—OH-15C:0, βOH-15C:0 | 643 | 37.42 | 163, C6H15N2O3 | 0.069 | 145, C6H13N2O2 | 0.136 | 128, C6H10NO2 | 0.135 |
Retention time, RT; Accuracy, Δmmu*; OH, hydroxy group attached to fatty acids; Gln, glutamine; I, proposed lysine lipid (LL); I', proposed lysine lipid with hydroxylated fatty acid (LLHFA); I, proposed hydroxylysine lipid (HLL); II', proposed hydroxylysine lipid with hydroxylated fatty acid (HLLHFA).
*In general the 0.5 Δmmu (milli mass unit) range represented very high confidence molecular formula assignments and the 1.0 Δmmu range represented good confidence molecular formula assignments (Kostiainen et al., 1997; Sakayanagi et al., 2006; Calza et al., 2012).
IPLs of P. saltans
HPLC-ESI/IT/MS analysis of the lipid extract of P. saltans showed abundant PE, OL, and OLHFA IPLs (Figure 1B) making up 8, 10, and 12%, respectively, of HPLC-ESI/IT/MS chromatogram base peak area and four clusters with unknown IPLs, groups I, I', II, and II' making up 23, 28, 1.5, and 7% of HPLC-ESI/IT/MS chromatogram base peak area, respectively. OLHFA were hydroxylated on the ester linked fatty acids at the α-position. P. saltans group I produced MS3 fragmentation products of m/z 129, 130, and 147, identical to the group I LLs identified in F. johnsoniae. The relative retention time of group I IPLs in P. saltans are also similar to the retention time of the LLs in F. johnsoniae (Table 1), and were therefore also identified as LLs. This was confirmed by HPLC-HRAM/MS analysis (Table 1). P. saltans group I' IPLs exhibited the same MS3 fragmentation products (m/z 129, 130, and 147) as group I LLs, but MS2 fatty acid fragmentation losses were 16 Th larger than observed for group I, indicating the IPLs in group I' are potentially hydroxylated-fatty acid versions of group I LLs (LLHFA). This was also confirmed by HPLC-HRAM/MS analysis (Table 1). As with OLHFA, group I' LLHFA were hydroxylated on the ester linked fatty acid α-position. The most abundant P. saltans fatty acids were iso branched as previously observed (Steyn et al., 1998; Liolios et al., 2011), and the distribution of fatty acid chain lengths and double bond equivalents were identical between the LLs, LLHFA, OLs, and OLHFA, suggesting a biosynthetic link between these IPL classes (Table 1).
Group I LLs and group I' LLHFA were isolated from the P. saltans extract using preparatory HPLC to confirm the lysine head group structure. Approximately 0.6 mg of both LL and LLHFA were individually isolated for NMR analysis. For comparison, roughly 2.0 mg of known OL was also isolated from the F. johnsoniae extract. Results from the 1H-NMR, COSY, and 13C-NMR analysis of isolated LLs and LLHFA (Table 2; Supplementary Materials) revealed many similarities with NMR characterization reported for OLs (Okuyama and Monde, 1996; Linscheid et al., 1997; Moore et al., 2013). The fatty acid chain chemical shifts were almost identical between LLs, LLHFA, and OLs, except for the OH group attached to the hydroxylated fatty acids of LLHFA. There were additional 1H and 13C signals for the head group of LLs and LLHFA corresponding to the ε-position of the lysine head group, compared to the OL head group, in agreement with the fact that lysine has one more carbon than ornithine. The most compelling 2D-NMR evidence of a lysine head group in the proposed LLs and LLHFA was observed in the TOCSY interactions among the head group positions (Table 2). The same TOCSY interactions were previously described in the structural determinations of components containing an esterified lysine moiety (Anderson et al., 1995), and peptide-bound lysine (Rabenstein et al., 1997). Interactions observed by adjacent carbon and proton atoms in the HSQC spectra, and spatially close protons in NOESY spectra, further supported the lysine head group assignment of LLs and LLHFA. Amino acid analysis of each of the isolated LL, LLHFA, and OL components confirmed the presence of a lysine head group in the LL and LLHFA and an ornithine head group in the OL (Table 3).
Table 2.
Nuclear magnetic resonance (NMR) proton and carbon chemical shifts, and 2D NMR correlations (correlation spectroscopy: COSY; total correlation spectroscopy: TOCSY) of lysine lipid (LL) and lysine hydroxy fatty acid lipid (LLHFA).
