Abstract
DEAD-box helicases are conserved enzymes involved in nearly all aspects of RNA metabolism, but their mechanisms of action remain unclear. Here, we investigated the mechanism of the DEAD-box protein Mss116 on its natural substrate, the group II intron ai5γ. Group II introns are structurally complex catalytic RNAs considered evolutionarily related to the eukaryotic spliceosome, and an interesting paradigm for large RNA folding. We used single-molecule fluorescence to monitor the effect of Mss116 on folding dynamics of a minimal active construct, ai5γ–D135. The data show that Mss116 stimulates dynamic sampling between states along the folding pathway, an effect previously observed only with high Mg2+ concentrations. Furthermore, the data indicate that Mss116 promotes folding through discrete ATP-independent and ATP-dependent steps. We propose that Mss116 stimulates group II intron folding through a multi-step process that involves electrostatic stabilization of early intermediates and ATP hydrolysis during the final stages of native state assembly.
Keywords: Group II introns, Mss116 DEAD-box protein, single-molecule fluorescence
DEAD-box proteins are enzymes that play essential roles in cellular processes involving RNA1. Although these have been studied in vitro and in vivo, there are few examples of DEAD-box proteins whose mechanisms have been dissected using a natural substrate in vitro2. Mss116 is a DEAD-box protein that facilitates splicing of all Saccharomyces cerevisiae (Sc.) mitochondrial group I and II introns in vivo3. Mss116 exhibits RNA binding, unwinding, annealing and ATPase activities and has been shown to facilitate group II intron splicing in vitro under near-physiological conditions4–6.
Group II introns are large ribozymes that catalyze self-splicing7 and are thought to be evolutionarily related to nuclear splicing. They also function as mobile genetic elements in bacteria, which may have played an important role during evolution and genome diversification8. Group II introns require high ionic strength and elevated temperatures to function in vitro7, but splicing in vivo is facilitated by protein cofactors. Despite diversity in primary sequence, group II introns have highly conserved secondary and tertiary structures9,10. The secondary structure consists of six domains (D1–D6) radiating from a central core. D1, an assembly scaffold for the other domains11,12, and D5, the catalytic core, are the only absolutely essential domains for minimal catalytic activity in group II introns13,14. One of the best characterized active forms of the Sc.ai5γ group II intron is D135, which contains D1, D3, a catalytic effector, and D5 (Fig. 1a)15–18. Folding studies have shown that D135 folds slowly but directly to the native state15–18, making it an interesting model system.
Figure 1. Single-molecule fluorescence detection of group II intron folding with its natural cofactor Mss116.
a, Secondary structure of D135-L14 ribozyme. Fluorophore-labeled DNA oligonucleotides (Cy3 - green, Cy5 - red) and biotin (yellow) labeled DNA oligonucleotide are shown. b, Single-molecule detection of D135-L14 folding in the presence of Mss116 (pink). The folding pathway consists of three states: the extended intermediate state (I), the folded intermediate state (F) and the native state (N).
Mss116 has been proposed to promote group II intron splicing by stabilizing on-pathway intermediates and/or by disrupting off-pathway misfolded structures4,5. However, the mechanism by which Mss116 mediates group II intron splicing remains highly debated4–6. Previous studies on other systems have shown that both mechanisms are possible. For example, the LtrA protein encoded by a group IIA intron promotes self-splicing by binding the intron RNA19. RNA chaperones, such as StpA, bind nonspecifically and unfold RNA to allow refolding into the active conformation20,21.
Previous work has used group II intron catalytic activity to report on Mss116 function. However, to further understand how DEAD-box proteins facilitate RNA-folding, it is necessary to observe RNA folding directly22. Therefore, we have characterized the effect of Mss116 on the folding dynamics of a fluorophore-labeled D135 ribozyme (D135-L14) using single-molecule Fluorescence Resonance Energy Transfer (smFRET, Fig. 1b)17,18.
