Abstract
The evolutionarily conserved CD1 family of antigen-presenting molecules presents lipid antigens rather than peptide antigens to T cells. CD1 molecules, unlike classical MHC molecules, display limited polymorphism, making CD1-restricted lipid antigens attractive vaccine targets that could be recognized in a genetically diverse human population. Group 1 CD1 (CD1a, CD1b, and CD1c)-restricted T cells have been implicated to play critical roles in a variety of autoimmune and infectious diseases. In this review, we summarize current knowledge and recent discoveries on the development of group 1 CD1-restricted T cells and their function in different infection models. In particular, we focus on (1) newly identified microbial and self-lipid antigens, (2) kinetics, phenotype, and unique properties of group 1 CD1-restricted T cells during infection, and (3) the similarities of group 1 CD1-restricted T cells to the closely related group 2 CD1-restricted T cells.
Keywords: Mycobacterium tuberculosis, CD1, antigen presentation, T cells, NKT cells, animal models
Introduction
The CD1 gene family encodes several MHC class I-like antigen-presenting molecules, which are specialized to present lipid antigens to T cells. The lipid antigens presented by CD1 include a diverse array of lipids/glycolipids, ranging from foreign lipids unique to specific microorganisms to common mammalian self-lipids. Three major groups of CD1 isoforms have been identified in humans: group 1 CD1 (CD1a, CD1b, and CD1c), group 2 CD1 (CD1d), and group 3 CD1 (CD1e) (1). Although homologs exist in the guinea pigs and other vertebrates, mice, and rats do not express group 1 CD1 (2, 3), making the in vivo functional study of group 1 CD1-restricted T cells difficult until the recent development of small animal models (4–6). This review will outline group 1 CD1 antigen binding and presentation, newly identified group 1 CD1-restricted self- and microbial lipid antigens, and the interaction of group 1 CD1-expressing antigen-presenting cells (APCs) with their corresponding T cell subsets. Additionally, the development and function of group 1 CD1-restricted T cells will be discussed, with special emphasis placed on small animal models, dynamics of these T cells during Mycobacterium tuberculosis (Mtb) infection, and the contribution of microbial antigen-specific and autoreactive group 1 CD1-restricted T cells to host defense against infection. Finally, the expansion of our knowledge of group 1 CD1-restricted T cell responses allows for their comparison with a more well-studied and related subset of T cells, the group 2 CD1-restricted NKT cells. The last section will focus on comparing and contrasting the properties of group 1 vs. group 2 CD1-restricted T cells.
Expression of Group 1 CD1 Molecules
Unlike MHC class Ia, which is expressed on all nucleated cells, the expression of group 1 CD1 is mainly limited to APCs and double-positive (CD4+CD8+) cortical thymocytes (1). CD1a molecules are highly expressed on skin resident DCs, or Langerhans cells (7). CD1b is most highly expressed on a subset of migrating lymph dendritic cells and myeloid-derived dendritic cells (8). CD1c is the most ubiquitously expressed group 1 CD1 molecule, being found on monocyte-derived DCs, B cells, and Langerhans cells under steady-state conditions (9, 10). CD1e is the only CD1 isoform that is not expressed on the surface of APCs (11). In immature DCs, CD1e is mainly localized in the Golgi apparatus and in mature DCs, it is detected in late endosomes, where it is cleaved into a functional soluble form (12). CD1e is thought to facilitate the processing and presentation of certain lipid antigens presented by CD1b (13).
Group 1 CD1 expression can also be regulated by various cytokines. Treatment of activated monocytes with GM-CSF and IL-4, which leads to dendritic cell differentiation, results in increased levels of group 1 CD1 expression (14). Group 1 CD1 expression is also altered in different disease models, implying a role for group 1 CD1-restricted T cells in the pathology and/or repression of the disease. Patients with the tuberculoid form of leprosy, a clinical pattern associated with active cellular immunity toward Mycobacterium leprae, show induction of CD1a, CD1b, and CD1c on DCs in dermal granulomas (15). Increased CD1b and CD1c expression has also been reported in the human skin upon Borrelia burgdorferi infection (16). CD1a expression on Langerhans cells has been correlated with a variety of autoimmune skin diseases, including atopic eczema (7, 17, 18) and psoriasis (19, 20). In addition, some leukemias and lymphomas express one or all group 1 CD1 molecules (21). One recent study showed that CD1c+ B cell leukemia precursors are efficiently targeted and lysed by CD1c autoreactive T cells, highlighting a role of group 1 CD1-restrcited T cells in anti-tumor immunity (22).
Binding and Presentation of Lipid Antigens by Group 1 CD1 Molecules
Group 1 CD1 isoforms have structurally diverse antigen-binding grooves that allow them to bind very different lipid classes. Group 1 CD1 isoforms also differ in their stability when lipids are bound in the binding groove vs. when the groove lies empty, which is consistent with their intracellular trafficking patterns and consequently the types of lipids they are exposed to (23, 24). CD1b and CD1c molecules traffic through and load lipids in the late endosome and the lysosome, and thus, are exposed to a low pH that may facilitate lipid exchange (25). However, CD1a molecules are unique in that under steady-state conditions, they do not accumulate or traffic through the late endosomes or lysosomes, and thus, are not exposed to the same low pH conditions as CD1b and -c molecules (26). Instead, modeling simulations suggest that CD1a molecules can be stably expressed on the cell surface in the absence of loaded lipid antigens (27), and other studies have shown that the CD1a binding groove can be stabilized by exogenously added lipids after being expressed on the cell surface (28).
Group 1 CD1 molecules present both microbial and self-lipid antigens to T cells. Historically, microbial antigens that were found to bind to group 1 CD1 molecules were derived from the cell wall of Mycobacteria species. Below, we will describe how the various group 1 CD1 molecules interact with different types of lipids, and what some of the functional consequences for T cell activation might be.
Lipid antigen presentation by CD1a
CD1a molecules can bind the mycobacterial lipopeptide didehydroxymycobactin (DDM), a biosynthetic precursor of the iron-chelating siderophore mycobactin, which is essential for Mtb growth within macrophages (29). CD1a presentation of DDM and subsequent TCR recognition of antigen have features reminiscent of both CD1 and MHC-like binding modes in that the CD1a-restricted TCR is capable of responding in an amino-acid sequence-specific manner to the solvent-exposed portion of the DDM lipopeptide (30). DDM-specific CD1a-restricted T cells were found to secrete varying levels of IL-2 in response to different synthetic DDM analogs, with the largest amount of IL-2 being produced when the DDM antigen was in its native state (31). This showed that CD1a-restricted T cells could specifically respond to the structural components of an antigen that are responsible for virulence (31).
The crystal structure of CD1a–DDM complex showed that A′ pocket of CD1a antigen-binding groove binds alkyl chains of discrete length, while the F′ pocket is less rigid and allows for different chemical head groups of lipid antigens to protrude from the groove (30). The crystal structure of CD1a with the bound self-antigen sulfatide provides another example of this type of binding mode wherein the sphingosine fits into the A′ pocket and the polar headgroup of sulfatide protrudes from the middle of the groove, allowing for optimal TCR recognition (32). However, despite the body of work showing that CD1a lipid antigens generally contain protruding polar headgroups and hydrophobic tails, a recent study showed that CD1a molecules could also bind and present “headless” lipid antigens to T cells (33). In fact, synthetic squalene, a lipid containing no headgroup, was able to activate the BC2 CD1a-restricted T cell line to the same extent as what was seen from challenge with sebaceous gland extract containing a mixture of CD1a autoantigens (33). This suggests that CD1a has the ability to recognize “headless” lipid antigens that lack a polar moiety available to be recognized by the TCR. A recent finding by Rossjohn’s group provided a potential explanation for this (34). This study first solved the crystal structure of a TCR bound to CD1a and mechanistically showed that the TCR can directly contact CD1a in the absence of lipid interaction. This interaction is sufficient for T cell activation. However, if CD1a is loaded with non-permissive lipids, there is a disruption of the TCR–CD1a docking site (34). Therefore, CD1a autoreactivity can result from direct contact of the TCR with CD1a, but only when the surface of CD1a is not disrupted by non-permissive ligands. Furthermore, a recent study showed that bee venom-derived phospholipase 2 could activate CD1a-autoreactive T cells through the generation of small neo-antigens, such as free fatty acids and lysophopholipids, from common phosphodiacyl glycerides (35). This finding provides a potential mechanism underlying phospholipase-dependent inflammatory skin disease.
