Abstract
The schweinfurthins, a family of natural products derived from the isoprenoid biosynthetic pathway (IBP), have marked growth inhibitory activity. However, the biochemical basis for the schweinfurthins cellular effects has remained ill-defined. Here, the effects of the synthetic schweinfurthin, 3-deoxyschweinfurthin (3dSB) on multiple aspects of isoprenoid homeostasis are explored. Cytotoxicity assays demonstrate a synergistic interaction between 3dSB and the HMG-CoA reductase inhibitor lovastatin but not with other IBP inhibitors in a variety of human cancer cell lines. The cytotoxic effects of 3dSB were enhanced in cells incubated in lipid-depleted serum. 3dSB was found to enhance the lovastatin-induced decrease in protein prenylation. In addition, 3dSB decreases intracellular farnesyl pyrophosphate and geranylgeranyl pyrophosphate levels in both established cell lines and primary cells. To determine whether 3dSB alters the regulation of expression of genes involved in isoprenoid homeostasis, real-time PCR studies were performed in human cell lines cultured in either lipid-replete or -deplete conditions. These studies demonstrate that 3dSB abrogates lovastatin-induced upregulation of sterol regulatory element-containing genes and lovastatin-induced downregulation of ABCA1. In aggregate, these studies are the first to demonstrate that a schweinfurthin exerts pleiotropic effects on isoprenoid homeostasis.
Keywords: Isoprenoid, Schweinfurthin, Lovastatin, Farnesyl pyrophosphate, Geranylgeranyl pyrophosphate, Sterol, ABCA1, Prenylation
Introduction
Schweinfurthins are a family of natural compounds that were originally isolated from the leaves of the flowering tree Macaranga schweinfurthii in Western Cameroon [1]. The extracts from these leaves were noted to have potent growth inhibitory activity in the NCI 60-cell line cancer screen and subsequent isolation of the active components identified schweinfurthins A and B with mean GI50’s of 0.36 and 0.81 μM, respectively [1]. Extensive structure-function analysis has provided insight into the key structural features which are necessary to retain schweinfurthin-like activity [2–7], but the mechanism of action has yet to be determined. Given the difficulty in isolating sufficient quantities of these compounds to allow for further biological exploration, a number of analogues have been prepared, including 3-deoxyschweinfurthin B (3dSB) (Fig. 1), which has a mean GI50 of 0.74 μM and has been shown to retain schweinfurthin-like activity [8].
Fig. 1.
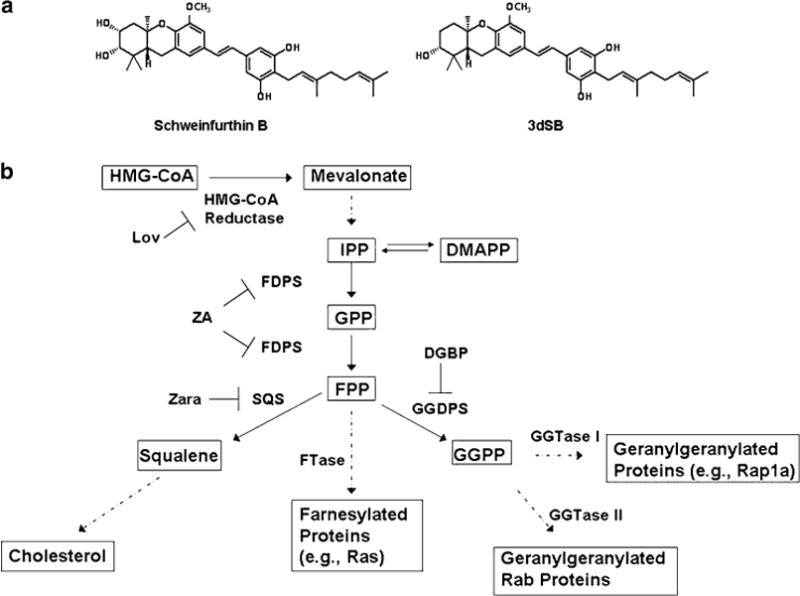
a Structures of schweinfurthin B and 3-deoxyschweinfurthin B (3dSB). b The isoprenoid biosynthetic pathway (IBP) and pharmacological inhibitors. Abbreviations: Lov lovastatin, ZA zoledronic acid, Zara zaragozic acid, DGBP digeranyl bisphosphonate
The schweinfurthin family shares the common structural feature of having one or more prenyl side chains. The prenyl groups are derived from the isoprenoid biosynthetic pathway (IBP) which is responsible for the synthesis of all sterol and non-sterol isoprenoids (Fig. 1). In addition to the products displayed in Fig. 1, this pathway also leads to the synthesis of other biologically important molecules such as the ubiquinones, dolichols, and retinoids [9]. There is an extraordinary degree of complexity with regards to regulation of the IBP and sterol homeostasis [10]. The expression of the majority of the enzymes in the IBP are regulated through sterol regulatory element-binding protein (SREBP) transcription factors [11, 12]. In conditions of sterol depletion, SREBPs upregulate the expression of genes with SRE-containing promoters, leading to increased de novo synthesis of sterols and nonsterols as well as increased cellular uptake of sterols via the LDL receptor. Conversely, in conditions of sterol excess, SREBPs are sequestered in the endoplasmic reticulum (ER) via Insigs and there is upregulation of sterol efflux proteins such as ATP-binding cassette transporter 1 (ABCA1) via liver X receptor (LXR)-mediated regulation [13–15].
Sterols (including oxysterols, cholesterol, and lanosterol), nonsterols [e.g., farnesyl pyrophosphate (FPP) and geranylgeranyl pyrophosphate (GGPP)], and miRNA (MiR-33) have all been shown to be important regulators of isoprenoid homeostasis [16–21]. Furthermore, a multitude of natural products derived from the IBP have been demonstrated to affect IBP regulation and sterol homeostasis through a variety of mechanisms, including the diterpenes cafestol, Daphnetoxin and Gniditrin [22, 23], monoterpenes [24, 25], tocotrienols [26], and triterpenoids [27–29], further highlighting the extraordinarily complex relationship between the IBP and its products. Finally, select inhibitors of the IBP also alter regulation of the IBP and affect the expression of derivatives of the IBP [30–35]. Thus, given the extent to which IBP products and inhibitors can influence IBP regulation, we were interested in exploring the potential interaction between schweinfurthins and IBP inhibitors, determining the effects of schweinfurthins on isoprenoid homeostasis, and providing insight into the basis for schweinfurthins’ biological activity. Here we demonstrate that the synthetic schweinfurthin 3dSB enhances inhibition of protein prenylation and induction of cytotoxicity in the setting of mevalonate depletion and reveal that 3dSB disrupts regulation of isoprenoid homeostasis.
Materials and Methods
Reagents
Lovastatin (converted to the dihydroxy-open acid form prior to use), DL-mevalonic acid lactone (converted to mevalonate prior to use), FPP, GGPP, and zaragozic acid were obtained from Sigma (St. Louis, MO). Zoledronic acid was purchased from Novartis (East Hanover, NJ). Digeranyl bisphosphonate (DGBP) [36] and 3-deoxyschweinfurthin [5] were kindly supplied by Professor David Wiemer, Department of Chemistry, University of Iowa. 3dSB was dissolved in DMSO. Anti-pan-Ras was obtained from Inter Biotechnology (Tokyo, Japan). Anti-β-tubulin, anti-Rap1a, anti-Rab6, and anti-goat IgG HRP antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-mouse and anti-rabbit HRP-linked antibodies were obtained from Amersham (GE Healthcare, Piscataway, NJ). D*-GCVLS and D*-GCVLL (dansyl-labeled peptides) were obtained from Bio-Synthesis (Lewisville, TX). Rat recombinant FTase and GGTase I were purchased from Jena Biosciences (Jena, Germany). HPLC-grade water was prepared with a Milli-Q system (Millipore, Bedford, MA). All solvents were optima or HPLC grade. Lipoprotein deficient serum was obtained from Hyclone (Thermo Scientific, Logan, UT).
Cell Cultures
Human multiple myeloma (RPMI-8226 and U266) and human glioblastoma multiforme (SF-295) cell lines were purchased from American Type Culture Collection (Manassas, VA). A549 cells, a human non-small cell lung cancer cell line, were obtained from the NCI (Frederick, MD). Cells were grown in RPMI-1640 media with 10% (RPMI-8226, SF-295) or 15% (U266) heat-inactivated fetal calf serum supplemented with glutamine and penicillin–streptomycin at 37 °C and 5% CO2. A549 cells were maintained in F-12 media supplemented with 10% fetal calf serum, glutamine, and penicillin–streptomycin. For the studies involving primary cells, after informed consent, peripheral blood samples were obtained from patients with acute leukemia. The protocol (IRB ID # 200610770) was approved by our Institutional Review Board for human subjects. Mononuclear cells were isolated from peripheral blood of patients with newly diagnosed acute leukemia and incubated ex vivo with 3dSB or lovastatin for 24 h. Formal pathological review of the bone marrow samples as well as cytogenetic analysis was performed by UIHC Hematopathology.