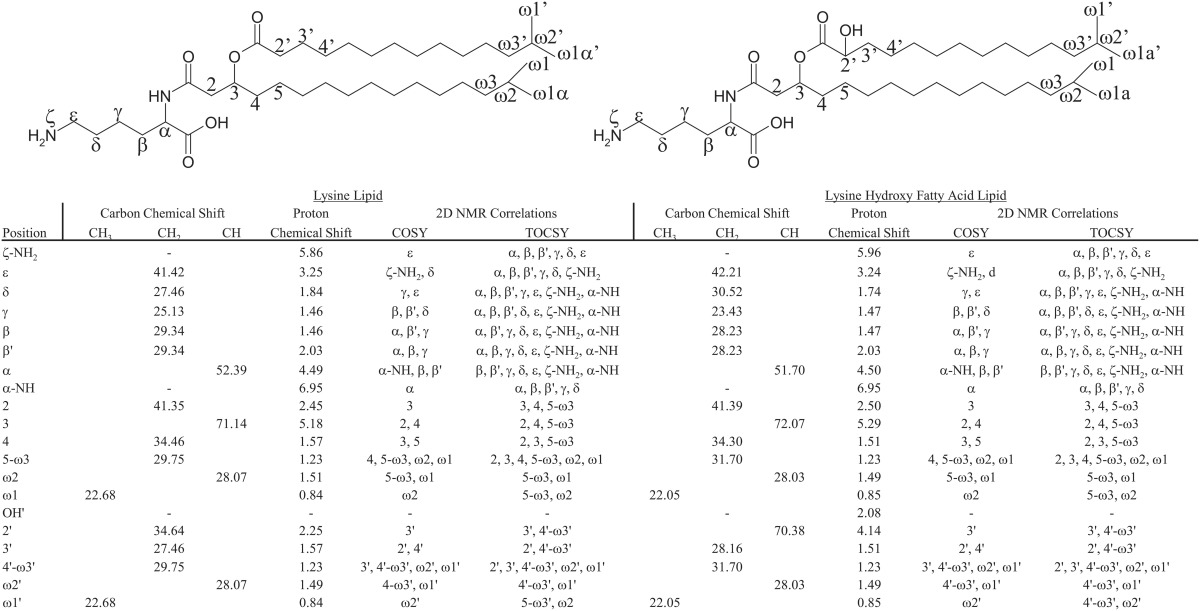
Table 3.
Amino acid analysis retention times (RT) of amino acids released from ornithine lipid, proposed lysine lipid (group I, LL), proposed lysine lipid with hydroxylated fatty acid (group I', LLHFA), and proposed hydroxylysine lipid with hydroxylated fatty acid (group II', HLLHFA) hydrolysates.
| Amino acid standard | Lipid head group | Amino acid standard RT (min) | Lipid hydrolysate head group RT (min) |
|---|---|---|---|
| Ornithine | Ornithine | 100.11 | 100.11 |
| Lysine | I, I' | 103.58 | 103.59, 103.61 |
| Hydroxylysine | II' | 95.99 | 95.63 |
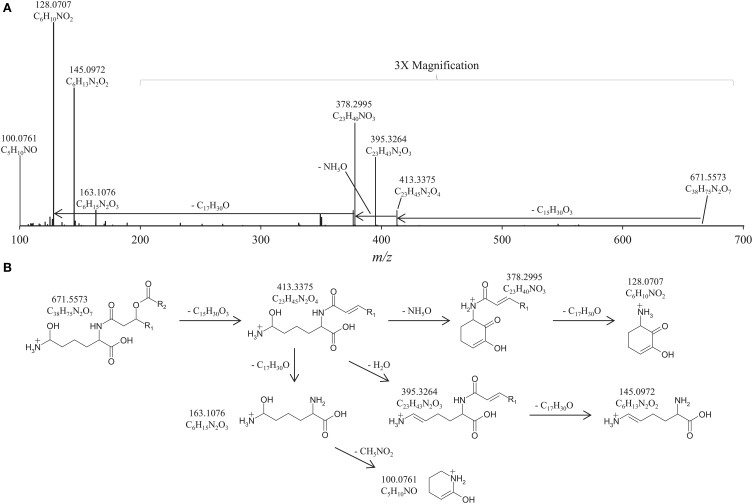
Another pair of unknown IPLs (groups II and II') observed in the P. saltans lipid extract eluted at 29.2 and 34.9 min, respectively (Figure 1B). The lipids in both of these groups exhibited identical OL-like fragmentation with ESI/IT/MS MS3 products at m/z 129 and 145. As observed before for OLHFA and LLHFA compared to OLs and LLs, respectively, the later eluting group II' IPLs displayed MS2 fatty acid fragmentation losses m/z 16 larger than group II, indicating groups II and II' contain the same polar head group but group II' is potentially hydroxylated on one of the fatty acids chains. HPLC-HRAM/MS analysis confirmed the presence of hydroxylated fatty acids in group II', and revealed that in both groups II and II', fragmentation losses of fatty acids results in the m/z 163.1076 product, which has the exact elemental composition of protonated hydroxylysine, C6H15N2O3 (Figure 3). Further fragmentation (loss of H2O) results in the m/z 145.0972 product (C6H13N2O2). The loss of NH5O from the C6H15N2O3 head group results in the abundant proposed cyclic m/z 128.0707 product with elemental composition C6H10NO2, and the loss of CH5NO2 from the C6H15N2O3 head group results in the proposed cyclic m/z 100.0761 product (C5H10NO). These observed elemental compositions are similar to those for LL and LLHFA but all with an additional oxygen (Table 1, Figure 3). We propose that these lipids contain hydroxylysine as the amino acid head group, which would explain the additional oxygen in the elemental composition, the observed fragments, and higher retention times (due to its more polar nature) compared to LL (Figure 1B). The fatty acid composition of the proposed hydroxylysine IPL (HLL) and fatty acid hydroxylated hydroxylysine lipids (HLLHFA) in groups II and II', respectively, are also the same as the OLs and LLs, including fatty acid hydroxylation (Table 1). The retention time of the amino acid released by acid hydrolysis from the isolated HLLHFA (group II') was closer to the 5-hydroxylysine standard than any other amino acid standard, but the retention times were still 18 s apart (Table 3). It is therefore likely that the proposed hydroxylysine head group is hydroxylated in a different position than the hydroxylysine standard resulting in a slightly different retention time. This is in agreement with the study by Morin et al. (1998), which found different hydroxylation positions on lysine resulted in different amino acid analysis retention times.