Folding requires high salt
Consistent with previous studies17,18, in high ionic strength (Methods Summary), the intron exhibits repeated stochastic transitions between three distinct conformational states (Fig. 2a)17: an extended intermediate (I, FRET ~0.1), a folded intermediate (F, FRET ~0.4) and the native state (N, FRET ~0.6). Among all transitions, fewer than 2% of molecules show direct transitions between I and N, indicating that F is an obligatory folding intermediate. In single-molecule experiments the unfolded state (U) is indistinguishable from donor-only species17,18. These experiments are complementary to gel-shift assays in which U to I transitions are readily observable, but I, F and N co-migrate as a single ‘compact’ species, and N can only be differentiated by the presence of catalytic activity22–24
Figure 2. Effect of ionic strength on the folding dynamics of D135-L14 ribozyme.
a, Typical FRET trajectory (left) showing the dynamics of a single D135-L14 ribozyme under high ionic strength conditions, and histogram representing the distribution of FRET states of ~100 trajectories (right). Three different conformations are observed: I (FRET~0.1), F (FRET~0.4) and N (FRET~0.6). Black line represents idealized HMM and arrows indicate rapid transient excursions to the N state. b, Typical FRET trajectory (left) showing the dynamics of a single D135-L14 ribozyme under near-physiological conditions, and histogram representing the FRET states distribution of ~100 trajectories (right).
Under near-physiological conditions (Methods Summary), D135-L14 displays only one FRET distribution (Fig. 2b, FRET ~0.1), indicating that the vast majority of molecules reside in the extended intermediate state, consistent with gel-shift experiments17,24. Most molecules in low ionic strength (≥95%) appear static and cannot make even transient excursions to N.
Mss116 mediates folding
We sought to characterize the effect of active Mss116 (Supplementary Fig. 1) on the folding of D135-L14 under near-physiological conditions. Since Mss116 is ATP-dependent, initial experiments included ATP. The observed smFRET trajectories and distributions of D135-L14 in the presence of Mss116 exhibit transitions between all conformational states (I, F and N, Fig. 3a–c), in contrast to the smFRET distribution in the absence of protein (Fig. 2b). The appearance of F and N with Mss116 indicates that it facilitates RNA folding. Approximately 30% of molecules reach N transiently or stably, including a minor population of molecules (~9%) that remains static in N (Supplementary Fig. 2). As a control, we monitored the FRET ratio of a single D135-L14 ribozyme before and after addition of Mss116 and ATP (Fig. 3d). Initially, D135-L14 appears static in I (FRET ~0.1). After the addition, transitions from I into the higher FRET conformations were observed, providing direct evidence of Mss116-mediated RNA folding (Fig. 3d).
Figure 3. Mss116 promotes folding of group II introns at near-physiological conditions.
a and b, FRET trajectories of a single D135-L14 ribozyme in the presence of Mss116 and ATP. Black lines represent idealized HMM and arrows indicate transient excursions to the native state. c, Histogram showing the distribution of three structural conformations. d, FRET trajectory showing dynamic behavior of a single ribozyme upon addition of Mss116 and ATP at 15–20 seconds. e, FRET state distribution in the presence of the indicated non-specific RNA binding proteins.
Lowering the Mg2+ concentration to 1 mM yielded similar results (Supplementary Fig. 3), showing that Mss116 can also help D135-L14 form F and N under physiological conditions. However, we also observed a higher number of molecules in the 0 FRET state, suggesting that the number of molecules in the unfolded state is higher at 1 mM [Mg2+]. Interestingly, at both 8 and 1 mM Mg2+, we find that the average FRET ratio of I increases to ~0.18 in the presence of Mss116, indicating a slight protein-induced compaction of this conformer.
In high ionic strength, exon-based substrates (17/7 or 17/7-dC) stabilize the native state17. Here we show that during protein-facilitated folding, the substrate 17/7 stabilizes N, increasing its population by ~30% (Supplementary Fig. 4 and Supplementary Table 1). Similar behavior was also observed with a slow-cleaving substrate (17/7-dC). However, the substrate alone does not promote folding into the native state under near-physiological conditions (Supplementary Fig. 5).
To evaluate the specificity of Mss116-induced folding, we examined the folding dynamics of D135-L14 in the presence of three different basic RNA-binding proteins: the HIV nucleocapsid (NC)25, Polypyrimidine tract binding (PTB)26 and Hfq proteins27. The corresponding smFRET histograms show all three proteins can populate the folded intermediate to a lesser extent than Mss116, and none promotes folding into the native state (Fig. 3e). Among the molecules observed in the presence of each non-specific protein, more than 90% appear static in either I or F (Supplementary Fig. 6). Unlike Mss116, none of these proteins affects the average FRET ratio of I, indicating that Mss116 may recognize this conformation more specifically. These data show that the formation of the folded intermediate can be facilitated by a diverse set of basic RNA binding proteins. However, the native state is formed only in the presence of Mss116 and ATP. Thus, Mss116 plays a specific role during the final, rapid stages of ai5γ folding.