MTT Assay
For the suspension cell lines (RPMI-8226 and U266), cells were seeded (5 × 10/150 μL per well) in 96-well flat-bottom plates and incubated for 48 h at 37 °C and 5% CO2 in the presence or absence of compounds. For the adherent cell lines (SF-295 and A549), cells were plated in 24-well plates, allowed to reach 65% confluency, and then incubated in the presence or absence of compounds for 48 h. MTT assays were performed as previously described [3, 43]. The absorbance for control untreated cells was defined as an MTT activity of 100%. DMSO treatment did not alter MTT activity by more than 5%.
Western Blot Analysis
Cells (5 × 106/5 mL) were incubated in the presence or absence of drugs. At the conclusion of the incubations, cells were collected, washed with PBS, and lysed in RIPA buffer (0.15 M NaCl, 1% sodium deoxycholate, 0.1% SDS, 1% Triton (v/v) X-100, 0.05 M Tris–HCl) containing protease and phosphatase inhibitors. Protein content was determined using the bicinchoninic acid method (Pierce Chemical, Rockford, IL). Equivalent amounts of cell lysate were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membrane, probed with the appropriate primary antibodies, and detected using HRP-linked secondary antibodies and Amersham Pharmacia Biotech ECL Western blotting reagents per manufacturer’s protocols.
Intracellular FPP and GGPP Measurements
Intracellular FPP and GGPP levels were measured using the previously reported reversed phase HPLC methodology [38]. Briefly, following incubation with drugs, cells were collected and counted using a hemocytometer. Isoprenoid pyrophosphates were extracted from cell pellets with extraction solvent (butanol/75 mM ammonium hydroxide/ethanol 1:1.25:2.75). Following drying down by nitrogen gas, the FPP and GGPP in the residue were incorporated into fluorescent GCVLS or GCVLL peptides by farnesyl-transferase or geranylgeranyl transferase I, respectively. The prenylated fluorescent peptides were separated and quantified by reversed phase HPLC with fluorescence detection.
HMGR Activity Assay
The HMG-CoA reductase assay kit was purchased from Sigma (St. Louis, MO) and the assay was performed according to manufacturer’s specifications. This assay detects the oxidation of NADPH by HMGR in the presence of HMG-CoA. The decrease in absorbance at 340 nm over time was measured using a Molecular Devices SpectraMax M2e microplate reader. Measurements were taken every 25 s. Pravastatin, which was included in the kit, was used as the positive control. The reaction mixture was incubated in the presence or absence of varying concentrations of 3dSB.
Real-Time PCR
RPMI-8226 or U266 cells were grown in the presence or absence of drugs in media containing either standard FCS or 10% LPDS for 24 h. Each condition was performed in triplicate. RNA was isolated using an RNeasy kit (Qiagen, Valencia, CA) and a BioRad (Hercules, CA) cDNA synthesis kit was used to prepare cDNA. Primers for HMGR, FDPS, SS, GGDPS, SREBP-2, Insig-1, LXRα, ABCA1 and β-actin (Supplementary Table 1) were designed using PrimerQuest. Real-time PCR was performed on an Applied Biosystems Model 7900HT using an Applied Biosystems (Carlsbad, CA) reaction kit with SYBR green. Data were analyzed using ABI SDS 2.3 software and normalized to β-actin RNA. Quantities were determined using the relative standard curve method. Each sample was run in triplicate.
Statistical Analysis
Two-tailed t testing was used to calculate statistical significance for the studies involving measurement of FPP and GGPP levels. An α of 0.05 was set as the level of significance. Isobologram analysis was used to evaluate the data from the MTT assays via CalcuSyn software (Biosoft). The software analyzes drug interactions based on the method of Chou and Talalay [26]. Combination indices (CI) ≥ 0.8 were deemed synergistic, CI > 1.2 antagonistic, and CI > 0.8 but less than 1.2 were deemed additive. ANOVA was used to compare changes in gene expression level following treatment with two drugs (lovastatin alone, 3dSB alone, and the two in combination) under four sets of conditions (two cell lines, two media conditions). The gene expression levels were compared separately for each of the four conditions. Expression levels were log-transformed to satisfy the ANOVA assumptions of normality and equal variances. Differences in expression levels were evaluated with the two-sided pair-wise comparisons of the treatment groups. The False Discovery Rate approach using q values was applied to adjust for multiple comparisons [39, 40] (Supplementary Table 2). Pair-wise comparisons for which the q values were less than 0.05 were declared to be statistically significant. This criterion ensures that the expected proportion of incorrectly rejected pair-wise comparisons is no greater than 5%. SAS version 9.2 and R 2.11.1 were used for the analysis.
Results
The Combination of 3dSB and Lovastatin Induces Synergistic Cytotoxic Effects in Human Malignant Cell Lines
The cytotoxic effects of 3dSB, both alone and in combination with the HMGR inhibitor lovastatin, in MTT assays were assessed in two human multiple myeloma cell lines (RPMI-8226 and U266), a human glioblastoma multiforme cell line (SF-295) and a human non-small cell lung cancer cell line (A549) (Fig. 2a). The RPMI-8226 and SF-295 cells have been previously noted to be particularly sensitive to schweinfurthins with mean GI50’s for schweinfurthin B of 15 and 24 nM, respectively [1]. The A549 cells, on the other hand, represent a relatively more resistant cell line, with a mean GI50 of 0.34 μM [1]. The U266 cell line was not part of the NCI 60-cell line panel, but was chosen for these studies based on our prior work which demonstrated resistance to select IBP inhibitors as a consequence of markedly elevated basal isoprenoid levels [41]. Consistent with the NCI findings for schweinfurthin B, the RPMI-8226 cells were noted to be the most sensitive to 3dSB with an IC50 of ~150 nM and the A549 cells the least sensitive with an IC50 of 5 μM at 48 h. The SF-295 and U266 cells displayed intermediate sensitivity to 3dSB alone with IC50 values of 0.5 and 1 μM, respectively. Interestingly, the addition of lovastatin to 3dSB resulted in enhanced cytotoxicity in all four cells. The nature of this interaction was evaluated by isobologram analysis and determined to be synergistic (Table 1). Furthermore, when RPMI-8226 or U266 cells were incubated in media containing lipoprotein deficient serum (LPDS), the cytotoxicity of 3dSB, both alone (3dSB IC50 of 80 nM for RPMI-8226 and 0.3 μM for U266) and in combination with lovastatin, was increased (Fig. 2b). This suggested a dependence on sterol availability for 3dSB cytotoxicity.
Fig. 2.
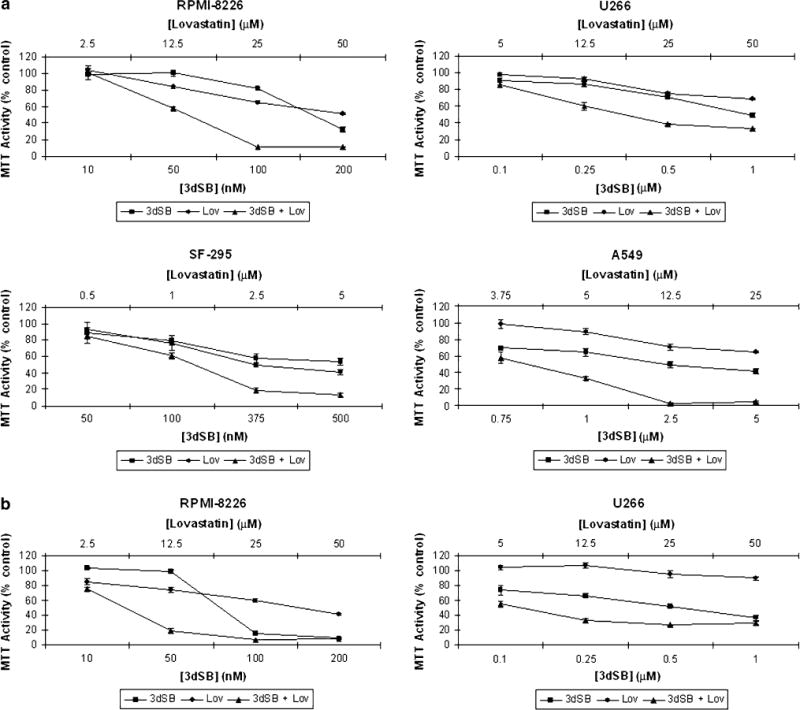
The combination of lovastatin and 3dSB induces a synergistic cytotoxic effect in malignant cell lines. Cells were incubated for 48 h in the presence or absence of drugs and the MTT assay was performed as described in the “Materials and methods”. a RPMI-8226, U266, SF-295, and A549 cells were incubated in standard FCS. b RPMI-8226 and U266 cells were incubated in LPDS. Data are expressed as a percentage of control (mean ± SD, n = 4) and are representative of at least two independent experiments
Table 1.
