Abstract
Background
Despite considerable knowledge that prenatal ethanol exposure can lead to devastating effects on the developing fetus, alcohol consumption by pregnant women remains strikingly prevalent. Both clinical and basic research has suggested that, in addition to possible physical, behavioral, and cognitive deficits, gestational exposure to alcohol may lead to an increased risk for the development of later alcohol-related use and abuse disorders. The current work sought to characterize alterations in endogenous opioid signaling peptides and gene expression produced by ethanol exposure during the last days of gestation.
Methods
Experimental subjects were 4-, 8-, and 12-day old infant rats obtained from pregnant females that were given daily intubations of 0, 1, or 2 g/kg ethanol during the last few days of gestation (GD17-20). Using real-time RT-PCR, western blotting analysis, and enzyme immunoassays, we examined mRNA and protein for three opioid receptors and ligands in the nucleus accumbens, ventral tegmental area, and hypothalamus.
Results
Three main trends emerged - (1) mRNA for the majority of factors were found to upregulate across each of the three postnatal ages assessed, indicative of escalating ontogenetic expression of opioid-related genes; (2) prenatal ethanol significantly reduced many opioid peptides, suggesting a possible mechanism by which prenatal exposure can affect future responsiveness towards ethanol; and (3) the nucleus accumbens emerged as a key site for ethanol-dependent effects, suggesting a potential target for additional assessment and intervention towards understanding the ethanol's ability to program the developing brain.
Conclusion
We provide a global assessment of relatively long-term changes in both opioid gene expression and protein following exposure to only moderate amounts of ethanol during a relatively short window in the prenatal period. These results suggest that, while continuing to undergo ontogenetic changes, the infant brain is sensitive to prenatal ethanol exposure and that such exposure may lead to relatively long-lasting changes in the endogenous opioid system within the reward circuitry. These data indicate a potential mechanism and target for additional assessments of ethanol's ability to program the brain, affecting later responsiveness towards the drug.
Keywords: Ethanol, ontogeny, opioids, rat, protein, mRNA
1. Introduction
Despite considerable knowledge that drinking ethanol during gestation leads to devastating effects to the developing fetus, alcohol consumption by pregnant women remains strikingly prevalent. Consequences of ethanol consumption during gestation depend, in part, on the overall amount of alcohol consumed per intake session and the gestational period in which the drug was administered (e.g., [1], [2], and [3]). For example, intake of large amounts of alcohol throughout gestation often lead to severe developmental consequences, sometimes including mental retardation and the hallmark cranial-facial malformations associated with Fetal Alcohol Syndrome [1] and [4]. Consumption of smaller amounts of ethanol during this critical period of development, however, may result in less obvious, yet equally devastating consequences. Recent research suggests that even moderate exposure to ethanol during the last portion of gestation or early postnatal life may enhance ethanol intake [5], [6], [7], [8], [9], [10], and [11], preference [6], [12], and [13], and reinforcement [14], [15], [16], and [17] throughout life. The neurochemical and neuroanatomical mechanisms mediating these effects, however, are currently unknown.
Studies examining alcohol intake and reinforcement have indicated that the endogenous opioid system plays an important role in many prenatal ethanol effects. Known to be involved in ethanol intake and reinforcement during adulthood, the endogenous opioid system has also been implicated in ethanol's appetitive effects during early postnatal life (e.g., [18], [9], [19], [20], and [14]). Administration of a mu or kappa opioid receptor antagonist, for example, was sufficient to eliminate ethanol reinforcement normally observed in the infant rat [19] and [20]. Additionally, naloxone, a nonselective opioid antagonist, administered in combination with ethanol to pregnant females eliminated evidence of augmented alcohol consumption typically found following prenatal exposure to ethanol alone [5], and [7]. Taken together, these results imply activation of the endogenous opioid system following ethanol administration and, perhaps, an obligatory role of opioid activity in order for ethanol to function as an effective positive reinforcer during early infancy.
Multiple groups have already reported a relationship between ethanol-induced opioid activity and increased ethanol preference and/or intake [21], [22], [23], [24], [25], and [26]. It appears that, in alcohol preferring strains of rodents and individuals considered to be at high risk of developing alcoholism, the endogenous opioid system is particularly sensitive to ethanol-induced activation. This is in contrast to low-risk individuals and animals genetically selected to avoid alcohol, for whom ethanol does not seem to be particularly effective at stimulating opioid peptide release [27] and [22]. Ethanol-induced enhanced sensitivity seems to be particular to the mu-opioid system in which beta-endorphin release, the endogenous ligand for the mu-opioid receptor, is especially responsive to ethanol administration [22]. Augmented opioid signaling contributes to enhanced ethanol reinforcement and/or intake through the release of dopaminergic cell bodies from GABAergic inhibition [22] and [23]. Similar work in animal models has implicated not only mu, but the delta-opioid system as well [22]. In contrast, the kappa-opioid receptor system is typically thought to convey aversive aspects of ethanol administration. While this generally seems to be the case for older infants [28] and adult animals, KOR activation is thought to convey appetitive and not aversive information during early postnatal life [20], [53], and [54]. What remains to be determined, however, is the consequence of prenatal ethanol administration on the basal development or activity of the endogenous opioid system (i.e., in the absence of a subsequent post-natal ethanol challenge).
Given that increases in alcohol intake are reported following prenatal exposure to the drug, it would be especially useful to examine the molecular and neurochemical mechanisms associated with prenatal exposure to ethanol during early infancy. Certainly, prenatal ethanol-induced changes in the neural mechanisms thought to mediate alcohol addiction may serve as one possible means by which fetal exposure to ethanol might increase the likelihood of future alcohol-related use and abuse disorders.
The current body of work sought to do just this by examining the expression of some of the key members of the opioid signaling family (including both ligands and receptors) in infant rats born to females intubated with ethanol during the last days of gestation. Specifically, we used real time RT-PCR to examine mRNA for the opioid receptors, mu, kappa, and delta, along with precursors for their endogenous ligands - proopiomelanocortin (POMC), preprodynorphin (PPD), and preproenkephalin (PPE), and western blotting analysis or enzyme immunoassay to assess protein for these same receptors and endogenous ligands (i.e., endorphin, dynorphin, enkephalin). All factors were assessed in the ventral tegmental area and nucleus accumbens, key mesolimbic structures known to be involved in ethanol intake and reinforcement. Additionally, we chose to assess these same factors in a third, offsite structure - the hypothalamus, due to its rich opioid activity, involvement in consummatory behavior, and connection to ethanol intake [31] and reinforcement [22]. Brain tissue was collected across several days during early postnatal life (PDs 4, 8 and 12), focusing on ages at which ethanol intake is known to be relatively low, moderate, and high, respectively [30] and [31]. To the extent that ethanol intake is mediated by the endogenous opioid system (e.g., [32], but see [33]) and differs across early ontogeny, and prenatal exposure to the drug alters subsequent consumption (e.g., [5], [6], [7], [8], [9], [10], [11], and [34] and reinforcement [14], [15], [16], and [17] of the drug, we expected to see differences in basal levels of opioid mRNA and/or protein as a function of both ontogeny and prenatal alcohol exposure (PAE). More specifically, we expected to see ontogenetic-dependent upregulation of opioid systems, indicative of ongoing developmental changes within this system. Additionally, based on previous work [35] and others [34], we expected to see ethanol-induced changes in mu-, kappa, and possibly delta- opioid receptors that reflect site-specific responsiveness to PAE. We anticipated that KOR expression would be reduced, while MOR may, depending upon the structure, be found in greater concentration following PAE. Lastly, we predicted that the majority of ethanol-dependent effects would be observed within the nucleus accumbens, a structure that seems especially responsive to ethanol-dependent effects.
2. Materials and Methods
2.1.1 Breeding
Rat pups derived from experimentally naïve Sprague-Dawley rats (Taconic, Germantown, NY) were used as experimental subjects. For breeding, a single male and female were housed in a wire-hanging cage and the droppings below each cage were checked daily for the appearance of a waxy sperm-plug. On the day detected, deemed as gestational day (GD) 0, females were removed and re-housed in standard maternity tubs with at least one other female impregnated on the same day. On GD 20, females were singly housed and observed daily for parturition; the day of delivery was deemed as postnatal day (PD) 0. On PD 1, litters were culled to a total of 10 animals, maintaining equal sex ratios whenever possible [36]. All animals were maintained in a temperature controlled environment (22° C), on a 14:10 light-dark cycle with lights on at 0700 hours and both food and water available ad libitum (Purina “Formulab Diet”, 5008, breeding formula, Ralston-Purina, St. Louis, MO). All animals were treated in accordance with the guidelines set forth by the National Institute of Health (1986) and the protocols approved by the IACUC of Binghamton University.