ATP is required for efficient folding
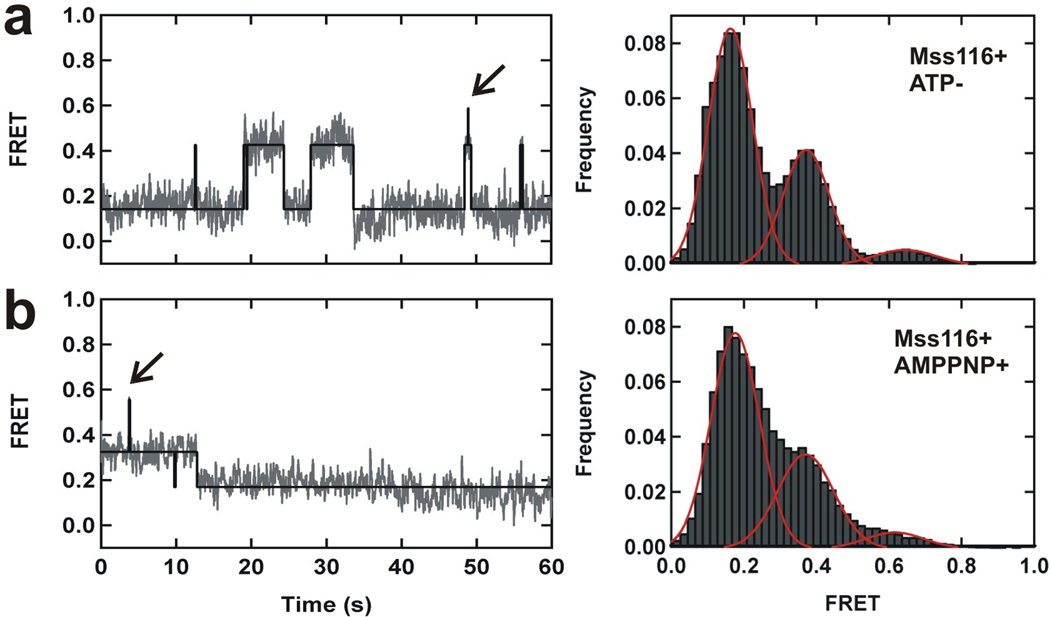
To investigate the role of ATP on Mss116-mediated folding of D135, we monitored the folding dynamics without ATP and with the non-hydrolysable ATP analog, AMPPNP (Fig. 4). The smFRET trajectories show transitions between all three conformations of the D135 ribozyme, as observed in the presence of ATP (compare Fig. 3c with Fig. 4). However, the population of N was ~2.5-fold lower, showing that ATP hydrolysis enhances formation of N (Fig. 4). This small population may be due to trace ATP that co-purifies with Mss116 or D135-L14. However, control experiments in the presence of glucose and hexokinase, which remove trace ATP, still yield low levels of the native state population (Supplementary Fig. 7). These results show that Mss116 facilitates folding into N in an ATP-dependent manner, but N can also be populated without ATP, albeit inefficiently. In the absence of Mss116, ATP or AMPPNP alone are not sufficient to form F or N (Supplementary Fig. 8). These data show that ATP binding and hydrolysis by Mss116 are important for the efficient formation of native D135, but hydrolysis is not strictly required.
Figure 4. Role of ATP in Mss116-mediated folding of group II introns.
a, smFRET time trace (left) and histogram (right) showing the distribution of different conformations of the Mss116-mediated folding pathway in the presence of wild type Mss116 protein without ATP. b, smFRET time trace (left) and histogram (right) showing the distribution of different conformations in the presence of wild type Mss116 protein and the non-hydrolysable ATP analog, AMPPNP. Black lines represent idealized HMM and arrows indicate rapid transient excursions to the native state. All experiments were performed under near-physiological conditions.