Figure 3.
(A) High Resolution Accurate Mass/Mass Spectrometer (HRAM/MS) fragmentation of Pseudopedobacter saltans IPL m/z 671.5573 at retention time 35.97 min; (B) Proposed MS fragmentation of IPL with hydroxylysine head group (HLL). The relative abundance of the m/z 200–700 range is magnified 3×. The positive-ion ESI settings were as follows: capillary temperature, 275°C; sheath gas (N2) pressure, 35 arbitrary units (AU); auxiliary gas (N2) pressure, 10 AU; spray voltage, 4.0 kV; probe heater temperature, 300°C; S-lens, 50 V. Target lipids were analyzed with a mass range of m/z 400–1000 (resolution, 70,000), followed by data dependent MS2 (resolution, 17,500), in which the five most abundant masses in the mass spectrum were fragmented successively (normalized collision energy, 35; isolation width, 1.0).
Biosynthetic pathway of lysine lipids
The synthesis pathway of LL has not yet been described but it appears to be linked to the OL synthesis. The first described synthetic pathway for OLs involves a N-acyltransferase coded by the OlsB gene transferring a 3-hydroxy acyl group from the constitutive acyl carrier protein (ACP) to the α-amino group of ornithine forming lyso-ornithine, and then an O-acyltransferase (encoded by the OlsA gene) transferring a fatty acyl chain from the acyl carrier protein to the 3-hydroxy group of the lyso-ornithine, forming OL (Weissenmayer et al., 2002; Gao et al., 2004). Vences-Guzmán et al. (2014) described the existence of a second pathway mediated by a bifunctional protein with two acyltransferase domains coded by the OlsF gene in some bacterial species, which have been described to form OL but lack the OlsAB genes. Vences-Guzmán et al. (2014) also described the formation of OL and LL, using MS analysis, in E. coli expressing the OlsF-coding protein from the F. johnsoniae strain UW101, but they did not confirm that LL were indeed produced by F. johnsoniae. However, our study confirms that F. johnsoniae synthesizes OL and LL. LLs have previously been reported in Agrobacterium tumefaciens by Tahara et al. (1976a) and in Mycobacterium phlei by Lerouge et al. (1988). The LL characterized in M. phlei is diacylglycerol based, while the structure of the A. tumefaciens LL is similar to the lysine lipids observed in this study by LC/MS and NMR, i.e., composed of an ester linked fatty acid moiety, and an amide linked β-OH-fatty acid.
Homologs of OlsF have been identified in genomes of γ-, δ-, and ε-Proteobacteria and in bacteria belonging to the CFB group (Vences-Guzmán et al., 2014). The genomes of F. johnsoniae and Pedobacter heparinus (both belonging to the CFB group) have been shown to harbor a homolog of OlsF (Vences-Guzmán et al., 2014). We searched for homologs of the OlsF-coding protein in genomes of Pedobacter species and P. saltans, whose lipid composition has been analyzed in our study. We detected putative homologs to OlsF in all Pedobacter genomes available (and in P. saltans) with an identity ≥70% to the P. heparinus putative OlsF (Vences-Guzmán et al., 2014), and an identity ≥40% to the OlsF protein of F. johnsoniae. These putative OlsF proteins in Pedobacter genomes have been previously annotated as hemolysines, as one of the genes of the hemolysin operon gene cluster, an acyltransferase catalyzing the transfer of a fatty-acyl group from acyl-ACP to α-amino groups of two lysine residues of the pro-toxin hemolysin converting it into a mature protein (Stanley et al., 1998). Considering this evidence, we speculate that the OlsF coding protein is involved not only in the synthesis of OL (Vences-Guzmán et al., 2014), but also in the synthesis of LL.