2.1.2 Subjects and procedures
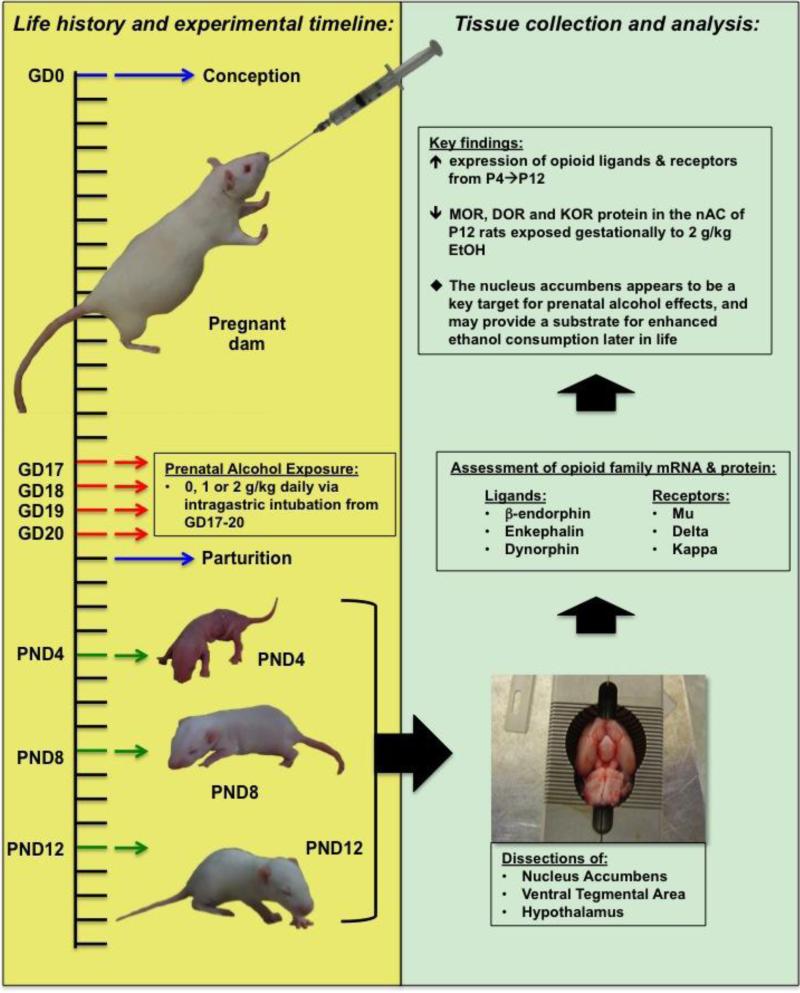
A total of 45 pregnant females, giving rise to 72 experimental subjects, were assigned to 1 of 3 prenatal treatment groups and given daily intubations (i.g.; GDs 17-20) of 0, 8.4, or 16.8% ethanol (v/v; total volume of 1.5% of body weight) to obtain a final dose of 0, 1, or 2 g/kg ethanol, respectively. Briefly, females were gently restrained in a soft towel and a stainless steel feeding tube, roughly 7.6 cm in length, was inserted through the intraoral cavity and into the stomach where fluid was infused. On PDs 4, 8, or 12, a total of 2 pups from each litter (1 male and 1 female) were removed from the dam and immediately killed for the assessment of mRNA for key members of the opioid family (Experiment 1). Four additional pups from each litter (2 male and 2 female) were removed from the dam and immediately killed for the assessment of basal opioid-related peptides (Experiment 2). The final design consisted of 3 prenatal conditions (0, 1, or 2 g/kg) and 3 postnatal ages (PD 4, 8, or 12), for which tissue was collected to assess for both mRNA (Exp 1) and protein (Exp 2) of 3 opioid receptors (i.e., mu, kappa, and delta) and 3 opioid ligands (i.e., POMC / β-endorphin, preproenkephalin / enkephalin, and preprodynorphin / dynorphin), in 3 separate brain regions (i.e., nucleus accumbens, ventral tegmental area, and hypothalamus) (n=8-10/group; see Figure 1 for summary and overview).
Figure 1.
Visual summary of experimental design and analysis. The timeline on the left corresponds to the both the prenatal and early postnatal period of experimental subjects. Ethanol was administered to pregnant dams on GD 17-20 and tissue was later collected from offspring on PDs 4, 8, or 12. Three brain regions were collected from each animal and examined for mRNA and protein for receptors and ligands in the opioid family. Results were indicative of a general upregulation of opioid factors and reduced opioid receptor expression in preweanlings given gestational exposure to ethanol (G17-20). The results also highlight the nucleus accumbens as a particularly vulnerable region for ethanol-induced effects on opioid-related genes.
2.2.1 Experiment 1 - RNA extraction and real time RT-PCR
For the examination of gene expression changes in Experiment 1, animals were quickly decapitated (unanesthetized) and brains were removed, placed in ice-cold physiological saline for roughly 30 sec, and then immediately transferred to a neonatal brain matrix. Tissue was then sectioned into 1 mm slices and incubated, at 4° C, in 350 μl of RNAlater (Qiagen) for 24 hr prior to storage at −20° C. At the time of dissection, two regions implicated in ethanol reinforcement (i.e., the ventral tegmental area (VTA) and nucleus accumbens (nAC)) along with a single offsite structure (i.e., hypothalamus (HYPO)), were identified according to the atlas of Altman and Bayer [37]. Tissue was microdissected and returned to RNAlater (Qiagen) at -20° C until the time of RNA extraction. Total RNA was extracted from tissue using a hand-held motorized homogenizer in the presence of 500 μl ice-cold Trizol® RNA reagent (Invitrogen). Tissue homogenate was then removed, passed through a QiaShredder (Qiagen) to shear residual genomic DNA and incubated with 100 μl chloroform for roughly 2 min before centrifugation at 12000 g (15 min). Following centrifugation, the aqueous phase of each sample was removed and added to 500 μl of 70% ethanol. Samples were then purified through RNeasy columns (Qiagen), according to manufacture protocol, and eluted with 30 μl of RNAse free water (65° C). RNA purity and yield were determined electrophoretically using the BIORAD Experion system and cDNA synthesis was performed on 0.1 μg of RNA in a total volume of 20 μl using a commercially available First-Strand cDNA synthesis kit containing a blend of random hexamers and oligoDT primers, and included a DNAse treatment step (QuantiTect Reverse Transcription Kit; Qiagen). PCR was performed on samples diluted 1:4 with RNAse free water.
For PCR reactions, a 20 μl volume containing 10 μl SYBR Green Supermix (Qiagen), 1 μl primer (final concentration of 250 nM; see Table 1), 1 μl cDNA template and 8 μl RNAse free water was pipetted into 96 wells (BioRad). Following a 3 min incubation period at 95° C, the PCR reaction consisted of 30 sec denaturation at 95° C, 30 sec annealing at 60° C, and 30 sec extension at 72° C for a total of 50 cycles. PCR was run on an iQ5, real time-PCR machine from BioRad. Following amplification, a melt-curve beginning at 55° C and increasing to 95° C in 0.5° C increments was used to ensure that only a single PCR product resulted. Primers used in Exp 1 (see Table 1) were designed to amplify mRNA for the opioid receptors – mu, kappa, and delta, along with the precursors for the endogenous ligands with greatest affinity for mu-, kappa- and delta-opioid receptors – proopiomelanocortin (POMC), preprodynorphin (PPD), and preproenkephalin (PPE), respectively.
Table 1.
Forward and reverse primer sequences along with corresponding amplicon size and accession numbers for real-time RT-PCR primers used in Experiment 1. Primers were designed to span an intron when possible.
| Primer | Sequence | Amplicon (bp) | Accession Number | |
|---|---|---|---|---|
| Ligands | POMC | Forward 5′ TCC ATA GAC GTG TGG AGC TG 3′ Reverse 5′ ACG TAC TTC CGG GGA TTT TC 3′ |
173 | NM_139326 |
| Preprodynorphin (PPD) | Forward 5′ GGG TTC GCT GGA TTC AAA TA 3′ Reverse 5′ TGT GTG GAG AGG GAC ACT CA 3′ |
83 | NM_019374 | |
| Preproenkephalin (PPE) | Forward 5′ AAA ATC TGG GAG ACC TGC AA 3′ Reverse 5′ CAT GAA ACC GCC ATA CCT CT 3′ |
197 | NM_017139 | |
| Receptors | Mu Receptor (MOR) | Forward 5′ ATC GTC AAC GTC TGC AAC TG 3′ Reverse 5′ CCC TGC CTG TAT TTT GTG GT 3′ |
81 | NM_001038598 |
| Kappa Receptor (KOR) | Forward 5′ CTT TGG CAG ATG CTT TG TT 3′ Reverse 5′ CAT CTC CAA AAG GCC AAG AA 3′ |
243 | NM_017167 | |
| Delta Receptor (DOR) | Forward 5′ AGC ATC TTC ACG CTC ACC AT 3′ Reverse 5′ CAA CAC CTG AAG CCA AGA CC 3′ |
136 | NM_012617 |
Previous work from our lab has demonstrated that administration of ethanol, either during pre- or postnatal life, may increase the overall amount of variability in mRNA for some housekeepers (unpublished results). For this reason, data were first analyzed using the qBASE system [38] and exported to geNORM [39], a software program designed to determine overall variability in each housekeeper. The application of geNORM allowed for the selection of those housekeepers that should be retained for subsequent comparison purposes. In all tissue compartments, 18s RNA was immediately discarded due to large amounts of variability across samples. Next, data were analyzed using the qBASE system with cyclophilin+beta-actin or cyclophilin+beta-actin+GAPDH as housekeepers. Analysis revealed similar findings with both methods, therefore, the inclusion of three housekeepers was chosen as the preferred method of analysis. All data are expressed using the qBASE method where relative expression levels are normalized using a set of reference primer pairs for three housekeeping genes (i.e., beta-actin, cyclophilin and GAPDH), and analyzed using separate between-groups ANOVA with any significant differences clarified with Fishers protected least significant difference (Fishers, PLSD).
2.2.2 Experiment 2 - Protein extraction and quantification via Western Blotting analysis and Enzyme Immuno Assay
For the examination of protein in Experiment 2, animals were quickly decapitated (unanesthetized) and brains were removed, placed in ice-cold physiological saline for roughly 30 sec and then transferred to a cold-plate for anatomical identification and isolation. As in Exp 1, several regions implicated in ethanol reinforcement and known to be high in opioid activity (e.g., VTA, nAC, and HYPO), were identified according to the atlas of Altman and Bayer [37], microdissected, and placed in a 1.5 ml eppendorf tube and flash-frozen for later protein extraction and assessment of opioid peptide.