Dynamics depend on Mss116 and ATP
To define the folding rate constants (k1, k−1, k2 and k−2), we analyzed the dynamic FRET trajectories using a Hidden Markov Model (HMM)28. Without Mss116 and under high ionic strength conditions, a large subpopulation (>75%) appears static in either I or F during the two-minute timescale of our observation window (Supplementary Fig. 2). Among the subpopulation of dynamic molecules (~24%), the observed folding rate constants between the extended and folded intermediates (k1 = 0.5 ± 0.3 s−1 and k−1 = 0.2 ± 0.1 s−1) agree closely with previous values17.
Under near-physiological conditions without Mss116, the dynamic subpopulation decreases to ≤5% (Supplementary Fig. 2a); therefore, folding rate constants could not be determined. However, upon addition of Mss116 and ATP, the fraction of dynamic molecules increases to ~34%, and transitions to both F and N were observed (Fig. 3a–c). The resulting folding rate constants (Supplementary Table 2) show that k1 and k−1 are comparable to the high ionic strength conditions, suggesting that Mss116 and Mg2+ stabilize these states similarly. However, in the presence of Mss116, k2 is ~3-fold higher and k−2 is ~2-fold lower than in 100 mM Mg2+,17 indicating that the protein stabilizes the native state more than Mg2+ ions.
In the presence of Mss116 without ATP or with AMPPNP, ~30% of molecules exhibited dynamics. The resulting folding rate constants k1 and k−1 are comparable to those with ATP present. However, k−2 is 3 to 6-fold higher than in the presence of ATP (Supplementary Table 2). This indicates that ATP hydrolysis, and not just binding, contributes to efficient formation of the native state. In the presence of Mss116 without ATP or with AMPPNP, the majority of molecules that dwell in F show transitions to I (~75%) rather than N (~25%, Supplementary Table 3). However, with both Mss116 and ATP, the percentage of F to I transitions decreases (58%) while the percentage of F to N transitions increases (42%) (Supplementary Table 3). Therefore, ATP hydrolysis also increases the probability that F molecules sample N instead of I.
Detailed analysis of static molecules in each state provides additional evidence for ATP-dependent Mss116 activity (Supplementary Fig. 2b–e). In the presence of Mss116 without ATP, the fraction of static molecules in I decreases from 95% to 53%, and they begin to appear in both F and N (15% and 4%, respectively). In the presence of both Mss116 and ATP, the fraction of static molecules in N increases significantly (2.5-fold, p = 0.02), while that in F does not change significantly (p = 0.16). These data support an important role for ATP hydrolysis for efficient Mss116-induced folding of the intron ai5γ, as proposed4,5.
Discussion
Based on previous biochemical and folding studies4–6,16,24, it has been proposed that Mss116 can promote group II intron splicing by stabilizing on-pathway intermediates and/or by disrupting off-pathway misfolded structures. Previous work has shown that ai5γ folds through a slow but smooth pathway devoid of kinetic traps16,24, and an Mss116 mutant with a significant helicase defect still retains the ability to promote splicing4,29. Other work supports a mechanism where Mss116 unwinds kinetic traps to promote splicing, even at low levels of helicase activity6. Both models are primarily based on indirect studies of ai5γ splicing, thus, any putative role for Mss116 in RNA folding is purely hypothetical. To specifically examine the effect of Mss116 on ai5γ folding, we have employed a well-characterized smFRET assay that enables us to directly monitor the role of Mss116 and ATP on the intron folding.
Previous experiments showed that under near-physiological conditions, group II introns alone do not stably form the native state24. Our data show that D135 cannot even transiently sample the native state. The large subpopulation of static molecules in I (~95%) indicates a high activation barrier between I and F (Fig. 5). The appearance of all three states in the presence of Mss116 shows that Mss116 can promote RNA folding by lowering the activation barriers between folded states, consistent with recent data22.
Figure 5. Mss116-mediated group II intron folding.
a, D135-L14 minimal folding pathway: unfolded (U), extended intermediate (I), folded intermediate (F) and native state (N). Folding rate constants of I, F and N are k1, k−1, k2 and k−2. b, Hypothetical free energy diagram based on this data and previous work17,22. In the absence of Mss116 and ATP (black), I is the most stable conformation. Mss116 alone (purple), results in formation of F. Addition of both Mss116 and ATP with and without trans exon-17/7 (red and green, respectively) enhances native state formation17. Cis exons (gray) further stabilize N22.