Additionally, in our study we report for the first time LLs that are modified by hydroxylation on one of its fatty acid chains and on the lysine head group. P. saltans produces a wide range of amino acid-containing IPLs including OL, OLHFA, LLs, LLHFA, HLL, and HLLHFA. All of the various ornithine and lysine lipids produced by P. saltans have the same fatty acid distribution (Table 1), suggesting a biosynthetic link. Hydroxylation of OL has been previously described and it is known to be mediated by the OL hydroxylase OlsC on ester-linked fatty acids (Rojas-Jimenez et al., 2005; Vences-Guzman et al., 2011), by the OlsD protein coding gene on the amide-linked fatty acid (González-Silva et al., 2011), or by the OlsE protein coding gene on the ornithine head group moiety (Rojas-Jimenez et al., 2005; Vences-Guzman et al., 2011). We performed protein blast searches for homologs of OlsC, OlsE of Rhizobium tropici (Rojas-Jimenez et al., 2005), and OlsD of Burkholderia cenocepacia but no putative homologs were detected in the currently available genomes of Pedobacter or P. saltans. However, we cannot rule out the presence of other putative OL hydroxylases in Pedobacter species genomes, which are expected to be quite divergent from the already described OL hydroxylases of Rhizobium and Burkholderia species, or that the hydroxylation of OL and LL in Pedobacter and Pseudopedobacter species is mediated by an alternative pathway.
Environmental controls on lysine IPLs in P. saltans
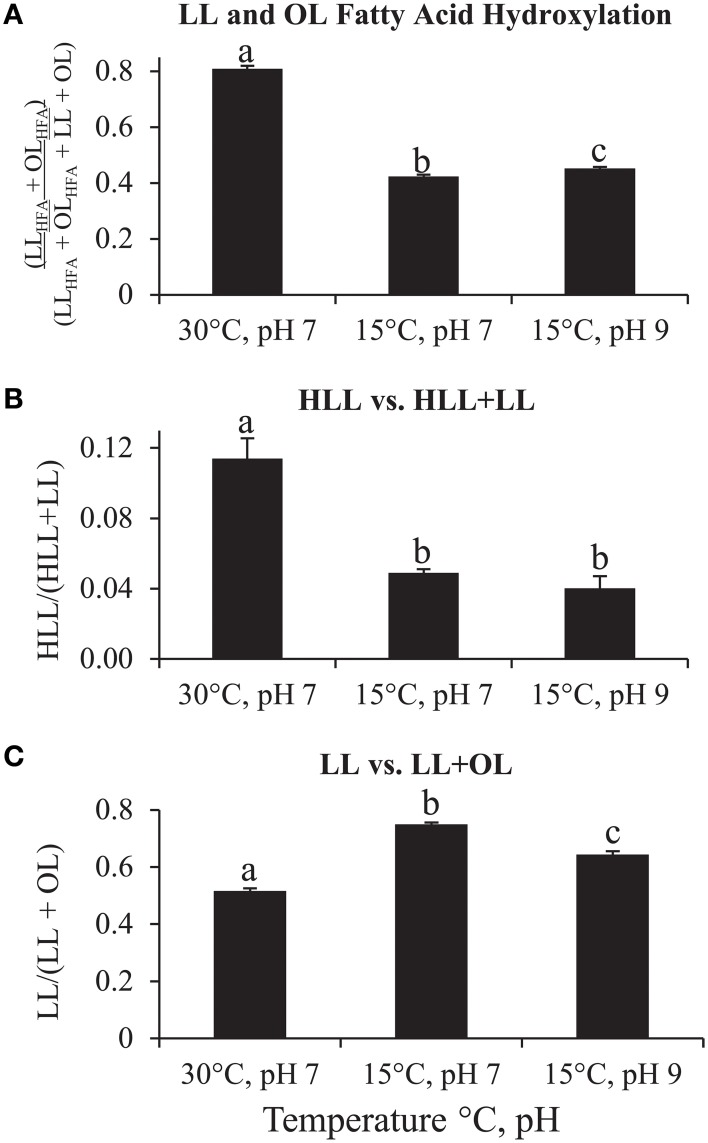
The novel characterization of HLL, and the high abundance of LLHFA and OLHFA in the lipid extract of P. saltans raises the question of their functional significance. Various studies have shown that certain bacteria increase fatty acid hydroxylation of OLs under temperature and pH stress (Taylor et al., 1998; Rojas-Jimenez et al., 2005; Vences-Guzman et al., 2011). The high abundance of LLHFA and OLHFA in P. saltans suggested that these lipids could be associated with in environmental stress response. We tested this hypothesis for LLs and OLs in P. saltans by growing triplicate cultures under three temperature/pH combinations: 30°C/pH 7, 15°C/pH 7, 15°C/pH 9. The ratios of LL and OL abundance, and the degree of fatty acid hydroxylation in P. saltans changed under different temperature and pH conditions (Figure 4). There was a statistically significant increase in the ratio of the sum of LLHFA and OLHFA (LLHFA + OLHFA) to total LLs and OLs (LLHFA + OLHFA + LL + OL) at higher temperatures and higher pH, with the largest ratio increase due to temperature (p < 0.0001) and a smaller increase due to pH rise (p = 0.0085, Figure 4A). There was also a significant increase in the ratio of HLL to the sum of HLL and LL at higher temperatures (p = 0.0115, Figure 4B). There were significant decreases in the ratio of total LLs vs. the sum of total LLs and OLs at higher temperatures (p < 0.0001) and higher pH (p = 0.0025), with the largest LL/(LL + OL) ratio decrease also due to temperature and a moderate decrease due to pH (Figure 4C).