Summary of 3dSB-IBP inhibitor interactions in MTT cytotoxicity assays
| Combination | Cell line |
|||
|---|---|---|---|---|
| RPMI-8226 | U266 | SF-295 | A549 | |
| 3dSB + Lov | Synergistica | Synergistica | Synergistica | Synergistica |
| 3dSB + ZA | Additiveb | Antagonisticd | Antagonistic/Additivef | ND |
| 3dSB + DGBP | Antagonisticc | Antagonisticd | Additiveb | ND |
| 3dSB + Zara | Antagonistice | Antagonistice | ND | ND |
Cells were incubated in standard FCS for 48 h in the presence of 3dSB and/or IBP inhibitors (Lov lovastatin, ZA zoledronic acid, DGBP digeranyl bisphosphonate, Zara zaragozic acid) prior to the addition of MTT salt. Combination indices (CI) for ED30 and ED50 (unless otherwise specified) were determined via isobologram analysis
ND Not determined
CI < 0.8 as determined by isobologram analysis
CI 0.8–1.2 as determined by isobologram analysis
CI > 1.2 as determined by isobologram analysis
MTT Activity of combination > 3dSB alone as determined by two-tailed t testing
CI’s for ED20 and ED30 calculated
CI antagonistic for ED30 and additive for ED50
To determine whether the synergistic interaction was unique to lovastatin, or was observed with other IBP inhibitors, additional MTT assays were performed in which cells were treated with 3dSB in the presence or absence of the FDPS inhibitor zoledronic acid (ZA), the GGDPS inhibitor digeranyl bisphosphonate (DGBP), or the squalene synthase inhibitor zaragozic acid (Zara). As shown in Table 1, none of these agents displayed a synergistic interaction with 3dSB. In particular, for the RPMI-8226 cells, both DGBP and ZA displayed an antagonistic interaction with 3dSB.
3dSB Enhances Lovastatin-Induced Decrease in Protein Prenylation
The pleiotropic effects of statins in malignant cells have generally been attributed to their ability to inhibit protein prenylation. To determine whether 3dSB, either alone or in combination with lovastatin, altered protein prenylation, immunoblot analysis was performed. Antibodies directed against Ras (substrate of FTase), Rap1a (substrate of GGTase I) and Rab6 (substrate of GGTase II) were utilized. For Ras and Rab6, the appearance of a more slowly migrating band represents unmodified protein, while for Rap1a, the antibody detects only unmodified protein. As shown in Fig. 3a, 3dSB alone has no effect on the prenylation status of the three proteins. As expected, lovastatin decreases both farnesylation and geranylgeranylation. Interestingly, 3dSB was noted to potentiate the effects of lovastatin in the RPMI-8226 cells as evidenced by the increase in the ratio of unmodified to modified protein for Ras and Rab6 and the increase in total unmodified protein for Rap1a. This potentiation occurred in a concentration-dependent manner and was observed for all three prenylated proteins. This effect was particularly evident in cells incubated with LPDS; disruption of protein prenylation was not detected in cells treated with 2.5 μM lovastatin alone, but was detected when cells were co-incubated with ≥ 50 nM 3dSB. Minimal potentiation was noted when cells were incubated with 3dSB and either zoledronic acid or DGBP (Fig. 3a). 3dSB was also found to potentiate lovastatin’s effects in U266 cells, particularly in cells treated with 2.5 μM lovastatin in either FCS or LPDS conditions (Fig. 3b). The effect of 10 μM lovastatin on disruption of protein prenylation in cells incubated with standard FCS was near maximal and the addition of 3dSB did not significantly alter the prenylation pattern.
Fig. 3.
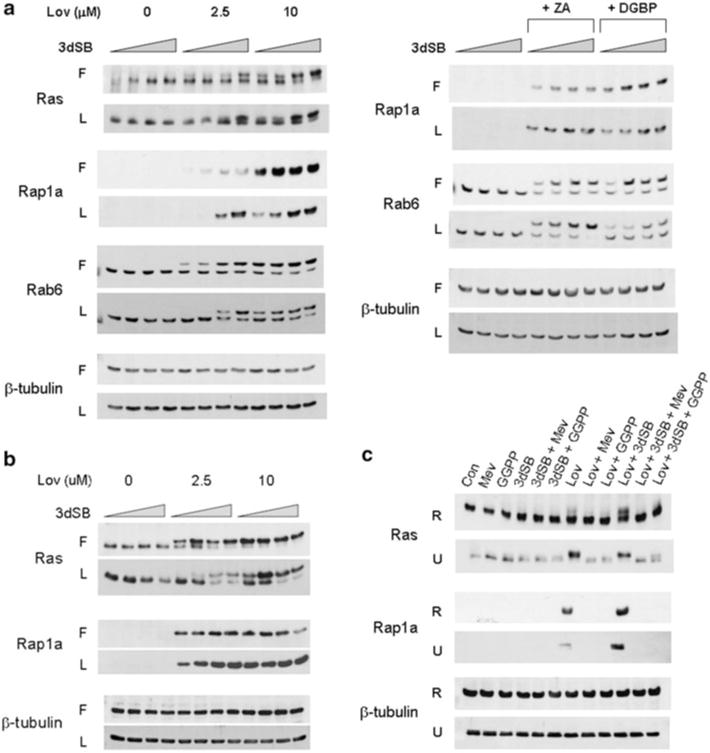
3dSB potentiates lovastatin-induced decrease in protein prenylation. a RPMI-8226 or b U266 cells were incubated in the presence or absence of 3dSB ± IBP inhibitor (Lov lovastatin, ZA zoledronic acid (50 μM) or DGBP digeranyl bisphosphonate (2.5 μM)) in either standard FCS (F) or LPDS (L) for 24 h. The wedge indicates increasing concentrations of 3dSB (0, 10, 50, 100 nM for RPMI-8226 cells and 0, 0.1, 0.5, 1 μM for U266 cells). c Lovastatin- or lovastatin + 3dSB-induced decrease in protein prenylation is prevented by co-incubation with select isoprenoid intermediates. RPMI-8226 (R) and U266 (U) cells were incubated for 24 h in media containing standard FCS in the presence or absence of lovastatin (10 μM), 3dSB (50 nM for RPMI-8226 and 1 μM for U266), mevalonate (1 mM), or GGPP (10 μM). Western blot analysis of Ras (more slowly migrating band represents unmodified Ras), Rap1a (antibody detects only unmodified Rap1a), Rab6 (more slowly migrating band represents unmodified Rab6) and β-tubulin (as a loading control) was performed. Data are representative of at least two independent experiments
To determine whether the induced changes in protein prenylation could be prevented by exogenous isoprenoid species, studies were performed in which RPMI-8226 or U266 cells incubated inmedia containing standard FCS were treated with 3dSB and/or lovastatin in the presence or absence of mevalonate or GGPP. As shown in Fig. 3c, addition of mevalonate or GGPP completely prevented the lovastatin- or lovastatin +3dSB-induced decrease in Rap1a geranylgeranylation. Mevalonate completely prevented the disruption of Ras prenylation while GGPP only partially restored Ras prenylation, consistent with the ability of normally farnesylated N- and K-Ras isoforms to undergo geranylgeranylation when farnesylation is disrupted [42].
Effects of 3dSB on Intracellular FPP and GGPP Levels
As 3dSB alone did not appear to inhibit protein prenyla-tion, but did potentiate lovastatin’s effects, it was hypothesized that 3dSB could be altering isoprenoid availability. Therefore, the effects of 3dSB on intracellular FPP and GGPP levels were determined. Lovastatin, which depletes cells of both FPP and GGPP, was used as a positive control. As shown in Fig. 4a, 3dSB decreased both FPP and GGPP levels in a concentration-dependent manner in all tested cell lines, with the exception of the RPMI-8226 cells. In addition, 3dSB also variably decreased FPP and GGPP levels in primary human acute myeloid leukemia cells which were incubated with the drug in an ex vivo manner (Fig. 5).
Fig. 4.