We observe two distinct effects on folding (Fig. 5). First, F state folding is promoted by Mss116 and other RNA-binding proteins even without ATP (Figs. 3 and 4). The fact that diverse, basic RNA-binding proteins promote this stage of folding suggests that formation of F is contingent on electrostatic stabilization or annealing. The lack of ATP dependence also shows that mechanical events such as translocation or duplex unwinding are not involved in the obligate early steps of ai5γ folding. Although diverse proteins stimulate the I→F transition, only Mss116 increases the FRET ratio of I (~0.12 to ~0.18), indicating a specific interaction with the extended intermediate. This suggests a slightly more collapsed I state in the presence of Mss116, and that this protein is ideally suited to folding of ai5γ. Second, Mss116 promotes folding to the native state through a mechanism involving ATP hydrolysis. Without ATP or with AMPPNP, only a small fraction of molecules reach N, indicating that ATP hydrolysis contributes to the function of Mss116 specifically during native state formation.
Several possible models explain the function of Mss116 during native state formation. In a stabilization model, Mss116 may bind and stabilize the κ–ζ region, a substructure of domain 1 that must form before the intron can fold productively23. This same substructure contains a binding site for the catalytic domain 5, which docks late in the assembly pathway24. If Mss116 stimulates folding by stabilizing the κ–ζ element, it must dissociate before D5 can stably dock within the core. Thus, ATP hydrolysis may stimulate a conformational change in Mss116 allowing protein release during the final, rapid stage of folding, consistent with previous data linking ATP hydrolysis to Mss116 turnover and dissociation29,30. The small fraction of the native population in the absence of ATP is consistent with this model, as Mss116 likely has a finite off-rate. In vivo, efficient recycling of Mss116 may be necessary to maintain a pool of active Mss116 enzymes and may allow the cell to overcome non-functional binding events while preventing inhibition of properly folded RNAs29. However, other models, such as unwinding during the final transition from the folded intermediate to the native state, also remain possible. According to this model, the protein would first facilitate the slow, early stages of folding (including D1 collapse) through electrostatic interactions, but formation of the final native state may be impeded by one or more small misfolded structures. To form the correct folded structure, the protein would resolve the misfolded structures through strand exchange or unwinding by local strand separation. This ATP-dependant remodeling event would provide an opportunity for the native contacts to form. Although we do not have direct evidence for the presence of misfolded structures, we have previously shown that I, F and N are likely heterogeneous17. Thus, it is possible that misfolded intermediates with FRET ratios similar to on-pathway intermediates are present but not readily distinguishable. However, recent in vivo experiments show that the ability of Mss116 to hydrolyze ATP correlates with function even with different degrees of helicase activity, in agreement with a recycling function for ATPase activity29. Helicase activity may play additional roles in the presence of long exons31.
In summary, we observe multiple roles for Mss116 in the folding pathway of ai5γ and propose that Mss116 mediates group II intron folding by stabilizing on-pathway intermediates and transition states. We also observe that efficient transition from the folded intermediate to the native state requires ATP hydrolysis, possibly for Mss116 release and recycling. The substrate further stabilizes the native state, raising the interesting possibility that in vivo and in the presence of all the group II intron domains and Mss116, the native state becomes the most stable conformation.
METHODS
Preparation of DNA, RNA and protein samples
The D135-L14 RNA (638 nt) was obtained by in vitro transcription of the Hind III digested pT7D135-L14 plasmid DNA with T7 RNA polymerase under standard conditions17. Biotinylated DNA strand (5’-Biotin-TGC ATG CCT GCA GGT CGA CTC TA-3’) Cy3-DNA strand (5’-Cy3-ACC AAG AGC GTT ATT AAT-5’), Cy5-DNA strand (5’-Cy5-ACG TAG TCC GAA ATA TAT-3’), wild type RNA substrate (17/7. 5’-CGU GGU GGG ACA UUU UCG AGC GGU-3’) and slower cleaving RNA substrate (17/7-dC, 5’-CGU GGU GGG ACA UUU UdC GAG CGG U-3’) were purchased from Howard Hughes Medical Institution Biopolymer/Keck Foundation Biotechnology Resource Laboratory (Yale University, New Haven, CT). The 2’-OH positions were deprotected following the manufacturer’s protocol. All samples were purified by denaturing 18% PAGE and subsequent C8 reversed-phase HPLC, as described17.