For assessment of the opioid receptors (i.e., mu, kappa, and delta) using Western Blotting analysis, tissue from 2 of the 4 animals in each litter (1 male and 1 female) was incubated with 250 μl ice-cold homogenization buffer (pH 7.2, 4° C) containing 50 mM Tris, 1 mM EDTA, 6 mM MgCl2, and Complete Mini Protease Inhibitor Cocktail Tablets (Roche Applied Science). Tissue was then sonicated for 10 sec using an ultrasonic dismembrator (Fisher, model 100) and centrifuged for 15 min at 14000 rcf (4° C). Supernatants were collected and total protein content was measured using the method of [40]. Samples were then adjusted to a final concentration of 1 μg/μl in a total volume of 50 μl. An equal volume of Laemmeli's buffer was then added to each tube prior to a 5 min boil. Samples were loaded onto a 12% Tris-glycine acrylamide gel and separated by SDS polyacrylamide gel electrophoresis (SDS-PAGE). Separated proteins were electrophoretically transferred from gels to PVDF membrane. Membranes were blocked using 5% BSA in 1X TBST for 1 hr and incubated overnight (4° C) with primary antibody specific for mu- (MOR-1 (H-80) sc-15310), kappa- (KOR-1 (H-70) sc-9112) or delta- (DOR-1 (H-60) sc-9111) opioid receptors, diluted 1:500 in 5% BSA. Secondary antibody (anti-rabbit, diluted 1:5000 in 5% BSA; Santa Cruz, CA) was applied the following day and immunopositive bands were visualized by chemiluminescence (Western Lightening® Plus; PerkinElmer). Optical density of immunopositive bands on X-ray film was measured with an image analysis system using Quantity One software (BioRad). Although we recognize the limitation of not examining a standard housekeeper protein in these samples, our pilot studies indicated degradation of signal after 2 cycles of stripping and re-probing. Thus, it was deemed higher priority to analyze all 3 receptor targets in the same samples than to include a housekeeper protein. To partially account for this limitation, PVDF membranes were incubated with each of the three antibodies in counter-balanced order. Prior to re-incubation with subsequent antibodies, membranes were stripped of primary and secondary antibodies with a stripping buffer (Restore PLUS Western Blot Stripping Buffer, Pierce Protein) for 15 min and re-blocked for 1 hr. Effectiveness of the stripping procedure was confirmed by the absence of bands in a subset of PVDF membranes in which stripped blots were incubated with Western Lightening® Plus and exposed to X-ray film. Due to the low sample volume available from neonatal pups, it was not possible to utilize a housekeeper approach to control for (i) specificity of experimental effects and (ii) potential loading errors. However, the examination of multiple proteins from the same samples on reprobed blots essentially serves the same purposes, and was viewed as a viable alternative to the (somewhat arbitrary) selection of an unrelated housekeeper protein.
Tissue processing for assessment of β-endorphin, enkephalin and dynorphin through Enzyme Immuno Assay was similar to that just described. The remaining samples (2 from each litter; 1 male and 1 female) were incubated with 250 μl ice-cold homogenization buffer and sonicated, as described above. Following centrifugation (14000 rcf for 15 min at 4° C), supernatants were transferred to 1.5 ml tube and stored (-80° C) for later sample purification and concentration. Samples were purified via column separation using Strata C18 Separation Columns according to manufacturer protocol (Phoenix Pharmaceuticals, CA). Following purification, samples were dehydrated (using a SpeedVac) and reconstituted in 250 μl assay buffer (Phoenix Pharmaceuticals, CA). Total protein content was determined using Experion protein analysis chips (BioRad), according to manufacturer protocol (BioRad). A total of 80 samples were included for the assessment of opioid ligands using EIA kits according to manufacturer protocol (EK-022-33; EK-021-03; EK-024-21; Phoenix Pharmaceuticals, CA).
3. Results
3.1.1 Experiment 1 - Ontogenetic effects on opioid-related mRNA
Based on the ontogenetic profile of ethanol intake and reinforcement across early infancy [41], [42], [30], and [31], we hypothesized that opioid receptors and/or ligands would be differentially expressed across the three postnatal ages assessed. Indeed, initial analyses examining Age x PAE revealed that nearly every factor assessed changed significantly across the three postnatal days under examination. Upon further investigation of PAE effects, however, it became apparent that the traditional multifactorial approach to these analyses would lead to two major limitations, namely that (1) ontogenetic effects would be artificially inflated by PAE and (2) larger ontogenetic changes in the endogenous opioid system would preclude our ability to observe smaller ethanol-dependent effects. Therefore, we chose a more conservative approach - to analyze and discuss our data using very focused, a priori, comparisons examining the consequences of both ontogeny and PAE separately. As is tradition, however, multifactorial ANOVAs were performed and are included, along with all other results, in Tables 2-4. It should also be noted that, since both male and female offspring were included in the experimental design, initial comparisons examined for potential sex differences. Since no sex-differences in opioid related mRNA were observed, we collapsed across sex for all remaining analyses.
Table 2.
Results summary for examination of mRNA (Exp 1) and protein (Exp 2) within the nucleus accumbens of rat pups on PD4, 8 or 12 following prenatal exposure to 0, 1 or 2 g/kg EtOH on GD17-20.
| Nucleus Accumbens | mRNA | Protein | |||||||
|---|---|---|---|---|---|---|---|---|---|
| PD4 | PD8 | PD12 | PD4 | PD8 | PD12 | ||||
| MOR | 0g/kg 1g/kg 2g/kg |
2.62 ± 0.52 2.69 ± 0.36 3.01 ± 0.53 |
2.33 ± 0.23 2.20 ± 0.18 1.73 ± 0.12 |
1.60 ± 0.13 1.56 ± 0.06 1.38 ± 0.06 |
Age: F(2,60)=14.006, p<0.0001 Dose: F(2,60)=0.196, p=.822 Age × Dose: F(4,60)=0.761, p=.555 |
124858.16 ± 3827.70 107210.42 ± 8433.81 123952.91 ± 3160.56 |
115867.83 ± 7392.51 115815.40 ± 6803.24 120808.35 ± 6950.47 |
135626.92 ± 2478.19 127742.67 ± 3277.05 91364.74 ± 4315.04 |
Age: F(2,68)=0.032, p=.969 Dose: F(2,68)=3.571, p<0.05 Age × Dose: F(4,68)=6.23, p<.0005 |
| POMC / β-endorphin | 0g/kg 1g/kg 2g/kg |
121.94 ± 41.36 52.91 ± 12.36 214.14 ± 66.49 |
404.84 ± 124.38 157.24 ± 42.92 118.15 ± 18.24 |
308.19 ± 54.38 406.20 ± 64.94 385.42 ± 47.08 |
Age: F(2,58)=11.053, p<0.0001 Dose: F(2,58)=1.066, p=.351 Age × Dose: F(4,58)=3.792, p<0.05 |
0.112 ± 0.025 0.093 ± 0.017 0.096 ± 0.017 |
0.082 ± 0.016 0.120 ± 0.019 0.064 ± 0.010 |
0.070 ± 0.012 0.074 ± 0.012 0.083 ± 0.011 |
Age: F(2,64)=2.297, p=.109 Dose: F(2,64)=.675, p=.513 Age × Dose: F(4,64)=1.857, p=.129 |
| KOR | 0g/kg 1g/kg 2g/kg |
2.56 ± 0.29 3.57 ± 0.35 3.44 ± 0.44 |
3.39 ± 0.44 4.11 ± 0.39 5.92 ± 0.30 |
4.49 ± 0.28 4.30 ± 0.21 4.04 ± 0.37 |
Age: F(2,56)=10.577, p<0.0005 Dose: F(2,56)=5.554, p<0.01 Age × Dose: F(4,56)=5.65, p<0.001 |
100811.18 ± 3915.24 98333.27 ± 3074.02 90815.98 ± 4211.37 |
96215.92 ± 5799.11 97593.04 ± 2701.47 105432.80 ± 4372.21 |
104894.08 ± 1299.38 100192.23 ± 2734.94 93432.43 ± 3859.63 |
Age: F(2,66)=0.631, p=.535 Dose: F(2,66)=0.875, p=.421 Age × Dose: F(4,66)=2.527, p<0.05 |
| PPD / dynorphin | 0g/kg 1g/kg 2g/kg |
5.04 ± 0.46 7.28 ± 1.20 7.98 ± 0.92 |
12.72 ± 1.32 12.88 ± 1.34 18.44 ± 2.21 |
20.98 ± 3.85 21.58 ± 0.82 25.09 ± 3.48 |
Age: F(2,60)=38.88, p<0.0001 Dose: F(2,60)=3.109, p=052 Age × Dose: F(4,60)=0.315, p=.867 |
0.968 ± 0.409 1.279 ± 0.310 1.467 ± 0.538 |
2.287 ± 0.680 0.977 ± 0.283 0.330 ± 0.109 |
0.689 ± 0.163 0.554 ± 0.283 0.785 ± 0.272 |
Age: F(2,64)=1.772, p=.178 Dose: F(2,64)=1.134, p=.328 Age × Dose: F(4,64)=2.788, p<0.05 |
| DOR | 0g/kg 1g/kg 2g/kg |
3.74 ± 0.62 5.92 ± 0.78 6.48 ± 1.22 |
5.91 ± 0.76 7.50 ± 0.75 7.77 ± 0.75 |
9.19 ± 1.35 8.01 ± 0.68 9.79 ± 1.68 |
Age: F(2,59)=9.281, p<0.001 Dose: F(2,59)=2.135, p=.127 Age × Dose: F(4,59)=0.807, p=.523 |
111173.68 ± 4365.36 96218.83 ± 5441.77 114707.86 ± 6736.15 |
108291.47 ± 5026.43 118189.99 ± 4077.11 114695.44 ± 5618.89 |
107695.35 ± 4391.24 105273.05 ± 2840.05 96313.74 ± 2200.25 |
Age: F(2,65)=3.59, p<0.05 Dose: F(2,65)=0.220, p=.726 Age × Dose: F(4,65)=3.171, p<0.05 |
| PPE / enkephalin | 0g/kg 1g/kg 2g/kg |
4.63 ± 1.39 5.70 ± 1.25 7.18 ± 0.44 |
10.78 ± 0.83 10.40 ± 1.93 15.80 ± 1.73 |
22.64 ± 3.49 25.53 ± 1.72 23.86 ± 4.13 |
Age: F(2,56)=42.604, p<0.0001 Dose: F(2,56)=1.096, p=.341 Age × Dose: F(4,56)=.570, p=.685 |
0.039 ± 0.014 0.042 ± 0.009 0.049 ± 0.015 |
0.053 ± 0.012 0.084 ± 0.019 0.052 ± 0.018 |
0.147 ± 0.038 0.083 ± 0.026 0.170 ± 0.062 |
Age: F(2,63)=8.831, p<0.0005 Dose: F(2,63)=.399, p=.672 Age x Dose: F(4,63)1.3063, p=.277 |
Data represent means and SEM for designated factors. Significant differences from water-intubated controls (i.e., 0 g/kg) are represented by bold font (p<0.05).