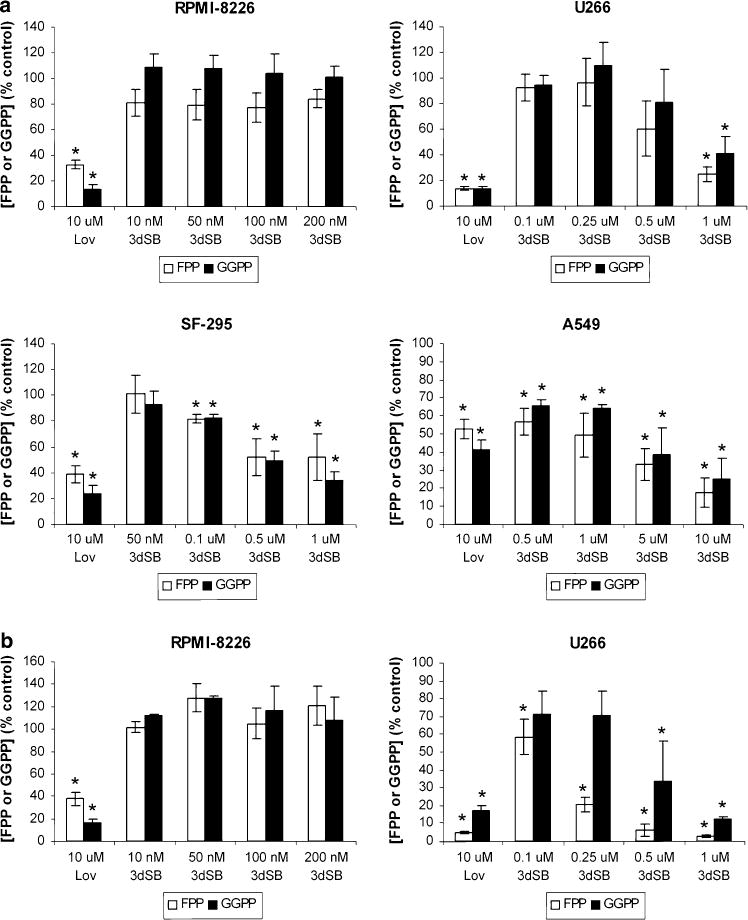
Effects of 3dSB on intracellular FPP and GGPP levels. Cells were incubated in the presence of 3dSB or lovastatin (Lov) for 24 h in media containing either standard FCS (a) or LPDS (b). Intracellular FPP and GGPP levels were determined as described in the “Materials and methods” and are expressed as a percent of control (mean ± SD of three independent experiments). * Denotes p < 0.05 per unpaired two-tailed t test and compares the treated cells to the control untreated cells
Fig. 5.
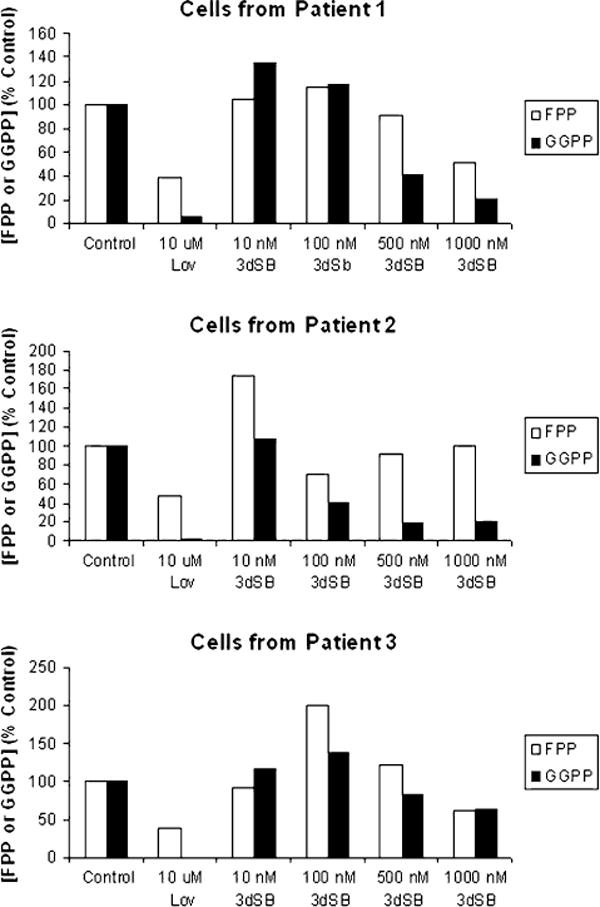
Effect of 3dSB on FPP and GGPP levels in primary acute leukemia cells. Mononuclear cells were isolated from the peripheral blood of patients with newly diagnosed acute leukemia and incubated ex vivo with 3dSB or lovastatin for 24 h. All patients had greater than 40,000 K/mm3 circulating blasts. Formal pathological review of the patient bone marrow samples revealed acute myelomonocytic leukemia for patient 1, acute myeloid leukemia with t(11;17) translocation for patient 2, and acute myeloid leukemia with aberrant CD7 expression for patient 3
The effects of 3dSB on FPP and GGPP levels were also evaluated in RPMI-8226 and U266 cells incubated with media containing LPDS. As shown in Fig. 4b, little effect was seen in the RPMI-8226 cells, but an enhanced depletion of FPP and GGPP levels was observed in the U266 cell line. As described previously [41], basal FPP levels are significantly higher in the U266 cell line compared with the RPMI-8226 line. Of note, incubation in LPDS-containing media significantly increased basal FPP (8.3-fold), but not GGPP levels in the U266 cells (Table 2), while basal levels of FPP and GGPP did not significantly change in RPMI-8226 cells. The LPDS-induced increase in FPP observed in the U266 cells could be overcome by either lovastatin or 3dSB (Fig. 4b). Incubation of SF-295 and A549 cells in LPDS resulted in an approximately 1.5-fold increase in FPP and GGPP levels compared with cells incubated in standard FCS (Table 2).
Table 2.
Effects of serum composition on basal FPP and GGPP levels
| FPP (pmol/106 cells) (Mean ± SD) |
GGPP (pmol/106 cells) (Mean ± SD) |
|
|---|---|---|
| RPMI-8226, FCS | 0.168 ± 0.030 | 0.273 ± 0.022 |
| RPMI-8226, LPDS | 0.122 ± 0.033 | 0.235 ± 0.096 |
| U266, FCS | 1.87 ± 0.271 | 0.682 ± 0.193 |
| U266, LPDS | 15.5 ± 1.25a | 1.115 ± 0.223 |
| SF-295, FCS | 0.088 ± 0.002 | 0.367 ± 0.023 |
| SF-295, LPDS | 0.155 ± 0.005a | 0.534 ± 0.032a |
| A549, FCS | 0.146 ± 0.003 | 0.291 ± 0.031 |
| A549, LPDS | 0.242 ± 0.026a | 0.460 ± 0.048a |
Cells were incubated for 24 h in media containing either standard FCS or LPDS. FPP and GGPP levels were measured as described in the section “Materials and methods”. The means and standard deviations of the results of three independent experiments are shown
p < 0.05 by two-tailed t testing comparing FCS to LPDS levels
To investigate the effect of combining 3dSB on IBP inhibitor-induced changes in FPP and GGPP levels, studies were performed in which U266 cells were treated with 3dSB and/or lovastatin, ZA, or DGBP. Consistent with our prior studies, treatment with ZA or DGBP only partially depletes FPP or GGPP levels, respectively, in U266 cells incubated in standard FCS [43]. Interestingly, however, the addition of 3dSB to either ZA or DGBP results in a significant decrease in both FPP and GGPP levels. Similar results were observed when cells were incubated in media containing LPDS (Fig. 6). The lovastatin-induced depletion of FPP and GGPP was near maximal in U266 cells incubated with standard FCS and the addition of 3dSB did not significantly alter FPP or GGPP levels. In contrast, when cells were incubated with LPDS, the addition of 3dSB to lovastatin resulted in a further decrease in GGPP levels. In aggregate these studies demonstrate the ability of 3dSB to lower intracellular FPP and GGPP levels irrespective of the presence of agents which disrupt the IBP. Finally, the ability of 3dSB to directly inhibit HMGR was evaluated in an in vitro enzyme assay and inhibitory activity was not observed (Fig. 7).
Fig. 6.