Figure 4.
Changes in amino acid-containing IPL abundance ratios based on HPLC-ESI/IT/MS chromatogram base peak area of each lipid group in Pseudopedobacter saltans lipid extract under 30°C/pH 7, 15°C/pH 7, and 15°C/pH 9 growth conditions. (A) Ratio of total fatty acid hydroxylated lysine and ornithine lipids (LLHFA + OLHFA) to total hydroxylated plus total unhydroxylated fatty acid lysine and ornithine lipids (LLHFA + OLHFA + LL + OL); (B) Ratio of total hydroxylysine lipids (HLL) to total HLL plus total LL; (C) Ratio of total LL to total LL plus total OL. Student's t-test statistically significant differences (p < 0.05) of IPL ratios between different growth conditions are represented by letters (a, b, c) over each bar (different letters represent statistically significant differences between bar graph values within each graph, values with the same letter within each graph are not statistically different).
There appears to be an OL and LL fatty acid hydroxylation stress response to temperature and pH in P. saltans, similar to the fatty acid hydroxylation stress response to temperature and pH observed in various other bacteria, which is proposed to increase membrane hydrogen bonding and stability (Taylor et al., 1998; Rojas-Jimenez et al., 2005; Vences-Guzman et al., 2011). In this case, there is a greater response to temperature than pH for P. saltans. The lysine head group hydroxylation also follows the same trend as fatty acid hydroxylation with more hydroxylation at higher temperature, as also observed in OL head group hydroxylation in R. tropici (Rojas-Jimenez et al., 2005; Vences-Guzman et al., 2011). OLs may also be preferentially produced over LLs at higher temperatures or pH levels in order to provide membrane stability due to the higher water solubility of ornithine than lysine. The various IPL composition changes due to temperature and pH show that the cell membrane of P. saltans is very dynamic in response to changing environmental conditions.
Conclusions
This is the first study to identify LL fatty acid hydroxylation and LL head group hydroxylation in membrane lipids. Confirmation of OL and LL formation in F. johnsoniae and P. saltans and the presence of OlsF putative homologs in P. saltans suggest the OlsF gene coding protein is possibly involved in OL and LL biosynthesis in both species, however, potential pathways of OL and LL hydroxylation in P. saltans are still undetermined.
LL and OL fatty acid hydroxylation and LL vs. OL fractional abundance changes due to temperature and pH in microbial culture experiments were observed, thus expanding knowledge of microbial response to environmental change. The IPLs of related soil bacteria (i.e., Pedobacter sp.) should be characterized as well to investigate stress biomarker potential of environmental conditions such as temperature or pH. Analysis of soil samples is needed as well to reveal if these membrane lipid responses to temperature and pH changes can be observed in the environment.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the Darwin Center for Biogeosciences grant # 142.16.3082. JD and AS were supported by Gravitation grant SIAM (24002002) of the Netherlands Ministry of Education, Culture and Science and the Netherlands Science Foundation (NWO). AS and ISA were also supported by ERC grant project 323009. We thank Rozelin Aydin for culturing one of the initial cultures of Pseudopedobacter saltans.
Footnotes
1Commission Regulation (EC) no., 152/2009 of January 27 2009. Laying down the methods of sampling and analysis for the official control of feed. Official Journal of the European Union L 54/28 Part F.