Table 4.
Results summary for examination of mRNA (Exp 1) and protein (Exp 2) within the hypothalamus of rat pups on PD4, 8 or 12 following prenatal exposure to 0, 1 or 2 g/kg EtOH on GD17-20.
| Hypothalamus | mRNA | Protein | |||||||
|---|---|---|---|---|---|---|---|---|---|
| PD4 | PD8 | PD12 | PD4 | PD8 | PD12 | ||||
| MOR | 0g/kg 1g/kg 2g/kg |
1.8 ± 0.18 1.71 ± 0.25 1.66 ± 0.08 |
2.26 ± 0.25 1.85 ± 0.23 1.59 ± 0.03 |
1.93 ± 0.19 1.56 ± 0.12 1.66 ± 0.13 |
Age: F(2,60)=.945, p=.394 Dose: F(2,60)=4.084, p<0.05 Age × Dose: F(4,60)=.607, p=.659 |
64151.35 ± 4930.75 61000.29 ± 4311.21 92867.91 ± 12570.87 |
63913.68 ± 5117.81 57288.82 ± 2368.14 65215.05 ± 6266.99 |
68125.98 ± 6261.64 68592.08 ± 4463.47 57014.46 ± 4734.72 |
Age: F(2,60)=1.887, p=.160 Dose: F(2,60)=1.576, p=.215 Age × Dose: F(4,60)=3.650, p<0.05 |
| POMC / β-endorphin | 0g/kg 1g/kg 2g/kg |
65.48 ± 10.10 97.15 ± 20.38 62.11 ± 17.16 |
170.11 ± 41.41 99.80 ± 38.21 52.35 ± 16.90 |
204.26 ± 39.28 172.68 ± 36.87 250.38 ± 52.47 |
Age: F(2,60)=13.821, p<0.0001 Dose: F(2,60)=.535, p=.589 Age × Dose: F(4,60)=2.148, p=.086 |
0.024 ± 0.003 0.056 ± 0.020 0.023 ± 0.004 |
0.027 ± 0.007 0.020 ± 0.051 0.027 ± 0.005 |
0.021 ± 0.005 0.019 ± 0.003 0.020 ± 0.003 |
Age: F(2,66)=3.159, p<0.05 Dose: F(2,66)=1.336, p=.270 Age × Dose: F(4,66)=2.928, p<0.05 |
| KOR | 0g/kg 1g/kg 2g/kg |
2.06 ± 0.12 1.46 ± 0.14 1.50 ± 0.10 |
2.88 ± 0.15 2.38 ± 0.33 2.37 ± 0.20 |
2.36 ± 0.30 1.67 ± 0.06 2.49 ± 0.40 |
Age: F(2,59)=10.402, p<0.0001 Dose: F(2,59)=4.766, p<0.05 Age × Dose: F(4,59)=1.179, p=.329 |
105399.84 ± 2932.96 103420.41 ± 4391.55 102925.07 ± 2642.93 |
100129.41 ± 5861.46 92810.74 ± 2037.09 112389.56 ± 5376.53 |
108459.73 ± 2554.49 105566.13 ± 4434.60 102501.14 ± 7152.48 |
Age: F(2,64)=.513, p=.601 Dose: F(2,64)=1.194, p=.310 Age × Dose: F(4,64)=2.229, p=.076 |
| PPD / dynorphin | 0g/kg 1g/kg 2g/kg |
2.44 ± 0.43 2.76 ± 0.26 4.50 ± 1.24 |
3.39 ± 0.21 2.92 ± 0.36 2.40 ± 0.39 |
4.89 ± 0.55 4.22 ± 0.52 4.53 ± 0.19 |
Age: F(2,56)=29.571, p<0.0001 Dose: F(2,56)=2.637, p=.081 Age × Dose: F(4,56)=.990, p=.421 |
0.024 ± 0.009 0.048 ± 0.019 0.012 ± 0.002 |
0.016 ± 0.004 0.061 ± 0.027 0.032 ± 0.012 |
0.023 ± 0.011 0.066 ± 0.023 0.011 ± 0.023 |
Age: F(2,66)=.167, p=.846 Dose: F(2,66)=5.434, p<0.05 Age × Dose: F(4,66)=.351, p=.842 |
| DOR | 0g/kg 1g/kg 2g/kg |
4.21 ± 0.73 2.60 ± 0.54 4.50 ± 1.24 |
11.25 ± 4.18 13.30 ± 5.79 9.05 ± 2.32 |
8.92 ± 1.79 8.71 ± 2.43 8.09 ± 1.77 |
Age: F(2,57)=5.399, p<0.01 Dose: F(2,57)=.120, p=.887 Age × Dose: F(4,57)=.307, p=.872 |
128240.89 ± 2783.10 122809.33 ± 7848.01 122198.46 ± 3848.20 |
125663.78 ± 7026.46 121646.89 ± 4087.73 125232.89 ± 3419.73 |
123513.02 ± 4056.04 127424.89 ± 3834.52 116552.02 ± 4605.46 |
Age: F(2,64)=.175, p=.840 Dose: F(2,64)=.752, p=.476 Age × Dose: F(4,64)=.871, p=.486 |
| PPE / enkephalin | 0g/kg 1g/kg 2g/kg |
2.68 ± 0.30 2.38 ± 0.29 2.58 ± 0.45 |
4.94 ± 0.41 4.19 ± 0.52 5.20 ± 0.80 |
8.46 ± 0.88 7.21 ± 0.86 8.24 ± 0.51 |
Age: F(2,61)=60.418, p<0.0001 Dose: F(2,61)=1.504, p=.230 Age × Dose: F(4,61)=.229, p=.921 |
0.018 ± 0.004 0.036 ± 0.009 0.025 ± 0.006 |
0.027 ± 0.005 0.027 ± 0.007 0.025 ± 0.004 |
0.090 ± 0.021 0.043 ± 0.006 0.039 ± 0.005 |
Age: F(2,68)=10.125, p<0.0005 Dose: F(2,68)=1.844, p=.166 Age × Dose: F(4,68)=3.877, p<0.05 |
Data represent means and SEM for designated factors. Significant differences from water-intubated controls (i.e., 0 g/kg) are represented by bold font (p<0.05).
Firstly, we sought to examine age-dependent effects on opioid gene expression. For this comparison, only those animals receiving water during gestation (i.e., 0 g/kg), and not ethanol, were examined for developmental changes in the amount of mRNA for opioid-related factors in the ventral tegmental area (VTA), nucleus accumbens (nAC), and hypothalamus (HYPO). Analysis of factors within the hypothalamus revealed significant main effects of age on mRNA for POMC [F(2,20) = 5.062, p<0.05], preprodynorphin [F(2,21) = 4.403, p<0.05], and preproenkephalin [F(2,21) = 24.587, p<0.0001], precursors for the endogenous ligands with greatest affinity for mu-, kappa- and delta-opioid receptors, respectively. Subsequent post hoc analyses revealed that mRNA for POMC was significantly greater on both PDs 8 and 12 than on PD 4 (Table 4). Similarly, mRNA for PPD was significantly greater on PD 12 than PD 4 (Table 4). Lastly, PPE mRNA was significantly greater on PD 8 than PD 4 and again on PD 12 than both PDs 4 and 8 (Table 4). There were no significant differences in mRNA for any of the receptors across the three postnatal days assessed.
Assessment of opioid-related mRNA in the VTA revealed fewer ontogenetic differences than in the hypothalamus. No differences in mRNA were present for opioid receptors as a function of postnatal age. Analyses of ligands revealed a main effect of age in POMC [F(2,18) = 6.629, p<0.01], but not PPE or PPD, in which POMC mRNA was significantly greater when examined on PD 12 compared to the two younger ages assessed (Table 3).
Table 3.
Results summary for examination of mRNA (Exp 1) and protein (Exp 2) within the ventral tegmental area of rat pups on PD4, 8 or 12 following prenatal exposure to 0, 1 or 2g/kg EtOH on GD17-20.