Lipid antigen presentation by CD1b
CD1b has the largest antigen-binding groove among the CD1 proteins described to date. As such, CD1b binds and presents the most diverse array of mycobacterial lipids, which include mycolic acid (36), glucose monomycolate (GMM) (37), glycerol monomycolate (38), phosphatidylinositol mannosides (PIMs) (39, 40), lipoarabinomannans (LAM) (41–43), and sulfoglycolipids (44). The CD1b binding groove is composed of four interconnected pockets, which enable binding of the long hydrophobic tails of lipid antigens (45). The structure of CD1b–GMM complex showed that the A′, C′, F′, and T′ pockets in the CD1b binding groove form a deep “maze” that allows them to present large mycobacterial lipid antigens to T cells, and the close association of lipid ligands with CD1b molecules is suggestive of this complex having a long half-life (45). These studies also show that the pleiotropic binding of structurally diverse lipids to CD1b molecules can occur due to critical amino-acid residues in the CD1b binding groove having the ability to change their conformation to induce a tighter fit for different lipids (45, 46). However, the finding that CD1b molecules bind and are stabilized by such large microbial lipid ligands led to the question of how unloaded CD1b molecules retain their structure in the absence of endosomally loaded lipid. Operating from the hypothesis that endogenous self-lipids could act as stabilizers of the Ag-binding groove for CD1b in the absence of foreign ligands, Garcia-Alles et al. showed that CD1b associates with phosphatidylcholine (PC) and a long 40+ carbon spacer molecule prior to endosomal lipid loading (47). The combined number of carbons shared between PC and the spacer molecule is greater than the C65–C70 capacity of the CD1b binding groove, but this “overloading” of the groove may actually facilitate lipid exchange in the endolysosome (47).
Mycolic acids were the first CD1b-restricted lipids identified (36). Mycolic acids are the major Mtb cell wall lipid components and are crucial for Mtb virulence (48). Studies in untreated human Mtb patients showed that many patients contained IFN-γ producing, mycolic acid-specific CD1b-restricted T cells in their PBMCs (49), suggesting that mycolic acids are immunodominant CD1b-restricted antigens relevant during Mtb infection. While free mycolic acids can activate some CD1b-restricted T cells; one recent study suggested that mycolic acids may act as scaffolds for other lipid antigens, with the tight association of the long hydrophobic tails in the CD1b binding pockets leading to proper positioning of various lipid antigenic headgroup for TCR recognition (38).
Diacylated sulfoglycolipids (Ac2SGL) are produced by virulent Mtb strains upon infection of the human host (44, 50). Studies using human CD1b-restricted T cell lines specific for Ac2SGL showed that the aliphatic tail region is responsible for anchoring the molecule deep in the CD1b antigen-binding groove, while the trehalose headgroup is responsible for TCR recognition (50). However, the antigen specificity of particular T cell lines could be influenced by the length and isomerization of the hydrophobic tail region of Ac2SGL (50). This corroborates the study described above, which showed that the long hydrophobic lipid tails of CD1b lipid antigens, including Ac2SGLs, can act to position the antigenic headgroups optimally for TCR recognition (38, 47).
Mycobacterium tuberculosis PIMs and their polyglycosylated extensions lipomannans (LM) and LAM (51) are components of the Mtb cell envelope (52). Many forms of LM and LAM are presented by CD1b (42, 43). Biochemical and functional characterization of structurally diverse forms of LM showed that the pathogenic Mtb strain H37RV contained LM moieties with longer and more branched mannan polymer domains than those from the avirulent strain Mycobacterium smegmatis (43). Thus, it is possible that virulent Mtb strains evolved to use their LM lipids as immune evasion factors, as CD1b–LM complexes from H37Rv were less potent activators of CD1b-restricted T cell responses than those from M. smegmatis (43).
Lipid antigen presentation by CD1c
The only known Mtb antigen presented by CD1c is phosphomycoketide (PM), a lipid that contains a single fully saturated alkyl chain with methyl branches at every fourth carbon (53, 54). The production of Mtb mannose-1-β-phosphomycoketide (MPM) occurs via enzymatic catalysis by Pks12, which is only found in slow-growing pathogenic strains of Mtb (55), suggesting that MPM is a potential virulence factor in Mtb pathogenesis. Recent studies showed that PM, a putative biosynthetic precursor for MPM can also bind to CD1c and activate CD1c-restricted T cells (56).
CD1c molecules have a unique antigen-binding mode that allows for presentation of diverse antigens, such as highly branched lipid species to T cells (57). Whereas lipid antigens bind in both the A′ and F′ pockets in CD1a and CD1b molecules (32, 45), MPM binds only to the A′ pocket in CD1c molecules, allowing for the branched side chains to protrude out from the groove (57). Like CD1a, CD1c can also present lipopeptide antigens to T cells (58). In addition, CD1c has been shown to bind sulfatide and other diacylated lipids similar to CD1a and CD1b (50, 59), suggesting that the CD1c binding groove has evolved to present a diverse array of lipid species.
A recent paper also helped shed light on a potential role for autoreactive CD1c-restricted T cells (22). Autoreactive CD1c-restricted T cells were found to recognize methyl-lysophosphotidic acid, a novel class of self-lipids, which accumulate in leukemia cells (22). While these autoreactive CD1c-restricted T cells poorly recognize non-tumor CD1c-expressing cells, they killed CD1c+ acute leukemia cells and protected immunodeficient mice against CD1c+ human leukemia cells (22). This study suggests that autoreactive CD1c-restricted T cells may play a role in anti-tumor immunity (22). However, their role in anti-microbial immunity remains unknown.
Kinetics, Phenotype, and Unique Properties of Group 1 CD1-Restricted T Cells
Much of the information about group 1 CD1-restricted T cells comes from in vitro assays using human T cell clones. Recent discoveries in tetramer development and improved cell isolation technology have greatly enhanced the study of group 1 CD1 molecules in humans, though animal models are critical to fully understand the dynamic biology of group 1 CD1-restricted T cells. A recent study using rhesus macaques identified a GMM-specific T cell population in BCG-immunized monkeys (60). However, unlike humans, GMM-specific T cells were found to be CD1c restricted (61). Though interesting, following up on such findings in primates can be challenging due to cost constraints. As an alternative to studying non-human primates, small animal models using transgenic mice have proven invaluable for the study of group 1 CD1-restricted T cells. In the following sections, we will review recent discoveries about group 1 CD1-restricted T cells and the role they play in host immunity in vivo.
Small animal models to study the In vivo function of group 1 CD1-restricted T cells
There have been a number of small animal models used to study the in vivo function of group 1 CD1-restricted T cell responses during Mtb infection, including the guinea pig model and humanized mouse model. Based on comparison with human CD1 isoforms, the nine guinea pig CD1 genes encode four CD1b-like, three CD1c-like, one CD1d-like, and one CD1e-like molecule (2, 62). Immunization of guinea pig with mycobacterial lipids has been shown to elicit antigen-specific proliferation and cytolytic capacity of group 1 CD1-restricted T cells (63). In addition, Mtb lipid-vaccinated guinea pigs exhibit reduced lung pathology after subsequent Mtb challenge (64). However, it has been difficult to show that group 1 CD1-restricted T cells mediate these protective effects due to limited experimental tools and reagents. Moreover, guinea pigs lack CD1a and have multiple isoforms of the CD1b and CD1c genes, suggesting that the group 1 CD1-restricted T cells response may be different in guinea pigs and humans.
A humanized mouse model developed by Gumperz’s group was observed to develop a functional CD1 compartment (6). This model involved grafting immunodeficient mice with human fetal thymus, liver, and CD34+ hematopoietic stem cells (6). While these mice were observed to express group 1 CD1 molecules, CD1a tissue expression patterns differed from humans (6). For instance, CD1a-expressing cells, which are normally abundant in human skin, are largely absent in humanized mice. It remains to be seen whether group 1 CD1-restrcited T cell responses can be elicited in these humanized mice upon immunization or during infection.
A transgenic mouse model that expresses the human group 1 CD1 genes under their endogenous human promoters has been generated (4). Human group 1 CD1 transgenic (hCD1Tg) mice express CD1a, CD1b, and CD1c in a pattern similar to that seen in humans and support the development of group 1 CD1-restricted T cells (4). These T cells from hCD1Tg mice also share many phenotypic and functional characteristics with CD1-restricted T cells found in humans (4). These data demonstrate the viability of hCD1Tg mice as an animal model to study the in vivo function of group 1 CD1-restricted T cells. Indeed, both infection with Mtb and immunization with Mtb lipids elicit group 1 CD1-restricted T cell responses in hCD1Tg mice (4).