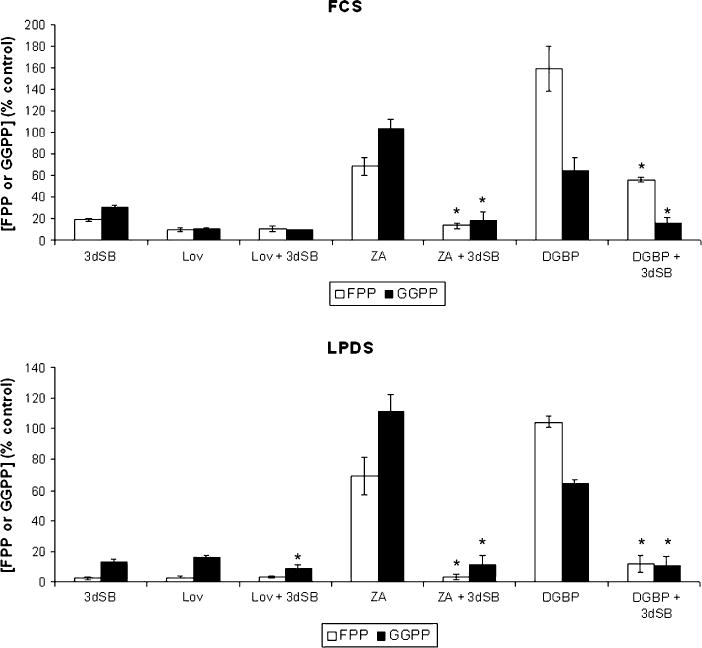
Effects of 3dSB on IBP inhibitor-induced changes in FPP and GGPP levels in U266 cells. Cells were incubated in the presence or absence of 1 μM 3dSB and/or 10 μM lovastatin, 50 μM ZA, or 10 μM DGBP for 24 h in media containing either standard FCS or LPDS. Intracellular FPP and GGPP levels were determined as described in the section “Materials and methods” and are expressed as a percentage of the control (mean ± SD of three independent experiments). The * denotes p < 0.05 per unpaired two-tailed t test and compares the IBP inhibitor + 3dSB combination to the IBP inhibitor alone
Fig. 7.
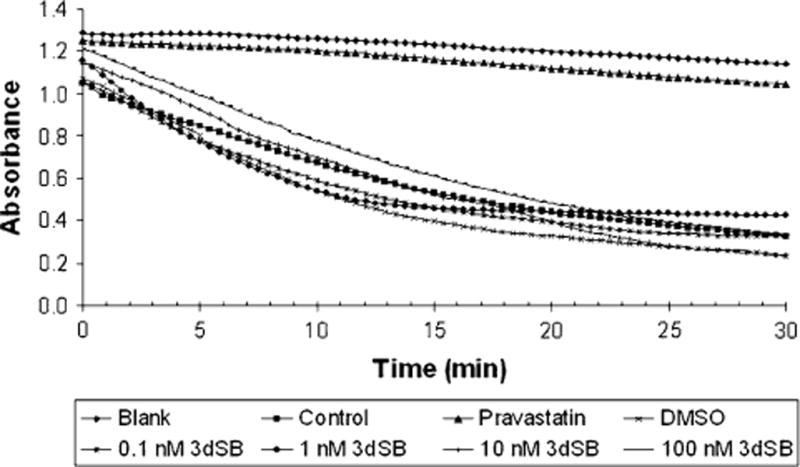
3dSB does not inhibit HMGR in an in vitro enzyme assay. The enzyme assay was performed as described in the “Materials and methods” section. Blank represents reaction mixture lacking HMGR while Control represents the complete reaction mixture. Pravastatin was used as a positive control
The Effects of Exogenous Isoprenoids on 3dSB-Induced Cytotoxicity and FPP and GGPP Levels
Lovastatin-induced cytotoxicity has previously been shown to be prevented by co-incubation with select isoprenoid species [43]. To determine whether 3dSB-induced cytotoxicity could be similarly prevented, MTT experiments were performed in which RPMI-8226 and U266 cells were treated with 3dSB and/or lovastatin in the presence or absence of select isoprenoids (mevalonate, FPP, or GGPP). As shown in Fig. 8a, mevalonate or GGPP, and to a lesser extent FPP, could prevent lovastatin-induced cytotoxicity. However, none of the isoprenoid species prevented 3dSB-induced cytotoxicity. The enhanced cytotoxicity observed with the combination of 3dSB and lovastatin was only partially reversed by co-incubation with the isoprenoids. Co-incubation with either mevalonate or GGPP induced a level of cytotoxicity equivalent to 3dSB alone in the drug-combination treatment. To determine whether the add-back of isoprenoid species was effective in repleting FPP and GGPP levels in 3dSB and/or lovastatin-treated cells, levels of FPP and GGPP were measured in U266 cells. As shown in Fig. 8b, mevalonate most efficiently restored both FPP and GGPP levels in drug-treated cells. Incubation with exogenous FPP only partially restored FPP levels in drug-treated cells and had no effect on GGPP levels. Incubation with GGPP completely restored GGPP levels in either 3dSB- or lovastatin-treated cells and partially restored levels in 3dSB + lovastatin-treated cells. Thus, although incubation with exogenous isoprenoids (mevalonate/GGPP > FPP) restores isoprenoid levels in 3dSB-treated cells, this agent’s cytotoxic effects are not significantly altered. In aggregate these data suggest that the mechanism underlying 3dSB’s cytotoxic effects extends beyond this agent’s ability to alter FPP and GGPP levels.
Fig. 8.
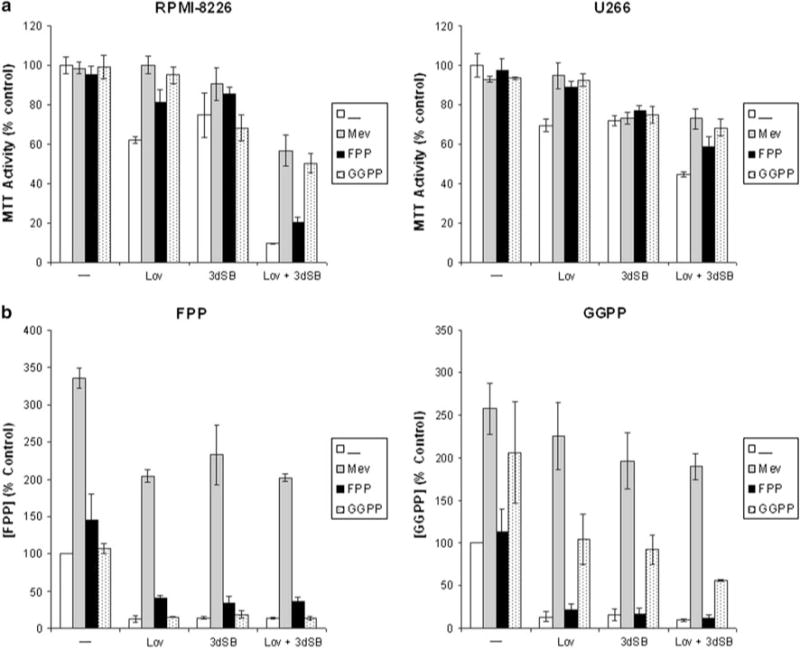
a Add-back of select IBP intermediates prevents lovastatin-, but not 3dSB-induced cytotoxicity in RPMI-8226 and U266 cells. Cells were incubated for 48 h in the presence or absence of 25 μM lovastatin (Lov), 3dSB (0.1 μM for RPMI-8226 and 0.5 μM for U266), 1 mM mevalonate (Mev), 10 μM FPP or 10 μM GGPP. MTT assays were performed as described in the “Materials and methods”. Data are expressed as a percent of control (mean ± SD, n = 4) and are representative of two independent experiments. b Effects of exogenous IBP intermediates on intracellular FPP and GGPP levels in U266 cells treated with 1 μM 3dSB and/or lovastatin for 24 h. Data are expressed as a percent of control (mean ± SD, n = 3 independent experiments)
3dSB Disrupts Regulation of Isoprenoid Homeostasis
While lovastatin disrupts sterol homeostasis by targeting HMGR and inhibiting isoprenoid biosynthesis, it appeared that 3dSB might be more indirectly disrupting the IBP. To address this hypothesis, real-time PCR was used to assess the effects of 3dSB, either alone or in combination with lovastatin, on the expression of key components of the IBP (HMGR, FDPS, SS, GGDPS) and regulators of sterol homeostasis (SREBP-2, Insig-1, LXRα, ABCA1). Treatment with lovastatin resulted in an increase in expression (1.5- to 4-fold) of the SRE-containing genes HMGR, FDPS, SS, SREBP-2, and Insig-1 in the RPMI-8226 and U266 cells under standard serum conditions while decreasing ABCA1 expression (Fig. 9). With the exception of HMGR (increase) and ABCA1 (decrease), 3dSB had little effect on any of the tested genes in the RPMI-8226 cell line, while in the U266 cells, statistically significant increases in HMGR, SS, SREBP-2, and Insig-1 levels were observed. Interestingly, 3dSB abrogated lovastatin-induced upregulation of HMGR, FDPS, SS, and Insig-1 in the RPMI-8226 cell line while potentiating the upregulation of GGDPS. In contrast, in the U266 cell line, 3dSB minimally altered lovastatin-induced upregulation of HMGR, FDPS, SS, or SREBP-2 but enhanced the upregulation of GGDPS, LXR, and ABCA1 compared with either drug alone.
Fig. 9.