| Ventral Tegmental Area | mRNA | Protein | |||||||
|---|---|---|---|---|---|---|---|---|---|
| PD4 | PD8 | PD12 | PD4 | PD8 | PD12 | ||||
| MOR | 0g/kg 1g/kg 2g/kg |
2.46 ± 0.22 3.67 ± 0.47 2.66 ± 0.31 |
2.09 ± 0.23 2.37 ± 0.42 3.33 ± 0.76 |
2.29 ± 0.48 1.81 ± 0.14 2.24 ± 0.37 |
Age: F(2,56)=2.070, p=.136 Dose: F(2,56)=.857, p=.430 Age × Dose: F(4,56)=1.500, p=.215 |
117787.60 ± 4531.00 128330.09 ± 2534.83 113722.84 ± 5417.27 |
111168.49 ± 8221.46 108331.76 ± 6717.11 113217.77 ± 4807.15 |
120219.20 ± 4071.92 125140.32 ± 6091.26 123954.48 ± 4056.12 |
Age: F(2,66)=3.897, p<0.05 Dose: F(2,66)=.514, p=.600 Age × Dose: F(4,66)=.895, p=.472 |
| POMC / β-endorphin | 0g/kg 1g/kg 2g/kg |
3.97 ± 0.80 4.38 ± 0.87 5.63 ± 1.36 |
6.13 ± 1.33 6.27 ± 2.03 9.09 ± 1.20 |
10.35 ± 1.38 11.36 ± 1.69 12.58 ± 1.99 |
Age: F(2,55)=14.759, p<0.0001 Dose: F(2,55)=1.747, p=.184 Age × Dose: F(4,55)=.106, p=.980 |
0.387 ± 0.197 tissue destroyed 0.264 ± 0.097 |
0.975 ± 0.512 0.809 ± 0.343 0.838 ± 0.555 |
0.921 ± 0.525 1.051 ± 0.386 0.959 ± 0.501 |
Age: F(2,28)=2.701, p=.085 Dose: F(2,28)=.085, p=.919 Age × Dose: F(4,28)=.094, p=.984 |
| KOR | 0g/kg 1g/kg 2g/kg |
2.23 ± 0.13 2.32 ± 0.19 2.10 ± 0.21 |
1.90 ± 0.11 2.30 ± 0.21 3.13 ± 0.62 |
1.73 ± 0.16 1.97 ± 0.23 1.58 ± 0.25 |
Age: F(2,55)=4.145, p<0.05 Dose: F(2,55)=.838, p=.438 Age × Dose: F(4,55)=2.085, p=.095 |
94981.68 ± 2480.91 84392.65 ± 3908.53 91051.88 ± 3255.87 |
79846.06 ± 5409.50 78766.42 ± 2952.21 97012.18 ± 5168.83 |
91909.90 ± 2402.59 82291.56 ± 2544.92 78804.42 ± 2781.36 |
Age: F(2,66)=2.687, p=.076 Dose: F(2,66)=4.310, p<0.05 Age × Dose: F(4,66)=5.188, p<0.005 |
| PPD / dynorphin | 0g/kg 1g/kg 2g/kg |
6.33 ± 0.47 6.61 ± 1.03 9.50 ± 0.72 |
8.15 ± 0.54 8.36 ± 0.74 9.68 ± 0.96 |
8.38 ± 1.09 8.20 ± 1.47 11.18 ± 1.81 |
Age: F(2,53)=1.964, p=.150 Dose: F(2,53)=4.759, p<0.05 Age × Dose: F(4,53)=.225, p=.923 |
2.10 ± 1.70 tissue destroyed 0.70 ± 0.40 |
4.80 ± 2.00 6.70 ± 3.30 4.50 ± 2.60 |
5.30 ± 3.10 7.50 ± 4.30 4.60 ± 3.70 |
Age: F(2,28)=2.500, p=.100 Dose: F(2,28)=.188, p=.830 Age × Dose: F(4,28)=.178, p=.948 |
| DOR | 0g/kg 1g/kg 2g/kg |
2.25 ± 0.31 2.82 ± 0.82 2.77 ± 0.59 |
3.93 ± 1.26 4.87 ± 1.33 3.88 ± 0.74 |
2.99 ± 0.28 7.03 ± 1.77 6.49 ± 1.24 |
Age: F(2,55)=4.469, p<0.05 Dose: F(2,55)=1.99, p=.147 Age × Dose: F(4,55)=.825, p=.515 |
64483.55 ± 8041.58 69866.27 ± 5914.94 61269.89 ± 9288.83 |
65229.18 ± 8432.09 58412.89 ± 6015.43 54109.21 ± 5022.88 |
71046.83 ± 6438.47 61659.10 ± 2866.75 67567.03 ± 7230.39 |
Age: F(2,66)=.813, p=.448 Dose: F(2,66)=.508, p=.604 Age × Dose: F(4,66)=.425, p=.790 |
| PPE / enkephalin | 0g/kg 1g/kg 2g/kg |
3.33 ± 0.42 4.01 ± 1.16 7.34 ± 1.79 |
7.30 ± 0.84 6.60 ± 0.46 8.20 ± 1.21 |
6.62 ± 0.64 12.74 ± 2.52 8.01 ± 1.25 |
Age: F(2,53)=6.713, p<0.01 Dose: F(2,53)=2.135, p=.128 Age × Dose: F(4,53)=3.043, p<0.05 |
0.011 ± 0.005 tissue destroyed 0.005 ± 0.003 |
0.028 ± 0.009 0.044 ± 0.023 0.017 ± 0.006 |
0.027 ± 0.010 0.116 ± 0.089 0.058 ± 0.053 |
Age: F(2,28)=2.188, p=.131 Dose: F(2,28)=.668, p=.521 Age × Dose: F(4,28)=.527, p=.717 |
Data represent means and SEM for designated factors. Significant differences from water-intubated controls (i.e., 0g/kg) are represented by bold font (p<0.05).
In contrast to the VTA and HYPO, far more age-dependent alterations in mRNA for opioid-related factors were observed within the nucleus accumbens. Analysis of opioid-receptor expression in this structure revealed a significant main effect of age in DOR mRNA [F(2,21) = 5.123, p<0.05]. Subsequent post hoc analyses revealed that DOR mRNA was significantly greater on PD 12 than PD 4; mRNA for DOR on PD 8 did not differ from that on either PD 4 or 12 (Table 2). Examination of mRNA for opioid ligands revealed significant increases in both PPE [F(2, 21) = 8.249, p<0.01] and PPD [F(2,21) = 11.354, p<0.001] as a function of ontogeny. Subsequent post hoc analyses revealed that, for PPD, mRNA was significantly greater on PD8 than 4, and greater on PD 12 than both PDs 4 and 8 (Table 2). Similarly, PPE mRNA was significantly greater on PD 12 than PDs 4 and 8; PPE mRNA did not differ between PDs 4 and 8 (Table 2).
3.1.2 Experiment 1 - Ethanol effects on opioid related mRNA
As mentioned previously (see 3.1.1), age-related differences in the endogenous opioid system substantially limited our ability to observe ethanol-dependent effects. Therefore, we again chose to utilize focused, a priori, comparisons examining the effects of PAE separately at each age at assessment. Again, since both male and female offspring were included in the experimental design, initial analyses accounted for potential sex differences. Since no sex-differences in opioid related mRNA were observed, we collapsed across sex for all remaining analyses. A summary of results is presented in Tables 2-4.
Analysis of mRNA within the hypothalamus revealed a main effect of prenatal ethanol treatment on KOR mRNA [F(2, 21) = 8.49, p<0.005] at PD 4, but not PDs 8 or 12. Kappa-receptor mRNA was significantly reduced in infants born to females intubated with either 1 or 2 g/kg ethanol during gestation compared to controls. No other factors were found to differ as a function of prenatal ethanol treatment within the hypothalamus (Table 4). Analysis of opioid mRNA within the VTA revealed a significant main effect of prenatal treatment on PPD mRNA, again on PD 4 [F(2,17) = 4.041, p < 0.05] - infants born to females intubated with 2 g/kg during the last day of gestation exhibited an increase in mRNA for preprodynorphin within this structure (Table 3). There were no differences in VTA opioid-related mRNA on PDs 8 or 12.
Again, the majority of gene expression changes were observed in the nucleus accumbens. In particular, analyses revealed a significant main effect of prenatal treatment on mRNA for kappa-receptor [F(2,20) = 11.821, p < 0.0005], POMC [F(2,19) = 3.657, p < 0.05], PPD [F(2,21) = 3.711, p < 0.05] and PPE [F(2,20) = 3.508, p < 0.05] within this same structure. In the case of KOR, PPE and PPD, mRNA from infants born to females intubated with 2 g/kg ethanol during the last days of gestation was significantly greater than from pups in the remaining two groups (Table 2). POMC mRNA, on the other hand, was found to be significantly less in ethanol-treated infants compared to water-intubated controls (Table 2). There were no effects of prenatal alcohol exposure on opioid-related gene expression apparent in the nAC on PD12.
3.2.1 Experiment 2 - Ontogenetic effects on opioid related protein
As in Exp 1 (see 3.1.1), assessments of age-dependent effects in opioid peptides were restricted to the analysis of water-intubated control animals (i.e., 0 g/kg). In doing so, we observed a significant main effect of age on MOR within the nAC [F(2,20) = 4.501, p < 0.05] (Table 2) and KOR within the VTA [F(2, 20) = 5.36, p < 0.05] (Table 3). Subsequent post hoc analyses revealed that, in both cases, protein for the respective receptors was significantly greater on PD 12 than PD 8. In addition, KOR in the VTA was greater on PD 4 than on PD 8. No other differences in opioid-receptor protein were observed with respect to ontogeny.
With respect to endogenous opioid ligands, there was a significant age-related increase in hypothalamic enkephalin [F(2,25) = 8.917, p < 0.005] in which tissue obtained from infants on PD 12 revealed significantly greater enkephalin compared to animals from the two previous age groups (Table 4). There were no developmental differences in ligand protein within the VTA. Like the previous results, the majority of changes were observed within the nucleus accumbens. In particular, β-endorphin [F(2,23) = 8.145, p < 0.005] and dynorphin [F(2,24) = 4.269, p < 0.05] were found to differ significantly as a function of ontogeny. Dynorphin was significantly greater on PD 8 than PD 12 (Table 2) and β-endorphin was greatest on PD 4 compared to the later two ages assessed (Table 2).
3.2.2 Experiment 2 - Ethanol effects on opioid related protein
Analysis of opioid receptors within the hypothalamus revealed a significant main effect of prenatal ethanol administration on MOR on PD 4 [F(2,20) = 3.75, p < 0.05]. In particular, hypothalamic MOR protein was significantly greater in four-day old animals born to females intubated with 2 g/kg ethanol during the last days of gestation than in pups exposed prenatally to lower doses of ethanol (Table 4). Like Experiment 1, relatively few ethanol-induced effects were observed in the ventral tegmental area. The lone exception was a significant main effect of KOR within the VTA when assessed on PD 12 [F(2, 24) = 6.43, p < 0.05] in which infants exposed to ethanol during the last days of gestation expressed less kappa receptor protein than those born to water intubated controls (Table 3).
Assessment of opioid-receptor protein in the nucleus accumbens revealed a significant main effect of prenatal treatment on MOR when assessed on PD 4 [F(2,23) = 3.519, p < 0.05] in which MOR protein was significantly less in infants born to females intubated with 1g/kg compared to infants from the remaining groups (Table 2). Additionally, analysis of nAC opioid-receptor protein revealed a significant main effect of prenatal treatment on MOR [F(2,22) = 45.866, p < 0.0001], KOR [F(2,22) = 3.498, p < 0.05] and DOR [F(2,23) = 3.826, p < 0.05] on PD 12 (Table 2). In all cases, immunopositive banding was significantly less in 12-day old infants born to females intubated with 2 g/kg during the last days of gestation compared to water-intubated controls. MOR from infants given the highest ethanol dose during gestation was also significantly less compared to pups given 1 g/kg during this same prenatal period.