Group 1 CD1-restricted T cell responses during Mtb infection
The presentation of mycobacterial lipid antigens by group 1 CD1 molecules has been well characterized. However, our understanding of the phenotype and function of responding T cells was mostly limited to in vitro assays with T cell clones until recently. Nevertheless, in vitro studies have demonstrated that group 1 CD1-restricted T cells are cytotoxic and produce IFN-γ and TNF-α upon encountering mycobacterial antigens (65, 66). Moreover, group 1 CD1-restricted Mtb lipid antigen-specific T cells are found in higher frequencies in individuals exposed to Mtb compared with a control population, suggesting that Mtb-specific CD1-restricted T cells are activated following infection with Mtb (38, 44, 65–67). The discovery of CD1-tetramer technology has made it easier to study T cell reactivity by enabling the direct isolation of antigen-specific T cells from the blood and tissue (56, 68). Moreover, the establishment of a group 1 CD1 transgenic mouse model has enabled the study of tissue distribution, activation kinetics, and phenotype of group 1 CD1-restricted T cells.
CD1a dextramers loaded with the mycobacterial lipopeptide DDM stained CD1a-restricted T cells in individuals with active TB and tuberculin-positive individuals, suggesting an expansion had occurred upon exposure to Mtb (69). CD1a/DDM dextramer+ T cells were found to be either CD4+ or CD8+, and produced IFN-γ and TNF-α in response to CD1a-expressing APCs treated with DDM. Studies with CD1b/GMM tetramers revealed the expansion of CD1b/GMM-specific T cells in individuals infected with Mtb, but not healthy individuals (68). These cells were found to be CD4+ TCRαβ+ and appeared to have a conserved TCR repertoire. CD1b/GMM-specific T cells were later separated into two types based on their avidity to CD1b/GMM. GMM-specific T cells with a high avidity to CD1b/GMM termed GEM (Germline-encoded, mycolyl-reactive) T cells expressed highly conserved TCRs composed of a TRAV1-2 (Vα 7.2) V segment rearranged with TRAJ9, with limited CDR3α diversity (70). GEM T cells were found at a very low frequency in Mtb naïve individuals. A second population with an intermediate avidity to CD1b/GMM was also observed and this population showed diverse TCR usage (71). Recently, Moody and colleagues described that CD1c-restricted T cells respond to glycosylated and unglycosylated forms of mycoketide presented by DCs or B cells (56). However, in cell-free systems, CD1c-restricted T cells responded only to unglycosylated forms of mycoketide (PM). Moreover, PM-loaded CD1c tetramers could detect antigen-specific T cells from PBMC of donors with latent TB. A comprehensive molecular analysis of TCR recognition of CD1c-mediated PM/MPM presentation revealed that some T cell clones recognize both PM and MPM while others are specific to either PM or MPM (72).
Studies infecting group 1 CD1-expressing hCD1Tg mice with BCG or Mtb revealed that the kinetics of group 1 CD1-restricted T cell responses were different from group 2 CD1d-restricted iNKT cells (4). Group 1 CD1-restricted T cells were detected at week 3 or 4 post infection, suggesting that they follow similar activation kinetics as peptide-specific conventional T cells. This study provided the first direct in vivo evidence that group 1 CD1-restricted T cell responses are induced by Mtb infection (4). Immunizing mice with BMDCs pulsed with Mtb lipid antigens followed by secondary challenge accelerated group 1 CD1-restricted T cell responses, indicating that group 1 CD1-restricted T cells can mount a memory response (4). Taken together, these studies support the potential use of group 1 CD1-restricted Mtb lipid antigens as components of subunit vaccines.
Autoreactive group 1 CD1-restricted T cells
Early reports of T cell recognition of group 1 CD1 molecules suggested that many CD1-restricted T cells are autoreactive (73, 74). A recent analysis of T cell clones from human PBMC showed that CD1a self-reactive T cells are present at high frequencies in the blood of healthy individuals (75). CD1a-restricted autoreactive T cells expressed diverse TCRs and skin homing receptors CCR4, CCR6, CCR10, and secreted IL-22. Given the properties of CD1a-autoreactive T cells and the fact that CD1a recognizes skin lipid antigens and is highly expressed on Langerhans cells implies that these T cells play a role in dermal immunity (33, 75). Another study investigated that the frequency and effector phenotype of autoreactive group 1 CD1-restricted T cells in human blood and cord blood using C1R transfectants expressing CD1a, CD1b, and CD1c (76). They found that the frequencies of group 1 CD1 autoreactive T cells were in the range of 1/10–1/300 circulating T cells. CD1a- and CD1c-restricted T cells comprised the most abundant autoreactive T cells. These group 1 CD1 autoreactive T cells had a diverse phenotype and exhibited mostly Th1 and Th0 functional activities (76).
Most of the CTLs derived from hCD1Tg mice are autoreactive (4). While the endogenous lipid antigens that activate these CTLs were not identified in this study, these clones were found to lyse hCD1Tg DC and group 1 CD1-expressing cells from humans. This suggests that the self-antigens recognized by these CTLs are conserved between mice and humans. Autoreactive T cells can also be detected from Mtb-infected mice. A transgenic mouse model, HJ1Tg, expressing an autoreactive CD1b-restricted TCR has been generated to study the development and function of group 1 CD1 autoreactive T cells (5). Similar to CD1d-restricted iNKT cells, HJ1 T cells exhibited an activated phenotype (CD44+CD69+CD122+) and a subset of HJ1 T cells expressed NK1.1 and was enriched in the liver (5). This finding raises the possibility that autoreactive group 1 CD1-restricted T cells may also reside within the NKT cell compartment of the human liver. HJ1 T cells were cytolytic and secreted proinflammatory cytokines (IFN-γ, IL-17A, and TNF-α) in response to CD1b-expressing DCs. TLR agonists (Pam3Cys and LPS) enhanced the autoreactivity of HJ1 T cells by stimulating DCs to produce IL12/IL23. Furthermore, HJ1 T cells were activated early during Listeria infection and played an immunoprotective role (5). The potential role of group 1 CD1 autoreactive T cells in other infections remains to be defined.
Developmental requirements of group 1 CD1-restricted T cells
The development of group 1 CD1-restricted T cells has not been well-studied due to a lack of a suitable small animal model, but some progress has been made in this regard. Studies using HJ1Tg mice showed that CD1b-expressing hematopoietic cells are necessary and sufficient to mediate the positive selection of HJ1 T cells (5). Cortical thymocytes are most likely the main cell type involved in the selection of HJ1 T cells as CD1b is not expressed on mature thymocytes. HJ1 T cells that developed in the hCD1Tg background expressed similar levels of the transcription factor PLZF as iNKT cells. PLZF is expressed by several non-conventional T cell subsets that display an activated phenotype and is responsible for their innate-like effector program (77–80). However, studies of group 1 CD1 autoreactive T cells in humans showed that autoreactive group 1 CD1-restricted T cells were present in both naive (CD45RA+) and effector/memory (CD45RO+) compartments (75, 76). It is unclear whether TCR–CD1 avidity and the nature of the selecting lipid antigen(s) may play a role in determining the phenotype of group 1 CD1 autoreactive T cells. Moreover, the developmental selection program for microbial lipid antigen-specific group 1 CD1-restricted T cells has not yet been investigated.
Group 1 vs. Group 2 CD1-Restricted T Cells: Some Similarities, Many Differences
Group 1 and group 2 CD1 molecules share many similarities beyond the presentation of lipid antigens to T cells, including molecular assembly mechanisms and sharing self-lipid moieties that stabilize the CD1 binding groove in the absence of foreign antigen (81, 82). However, many differences also exist regarding the functional capacity of their cognate T cells and APC–T cell interactions. The following sections will focus on comparing and contrasting group 1 and group 2 CD1-restricted T cells in terms of TCR diversity, lipid recognition, cytokine production, and APC–T cell interactions. Table 1 highlights the properties of these two groups of CD1-restricted T cells.
Table 1.