2European Pharmacopoeia chapter 2.2.56, USP chapter 1052 (2005) Method 1 for Analysis.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00637
References
- Anderson R. C., Stokes J. P., Shapiro M. J. (1995). Structure determination in combinatorial chemistry: utilization of magic angle spinning HMQC and TOCSY NMR spectra in the structure determination of Wang-bound lysine. Tetrahedr. Lett. 36, 5311–5314. [Google Scholar]
- Aygun-Sunar S., Mandaci S., Koch H. G., Murray I. V. J., Goldfine H., Daldal F. (2006). Ornithine lipid is required for optimal steady-state amounts of c-type cytochromes in Rhodobacter capsulatus. Mol. Microbiol. 61, 418–435. 10.1111/j.1365-2958.2006.05253.x [DOI] [PubMed] [Google Scholar]
- Bardgett R. D., Freeman C., Ostle N. J. (2008). Microbial contribution to climate change through carbon cycle feedbacks. ISME J. 2, 805–8014. 10.1038/ismej.2008.58 [DOI] [PubMed] [Google Scholar]
- Batrakov S. G., Nikitin D. I., Mosezhnyi A. E., Ruzhitsky A. O. (1999). A glycine containing phosphorus-free lipoaminoacid from the gram-negative marine bacterium Cyclobacterium marinus WH. Chem. Phys. Lipids 99, 139–143. [Google Scholar]
- Boumann H. A., Longo M. L., Stroeve P., Poolman B., Hopmans E. C., Stuart M. C. A., et al. (2009). Biophysical properties of membrane lipids of anammox bacteria. I. Ladderane phospholipids form highly organized fluid membranes. Biochim. Biophys. Acta 1788, 1444–1451. 10.1016/j.bbamem.2009.04.008 [DOI] [PubMed] [Google Scholar]
- Calza P., Medana C., Padovano E., Dal Bello F., Baiocchi C. (2012). Identification of the unknown transformation products derived from lincomycin using LC-HRMS technique. J. Mass Spectrom. 47, 751–759. 10.1002/jms.3012 [DOI] [PubMed] [Google Scholar]
- Cao J., Lai Q., Li G., Shao Z. (2014). Pseudopedobacter beijingensis gen. nov., sp. nov., isolated from coking wastewater activated sludge, and reclassification of Pedobacter saltans as Pseudopedobacter saltans comb. nov. Int. J. Syst. Evol. Micr. 64, 1853–1858. 10.1099/ijs.0.053991-0 [DOI] [PubMed] [Google Scholar]
- Cerny R. L., Tomer K. B., Gross M. L. (1986). Desorption ionization combined with tandem mass spectrometry: advantages for investigation complex lipids, disaccharides, and organometallic complexes. Org. Mass Spectrom. 21, 655–660. [Google Scholar]
- Freer E., Moreno E., Moriyon I., Pizarro-Cerda J., Weintraub A., Gorvel J. P. (1996). Brucella-Salmonella lipopolysaccharide chimeras are less permeable to hydrophobic probes and more sensitive to cationic peptides and EDTA than are their native Brucella sp counterparts. J. Bacteriol. 178, 5867–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. L., Weissenmayer B., Taylor A. M., Thomas-Oates J., Lopez-Lara I. M., Geiger O. (2004). Identification of a gene required for the formation of lyso-ornithine lipid, an intermediate in the biosynthesis of ornithine-containing lipids. Mol. Microbiol. 53, 1757–1770. 10.1111/j.1365-2958.2004.04240.x [DOI] [PubMed] [Google Scholar]
- Geiger O., Gonzalez-Silva N., Lopez-Lara I. M., Sohlenkamp C. (2010). Amino acid-containing membrane lipids in bacteria. Prog. Lipid Res. 49, 46–60. 10.1016/j.plipres.2009.08.002 [DOI] [PubMed] [Google Scholar]
- González-Silva N., López-Lara I. M., Reyes-Lamothe R., Taylor A. M., Sumpton D., Thomas-Oates J., et al. (2011). The dioxygenase-encoding olsD gene from Burkholderia cenocepacia causes the hydroxylation of the amide-linked fatty acyl moiety of ornithine-containing membrane lipids. Biochemistry 50, 6396–6408. 10.1021/bi200706v [DOI] [PubMed] [Google Scholar]
- Hilker D. R., Gross M. L., Knoche H. W., Shively J. M. (1978). Interpretation of mass-spectrum of an ornithine-containing lipid from Thiobacillus thiooxidans. Biomed. Mass Spectrom. 5, 64–71. 10.1002/bms.1200050112 [DOI] [PubMed] [Google Scholar]
- Hilker D. R., Knoche H. W., Gross M. L. (1979). Thermolysis chemical ionization of a complex polar lipid. Biomed. Mass Spectrom. 6, 356–358. 10.1002/bms.1200060810 [DOI] [PubMed] [Google Scholar]
- Kawazoe R., Monde K., Reichardt W. (1992). Lipoamino acids and sulfonolipids in Cytophaga johnsonae Stanier strain C21 and 6 related species of Cytophaga. Archiv. Microbiol. 158, 171–175. [Google Scholar]
- Kawazoe R., Okuyama H., Reichardt W., Sasaki S. (1991). Phospholipids and a novel glycine-containing lipoamino acid in Cytophaga johnsonae Stanier strain C21. J. Bacteriol. 173, 5470–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostiainen R., Tuominen J., Luukanen L., Taskinen J., Green B. N. (1997). Accurate mass measurements of some glucuronide derivatives by electrospray low resolution quadrupole mass spectrometry. Rapid Commun. Mass Spectrom. 11, 283–285. [Google Scholar]