Relatively few ethanol-dependent effects were observed in assessment of endogenous opioid ligands. As mentioned previously, effects of prenatal ethanol administration on opioid peptides was assessed independently at each age. Analyses of dynorphin within the nucleus accumbens revealed a significant main effect of prenatal treatment on PD 8 [F(2,23) = 3.062, p < 0.05] (Table 2). Similarly, hypothalamic enkephalin differed significantly as a function of prenatal ethanol administration when assessed on PD 12 [F(2,23) = 4.014, p < 0.05] (Table 2). In both cases, protein for the respective ligand was significantly reduced in infants born to ethanol-exposed dams compared to water intubated controls. There were no effects of PAE on opioid ligands in PD 4 tissue.
4. Discussion
The current body of work sought to characterize basal levels of opioid-related mRNA and protein in infant rats exposed to ethanol while in utero. In contrast to studies examining consequences of fetal alcohol exposure following the administration of large doses of alcohol (e.g., liquid diets contributing 25-36% of daily calories from ethanol) throughout the majority of gestation, the current work focused on consequences of moderate ethanol exposure (i.e., 1 and 2 g/kg) given during only a small portion of the gestational period (GD 17-20). The days in which alcohol was administered are not only those in which the fetus can successfully detect ethanol within the amniotic fluid (e.g., [43] and [44]) but, importantly, are also those in which ethanol exposure is capable of modifying subsequent intake of and responsiveness towards the drug (see [8] and [12] for reviews).
We chose to examine the effects of both age and prenatal alcohol exposure (PAE) on opioid protein and mRNA for the opioid receptors mu, kappa, and delta, along with their endogenous ligands, β-endorphin, dynorphin, and enkephalin. These factors were carefully selected based on their known involvement in both ethanol intake and reinforcement throughout early development (e.g., [9], [14], [18], [19], [20], and [35]) and adulthood (e.g., [21], [22], [23], [24], [25], and [26]). Each factor was examined in three brain regions - the nucleus accumbens, ventral tegmental area, and hypothalamus - sites known to be both high in opioid-containing neurons and implicated in general consummatory behavior or ethanol's appetitive effects. Tissue from animals prenatally exposed to either ethanol (1 or 2 g/kg) or water (0 g/kg) was later collected on one of three postnatal days - PD 4, 8, or 12 - days in which infants have been known to exhibit relatively low, moderate, and high ethanol consumption, respectively [30], and [31]. See Figure 1 for a summary of the entire experimental design.
The results presented here revealed a complex series of interactions between ontogeny, prenatal ethanol administration, brain structure, and factor of interest (see Tables 2-4 for a summary of results). After careful consideration of both age- and PAE- dependent effects, three main trends began to emerge. Firstly, age-dependent differences in opioid mRNA appear to be indicative of general upregulation of opioid signaling across the three postnatal ages assessed. Similar, albeit somewhat mixed, results were observed at the level of protein. Previous evaluations of opioid development have determined that, at the time of birth, opioid receptor density and localization within the CNS only partially resembles that of the adult [45]. Using in situ hybridization, both mu and kappa opioid receptor mRNA have been detected in the CNS as early as GD 13 - well before the age of assessment used here. In contrast to early developing mu- and kappa-opioid receptors, delta opioid receptor mRNA is not detected until GD 21 [45]. Delta-receptor mRNA first appears in, among other structures, the nucleus accumbens and soon thereafter, at birth, resembles the patterns (but not the density) of expression found in the adult [45]. Although we are unsure of the exact age at which adult-like opioid expression is apparent, one might suspect that the pattern of results observed here, in which the majority of factors were found to upregulate across early postnatal life, likely reflects continuing ontogenetic development and organization of this complicated system. Importantly, as we will now note, this developmental organization can be significantly affected by PAE.
Secondly, exposure to ethanol during the last days of gestation led to significant reductions in opioid-related protein, most notably within the nucleus accumbens. Importantly, one must be reminded that ethanol was administered only during the last days of gestation (GD 17-20) and not during postnatal life. Therefore, any effects of prenatal drug exposure observed at the time of assessment reflect relatively long-term alterations in basal opioid expression patterns and not acute activational responses to ethanol exposure at the time of tissue collection. Nevertheless, PAE effects on mRNA did not necessarily reflect these same alcohol-dependent reductions in protein. Consistent with other assessments of ethanol's effects on opioid gene expression (e.g., [46] and [47]), changes in mRNA produced by the prenatal treatment do not necessarily reflect alterations in protein for the same factors. Because there is a large divergence in the results obtained with mRNA and protein, it is especially difficult to interpret the ultimate consequence of ethanol's effects on the opioid system. Upon further investigation, however, one may notice that, at least within the nAC, the upregulation of mRNA for POMC, PPD and PPE present on PDs 4 or 8 are met by a reduction in protein for the mu-, kappa- and delta-opioid receptors on PD 12. Several interpretations can be generated to explain these findings. For example, early ontogeny and exposure to ethanol may both be associated with a reduction in the translational efficiency of mRNA and/or alterations in mRNA stability. A second interpretation of the present findings might be increased utilization of ligands as a result of PAE. Lastly, limitations in the sensitivity of our peptide quantification method could be responsible for the divergence between mRNA and protein observed here. Studies that have made use of radioimmunoassay have successfully found increases in met-enkephalin, the endogenous ligand for the DOR, within the nAC and hypo following a similar prenatal manipulation [34]. While future studies will be necessary to clarify the reasons for the dissociation between mRNA and protein observed here, such effects are common (e.g., [48] and [49], but see [50]) and underscore the importance of examining both mRNA and protein as a function of experimental treatment. Regardless, the present data provide important information regarding functional plasticity of the opioid system across early ontogeny and highlight the vulnerability of this system to prenatal programming effects of PAE. These results support the notion that dynamic changes in opioid signaling pathways are likely to play a role in age-related differences in ethanol reinforcement and, perhaps, long-term changes in responsiveness towards the drug.
Thirdly, ethanol-related effects were most abundant in the nucleus accumbens, a brain region that has been highly implicated in the consumption and dependence of nearly every drug of abuse. Among the main findings here were a significant reduction in accumbal mu-, kappa-, and delta-opioid receptors at 12 days of age. This seems especially important since mu- and delta-opioid receptor activation has been implicated in ethanol acceptance during adulthood (see [22], [23], [24], [51], and [52] for reviews), and kappa-receptor activation seems to be necessary to observe ethanol reinforcement during early infancy [20]. However, likely due to the relatively late emergence of delta-opioid receptors, this system has not yet been implicated in ethanol's appetitive effects during early infancy. Although the involvement of delta-opioid receptors in ethanol-related behavior during infancy was thought to be marginal at best, the current results suggest that delta-opioid receptors located within the nAC are susceptible to the programming influence of prenatal ethanol administration.
The kappa-opioid receptor system, on the other hand, in contrast to what is typically observed in the adult, has been found to be uniquely involved in ethanol-related behavior during early development. Previous work examining ethanol-related behaviors during early infancy have made mention of the unique ontogenetic shift in KOR activity (e.g., [20], [35], and [28]. Additional studies have led to the discovery that kappa-receptor activation may convey appetitive, and not aversive, information during the first few days of life [53] and [54] and, importantly, activation at the KOR seems necessary to observe ethanol reinforcement during early infancy [20]. More recently, we have found that prenatal exposure to ethanol, in doses similar to those used here, led to a reduction in kappa opioid receptor expression on PD 14 and alterations in the sensitivity of these receptors to KOR activation [35]. Interestingly, it is around this time when KOR activation first begins to convey some of the aversive-aspects of ethanol administration [28]. While it is not yet known precisely when the developmental shift in KOR activity occurs, nor whether the differences in DOR observed here are functionally involved in changing responsiveness towards the drug, these results highlight the unique sensitivity of the endogenous opioid system to early ethanol exposure. Certainly, the generalized reduction in all 3 opioid receptors produced by prenatal exposure to 2g/kg ethanol might reflect either (i) a generalized switch in phenotype of cells in the nAC incurred by prenatal ethanol exposure; or (ii) epigenetic modifications in the endogenous opioid system that could, potentially, contribute to long-term changes in ethanol acceptance and/or reinforcement. Though we cannot rule out the possibility that these effects might be reflective of ethanol-induced neurotoxicity and the result of general cell loss, it can be noted that cell loss typically requires substantially larger doses of ethanol [55] or longer ethanol exposure periods [56] than were employed in the present studies. Nevertheless, our data point to the Nucleus Accumbens as an excellent candidate site to target with future micro-injection studies to further delineate the functional role of the endogenous opioid system as a neurobiological substrate of ethanol reinforcement across early ontogeny.
One potential limitation worthy of discussion is that the present studies did not utilize a separate group of non-manipulated rats to control for the potential influence of the intubation procedures and/or dietary alterations produced by such manipulations. There are several reasons for this choice. First of all, water intubated controls effectively control for all experiential aspects of the primary experimental manipulation (ethanol exposure) and was deemed as the most appropriate control group for these experimental purposes. Second, ethanol was not delivered as part of the pregnant dam's diet (as many other studies do), but instead was delivered via intragastric intubation, which typically produces less dietary disturbance than ethanol diets per se, suggesting that dietary fluctuations as a result of gestational ethanol exposure should be minimal. Importantly, it should be noted that all dams (and offspring) were given ad libidum access to food and water at all times, and thus were able to self-regulate food and water consumption freely and without distress associated with food-restricted controls. Finally, adding another experimental group to the large, multi-factorial design would have adversely impacted the feasibility of the study, and threatened our ability to successfully execute such a large, multi-factorial project. With that said, the present results should be considered within the context of this minor limitation, and future studies might be necessary to take into account the potential influence of the intubation procedure on ethanol-related changes in opioid activity.