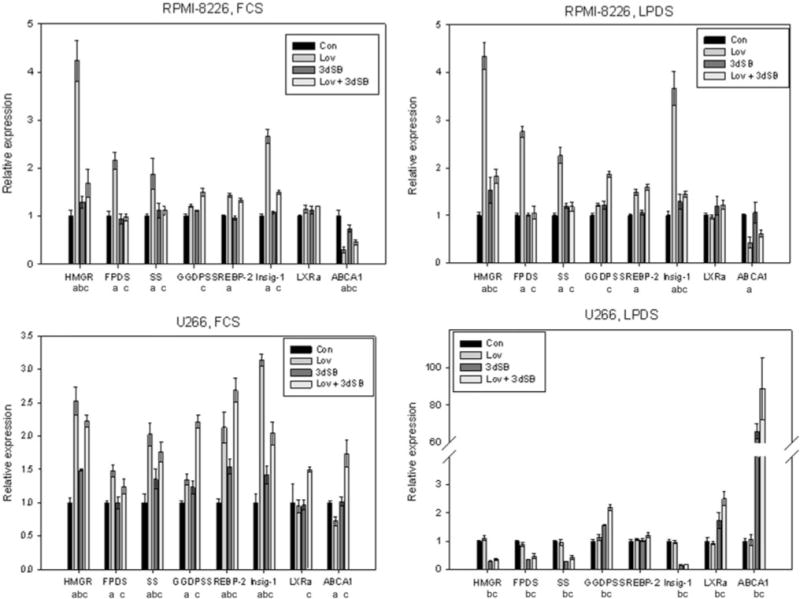
3dSB induces alteration in expression of genes involved with isoprenoid homeostasis. RPMI-8226 or U266 cells were incubated for 24 h in the presence or absence of lovastatin (Lov, 10 μM) and/or 3dSB (0.1 μM for RPMI-8226 and 0.5 μM for U266) in either standard FCS or LPDS. Real-time PCR was performed using primers for HMG-CoA reductase (HMGR), farnesyl diphosphate synthase (FDPS), squalene synthase (SS), geranylgeranyl diphosphate synthase (GGDPS), SREBP-2, Insig-1, ABCA1 and LXRα. Data were normalized to β-actin levels and are expressed as relative to control untreated cells (mean ± SE, n = 3). Data are representative of two independent experiments. Statistical significance, as described in the “Materials and methods” (q value < 0.05), is represented by “a” (lovastatin compared to control), “b” (3dSB compared to control), and “c” (3dSB + lovastatin compared to lovastatin)
These studies were also performed with cells that had been incubated with LPDS (Fig. 9). Overall, there was little difference in effect on gene expression between the FCS- and LPDS-treated RPMI-8226 cells: lovastatin induced an increase in HMGR, FDPS, SS, SREBP-2, and Insig-1, had little effect on LXRα, and decreased ABCA1 expression. 3dSB induced minimal effects on HMGR and Insig-1 levels. Likewise, as seen in the FCS-treated cells, the combination of lovastatin and 3dSB resulted in suppression of the lovastatin-induced increase in the SRE-dependent genes and an increase in GGDPS expression. While the effects of the drugson the expression profile were concordant between FCS and LPDS conditions in the RPMI-8226 cells, there were profound differences noted between FCS and LPDS conditions in the U266 cell line. Notably, lovastatin failed to induce an increase in any of the tested SRE-containing genes. In addition, with the exception of SREBP-2, 3dSB markedly decreased the expression of the SRE-containing genes. Most striking was the greater than 60-fold increase in ABCA1 expression induced by 3dSB in the LPDS-treated U266 cells; an effect which was further enhanced by co-incubation with lovastatin.
Discussion
The basis for the schweinfurthins’ potent cellular activities has thus far not been determined. In the studies presented herein we demonstrate that the synthetic schweinfurthin 3dSB has a very complex relationship with the IBP. In particular, 3dSB: (1) exerts a synergistic cytotoxic effect with the HMGR inhibitor lovastatin but additive or antagonistic effects with other IBP inhibitors, (2) enhances lovastatin-induced disruption of prenylation, (3) induces changes in intracellular FPP and GGPP levels, and (4) disrupts both SRE- and LXR-mediated regulation of genes involved in the IBP and sterol homeostasis. That these effects are enhanced under conditions of limited sterol availability, provides further evidence for the link between 3dSB and isoprenoid homeostasis.
The synergistic interaction between 3dSB and lovastatin is a consequence of lovastatin’s ability to deplete cells of one or more down-stream isoprenoids such as FPP, GGPP, or sterols. Notably, an antagonistic interaction was observed between 3dSB and agents that increase FPP levels but have disparate effects on GGPP and sterol synthesis (the GGDPS inhibitor DGBP and the squalene synthase inhibitor Zara), suggesting that the lovastatin/3dSB interaction is dependent on depletion of both nonsterols and sterols down-stream of FPP. Furthermore, 3dSB-induced cytotoxicity is enhanced under conditions of sterol depletion, providing further evidence for a connection between 3dSB activity and isoprenoid levels. In fact, 3dSB was found to decrease cellular FPP/GGPP levels in a variety of cultured and primary cells (Figs. 4, 5, 6). It is interesting to note that although 3dSB decreased FPP and GGPP to levels equivalent to lovastatin in the U266 cell line (Fig. 4), 3dSB alone did not decrease protein prenylation (Fig. 3). In addition, while 3dSB did not significantly alter FPP and GGPP levels in the RPMI-8226 cells, this agent did enhance lovastatin-induced decrease of protein prenylation (Fig. 3). These results, in conjunction with the finding that 3dSB-mediated enhancement of lovastatin-induced decrease of protein prenylation is further augmented in the presence of LPDS, suggests that these effects are not only due to changes in steady state levels of isoprenoids, but may also reflect alterations in flux through the pathway. The FPP/GGPP assay that was employed measures steady state total cellular FPP/GGPP levels. It remains to be determined whether shunting of isoprenoids to various branches of the IBP is altered in the presence of 3dSB. In addition, it is unknown whether there are discrete pools of these isoprenoid intermediates within the cell, and if so, whether 3dSB alters specific pools, thereby affecting substrate availability for disparate processes.
Further understanding of the mechanism(s) for 3dSB activity may be gained from the marked increase in FPP levels in U266 cells, but not in RPMI-8226 cells in response to incubation with LPDS. We have previously demonstrated that the U266 cell line is less sensitive than the RPMI-8226 cells to lovastatin and is resistant to downstream IBP inhibitors such as ZA and DGBP, likely a consequence of the markedly elevated basal FPP levels [43]. It has been noted that statin sensitivity in myeloma cell lines may be a consequence of altered HMGR regulation [44]. However, our work, which demonstrates a loss of lovastatin-induced upregulation of SRE-regulated genes in U266 cells incubated in LPDS-containing media suggests that statin-sensitivity is dependent on factors that extend beyond the level of HMGR regulation. In a similar manner, 3dSB-sensitivity may be a consequence of variation in isoprenoid pathway regulation.
In addition to revealing novel activities of 3dSB, these studies also provide insight into the regulation of isoprenoid levels. In our studies, GGDPS expression was minimally induced by either lovastatin or 3dSB alone, but was upregulated to a greater extent when the two agents were used in combination (Fig. 9). This effect was particularly evident in the LPDS-treated cells. Analysis of FDPS has demonstrated that it is transcriptionally regulated by both SREBP and LXR [37, 45–47]. Less is known about the regulation of GGDPS. While one report in a transgenic system suggested that GGDPS is a target of SREBP, another demonstrated that GGDPS mRNA levels did not significantly change in HeLa cells incubated with either an HMGR inhibitor or with LPDS in the presence or absence of sterols [48, 49]. The product of this enzyme, GGPP, has been shown to serve as a negative regulator for LXR [21, 50]. Our results suggest that GGPP levels may in part regulate expression of GGDPS. Whether this is due to an effect on LXR activity or is via another mechanism remains to be determined.
The add-back experiments, which demonstrated that exogenous FPP/GGPP can restore isoprenoid levels in 3dSB-treated cells, but does not reverse the cytotoxic effects (Fig. 8), further suggest that 3dSB’s effects extend beyond altering FPP and GGPP levels. In fact, we found that in RPMI-8226 cells, 3dSB abrogates statin-induced upregulation of SREBP-2-dependent genes as well as statin-induced downregulation of ABCA1 in both sterol replete (FCS) and deplete (LPDS) conditions (Fig. 9). In aggregate these data suggest that 3dSB shifts the cell’s sterol regulatory pathways from a sterol-deficient state to a sterol-replete state. This hypothesis is further supported by the findings from the U266-LPDS studies. As noted in Table 2, U266 cells, in response to incubation with LPDS, markedly increase intracellular FPP levels, presumably in an effort to compensate for the exogenous depletion of sterols. In this setting, 3dSB alone significantly downregulates SREBP-2-dependent genes while markedly upregulating ABCA1 (a LXRα-dependent gene). This pattern implies sterol excess. Possible mechanisms by which 3dSB could induce these changes include interference by 3dSB of the SRE-SREBP-2 interaction, disruption of SREBP-2 trafficking, direct activation of LXR, RXR or PPAR, or interference with the function of oxysterol binding proteins (OSBPs). A variety of oxysterols have been shown to be ligands for LXR and to induce LXR-target gene expression [51]. For example, the dietary plant sterol, stigmasterol, has been shown to induce ABCA1 expression and to suppress the SREBP pathway by promoting the formation of a Scap-SREBP-Insig-1 complex in the ER, thereby decreasing expression of SREBP target genes [16]. Whether 3dSB shares a similar mechanism of action, changes sterol levels, or alters the sterol-sensing machinery is of considerable interest and studies addressing these hypotheses are underway.