Comparison of group 1 and group 2 CD1-restricted T cells.
| Group 1 CD1-restricted T cells | Group 2 CD1-restricted T cells | |||
|---|---|---|---|---|
| Types | Microbial antigen-specific | Autoreactive | Type I (iNKT) | Type II (vNKT) |
| Retricted by | CD1a, CD1b, CD1c | CD1d | ||
| Antigens recognized | Mtb lipid antigens | CD1a – squalene, wax esters, triacylglycerides, sulfatide | α-GalCer analogs, glycosphingolipids, α-Gal-diacylglycerols, phospholipids, gangliosides, isoglobosides | β-GluCer, sulfatide, lysosulfatide, lysophospholipids |
| CD1b – gangliosides, sulfatide | ||||
| CD1c – methy-lysophosphatidic acid, sulfatide | ||||
| Antigen capture | CD1a – cell surface and early endosomes | Endosomes and lysosomes | ||
| CD1b – late endosomes and lysosomes | ||||
| CD1c – endosomes | ||||
| Cell type that mediates selection | Unknown | Hematopoietic cells | Hematopoietic cells | |
| Subset | CD4+, CD8+, DN | CD4+, CD8+, DN | Human-CD4+, CD8+, DN | CD4+, CD8+, DN |
| Mouse-CD4+, DN | ||||
| TCR usage | Diverse TCRαβ chains, Vα7.2-Jα9 (GEM T cells) | Diverse TCRαβ chains | Human – Vα24- | Diverse TCRαβ chains (mouse – Vα3.2, Vα8, Vβ8 bias) |
| Jα18; Vβ11 | ||||
| Mouse – Vα14- | ||||
| Jα18; Vβ8.2, Vβ7, Vβ2, | ||||
| Effector functions | Cytotoxic, produce IFNγ, TNFα | Cytotoxic, produce IFNγ, TNFα, IL-17 | Cytotoxic, produce Th1, Th2, Th17 cytokines | Cytotoxic, produce Th1, Th2 cytokines |
TCR diversity in CD1-restricted T cells
CD1d-restricted NKT cells can be divided into two subsets based on TCR diversity (83). Type I NKT cells, also known as invariant NKT (iNKT) cells, express an invariant TCR α chain. Murine type I NKT cells express an invariant Vα14–Jα18 TCR α chain paired with a limited set of TCR β chains, including Vβ8.2, Vβ7, and Vβ2 (83). Like murine type I NKT cells, human type I NKT cells express the invariant Vα24–Jα18 TCR α chain paired with predominantly the Vβ11 chain (84). All type I NKT cells recognize the marine sponge-derived glycolipid, α-galactosylceramide (α-GalCer) (85). Other populations of CD1d-restricted NKT cells that respond to lipid antigens are broadly classified as type II NKT cells, which exhibit more TCR sequence diversity compared to type I NKT cells (86). Type II NKT cells do not respond to α-GalCer, and therefore, cannot be identified using α-GalCer/CD1d tetramers (86). Several studies have suggested that group 1 CD1-restricted T cells have diverse TCR usage (4, 33, 74, 76). However, CD1b-restricted GEM T cells were recently described to express highly conserved TCRs composed of a TRAV1-2V segment rearranged with TRAJ9, with limited CDR3α diversity (70).
Lipid antigen recognition by group 1 and group 2 CD1-restricted T cells
Both group 1 and group 2 CD1 have been shown to present self-lipid antigens to CD1 autoreactive T cells. However, each member of the CD1 family binds distinct self-lipids. CD1d binds to sphingomyelin, gangliosides (e.g., GD3), globo/isoglobosides, phospholipids, plasmalogens, and lysophospholipids (82, 87–93). However, it remains unclear which of these lipids stimulate iNKT cells under physiological conditions and which contribute to the activation of iNKT cells during microbial infection (94). Gangliosides (e.g., GM1, GD1) were also described to bind to CD1b and activate CD1b-restricted T cells (95). Diacylglycerophosphocholines and diacylglycerophoshoinositol were shown to be bound by CD1c (87). However, it is unclear whether these self-lipids can be recognized by CD1c-restricted T cells. Self-lipids bound by CD1a molecules include squalene, wax esters, and triacylglycerides derived from the skin (33). Interestingly, sulfatide can bind to all group 1 and group 2 CD1 proteins (CD1a, CD1b, CD1c, CD1d) and activate CD1-restricted T cells (59, 96).
Most studies examining microbial antigen presentation by group 1 CD1 molecules have focused on Mtb, but CD1d-restricted iNKT cells have been shown to bind microbial antigens from a variety of pathogens. Both CD1d and CD1b molecules bind and present PIM moieties from Mtb, though they bind different isoforms (40, 97). In addition, diacylglycerol lipid species from pathogens including B. burgdorferi and Streptococcus pneumoniae have been identified as CD1d-restricted iNKT cell ligands, with physiologically relevant effects on T cell responses during infection (98, 99). No comparable diversity in microbial ligands has been shown to exist in group 1 CD1-mediated antigen presentation. However, the biochemical nature of the group 1 CD1 antigen-binding groove suggests that diverse microbial antigens can be presented.
Effector functions of group 1 and group 2 CD1-restricted T cells
Group 1 and group 2 CD1-restricted T cells display antigen-specific cytotoxicity in the context of sterile cancer and infectious disease. Like group 1 CD1 autoreactive T cells, type I and type II NKT cells are capable of directly lysing tumor cells (100–103). Upon activation, type I NKT cells can produce both Th1 (IFN-γ, TNF-α) and Th2 (IL-4, IL-5, IL-13) cytokines, and this dual functionality enables them to play both pathogenic and immunoregulatory roles in various disease settings (104). Type I NKT cells were also shown to produce anti-inflammatory cytokines, such as IL-10 and TGF-β. Similar to type I NKT cells, type II NKT cells are also capable of producing multiple cytokines in response to polyclonal TCR stimulation, including IFN-γ, IL-4, GM-CSF, and IL-13 (105). These findings show that group 2 CD1-restricted NKT cells can produce a variety of Th1, Th2, and regulatory cytokines in response to different environmental cues.
Group 1 CD1-restricted T cells have also been shown to produce multiple cytokines in the context of infection, inflammation, and cancer. Autoreactive group 1 CD1-restricted T cell lines produce IFN-γ, TNF-α, IL-17, and IL-22 in response to self-antigenic stimulation (5). Both in vitro and in vivo assays demonstrated that group 1 CD1-restricted T cells produce IFN-γ and TNF-α upon encountering mycobacterial antigens (4, 65, 66). Unlike group 2 CD1-restricted T cells, most group 1 CD1-restricted T cells do not appear to produce Th2 and/or regulatory cytokines in response to antigenic stimulation. However, to date, all the studies examining group 1 CD1-restricted T cells do so in Mtb infection settings. Further experiments need to be conducted to ascertain whether group 1 CD1-restricted T cells produce Th2 and/or regulatory cytokines under other infectious or inflammatory conditions.
Antigen-presenting cell–T cell interactions
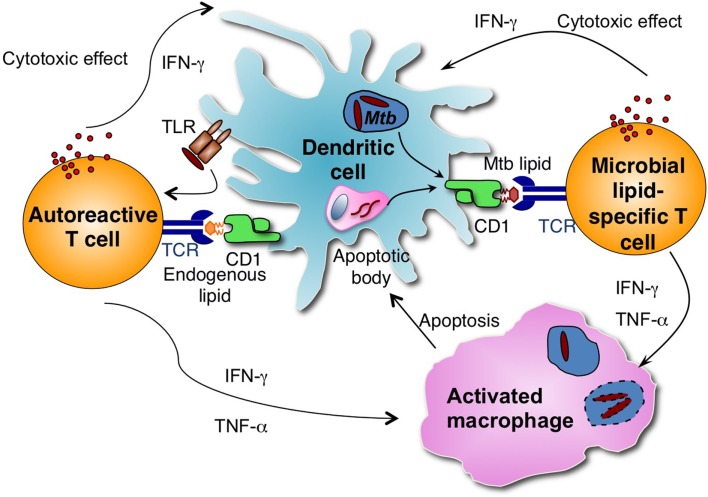
Although all APCs expressing CD1d have the potential to present lipid antigens to NKT cells, studies depleting specific types of APCs have revealed that APC subsets differ in their capacity to prime type I NKT cells with lipid antigens (106). A recent study demonstrated that CD8α+ DEC-205+ dendritic cells were the main cell type responsible for capturing and presenting multiple forms of α-GalCer and stimulated type I NKT cell responses in vivo (106). However, it is unclear which APC subset is responsible for presenting Mtb antigens to group 1 CD1-restricted T cells in vivo. Macrophages are the primary host cells for Mtb but do not express group 1 CD1 molecules. It was demonstrated that group 1 CD1-expressing DCs take up apoptotic vesicles from bystander infected cells and presented them to human CD8+ T cells in an in vitro co-culture experiment (107). Thus, group 1 CD1-expressing DCs may similarly cross-present Mtb lipid antigens to T cells during infection (Figure 1).