4. Conclusion
The endogenous opioid system has often been implicated in ethanol consumption and reinforcement (e.g., [19], [57], [58], [59], and [60]). Enhanced sensitivity of this system to alcohol administration is thought to underlie an increased vulnerability to high alcohol intake, at least in alcohol preferring animals and adult humans considered to be at high risk for the development of alcoholism [61], [62], [63], [64], [65], and [66]. While much is known regarding the opioid response to ethanol challenges during adulthood, considerably less is understood regarding this same response during early development or following prenatal exposure to the drug. The results presented here provide the first global assessment of the opioid family at the level of mRNA and protein across several brain regions and at three different ontogenetic time points. We suggest that exposure to moderate amounts of ethanol during only the last days of gestation are sufficient to produce relatively long-term alterations in basal opioid gene expression and protein. Importantly, these changes appear to be relatively site specific – being most robust in the nucleus accumbens - and differ as a function of early ontogeny. These results provide an early ontogenetic perspective on how opioid tone may influence alcohol intake as a function of age or prenatal ethanol treatment, thereby setting the stage for future studies to examine (a) programming effects of prenatal ethanol that persist into adulthood and (b) prenatal – postnatal ethanol interactions as a predictor for exposure-dependent enhancement of ethanol reinforcement.
Highlights.
Pregnant rats were exposed to moderate mounts of ethanol during late gestation.
Offspring were examined, as infants, for changes in opioid gene expression and protein.
Assessed factors in the nucleus accumbens, ventral tegmental area, and hypothalamus.
Results suggest relatively long-term changes stemming from prenatal exposure.
The nucleus accumbens emerged as a primary target for ethanol's effects.
Acknowledgements
The authors would like to acknowledge and honor Dr. Norman “Skip” Spear for his wisdom, generosity, patience, and support. Skip has made a lifetime of contributions to the field of psychology and has affected hundreds of lives through his science and his mentorship. Whether you've had the pleasure of sharing a cup of tea and a cookie in his office or presenting research at a meeting in which he attended, simply being in Skip's presence made you want to be a better scientist. We thank Skip for all that he has done and continues to do for the field and for the countless students and colleagues with which he's guided. Certainly, without him, none of this work would have been possible.
This work was supported by: NIH grants number AA016305 to T.D. and AA011960, AA01309, and AA017823 to Norman Spear, the Developmental Exposure Alcohol Research Center (DEARC; P50AA017823), and the Center for Development and Behavioral Neuroscience at Binghamton University. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the above stated funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interests:
The authors have no conflicts of interest to declare.
References
- 1.Clarren SK, Astley SJ, Bowden DM. Physical anomalies and developmental delays in nonhuman primate infants exposed to weekly doses of ethanol during gestation. Teratology. 1988;37(6):561–569. doi: 10.1002/tera.1420370605. [DOI] [PubMed] [Google Scholar]
- 2.Driscoll CD, Streissguth AP, Riley EP. Prenatal alcohol exposure: comparability of effects in humans and animal models. Neurotoxicol Tertol. 1990;12(3):231–237. doi: 10.1016/0892-0362(90)90094-s. [DOI] [PubMed] [Google Scholar]
- 3.Graham JM, Hanson JW, Darby BL, Barr HM, Streissguth AP. Independent dysmorphology evaluations at birth and 4 years of age for children exposed to varying amounts of alcohol in utero. Pediatrics. 1988;81(6):772–778. [PubMed] [Google Scholar]
- 4.Clarren SK, Astley SJ, Bowden DM, La H, Milam AH, Rudeen PK, Shoemaker WJ. Neuroanatomic and neurochemical abnormalities in nonhuman primate infants exposed to weekly doses of ethanol during gestation. Alcohol Clin Exp Res. 1990;14(5):674–683. doi: 10.1111/j.1530-0277.1990.tb01226.x. [DOI] [PubMed] [Google Scholar]
- 5.Arias C, Chotro MG. Increased palatability of ethanol after prenatal ethanol exposure is mediated by the opioid system. Pharmacology, Biochemistry, and Behavior. 2005;82(3):434–442. doi: 10.1016/j.pbb.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Arias C, Chotro MG. Increased preference for ethanol in the infant rat after prenatal exposure, expressed on intake and taste reactivity tests. Alcohol Clin Exp Res. 2005;29(3):337–346. doi: 10.1097/01.alc.0000156115.35817.21. [DOI] [PubMed] [Google Scholar]
- 7.Chotro MG, Arias C. Prenatal exposure to ethanol increases ethanol consumption: A conditioned response? Alcohol. 2003;30(1):19–28. doi: 10.1016/s0741-8329(03)00037-5. [DOI] [PubMed] [Google Scholar]
- 8.Chotro MG, Arias C, Laviola G. Increased ethanol intake after prenatal ethanol exposure: studies with animals. Neurosci Biobehav Rev. 2007;31(2):181–191. doi: 10.1016/j.neubiorev.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Cenzano E, Gaztanaga M, Chotro GM. Exposure to ethanol on prenatal days 19-20 increases ethanol intake and palatability in the infant rat: Involvement of kappa and mu opioid receptors. Developmental Psychobiology. 2014;56(6):1167–1178. doi: 10.1002/dev.21162. [DOI] [PubMed] [Google Scholar]
- 10.Dominguez HD, Lopez MF, Molina JC. Neonatal responsiveness to alcohol odor and infant alcohol intake as a function of alcohol experience during late gestation. Alcohol. 1998;16(2):109–117. doi: 10.1016/s0741-8329(97)00169-9. [DOI] [PubMed] [Google Scholar]
- 11.Fabio MC, March SM, Molina JC, Nizhniko ME, Spear NE, Pautassi RM. Prenatal ethanol exposure increases ethanol intake and reduces c-Fos expression in infralimbic cortex of adolescent rats. Pharmacol Biochem Behav. 2013;103(4):842–852. doi: 10.1016/j.pbb.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chotro MG, Arias C. Exposure to low and moderate doses of alcohol on late gestation modifies infantile response to and preference for alcohol in rats. Ann 1st Super Sanita. 2006;42(1):22–30. [PubMed] [Google Scholar]
- 13.Shea KM, Hewitt AJ, Olmstead MC, Brien JF, Reynolds JN. Maternal ethanol consumption by pregnant guinea pigs causes neurobehavioral deficits and increases ethanol preference in offspring. Behav Pharmacol. 2012;23(1):105–112. doi: 10.1097/FBP.0b013e32834ed866. [DOI] [PubMed] [Google Scholar]
- 14.Pautassi RM, Nizhnikov ME, Spear NE, Molina JC. Prenatal ethanol exposure leads to greater ethanol-induced appetitive reinforcement. Alcohol. 2012;46(6):585–593. doi: 10.1016/j.alcohol.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.March SM, Abate P, Spear NE, Molina JC. Fetal exposure to moderate ethanol doses: heightened operant responsiveness elicited by ethanol-related reinforcers. Alcohol Cllin Exp Res. 2009;33(11):1981–1993. doi: 10.1111/j.1530-0277.2009.01037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miranda-Morales RS, Nizhnikov ME, Spear NE. Prenatal exposure to ethanol during late gestation facilitates operant self-admnistration of the drug in 5-day-old rats. Alcohol. 2014;48(1):19–23. doi: 10.1016/j.alcohol.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nizhnikov ME, Molina JC, Varlinskaya EI, Spear NE. Prenatal ethanol exposure increases ethanol reinforcement in neonatal rats. Alcohol Cllin Exp Res. 2006;30(1):34–45. doi: 10.1111/j.1530-0277.2006.00009.x. [DOI] [PubMed] [Google Scholar]
- 18.Chotro MG, Arias C. Ontogenetic difference in ethanol reinforcing properties: the role of the opioid system. Behav Pharmacol. 2007;18(7):661–666. doi: 10.1097/FBP.0b013e3282f00754. [DOI] [PubMed] [Google Scholar]
- 19.Miranda-Morales RS, Spear NE, Nizhnikov ME, Molina JC, Abate P. Role of mu, delta and kappa opioid receptors in ethanol-reinforced operant responding in infant rats. Behav Brain Res. 2012;234(2):267–277. doi: 10.1016/j.bbr.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nizhnikov ME, Varlinskaya EI, Petrov ES, Spear NE. Reinforcing properties of ethanol in neonatal rats: Involvement of the opioid system. Behavioral Neuroscience. 2006;120(2):267–280. doi: 10.1037/0735-7044.120.2.267. [DOI] [PubMed] [Google Scholar]
- 21.Froehlich JC. Interactions between alcohol and the endogenous opioid system. In: Zakhari S, editor. Alcohol and the Endocrine System (National Institute on Alcohol Abuse and Alcoholism Research Monograph. 23. U. S. Department of Health and Human Services; Bethesda, MD: 1993. pp. 31–35. [Google Scholar]
- 22.Gianoulakis C. Implications of endogenous opioids and dopamine in alcoholism: Human and basic science studies. Alcohol and Alcoholism. 1996;31:33–42. [PubMed] [Google Scholar]
- 23.Gianoulakis C. Influence of the endogenous opioid system on high alcohol consumption and genetic predisposition to alcoholism. J Psychiatry Neurosci. 2001;26(4):304–318. [PMC free article] [PubMed] [Google Scholar]
- 24.Herz A. Endogenous opioid system and alcohol addiction. Psychopharmacology. 1997;129:99–111. doi: 10.1007/s002130050169. [DOI] [PubMed] [Google Scholar]
- 25.Oswald LM, Wand GS. Opioids and alcoholism. Physiology and Behavior. 2004;81:339–358. doi: 10.1016/j.physbeh.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Ulm RR, Volpicelli JR, Volpicelli LA. Opiates and alcohol self-administration in animals. Journal of Clinical Psychiatry. 1995;56:5–14. [PubMed] [Google Scholar]