Schweinfurthin A has recently been shown to induce reorganization of the actin cytoskeleton and to inhibit EGF-induced Rho activation in serum-starved astrocytoma cells [52]. It is interesting to note that a variety of interactions have been described between OSBPs and elements of the cytoskeleton. For example, overexpression of ORP4S induced collapse of the vimentin network and inhibited LDL-cholesterol esterification while silencing of ORP3 resulted in reorganization of the actin cytoskeleton [53, 54]. It remains to be determined whether schweinfurthins, either directly or indirectly, interact with OSBPs, thereby impairing sterol sensing/trafficking and disrupting the cytoskeleton.
In conclusion, these studies are the first to demonstrate that the synthetic schweinfurthin 3dSB exerts pleiotropic effects on isoprenoid homeostasis. Specifically, 3dSB has a synergistic interaction with lovastatin, can alter steady state isoprenoid levels, and can disrupt multiple aspects of the regulatory elements controlling the IBP and sterol homeostasis. These studies, which advance understanding the basis for schweinfurthin activities, also indicate that there is a complexity to regulation of the isoprenoid pathway that is as yet unknown but may depend upon isoprenoid flux as compared to absolute isoprenoid levels.
Supplementary Material
Acknowledgments
This project was supported by the Roy J. Carver Charitable Trust as a Research Program of Excellence and the Roland W. Holden Family Program for Experimental Cancer Therapeutics. S.A. Holstein was supported through a NIH T32 training [T32 HL07344] and a PhRMA Foundation Career Development Award. Brian Smith, PhD and Anna Button, MS from the University of Iowa Holden Comprehensive Cancer Center Biostatistics Core provided support with statistical analysis.
Abbreviations
- 3dSB
3-Deoxyschweinfurthin B
- ABCA1
ATP-binding cassette transporter 1
- DGBP
Digeranyl bisphosphonate
- ER
Endoplasmic reticulum
- FCS
Fetal calf serum
- FDPS
Farnesyl diphosphate synthase
- FPP
Farnesyl pyrophosphate
- FTase
Farnesyl transferase
- GGDPS
Geranylgeranyl diphosphate synthase
- GGPP
Geranylgeranyl pyrophosphate
- GGTase
Geranylgeranyl transferase
- GI50
50% growth inhibition
- HMGR
3-Hydroxy-3-methyl-glutaryl-CoA reductase
- IBP
Isoprenoid biosynthetic pathway
- LPDS
Lipoprotein deficient serum
- LXR
Liver X receptor
- MTT
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
- SRE
Sterol regulatory element
- SREBP
Sterol regulatory element-binding protein
- SS
Squalene synthase
- ZA
Zoledronic acid
- Zara
Zaragozic acid
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11745-011-3572-y) contains supplementary material, which is available to authorized users.
Conflict of interest Nothing to disclose.
Contributor Information
Sarah A. Holstein, Department of Internal Medicine, University of Iowa, Iowa City, IA 52242, USA
Craig H. Kuder, Department of Internal Medicine, University of Iowa, Iowa City, IA 52242, USA
Huaxiang Tong, Department of Internal Medicine, University of Iowa, Iowa City, IA 52242, USA.
Raymond J. Hohl, Department of Internal Medicine, University of Iowa, Iowa City, IA 52242, USA, raymond-hohl@uiowa.edu Department of Pharmacology, University of Iowa, 200 Hawkins Dr, SE 313 GH, Iowa City, IA 52242, USA.
References
- 1.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 2.Ulrich NC, Kodet JG, Mente NR, Kuder CH, Beutler JA, Hohl RJ, Wiemer DF. Structural analogues of schweinfurthin F: probing the steric, electronic, and hydrophobic properties of the D-ring substructure. Bioorg Med Chem. 2010;18:1676–1683. doi: 10.1016/j.bmc.2009.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuder CH, Neighbors JD, Hohl RJ, Wiemer DF. Synthesis and biological activity of a fluorescent schweinfurthin analogue. Bioorg Med Chem. 2009;17:4718–4723. doi: 10.1016/j.bmc.2009.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mente NR, Wiemer AJ, Neighbors JD, Beutler JA, Hohl RJ, Wiemer DF. Total synthesis of (R, R, R)- and (S, S, S)-schweinfurthin F: differences of bioactivity in the enantiomeric series. Bioorg Med Chem Lett. 2007;17:911–915. doi: 10.1016/j.bmcl.2006.11.096. [DOI] [PubMed] [Google Scholar]
- 5.Neighbors JD, Salnikova MS, Beutler JA, Wiemer DF. Synthesis and structure-activity studies of schweinfurthin B analogs: Evidence for the importance of a D-ring hydrogen bond donor in expression of differential cytotoxicity. Bioorg Med Chem. 2006;14:1771–1784. doi: 10.1016/j.bmc.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 6.Neighbors JD, Beutler JA, Wiemer DF. Synthesis of nonracemic 3-deoxyschweinfurthin B. J Org Chem. 2005;70:925–931. doi: 10.1021/jo048444r. [DOI] [PubMed] [Google Scholar]
- 7.Topczewski JJ, Kuder CH, Neighbors JD, Hohl RJ, Wiemer DF. Fluorescent schweinfurthin B and F analogs with anti-proliferative activity. Bioorg Med Chem. 2010;18:6734–6741. doi: 10.1016/j.bmc.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamadmad SN, Henry MK, Hohl RJ. Erythropoietin receptor signal transduction requires protein geranylgeranylation. J Pharmacol Exp Ther. 2006;316:403–409. doi: 10.1124/jpet.105.092510. [DOI] [PubMed] [Google Scholar]
- 9.Holstein SA, Hohl RJ. Isoprenoids: remarkable diversity of form and function. Lipids. 2004;39:293–309. doi: 10.1007/s11745-004-1233-3. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 11.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 13.Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 14.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci USA. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costet P, Luo Y, Wang N, Tall AR. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J Biol Chem. 2000;275:28240–28245. doi: 10.1074/jbc.M003337200. [DOI] [PubMed] [Google Scholar]
- 16.Yang C, McDonald JG, Patel A, Zhang Y, Umetani M, Xu F, Westover EJ, Covey DF, Mangelsdorf DJ, Cohen JC, Hobbs HH. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J Biol Chem. 2006;281:27816–27826. doi: 10.1074/jbc.M603781200. [DOI] [PubMed] [Google Scholar]
- 17.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci USA. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Correll CC, Ng L, Edwards PA. Identification of farnesol as the non-sterol derivative of mevalonic acid required for the accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 1994;269:17390–17393. [PubMed] [Google Scholar]
- 20.Song BL, Javitt NB, DeBose-Boyd RA. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 2005;1:179–189. doi: 10.1016/j.cmet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Gan X, Kaplan R, Menke JG, MacNaul K, Chen Y, Sparrow CP, Zhou G, Wright SD, Cai TQ. Dual mechanisms of ABCA1 regulation by geranylgeranyl pyrophosphate. J Biol Chem. 2001;276:48702–48708. doi: 10.1074/jbc.M109402200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Zhang H, Hua S, Ma L, Chen C, Liu X, Jiang L, Yang H, Zhang P, Yu D, Guo Y, Tan X, Liu J. Identification of two herbal compounds with potential cholesterol-lowering activity. Biochem Pharmacol. 2007;74:940–947. doi: 10.1016/j.bcp.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 23.Ricketts ML, Boekschoten MV, Kreeft AJ, Hooiveld GJ, Moen CJ, Muller M, Frants RR, Kasanmoentalib S, Post SM, Princen HM, Porter JG, Katan MB, Hofker MH, Moore DD. The cholesterol-raising factor from coffee beans, cafestol, as an agonist ligand for the farnesoid and pregnane X receptors. Mol Endocrinol. 2007;21:1603–1616. doi: 10.1210/me.2007-0133. [DOI] [PubMed] [Google Scholar]