Figure 1.
Group 1 CD1-restricted T cells consist of autoreactive and microbial antigen-specific T cells. Microbial lipid-specific T cells are activated by microbial lipids that are loaded on group 1 CD1 molecules. Autoreactive T cells are activated in combination by proinflammatory cytokines produced on TLR engagement and self-lipid antigens. During Mtb infection, group 1 CD1-expressing DCs can either be directly infected by Mtb or take up apoptotic vesicles from bystander infected cells and present Mtb-derived antigens to group 1 CD1-restricted Mtb lipid-specific T cells. Activated group 1 CD1-restricted T cells are cytotoxic and can directly lyse Mtb-infected cells. Activated group 1 CD1-restricted T cells also produce IFN-γ and TNF-α, which can activate infected macrophages and help to control Mtb growth.
Most of the studies involving CD1d have been performed in murine models. However, as humans possess both group 1 CD1 and group 2 CD1 molecules, it is possible that presence of group 1 CD1 molecules may affect CD1d antigen presentation. While there is some specialization in the antigen presentation functions of CD1 molecules, whereby they sample lipids from different intracellular compartments, sulfatide, a glycosphingolipid, is capable of promiscuously binding to CD1a, CD1b, CD1c, and CD1d (59). As CD1c tends to be co-expressed with CD1d on blood dendritic cells and a fraction of B cells, a recent study evaluated how the presence of CD1c influences the activation of CD1d-restricted iNKT cells (108). This study demonstrated that CD1c is able to present α-GalCer as a weak agonist to human iNKT cells, and the presence of CD1c enhances α-GalCer-dependent activation of iNKT cells by CD1d (108).
Role of group 1 and group 2 CD1-restricted T cells during infection
The studies described above support the likely role of group 1 CD1-restricted T cells in protective immunity against Mtb infection. However, the mechanisms by which they control Mtb infection remain to be elucidated. Aside from Mtb, the role of group 1 CD1-restricted T cells in other microbial infection has not been addressed. In contrast, the role of CD1d-restricted iNKT cells in host defense against various pathogens has been extensively studied (109). Studies examining iNKT cell dynamics in mice infected intravenously with BCG showed an early expansion of iNKT cells in the lung with a peak at day 8 after infection, followed by a subsequent decline likely due to programed cell death (110, 111). Other studies have suggested that iNKT cells may play a role in the formation of granulomas in response to mycobacterial cell wall components (112, 113). Furthermore, in vitro experiments have shown that iNKT cells possess the ability to kill Mtb-infected APCs through the production of GM-CSF (114). However, Mtb infection of CD1d−/− or Jα18−/− mice with Mtb resulted in no significant difference in mortality or bacterial burdens in the lung, suggesting that iNKT cells may play a redundant role in the control of Mtb (115–117).
Conclusion
We highlight several recent studies that have contributed to our understanding of group 1 CD1 antigen presentation and the role group 1 CD1-restricted T cells play during Mtb infection. Further studies are needed to characterize Mtb lipid-specific T cell memory responses and evaluate whether Mtb lipid antigens can effectively be used in a subunit vaccine. Future studies also need to expand on the role of autoreactive group 1 CD1-restricted T cells in Mtb infections, as it is unknown whether they have similar activation kinetics as antigen-specific T cells and how they affect host immunity to infection. Moreover, it remains unclear whether group 1 CD1 molecules present lipids from other microbial species and whether they play a role in other inflammatory diseases, presenting fertile ground for future investigation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely apologize to colleagues whose work we have not cited due to space constraints or oversight. We thank Sreya Bagchi for critical reading and helpful discussions. This work was supported by the National Institutes of Health R01 grants AI057460 and AI43407.
References
- 1.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol (2004) 22:817–90. 10.1146/annurev.immunol.22.012703.104608 [DOI] [PubMed] [Google Scholar]
- 2.Dascher CC, Hiromatsu K, Naylor JW, Brauer PP, Brown KA, Storey JR, et al. Conservation of a CD1 multigene family in the guinea pig. J Immunol (1999) 163(10):5478–88. [PubMed] [Google Scholar]
- 3.Martin LH, Calabi F, Lefebvre FA, Bilsland CA, Milstein C. Structure and expression of the human thymocyte antigens CD1a, CD1b, and CD1c. Proc Natl Acad Sci U S A (1987) 84(24):9189–93. 10.1073/pnas.84.24.9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felio K, Nguyen H, Dascher CC, Choi HJ, Li S, Zimmer MI, et al. CD1-restricted adaptive immune responses to mycobacteria in human group 1 CD1 transgenic mice. J Exp Med (2009) 206(11):2497–509. 10.1084/jem.20090898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S, Choi HJ, Felio K, Wang CR. Autoreactive CD1b-restricted T cells: a new innate-like T-cell population that contributes to immunity against infection. Blood (2011) 118(14):3870–8. 10.1182/blood-2011-03-341941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lockridge JL, Chen X, Zhou Y, Rajesh D, Roenneburg DA, Hegde S, et al. Analysis of the CD1 antigen presenting system in humanized SCID mice. PLoS One (2011) 6(6):e21701. 10.1371/journal.pone.0021701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wollenberg A, Kraft S, Hanau D, Bieber T. Immunomorphological and ultrastructural characterization of Langerhans cells and a novel, inflammatory dendritic epidermal cell (IDEC) population in lesional skin of atopic eczema. J Invest Dermatol (1996) 106(3):446–53. 10.1111/1523-1747.ep12343596 [DOI] [PubMed] [Google Scholar]
- 8.Olivier M, Foret B, Le Vern Y, Kerboeuf D, Guilloteau LA. Plasticity of migrating CD1b+ and CD1b- lymph dendritic cells in the promotion of Th1, Th2 and Th17 in response to Salmonella and helminth secretions. PLoS One (2013) 8(11):e79537. 10.1371/journal.pone.0079537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugita M, van Der Wel N, Rogers RA, Peters PJ, Brenner MB. CD1c molecules broadly survey the endocytic system. Proc Natl Acad Sci U S A (2000) 97(15):8445–50. 10.1073/pnas.150236797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milne P, Bigley V, Gunawan M, Haniffa M, Collin M. CD1c+ blood dendritic cells have Langerhans cell potential. Blood (2015) 125(3):470–3. 10.1182/blood-2014-08-593582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angenieux C, Salamero J, Fricker D, Cazenave JP, Goud B, Hanau D, et al. Characterization of CD1e, a third type of CD1 molecule expressed in dendritic cells. J Biol Chem (2000) 275(48):37757–64. 10.1074/jbc.M007082200 [DOI] [PubMed] [Google Scholar]
- 12.Angenieux C, Fraisier V, Maitre B, Racine V, van der Wel N, Fricker D, et al. The cellular pathway of CD1e in immature and maturing dendritic cells. Traffic (2005) 6(4):286–302. 10.1111/j.1600-0854.2005.00272.x [DOI] [PubMed] [Google Scholar]
- 13.de la Salle H, Mariotti S, Angenieux C, Gilleron M, Garcia-Alles LF, Malm D, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science (2005) 310(5752):1321–4. 10.1126/science.1115301 [DOI] [PubMed] [Google Scholar]
- 14.Kasinrerk W, Baumruker T, Majdic O, Knapp W, Stockinger H. CD1 molecule expression on human monocytes induced by granulocyte-macrophage colony-stimulating factor. J Immunol (1993) 150(2):579–84. [PubMed] [Google Scholar]
- 15.Sieling PA, Jullien D, Dahlem M, Tedder TF, Rea TH, Modlin RL, et al. CD1 expression by dendritic cells in human leprosy lesions: correlation with effective host immunity. J Immunol (1999) 162(3):1851–8. [PubMed] [Google Scholar]