- 27.Gianoulakis C, De Waele JP, Thavundayil J. Implication of the endogenous opioid system in excessive ethanol consumption. Alcohol. 1996;13(1):19–23. doi: 10.1016/0741-8329(95)02035-7. [DOI] [PubMed] [Google Scholar]
- 28.Pautassi RM, Nizhnikov ME, Acevedo MB, Spear NE. Early role of the k opioid receptor in ethanol-induced reinforcement. Physiol Behav. 2012;105(5):1231–1241. doi: 10.1016/j.physbeh.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barson JR, Carr AJ, Soun JE, Sobhani NC, Rada P, Leibowitz SF, Hoebel BG. Opioids in the hypothalamic paraventricular nucleus stimulate ethanol intake. Alcohol Clin Exp Res. 2010;34(2):214–222. doi: 10.1111/j.1530-0277.2009.01084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders SK, Spear NE. Ethanol acceptance is high during earlyfancy and becomes still higher after previous ethanol ingestion. Alcohol Clin Exp Res. 2007;31(7):1148–1158. doi: 10.1111/j.1530-0277.2007.00400.x. [DOI] [PubMed] [Google Scholar]
- 31.Spear NE, Molina JC. Fetal or infantile exposure to ethanol promotes ethanol ingestion in adolescence and adulthood: A theoretical review. Alcohol Clin Exp Res. 2005;29(6):909–929. doi: 10.1097/01.alc.0000171046.78556.66. [DOI] [PubMed] [Google Scholar]
- 32.Gilpin NW, Richardson HN, Koob GF. Effects of CRF1-receptor and opioid-receptor antagonists on depednece-induced increases in alcohol drinking by alcohol-preferring (P) rats. Alcohol Clin Exp Res. 2008;32(9):1535–1542. doi: 10.1111/j.1530-0277.2008.00745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juarez J, Barrios De Tomasi E. Naltrexone treatment produces dose-related effects on food and water intake but daily alcohol consumption is not affected. Nutr Neurosci. 2008;11(4):183–192. doi: 10.1179/147683008X301577. [DOI] [PubMed] [Google Scholar]
- 34.Abate P, Hernandez-Fonseca K, Reyes-Guzman AC, Barbosa-Luna IG, Mendez M. Prenatal ethanol exposure alters met-enkephalin expression in brain regions related with reinforcement: possible mechanism for ethanol consumption in offspring. Behav Brain Res. 2014 Nov 1;(274):194–204. doi: 10.1016/j.bbr.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 35.Nizhnikov ME, Pautassi RM, Carter JM, Landin JD, Varlinskaya EI, Bordner KA, Werner DF, Spear NE. Brief prenatal ethanol exposure alters behavioral sensitivity to the kappa opioid receptor agonist (U62,066E) and antagonist (Nor-BNI) and reduces kappa opioid receptor expression. Alcohol Clin Exp Res. 2014;38(6):1630–1638. doi: 10.1111/acer.12416. [DOI] [PubMed] [Google Scholar]
- 36.Agnish ND, Keller KA. The rationale for culling of rodent litters. Fundam Appl Toxicol. 1997;38(1):2–6. doi: 10.1006/faat.1997.2318. [DOI] [PubMed] [Google Scholar]
- 37.Altman J, Bayer SA. Atlas of Prenatal Rat Brain Development. CRC; Boca Raton: 1995. [Google Scholar]
- 38.Hellemans J, Mortier G, De Paepe A, Speeman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):1–12. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradford MM. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Annals of Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 41.Molina JC, Pautassi RM, Truxell E, Spear NE. Differential motivational properties of ethanol during early ontogeny as a function of dose and postadministration time. Alcohol. 2007;41(1):41–55. doi: 10.1016/j.alcohol.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molina JC, Ponce LF, Truxell E, Spear NE. Infantile sensitivity to ethanol's motivational effects: Ethanol reinforcement during the third postnatal week. Alcohol Clin Exp Res. 2006;30(9):1506–1519. doi: 10.1111/j.1530-0277.2006.00182.x. [DOI] [PubMed] [Google Scholar]
- 43.Arias C, Chotro MG. Amniotic fluid can act as an appetitive unconditioned stimulus in preweanling rats. Dev Psychobiol. 2007;49(2):139–149. doi: 10.1002/dev.20205. [DOI] [PubMed] [Google Scholar]
- 44.Molina JC, Chotro MG. Association between chemosensory stimuli and cesarean delivery in rat fetuses: neonatal presentation of similar stimuli increases motor activity. Behav Neural Biol. 1991;55(1):42–60. doi: 10.1016/0163-1047(91)80126-y. [DOI] [PubMed] [Google Scholar]
- 45.Georges F, Normand E, Bloch B, Le Moine C. Opioid receptor gene expression in the rat brain during ontogeny, with special reference to the mesostriatal system: An in situ hybridization study. Brain Res Dev Brain Res. 1998;109(2):187–199. doi: 10.1016/s0165-3806(98)00082-0. [DOI] [PubMed] [Google Scholar]
- 46.Chang GQ, Karatayev O, Ahsan R, Avena NM, Lee C, Lewis MJ, Hoebel BG, Leibowitz SF. Effects of ethanol on hypothalamic opioid peptides, enkephalin, and dynorphin: relationship with circulating triglycerides. Alcohol Clin Exp Res. 2007;31(2):249–259. doi: 10.1111/j.1530-0277.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- 47.Winkler A, Buzas B, Siems WE, Heder G, Cox BM. Effect of ethanol drinking on the gene expression of opioid receptors, enkephalinase, and antiotensin-converting enzyme in two inbred mice strains. Alcohol Clin Exp Res. 1998;22(6):1262–1271. [PubMed] [Google Scholar]
- 48.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19(3):1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nie L, Gang W, Zhang W. Correlation between mRNA and protein abundance in Desulfovibrio vulgaris: A multiple regression to identify sources of variations. Biochem Biophys Res Comm. 2006;339(2):603–610. doi: 10.1016/j.bbrc.2005.11.055. [DOI] [PubMed] [Google Scholar]
- 50.Doremus-Fitzwater TL, Buck HM, Bordner K, Richey L, Jones ME, Deak T. Intoxication- and withdrawal-dependent expression of central and peripheral cytokines following initial ethanol exposure. Alcohol Clin Exp Res. 2014;38(8):2186–2198. doi: 10.1111/acer.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cowen MS, Lawrence AJ. The role of opioid-dopamine interactions in the induction and maintenance of ethanol consumption. Prog Neuropsychopharmacol Biol Psychiatry. 1999;23(7):1171–1212. doi: 10.1016/s0278-5846(99)00060-3. [DOI] [PubMed] [Google Scholar]
- 52.Gianoulakis C. Endogenous opioids and excessive alcohol consumption. J Psychiatry Neurosci. 1993;18(4):148–156. [PMC free article] [PubMed] [Google Scholar]
- 53.Barr GA, Wang S, Carden S. Aversive properties of the kappa opioid agonist U50,488 in the week-old rat pup. Psychopharmacology. 1994;113(3-4):422–428. doi: 10.1007/BF02245218. [DOI] [PubMed] [Google Scholar]
- 54.Petrov ES, Nizhnikov ME, Varlinskaya EI, Spear NE. Dynorphin A (1-13) and responsiveness of the newborn rat to a surrogate nipple: immediate behavioral consequences and reinforcement effects in conditioning. Behav Brain Res. 2006;170(1):1–14. doi: 10.1016/j.bbr.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 55.Balaszczuk V, Bender C, Pereno GL, Beltramino CA. Alcohol-induced neuronal death in central extended amygdala and pyriform cortex during the postnatal period of the rat. Int J Dev Neurosci. 2011;29(7):733–742. doi: 10.1016/j.ijdevneu.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 56.Crews FT, Braun CJ, Hoplight B, Switzer RC, 3rd, Knapp DJ. Binge ethanol consumption causes differential brain damage in young adolescent rats compared with adult rats. Alcohol Clin Exp Res. 2000;24(11):1712–1723. [PubMed] [Google Scholar]
- 57.Camarini R, Pautassi RM, Mendez M, Quadros IM, Souza-Formigoni ML, Boerngen-Lacerda R. Behavioral and neurochemical studies in distinct animal models of ethanol's motivational effects. Curr Drug Abuse Rev. 2010;3(4):205–221. doi: 10.2174/1874473711003040205. [DOI] [PubMed] [Google Scholar]
- 58.Gianoulakis C. Endogenous opioids and addiction to alcohol and other drugs of abuse. Curr Top Med Chem. 2009;9(11):999–1015. doi: 10.2174/156802609789630956. [DOI] [PubMed] [Google Scholar]
- 59.Mendez M, Morales-Mulia M. Role of mu and delta opioid receptors in alcohol drinking behaviour. Curr Drug Abuse Rev. 2008;1(2):239–252. doi: 10.2174/1874473710801020239. [DOI] [PubMed] [Google Scholar]
- 60.Tseng A, Nguyen K, Hamid A, Garg M, Marquez P, Lutfy K. The role of endogenous beta-endorphin and enkephalins in ethanol reward. Neuropharmacology. 2013;73:290–300. doi: 10.1016/j.neuropharm.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Waele JP, Papachristou DN, Gianoulakis C. The alcohol-preferring C57BL/6 mice present an enhanced sensitivity to the hypothalamic beta-endorphin system to ethanol than the alcohol-avoiding DBA/2 mice. J Pharmacol Exp Ther. 1992;261(2):788–794. [PubMed] [Google Scholar]
- 62.Froehlich JC. Genetic factors in alcohol self-administration. Journal of Clinical Psychiatry. 1995;56(7):15–23. [PubMed] [Google Scholar]
- 63.Gianoulakis C, De Waele JP, Kiianmaa K. Differences in the brain and pituitary beta-endorphin system between the alcohol-preferring AA and alcohol-avoiding ANA rats. Alcohol Clin Exp Res. 1992;16(3):453–459. doi: 10.1111/j.1530-0277.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 64.Li XW, Li TK, Froehlich JC. Enhanced sensitivity of the nucleus accumbens proenkephalin system to alcohol in rats selectively bred for alcohol preference. Brain Res. 1998;794(1):35–47. doi: 10.1016/s0006-8993(98)00191-7. [DOI] [PubMed] [Google Scholar]
- 65.Mendez M, Morales-Mulia M. Ethanol exposure differentially alters pro-enkephalin mRNA expression in regions of the mesocorticolimbic system. Psychopharmacology (Berl) 2006;189(1):117–124. doi: 10.1007/s00213-006-0503-3. [DOI] [PubMed] [Google Scholar]
- 66.Ng GY, O'Dowd BF, George SR. Genotypic differences in mesolimbic enkephalin gene expression in DBA/2J and C57BL/6J inbred mice. Eur J Pharmacol. 1996;311(1):45–52. doi: 10.1016/0014-2999(96)00401-3. [DOI] [PubMed] [Google Scholar]