- Larkin J. M. (1989). Nonphotosynthetic, nonfruiting gliding bacteria, in Bergey's Manual of Systematic Bacteriology, eds Staley J. T., Bryant M. P., Pfennig N., Holt J. G. (Baltimore, MD: Williams and Wilkins; ), 2010–2138. [Google Scholar]
- Lerouge P., Lebas M. H., Agapakis-Caussé C., Promé J. C. (1988). Isolation and structural characterization of a new non-phosphorylated lipoamino acid from Mycobacterium phlei. Chem. Phys. Lipids. 49, 161–166. 10.1016/0009-3084(88)90003-5 [DOI] [PubMed] [Google Scholar]
- Linscheid M., Diehl B. W. K., Overmohle M., Riedl I., Heinz E. (1997). Membrane lipids of Rhodopseudomonas viridis. Biochim. Biophys. Acta 1347, 151–163. 10.1016/S0005-2760(97)00065-9 [DOI] [PubMed] [Google Scholar]
- Liolios K., Sikorski J., Lu M., Nolan M., Lapidus A., Lucas S., et al. (2011). Complete genome sequence of the gliding, heparinolytic Pedobacter saltans type strain (113T). Stand. Genom. Sci. 5, 30–40. 10.4056/sigs.2154937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lara I. M., Sohlenkamp C., Geiger O. (2003). Membrane lipids in plant-associated bacteria: their biosynthesis and possible functions. Mol. Plant Microb. Int. 16, 567–579. 10.1094/MPMI.2003.16.7.567 [DOI] [PubMed] [Google Scholar]
- McBride M. J. (2001). Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Ann. Rev. Microbiol. 55, 49–75. 10.1146/annurev.micro.55.1.49 [DOI] [PubMed] [Google Scholar]
- McBride M. J., Braun T. F., Brust J. L. (2003). Flavobacterium johnsoniae GldH is a lipoprotein that is required for gliding motility and chitin utilization. J. Bacteriol. 185, 6648–6657. 10.1128/JB.185.22.6648-6657.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore E. K., Hopmans E. C., Rijpstra W. I. C., Villanueva L., Dedysh S. N., Kulichevskaya I. S., et al. (2013). Novel Mono-, Di-, and trimethylornithine membrane lipids in Northern Wetland Planctomycetes. Appl. Environ. Microbiol. 79, 6874–6884. 10.1128/AEM.02169-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin B., Bubb W. A., Davies M. J., Dean R. T., Fu S. (1998). 3-Hydroxylysine, a potential marker for studying radical-induced protein oxidation. Chem. Res. Toxicol. 11, 1265–1273. 10.1021/tx980118h [DOI] [PubMed] [Google Scholar]
- Nelson S. S., McBride M. J. (2006). Mutations in Flavobacterium johsoniae secDF result in defects in gliding motility and chitin utilization. J. Bacteriol. 188, 348–351. 10.1128/JB.188.1.348-351.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama H., Monde K. (1996). Identification of an ornithine-containing lipid from Cytophaga johnsonae Stanier strain C21 by 1H-NMR. Chem. Phys. Lipids. 83, 169–173. [Google Scholar]
- Pitta T. P., Leadbetter E. R., Godchauz W. (1989). Increase of ornithine amino lipid-content in a sulfonolipid-deficient mutant of Cytophaga johnsonae. J. Bacteriol. 171, 952–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenstein D. L., Hari S. P., Kaerner A. (1997). Determination of acid dissociation constants of peptide side-chain functional groups by two-dimensional NMR. Anal. Chem. 69, 4310–4316. 10.1021/ac970675t [DOI] [PubMed] [Google Scholar]
- Rojas-Jimenez K., Sohlenkamp C., Geiger O., Martinez-Romero E., Werner D., Vinuesa P. (2005). A CIC chloride channel homolog and ornithine-containing membrane lipids of Rhizobium tropici CIAT899 are involved in symbiotic efficiency and acid tolerance. Mol. Plant Microbe. Int. 18, 1175–1185. 10.1094/MPMI-18-1175 [DOI] [PubMed] [Google Scholar]
- Rutters H., Sass H., Cypionka H., Rullkötter J. (2002). Phospholipid analysis as a tool to study complex microbial communities in marine sediments. J. Microbiol. Meth. 48, 149–160. 10.1016/S0167-7012(01)00319-0 [DOI] [PubMed] [Google Scholar]
- Sakayanagi M., Yamada Y., Sakabe C., Watanabe K., Harigaya Y. (2006). Identification of inorganic anions by gas chromatography/mass spectrometry. Forensic Sci. Int. 157, 134–143. 10.1016/j.forsciint.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Schubotz F., Wakeham S. G., Lipp J. S., Fredericks H. F., Hinrichs K. U. (2009). Detection of microbial biomass by intact polar membrane lipid analysis in the water column and surface sediments of the Black Sea. Environm. Microbiol. 11, 2720–2734. 10.1111/j.1462-2920.2009.01999.x [DOI] [PubMed] [Google Scholar]
- Seidel M., Rütters H., Rullkötter J., Sass H. (2013). Phosphate-free ornithine lipid contents in Desulfovibrio spp. respond to growth temperature. Org. Geochem. 59, 133–142. [Google Scholar]
- Singh B. K., Bardgett R. D., Smith P., Reay D. S. (2010). Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 8, 779–790. 10.1038/nrmicro2439 [DOI] [PubMed] [Google Scholar]