- 16.Yakimchuk K, Roura-Mir C, Magalhaes KG, de Jong A, Kasmar AG, Granter SR, et al. Borrelia burgdorferi infection regulates CD1 expression in human cells and tissues via IL1-beta. Eur J Immunol (2011) 41(3):694–705. 10.1002/eji.201040808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oji V, Seller N, Sandilands A, Gruber R, Gerss J, Huffmeier U, et al. Ichthyosis vulgaris: novel FLG mutations in the German population and high presence of CD1a+ cells in the epidermis of the atopic subgroup. Br J Dermatol (2009) 160(4):771–81. 10.1111/j.1365-2133.2008.08999.x [DOI] [PubMed] [Google Scholar]
- 18.Hussein MR. Evaluation of Langerhans’ cells in normal and eczematous dermatitis skin by CD1a protein immunohistochemistry: preliminary findings. J Cutan Pathol (2008) 35(6):554–8. 10.1111/j.1600-0560.2007.00861.x [DOI] [PubMed] [Google Scholar]
- 19.Abreu-Velez AM, Googe PB, Howard MS. Immune reactivity in psoriatic munro-saboureau microabscesses, stratum corneum and blood vessels. N Am J Med Sci (2012) 4(6):257–65. 10.4103/1947-2714.97204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alshenawy HA, Hasby EA. Immunophenotyping of dendritic cells in lesional, perilesional and distant skin of chronic plaque psoriasis. Cell Immunol (2011) 269(2):115–9. 10.1016/j.cellimm.2011.03.015 [DOI] [PubMed] [Google Scholar]
- 21.Aruffo A, Seed B. Expression of cDNA clones encoding the thymocyte antigens CD1a, b, c demonstrates a hierarchy of exclusion in fibroblasts. J Immunol (1989) 143(5):1723–30. [PubMed] [Google Scholar]
- 22.Lepore M, de Lalla C, Gundimeda SR, Gsellinger H, Consonni M, Garavaglia C, et al. A novel self-lipid antigen targets human T cells against CD1c(+) leukemias. J Exp Med (2014) 211(7):1363–77. 10.1084/jem.20140410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briken V, Jackman RM, Dasgupta S, Hoening S, Porcelli SA. Intracellular trafficking pathway of newly synthesized CD1b molecules. EMBO J (2002) 21(4):825–34. 10.1093/emboj/21.4.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odyniec AN, Barral DC, Garg S, Tatituri RV, Besra GS, Brenner MB. Regulation of CD1 antigen-presenting complex stability. J Biol Chem (2010) 285(16):11937–47. 10.1074/jbc.M109.077933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Relloso M, Cheng TY, Im JS, Parisini E, Roura-Mir C, DeBono C, et al. pH-dependent interdomain tethers of CD1b regulate its antigen capture. Immunity (2008) 28(6):774–86. 10.1016/j.immuni.2008.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cernadas M, Cavallari M, Watts G, Mori L, De Libero G, Brenner MB. Early recycling compartment trafficking of CD1a is essential for its intersection and presentation of lipid antigens. J Immunol (2010) 184(3):1235–41. 10.4049/jimmunol.0804140 [DOI] [PubMed] [Google Scholar]
- 27.Garzon D, Anselmi C, Bond PJ, Faraldo-Gomez JD. Dynamics of the antigen-binding grooves in CD1 proteins: reversible hydrophobic collapse in the lipid-free state. J Biol Chem (2013) 288(27):19528–36. 10.1074/jbc.M113.470179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manolova V, Kistowska M, Paoletti S, Baltariu GM, Bausinger H, Hanau D, et al. Functional CD1a is stabilized by exogenous lipids. Eur J Immunol (2006) 36(5):1083–92. 10.1002/eji.200535544 [DOI] [PubMed] [Google Scholar]
- 29.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O’Connor PB, et al. T cell activation by lipopeptide antigens. Science (2004) 303(5657):527–31. 10.1126/science.1089353 [DOI] [PubMed] [Google Scholar]
- 30.Zajonc DM, Crispin MD, Bowden TA, Young DC, Cheng TY, Hu J, et al. Molecular mechanism of lipopeptide presentation by CD1a. Immunity (2005) 22(2):209–19. 10.1016/j.immuni.2004.12.009 [DOI] [PubMed] [Google Scholar]
- 31.Young DC, Kasmar A, Moraski G, Cheng TY, Walz AJ, Hu J, et al. Synthesis of dideoxymycobactin antigens presented by CD1a reveals T cell fine specificity for natural lipopeptide structures. J Biol Chem (2009) 284(37):25087–96. 10.1074/jbc.M109.000802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol (2003) 4(8):808–15. 10.1038/ni948 [DOI] [PubMed] [Google Scholar]
- 33.de Jong A, Cheng TY, Huang S, Gras S, Birkinshaw RW, Kasmar AG, et al. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat Immunol (2014) 15(2):177–85. 10.1038/ni.2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birkinshaw RW, Pellicci DG, Cheng TY, Keller AN, Sandoval-Romero M, Gras S, et al. alphabeta T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat Immunol (2015) 16(3):258–66. 10.1038/ni.3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourgeois EA, Subramaniam S, Cheng TY, De Jong A, Layre E, Ly D, et al. Bee venom processes human skin lipids for presentation by CD1a. J Exp Med (2015) 212(2):149–63. 10.1084/jem.20141505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature (1994) 372(6507):691–4. 10.1038/372691a0 [DOI] [PubMed] [Google Scholar]
- 37.Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, et al. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science (1997) 278(5336):283–6. 10.1126/science.278.5336.283 [DOI] [PubMed] [Google Scholar]
- 38.Layre E, Collmann A, Bastian M, Mariotti S, Czaplicki J, Prandi J, et al. Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1-restricted T cells. Chem Biol (2009) 16(1):82–92. 10.1016/j.chembiol.2008.11.008 [DOI] [PubMed] [Google Scholar]
- 39.Ernst WA, Maher J, Cho S, Niazi KR, Chatterjee D, Moody DB, et al. Molecular interaction of CD1b with lipoglycan antigens. Immunity (1998) 8(3):331–40. 10.1016/S1074-7613(00)80538-5 [DOI] [PubMed] [Google Scholar]
- 40.Cala-De Paepe D, Layre E, Giacometti G, Garcia-Alles LF, Mori L, Hanau D, et al. Deciphering the role of CD1e protein in mycobacterial phosphatidyl-myo-inositol mannosides (PIM) processing for presentation by CD1b to T lymphocytes. J Biol Chem (2012) 287(37):31494–502. 10.1074/jbc.M112.386300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, et al. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science (1995) 269(5221):227–30. 10.1126/science.7542404 [DOI] [PubMed] [Google Scholar]
- 42.Torrelles JB, Sieling PA, Zhang N, Keen MA, McNeil MR, Belisle JT, et al. Isolation of a distinct Mycobacterium tuberculosis mannose-capped lipoarabinomannan isoform responsible for recognition by CD1b-restricted T cells. Glycobiology (2012) 22(8):1118–27. 10.1093/glycob/cws078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torrelles JB, Sieling PA, Arcos J, Knaup R, Bartling C, Rajaram MV, et al. Structural differences in lipomannans from pathogenic and nonpathogenic mycobacteria that impact CD1b-restricted T cell responses. J Biol Chem (2011) 286(41):35438–46. 10.1074/jbc.M111.232587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Bohmer G, et al. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J Exp Med (2004) 199(5):649–59. 10.1084/jem.20031097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gadola SD, Zaccai NR, Harlos K, Shepherd D, Castro-Palomino JC, Ritter G, et al. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat Immunol (2002) 3(8):721–6. 10.1038/ni821 [DOI] [PubMed] [Google Scholar]
- 46.Batuwangala T, Shepherd D, Gadola SD, Gibson KJ, Zaccai NR, Fersht AR, et al. The crystal structure of human CD1b with a bound bacterial glycolipid. J Immunol (2004) 172(4):2382–8. 10.4049/jimmunol.172.4.2382 [DOI] [PubMed] [Google Scholar]
- 47.Garcia-Alles LF, Collmann A, Versluis C, Lindner B, Guiard J, Maveyraud L, et al. Structural reorganization of the antigen-binding groove of human CD1b for presentation of mycobacterial sulfoglycolipids. Proc Natl Acad Sci U S A (2011) 108(43):17755–60. 10.1073/pnas.1110118108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barry CE, III, Lee RE, Mdluli K, Sampson AE, Schroeder BG, Slayden RA, et al. Mycolic acids: structure, biosynthesis and physiological functions. Prog Lipid Res (1998) 37(2–3):143–79. 10.1016/S0163-7827(98)00008-3 [DOI] [PubMed] [Google Scholar]