- 24.Clegg RJ, Middleton B, Bell GD, White DA. The mechanism of cyclic monoterpene inhibition of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase in vivo in the rat. J Biol Chem. 1982;257:2294–2299. [PubMed] [Google Scholar]
- 25.Clegg RJ, Middleton B, Bell GD, White DA. Inhibition of hepatic cholesterol synthesis and S-3-hydroxy-3-methylglutaryl-CoA reductase by mono and bicyclic monoterpenes administered in vivo. Biochem Pharmacol. 1980;29:2125–2127. doi: 10.1016/0006-2952(80)90183-5. [DOI] [PubMed] [Google Scholar]
- 26.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 27.Einbond LS, Soffritti M, Esposti DD, Park T, Cruz E, Su T, Wu HA, Wang X, Zhang YJ, Ham J, Goldberg IJ, Kronenberg F, Vladimirova A. Actein activates stress- and statin-associated responses and is bioavailable in Sprague-Dawley rats. Fundam Clin Pharmacol. 2009;23:311–321. doi: 10.1111/j.1472-8206.2009.00673.x. [DOI] [PubMed] [Google Scholar]
- 28.Ondeyka JG, Jayasuriya H, Herath KB, Guan Z, Schulman M, Collado J, Dombrowski AW, Kwon SS, McCallum C, Sharma N, MacNaul K, Hayes N, Menke JG, Singh SB. Steroidal and triterpenoidal fungal metabolites as ligands of liver X receptors. J Antibiot (Tokyo) 2005;58:559–565. doi: 10.1038/ja.2005.76. [DOI] [PubMed] [Google Scholar]
- 29.Jayasuriya H, Herath KB, Ondeyka JG, Guan Z, Borris RP, Tiwari S, de Jong W, Chavez F, Moss J, Stevenson DW, Beck HT, Slattery M, Zamora N, Schulman M, Ali A, Sharma N, MacNaul K, Hayes N, Menke JG, Singh SB. Diterpenoid, steroid, and triterpenoid agonists of liver X receptors from diversified terrestrial plants and marine sources. J Nat Prod. 2005;68:1247–1252. doi: 10.1021/np050182g. [DOI] [PubMed] [Google Scholar]
- 30.Brown MS, Faust JR, Goldstein JL, Kaneko I, Endo A. Induction of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in human fibroblasts incubated with compactin (ML-236B), a competitive inhibitor of the reductase. J Biol Chem. 1978;253:1121–1128. [PubMed] [Google Scholar]
- 31.Sever N, Song BL, Yabe D, Goldstein JL, Brown MS, DeBose-Boyd RA. Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J Biol Chem. 2003;278:52479–52490. doi: 10.1074/jbc.M310053200. [DOI] [PubMed] [Google Scholar]
- 32.Wong J, Quinn CM, Brown AJ. Statins inhibit synthesis of an oxysterol ligand for the liver X receptor in human macrophages with consequences for cholesterol flux. Arterioscler Thromb Vasc Biol. 2004;24:2365–2371. doi: 10.1161/01.ATV.0000148707.93054.7d. [DOI] [PubMed] [Google Scholar]
- 33.Holstein SA, Wohlford-Lenane CL, Hohl RJ. Consequences of mevalonate depletion. Differential transcriptional, translational, and post-translational up-regulation of Ras, Rap1a, RhoA, and RhoB. J Biol Chem. 2002;277:10678–10682. doi: 10.1074/jbc.M111369200. [DOI] [PubMed] [Google Scholar]
- 34.Holstein SA, Wohlford-Lenane CL, Hohl RJ. Isoprenoids influence expression of Ras and Ras-related proteins. Biochemistry. 2002;41:13698–13704. doi: 10.1021/bi026251x. [DOI] [PubMed] [Google Scholar]
- 35.Ness GC, Zhao Z, Keller RK. Effect of squalene synthase inhibition on the expression of hepatic cholesterol biosynthetic enzymes, LDL receptor, and cholesterol 7 alpha hydroxylase. Arch Biochem Biophys. 1994;311:277–285. doi: 10.1006/abbi.1994.1238. [DOI] [PubMed] [Google Scholar]
- 36.Shull LW, Wiemer AJ, Hohl RJ, Wiemer DF. Synthesis and biological activity of isoprenoid bisphosphonates. Bioorg Med Chem. 2006;14:4130–4136. doi: 10.1016/j.bmc.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Ericsson J, Jackson SM, Lee BC, Edwards PA. Sterol regulatory element binding protein binds to a cis element in the promoter of the farnesyl diphosphate synthase gene. Proc Natl Acad Sci USA. 1996;93:945–950. doi: 10.1073/pnas.93.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong H, Holstein SA, Hohl RJ. Simultaneous determination of farnesyl and geranylgeranyl pyrophosphate levels in cultured cells. Anal Biochem. 2005;336:51–59. doi: 10.1016/j.ab.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 39.Storey JD. A direct approach to false discovery rates. J Roy Stat Soc B. 2002;64:479–498. [Google Scholar]
- 40.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 42.Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 43.Holstein SA, Tong H, Hohl RJ. Differential activities of thalidomide and isoprenoid biosynthetic pathway inhibitors in multiple myeloma cells. Leuk Res. 2010;34:344–351. doi: 10.1016/j.leukres.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clendening JW, Pandyra A, Li Z, Boutros PC, Martirosyan A, Lehner R, Jurisica I, Trudel S, Penn LZ. Exploiting the mevalonate pathway to distinguish statin-sensitive multiple myeloma. Blood. 2010;115:4787–4797. doi: 10.1182/blood-2009-07-230508. [DOI] [PubMed] [Google Scholar]
- 45.Jackson SM, Ericsson J, Metherall JE, Edwards PA. Role for sterol regulatory element binding protein in the regulation of farnesyl diphosphate synthase and in the control of cellular levels of cholesterol and triglyceride: evidence from sterol regulation-defective cells. J Lipid Res. 1996;37:1712–1721. [PubMed] [Google Scholar]
- 46.Ericsson J, Jackson SM, Edwards PA. Synergistic binding of sterol regulatory element-binding protein and NF-Y to the farnesyl diphosphate synthase promoter is critical for sterol-regulated expression of the gene. J Biol Chem. 1996;271:24359–24364. doi: 10.1074/jbc.271.40.24359. [DOI] [PubMed] [Google Scholar]
- 47.Fukuchi J, Song C, Ko AL, Liao S. Transcriptional regulation of farnesyl pyrophosphate synthase by liver X receptors. Steroids. 2003;68:685–691. doi: 10.1016/s0039-128x(03)00100-4. [DOI] [PubMed] [Google Scholar]
- 48.Sakakura Y, Shimano H, Sone H, Takahashi A, Inoue N, Toyoshima H, Suzuki S, Yamada N. Sterol regulatory element-binding proteins induce an entire pathway of cholesterol synthesis. Biochem Biophys Res Commun. 2001;286:176–183. doi: 10.1006/bbrc.2001.5375. [DOI] [PubMed] [Google Scholar]
- 49.Ericsson J, Greene JM, Carter KC, Shell BK, Duan DR, Florence C, Edwards PA. Human geranylgeranyl diphosphate synthase: isolation of the cDNA, chromosomal mapping and tissue expression. J Lipid Res. 1998;39:1731–1739. [PubMed] [Google Scholar]
- 50.Forman BM, Ruan B, Chen J, Schroepfer GJ, Jr, Evans RM. The orphan nuclear receptor LXRalpha is positively and negatively regulated by distinct products of mevalonate metabolism. Proc Natl Acad Sci USA. 1997;94:10588–10593. doi: 10.1073/pnas.94.20.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lehmann JM, Kliewer SA, Moore LB, Smith-Oliver TA, Oliver BB, Su JL, Sundseth SS, Winegar DA, Blanchard DE, Spencer TA, Willson TM. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 52.Turbyville TJ, Gursel DB, Tuskan RG, Walrath JC, Lipschultz CA, Lockett SJ, Wiemer DF, Beutler JA, Reilly KM. Schweinfurthin A selectively inhibits proliferation and Rho signaling in glioma and neurofibromatosis type 1 tumor cells in a NF1-GRD-dependent manner. Mol Cancer Ther. 2010;9:1234–1243. doi: 10.1158/1535-7163.MCT-09-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C, JeBailey L, Ridgway ND. Oxysterol-binding-protein (OSBP)-related protein 4 binds 25-hydroxycholesterol and interacts with vimentin intermediate filaments. Biochem J. 2002;361:461–472. doi: 10.1042/0264-6021:3610461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lehto M, Mayranpaa MI, Pellinen T, Ihalmo P, Lehtonen S, Kovanen PT, Groop PH, Ivaska J, Olkkonen VM. The R-Ras interaction partner ORP3 regulates cell adhesion. J Cell Sci. 2008;121:695–705. doi: 10.1242/jcs.016964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