- Sinninghe Damsté J. S., Rijpstra W. I. C., Hopmans E. C., Weijers J. W. H., Foesel B. U., Overmann J., et al. (2011). 13,16-Dimethyl octacosanedioic acid (iso-diabolic acid), a common membrane-spanning lipid of Acidobacteria subdivisions 1 and 3. Appl. Environ. Microbiol. 77, 4147–4154. 10.1128/AEM.00466-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinninghe Damsté J. S., Rijpstra W. I. C., Schouten S., Fuerst J. A., Jetten M. S. M., Strous M. (2004). The occurrence of hopanoids in planctomycetes: implications for the sedimentary biomarker record. Org. Geochem. 35, 561–566. [Google Scholar]
- Smittenberg R. H., Hopmans E. C., Schouten S., Sinninghe Damsté J. S. (2002). Rapid isolation of biomarkers for compound specific radiocarbon dating using high performance liquid chromatography and flow injection analysis-atmospheric pressure chemical ionization mass spectrometry. J. Chromatogr. A 978, 129–140. 10.1016/S0021-9673(02)01427-9 [DOI] [PubMed] [Google Scholar]
- Spackman D. H., Stein W. H., Moore S. (1958). Automatic recording apparatus for use in the chromatography of amino acids. Anal. Chem. 30, 1190–1206. [PubMed] [Google Scholar]
- Stanier R. Y. (1947). Studies on non-fruiting myxobacteria. I. Cytophaga johnsonae, N. Sp., A chitin-decomposing myxobacterium. J. Bacteriol. 53, 297–315. [PMC free article] [PubMed] [Google Scholar]
- Stanley P., Koronakis V., Hughes C. (1998). Acylation of Escherichia coli Hemolysin: a unique protein lipidation mechanism underlying toxin function. Microbiol. Mol. Biol. R. 62, 309–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn P. L., Segers P., Vancanneyt M., Sandra P., Kersters K., Joubert J. J. (1998). Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. Proposal of the family Sphingobacteriaceae fam. nov. Int. J. System. Bacteriol. 48, 165–177. 10.1099/00207713-48-1-165 [DOI] [PubMed] [Google Scholar]
- Sturt H. F., Summons R. E., Smith K., Elvert M., Hinrichs K. U. (2004). Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry–new biomarkers for biogeochemistry and microbial ecology. Rap. Commun. Mass Spectrom. 18, 617–628. 10.1002/rcm.1378 [DOI] [PubMed] [Google Scholar]
- Tahara Y., Kameda M., Yamada Y., Kondo K. (1976b). A new lipid; the ornithine and taurine-containing “cerilipin.” Agric. Biol. Chem. 40, 243–244. [Google Scholar]
- Tahara Y., Yamada Y., Kondo K. (1976a). A new lysine-containing lipid isolated from Agrobacterium tumefaciens. Agric. Biol. Chem. 40, 1449–1450. [Google Scholar]
- Taylor C. J., Anderson A. J., Wilkinson S. G. (1998). Phenotypic variation of lipid composition in Burkholderia cepacia: a response to increased growth temperature is a greater content of 2-hydroxy acids in phosphatidylethanolamine and ornithine amide lipid. Microbiology 144, 1737–1745. 10.1099/00221287-144-7-1737 [DOI] [PubMed] [Google Scholar]
- Tomer K. B, Crow F. W., Knoche H. W., Gross M. L. (1983). Fast atom bombardment and mass spectrometry/mass spectrometry for analysis of a mixture of ornithine-containing lipids from Thiobacillus thiooxidans. Anal. Chem. 55, 1033–1036. [Google Scholar]
- Van Mooy B. A. S., Fredricks H. F., Pedler B. E., Dyhrman S. T., Karl D. M., KoblíŽek M., et al. (2009). Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458, 69–72. 10.1038/nature07659 [DOI] [PubMed] [Google Scholar]
- Vences-Guzmán M. A., Geiger O., Sohlenkamp C. (2012). Ornithine lipids and their structural modifications from A to E and beyond. FEMS Microbiol. Lett. 335, 1–10. 10.1111/j.1574-6968.2012.02623.x [DOI] [PubMed] [Google Scholar]
- Vences-Guzmán M. A., Guan Z., Escobedo-Hinojosa W. I., Bermúdez-Barrientos J. R., Geiger O., Sohlenkamp C. (2014). Discovery of a bifuctional acyltransferase responsible for ornithine lipid synthesis in Serratia proteamaculans. Environm. Microbiol. 17, 1487–1496. 10.1111/1462-2920.12562 [DOI] [PubMed] [Google Scholar]
- Vences-Guzman M. A., Guan Z., Ormeno-Orillo E., Gonzalez-Silva N., Geiger O., Sohlenkamp C. (2011). Hydroxylated ornithine lipids increase stress tolerance in Rhizobium tropici CIAT899. Mol. Biol. 79, 1496–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenmayer B., Gao J. L., Lopez-Lara I. M., Geiger O. (2002). Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol. Microbiol. 45, 721–733. 10.1046/j.1365-2958.2002.03043.x [DOI] [PubMed] [Google Scholar]
- Zhang X., Ferguson-Miller S. M., Reid G. E. (2009). Characterization of ornithine and glutamine lipids extracted from cell membranes of Rhodobacter sphaeroides. J. Am. Soc. Mass Spectrom. 20, 198–212. 10.1016/j.jasms.2008.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.