- 49.Montamat-Sicotte DJ, Millington KA, Willcox CR, Hingley-Wilson S, Hackforth S, Innes J, et al. A mycolic acid-specific CD1-restricted T cell population contributes to acute and memory immune responses in human tuberculosis infection. J Clin Invest (2011) 121(6):2493–503. 10.1172/JCI46216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guiard J, Collmann A, Garcia-Alles LF, Mourey L, Brando T, Mori L, et al. Fatty acyl structures of Mycobacterium tuberculosis sulfoglycolipid govern T cell response. J Immunol (2009) 182(11):7030–7. 10.4049/jimmunol.0804044 [DOI] [PubMed] [Google Scholar]
- 51.Willcox BE, Willcox CR, Dover LG, Besra G. Structures and functions of microbial lipid antigens presented by CD1. Curr Top Microbiol Immunol (2007) 314:73–110. 10.1007/978-3-540-69511-0_4 [DOI] [PubMed] [Google Scholar]
- 52.Fukuda T, Matsumura T, Ato M, Hamasaki M, Nishiuchi Y, Murakami Y, et al. Critical roles for lipomannan and lipoarabinomannan in cell wall integrity of mycobacteria and pathogenesis of tuberculosis. MBio (2013) 4(1):e472–412. 10.1128/mBio.00472-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moody DB, Ulrichs T, Muhlecker W, Young DC, Gurcha SS, Grant E, et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature (2000) 404(6780):884–8. 10.1038/35009119 [DOI] [PubMed] [Google Scholar]
- 54.Matsunaga I, Bhatt A, Young DC, Cheng TY, Eyles SJ, Besra GS, et al. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J Exp Med (2004) 200(12):1559–69. 10.1084/jem.20041429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsunaga I, Sugita M. Mycoketide: a CD1c-presented antigen with important implications in mycobacterial infection. Clin Dev Immunol (2012) 2012:981821. 10.1155/2012/981821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ly D, Kasmar AG, Cheng TY, de Jong A, Huang S, Roy S, et al. CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. J Exp Med (2013) 210(4):729–41. 10.1084/jem.20120624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scharf L, Li NS, Hawk AJ, Garzon D, Zhang T, Fox LM, et al. The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity (2010) 33(6):853–62. 10.1016/j.immuni.2010.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Rhijn I, Young DC, De Jong A, Vazquez J, Cheng TY, Talekar R, et al. CD1c bypasses lysosomes to present a lipopeptide antigen with 12 amino acids. J Exp Med (2009) 206(6):1409–22. 10.1084/jem.20082480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G. Presentation of the same glycolipid by different CD1 molecules. J Exp Med (2002) 195(8):1013–21. 10.1084/jem.20011963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morita D, Katoh K, Harada T, Nakagawa Y, Matsunaga I, Miura T, et al. Trans-species activation of human T cells by rhesus macaque CD1b molecules. Biochem Biophys Res Commun (2008) 377(3):889–93. 10.1016/j.bbrc.2008.10.075 [DOI] [PubMed] [Google Scholar]
- 61.Morita D, Hattori Y, Nakamura T, Igarashi T, Harashima H, Sugita M. Major T cell response to a mycolyl glycolipid is mediated by CD1c molecules in rhesus macaques. Infect Immun (2013) 81(1):311–6. 10.1128/IAI.00871-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Looringh van Beeck FA, Reinink P, Hermsen R, Zajonc DM, Laven MJ, Fun A, et al. Functional CD1d and/or NKT cell invariant chain transcript in horse, pig, African elephant and guinea pig, but not in ruminants. Mol Immunol (2009) 46(7):1424–31. 10.1016/j.molimm.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hiromatsu K, Dascher CC, LeClair KP, Sugita M, Furlong ST, Brenner MB, et al. Induction of CD1-restricted immune responses in guinea pigs by immunization with mycobacterial lipid antigens. J Immunol (2002) 169(1):330–9. 10.4049/jimmunol.169.1.330 [DOI] [PubMed] [Google Scholar]
- 64.Dascher CC, Hiromatsu K, Xiong X, Morehouse C, Watts G, Liu G, et al. Immunization with a mycobacterial lipid vaccine improves pulmonary pathology in the guinea pig model of tuberculosis. Int Immunol (2003) 15(8):915–25. 10.1093/intimm/dxg091 [DOI] [PubMed] [Google Scholar]
- 65.Ulrichs T, Moody DB, Grant E, Kaufmann SH, Porcelli SA. T-cell responses to CD1-presented lipid antigens in humans with Mycobacterium tuberculosis infection. Infect Immun (2003) 71(6):3076–87. 10.1128/IAI.71.6.3076-3087.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawashima T, Norose Y, Watanabe Y, Enomoto Y, Narazaki H, Watari E, et al. Cutting edge: major CD8 T cell response to live bacillus Calmette-Guerin is mediated by CD1 molecules. J Immunol (2003) 170(11):5345–8. 10.4049/jimmunol.170.11.5345 [DOI] [PubMed] [Google Scholar]
- 67.Moody DB, Guy MR, Grant E, Cheng TY, Brenner MB, Besra GS, et al. CD1b-mediated T cell recognition of a glycolipid antigen generated from mycobacterial lipid and host carbohydrate during infection. J Exp Med (2000) 192(7):965–76. 10.1084/jem.192.7.965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kasmar AG, van Rhijn I, Cheng TY, Turner M, Seshadri C, Schiefner A, et al. CD1b tetramers bind alphabeta T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. J Exp Med (2011) 208(9):1741–7. 10.1084/jem.20110665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kasmar AG, Van Rhijn I, Magalhaes KG, Young DC, Cheng TY, Turner MT, et al. Cutting edge: CD1a tetramers and dextramers identify human lipopeptide-specific T cells ex vivo. J Immunol (2013) 191(9):4499–503. 10.4049/jimmunol.1301660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Rhijn I, Kasmar A, de Jong A, Gras S, Bhati M, Doorenspleet ME, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol (2013) 14(7):706–13. 10.1038/ni.2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Rhijn I, Gherardin NA, Kasmar A, de Jager W, Pellicci DG, Kostenko L, et al. TCR bias and affinity define two compartments of the CD1b-glycolipid-specific T Cell repertoire. J Immunol (2014) 192(9):4054–60. 10.4049/jimmunol.1400158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roy S, Ly D, Li NS, Altman JD, Piccirilli JA, Moody DB, et al. Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by alphabeta T cells. Proc Natl Acad Sci U S A (2014) 111(43):E4648–57. 10.1073/pnas.1408549111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature (1989) 341(6241):447–50. 10.1038/341447a0 [DOI] [PubMed] [Google Scholar]
- 74.Vincent MS, Xiong X, Grant EP, Peng W, Brenner MB. CD1a-, b-, and c-restricted TCRs recognize both self and foreign antigens. J Immunol (2005) 175(10):6344–51. 10.4049/jimmunol.175.10.6344 [DOI] [PubMed] [Google Scholar]
- 75.de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nat Immunol (2010) 11(12):1102–9. 10.1038/ni.1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Lalla C, Lepore M, Piccolo FM, Rinaldi A, Scelfo A, Garavaglia C, et al. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. Eur J Immunol (2011) 41(3):602–10. 10.1002/eji.201041211 [DOI] [PubMed] [Google Scholar]
- 77.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol (2008) 9(9):1055–64. 10.1038/ni.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U S A (2009) 106(30):12453–8. 10.1073/pnas.0903895106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity (2008) 29(3):391–403. 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raberger J, Schebesta A, Sakaguchi S, Boucheron N, Blomberg KE, Berglof A, et al. The transcriptional regulator PLZF induces the development of CD44 high memory phenotype T cells. Proc Natl Acad Sci U S A (2008) 105(46):17919–24. 10.1073/pnas.0805733105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang S, Cheng TY, Young DC, Layre E, Madigan CA, Shires J, et al. Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proc Natl Acad Sci U S A (2011) 108(48):19335–40. 10.1073/pnas.1112969108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park JJ, Kang SJ, De Silva AD, Stanic AK, Casorati G, Hachey DL, et al. Lipid-protein interactions: biosynthetic assembly of CD1 with lipids in the endoplasmic reticulum is evolutionarily conserved. Proc Natl Acad Sci U S A (2004) 101(4):1022–6. 10.1073/pnas.0307847100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol (2005) 23:877–900. 10.1146/annurev.immunol.23.021704.115742 [DOI] [PubMed] [Google Scholar]
- 84.Spada FM, Koezuka Y, Porcelli SA. CD1d-restricted recognition of synthetic glycolipid antigens by human natural killer T cells. J Exp Med (1998) 188(8):1529–34. 10.1084/jem.188.8.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science (1997) 278(5343):1626–9. 10.1126/science.278.5343.1626 [DOI] [PubMed] [Google Scholar]
- 86.Blomqvist M, Rhost S, Teneberg S, Lofbom L, Osterbye T, Brigl M, et al. Multiple tissue-specific isoforms of sulfatide activate CD1d-restricted type II NKT cells. Eur J Immunol (2009) 39(7):1726–35. 10.1002/eji.200839001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haig NA, Guan Z, Li D, McMichael A, Raetz CR, Xu XN. Identification of self-lipids presented by CD1c and CD1d proteins. J Biol Chem (2011) 286(43):37692–701. 10.1074/jbc.M111.267948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Joyce S, Woods AS, Yewdell JW, Bennink JR, De Silva AD, Boesteanu A, et al. Natural ligand of mouse CD1d1: cellular glycosylphosphatidylinositol. Science (1998) 279(5356):1541–4. 10.1126/science.279.5356.1541 [DOI] [PubMed] [Google Scholar]
- 89.De Silva AD, Park JJ, Matsuki N, Stanic AK, Brutkiewicz RR, Medof ME, et al. Lipid protein interactions: the assembly of CD1d1 with cellular phospholipids occurs in the endoplasmic reticulum. J Immunol (2002) 168(2):723–33. 10.4049/jimmunol.168.2.723 [DOI] [PubMed] [Google Scholar]
- 90.Zhou D, Mattner J, Cantu C, III, Schrantz N, Yin N, Gao Y, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science (2004) 306(5702):1786–9. 10.1126/science.1103440 [DOI] [PubMed] [Google Scholar]
- 91.Giabbai B, Sidobre S, Crispin MD, Sanchez-Ruiz Y, Bachi A, Kronenberg M, et al. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J Immunol (2005) 175(2):977–84. 10.4049/jimmunol.175.2.977 [DOI] [PubMed] [Google Scholar]
- 92.Yuan W, Kang SJ, Evans JE, Cresswell P. Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J Immunol (2009) 182(8):4784–91. 10.4049/jimmunol.0803981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cox D, Fox L, Tian R, Bardet W, Skaley M, Mojsilovic D, et al. Determination of cellular lipids bound to human CD1d molecules. PLoS One (2009) 4(5):e5325. 10.1371/journal.pone.0005325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brigl M, Tatituri RV, Watts GF, Bhowruth V, Leadbetter EA, Barton N, et al. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med (2011) 208(6):1163–77. 10.1084/jem.20102555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shamshiev A, Donda A, Prigozy TI, Mori L, Chigorno V, Benedict CA, et al. The alphabeta T cell response to self-glycolipids shows a novel mechanism of CD1b loading and a requirement for complex oligosaccharides. Immunity (2000) 13(2):255–64. 10.1016/S1074-7613(00)00025-X [DOI] [PubMed] [Google Scholar]
- 96.Bai L, Picard D, Anderson B, Chaudhary V, Luoma A, Jabri B, et al. The majority of CD1d-sulfatide-specific T cells in human blood use a semiinvariant Vdelta1 TCR. Eur J Immunol (2012) 42(9):2505–10. 10.1002/eji.201242531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fischer K, Scotet E, Niemeyer M, Koebernick H, Zerrahn J, Maillet S, et al. Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d-restricted T cells. Proc Natl Acad Sci U S A (2004) 101(29):10685–90. 10.1073/pnas.0403787101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol (2011) 12(10):966–74. 10.1038/ni.2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol (2006) 7(9):978–86. 10.1038/ni1380 [DOI] [PubMed] [Google Scholar]
- 100.Metelitsa LS, Naidenko OV, Kant A, Wu HW, Loza MJ, Perussia B, et al. Human NKT cells mediate antitumor cytotoxicity directly by recognizing target cell CD1d with bound ligand or indirectly by producing IL-2 to activate NK cells. J Immunol (2001) 167(6):3114–22. 10.4049/jimmunol.167.6.3114 [DOI] [PubMed] [Google Scholar]
- 101.Fuss IJ, Heller F, Boirivant M, Leon F, Yoshida M, Fichtner-Feigl S, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest (2004) 113(10):1490–7. 10.1172/JCI19836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weng X, Liao CM, Bagchi S, Cardell SL, Stein PL, Wang CR. The adaptor protein SAP regulates type II NKT-cell development, cytokine production, and cytotoxicity against lymphoma. Eur J Immunol (2014) 44(12):3646–57. 10.1002/eji.201444848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zimmer MI, Nguyen HP, Wang B, Xu H, Colmone A, Felio K, et al. Polymorphisms in CD1d affect antigen presentation and the activation of CD1d-restricted T cells. Proc Natl Acad Sci U S A (2009) 106(6):1909–14. 10.1073/pnas.0808476106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol (2013) 13(2):101–17. 10.1038/nri3369 [DOI] [PubMed] [Google Scholar]
- 105.Zhao J, Weng X, Bagchi S, Wang CR. Polyclonal type II natural killer T cells require PLZF and SAP for their development and contribute to CpG-mediated antitumor response. Proc Natl Acad Sci U S A (2014) 111(7):2674–9. 10.1073/pnas.1323845111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Arora P, Baena A, Yu KO, Saini NK, Kharkwal SS, Goldberg MF, et al. A single subset of dendritic cells controls the cytokine bias of natural killer T cell responses to diverse glycolipid antigens. Immunity (2014) 40(1):105–16. 10.1016/j.immuni.2013.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schaible UE, Winau F, Sieling PA, Fischer K, Collins HL, Hagens K, et al. Apoptosis facilitates antigen presentation to T lymphocytes through MHC-I and CD1 in tuberculosis. Nat Med (2003) 9(8):1039–46. 10.1038/nm906 [DOI] [PubMed] [Google Scholar]
- 108.Fox LM, Miksanek J, May NA, Scharf L, Lockridge JL, Veerapen N, et al. Expression of CD1c enhances human invariant NKT cell activation by alpha-GalCer. Cancer Immun (2013) 13:9. [PMC free article] [PubMed] [Google Scholar]
- 109.Brigl M, Brenner MB. How invariant natural killer T cells respond to infection by recognizing microbial or endogenous lipid antigens. Semin Immunol (2010) 22(2):79–86. 10.1016/j.smim.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 110.Dieli F, Taniguchi M, Kronenberg M, Sidobre S, Ivanyi J, Fattorini L, et al. An anti-inflammatory role for V alpha 14 NK T cells in Mycobacterium bovis bacillus Calmette-Guerin-infected mice. J Immunol (2003) 171(4):1961–8. 10.4049/jimmunol.171.4.1961 [DOI] [PubMed] [Google Scholar]
- 111.Chiba A, Dascher CC, Besra GS, Brenner MB. Rapid NKT cell responses are self-terminating during the course of microbial infection. J Immunol (2008) 181(4):2292–302. 10.4049/jimmunol.181.4.2292 [DOI] [PubMed] [Google Scholar]
- 112.Apostolou I, Takahama Y, Belmant C, Kawano T, Huerre M, Marchal G, et al. Murine natural killer T(NKT) cells [correction of natural killer cells] contribute to the granulomatous reaction caused by mycobacterial cell walls. Proc Natl Acad Sci U S A (1999) 96(9):5141–6. 10.1073/pnas.96.9.5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guidry TV, Olsen M, Kil KS, Hunter RL, Jr, Geng YJ, Actor JK. Failure of CD1D-/- mice to elicit hypersensitive granulomas to mycobacterial cord factor trehalose 6,6’-dimycolate. J Interferon Cytokine Res (2004) 24(6):362–71. 10.1089/107999004323142222 [DOI] [PubMed] [Google Scholar]
- 114.Rothchild AC, Jayaraman P, Nunes-Alves C, Behar SM. iNKT cell production of GM-CSF controls Mycobacterium tuberculosis. PLoS Pathog (2014) 10(1):e1003805. 10.1371/journal.ppat.1003805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Behar SM, Dascher CC, Grusby MJ, Wang CR, Brenner MB. Susceptibility of mice deficient in CD1D or TAP1 to infection with Mycobacterium tuberculosis. J Exp Med (1999) 189(12):1973–80. 10.1084/jem.189.12.1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kawakami K, Kinjo Y, Uezu K, Yara S, Miyagi K, Koguchi Y, et al. Minimal contribution of Valpha14 natural killer T cells to Th1 response and host resistance against mycobacterial infection in mice. Microbiol Immunol (2002) 46(3):207–10. 10.1111/j.1348-0421.2002.tb02687.x [DOI] [PubMed] [Google Scholar]
- 117.Sugawara I, Yamada H, Mizuno S, Li CY, Nakayama T, Taniguchi M. Mycobacterial infection in natural killer T cell knockout mice. Tuberculosis (Edinb) (2002) 82(2–3):97–104. 10.1054/tube.2002.0331 [DOI] [PubMed] [Google Scholar]