Mss116 was overexpressed in Rosetta 2 cells (Novagen) and grown at 16°C overnight after induction. After purification of protein on a nickel column, it was cleaved with SUMO protease at 4°C overnight and purified using a gel filtration column. Untagged protein was concentrated by an Amicon ultra concentrator and stored at −80°C. The activity of Mss116 protein was confirmed by in vitro splicing, ATPase and unwinding assays.
Single-molecule FRET experiments
Protein-free single-molecule experiments were performed by heat-annealing (90°C for 45 seconds) Cy3-DNA, Cy5-DNA, biotin-DNA (10 µM each) and D135-L14 RNA (1 µM) in 100 mM KCl, 40 mM MOPS, pH 7.5 and 0.5% 2-mercaptoethanol. After the addition of 8 mM MgCl2, the reaction mixture was incubated at 30°C for 15–20 min. The fluorophore-labeled, biotinylated RNA/DNA complex was diluted to ~25 pM and bound to a streptavidin-coated quartz slide via a biotin-streptavidin linkage. Excess fluorophore-labeled and biotin-labeled DNA strands were removed from the slide by washing with reaction buffers. To minimize the non-specific binding of protein on the slide, all quartz slides were PEG-passivated. In order to reduce photobleaching of fluorophores, an oxygen-scavenging system (OSS) consisting of 10% (w/v) glucose, 2% (v/v) 2-mercaptoethanol, 50 µg/ml glucose oxidase and 10 µg/ml catalase was used in all experiments. The smFRET experiments with Mss116 were performed by introducing 25 nM Mss116 with and without 1 mM ATP/AMPPNP in OSS solution as described earlier. Similarly, smFRET experiments with the substrate (17/7 or 17/7-dC) were performed after addition of 25 nM substrate RNA into OSS solution containing 25 nM protein and 1 mM ATP. All TIRF-based smFRET experiments were performed with 33 ms time resolution at room temperature under different experimental conditions, as described32.
FRET histograms from different experimental conditions were constructed by collecting ~100 single-molecule trajectories showing the transitions between different structural conformations. Since we cannot distinguish molecules in the unfolded state (U, FRET ~ 0) from those containing only a donor fluorophore (FRET = 0, due to photobleaching, for example), we did not include them in the FRET histograms. Folding rate constants (k1, k−1, k2 and k−2) of the D135 ribozyme under different experimental conditions were obtained by analyzing FRET trajectories with a Hidden Markov Model (HMM)28. First, FRET trajectories were fitted using HMM and then transition density plots (TDP) were constructed to obtain folding rate constants as described28. Threshold FRET values 0.27 and 0.51 were used to separate transitions among the three different FRET states (Supplementary Fig 9). The effect of Mss116 without ATP was further determined in the presence of glucose and hexokinase. Incubation of the reaction mixture with glucose and hexokinase removes the trace amount of ATP associated with sample preparation.
Splicing assay
The body-labeled full-length ai5γ precursor RNA was incubated in 40 mM MOPS pH 7.5 at 90°C for 1 min followed by 3 min at 30°C. The RNA solution (1 nM final concentration) was added to a reaction mixture containing 20 nM Mss116 protein, 100 mM KCl, 40 mM MOPS, pH 7.5 and 8 mM MgCl2, 5% glycerol, 2 mM DTT and 2U/µL RNase inhibitor. After incubation for 10 min at 30°C, the splicing reaction was initiated with the addition of ATP (1 mM final concentration). The sample aliquots were removed at 0 and 120 min time points and detected using 5% native polyacrylamide gel.
Unwinding assay
250 nM Mss116 protein was pre-incubated with 0.1 nM end-labeled RNA duplex (12 bp) at 30°C for 10 min and unwinding reaction was initiated by the addition of ATP as described4. The unwinding reaction was performed in 100 mM KCl, 40 mM MOPS, pH 7.5, 4 mM MgCl2, 0.01% IGEPAL, 2 mM DTT, 2U/µL RNase inhibitor and 4 mM ATP. The sample aliquots were removed at 0 and 30 min time points and detected using 10% native PAGE.
ATPase assay
250 nM Mss116 protein was preincubated with or wthout 5 µM single-stranded RNA at 30°C for 10 min and all reactions were initiated by the addition of a mixture of P-32 labeled and unlabeled ATP. ATPase assays were performed according to standard protocol35 in 100 mM KCl, 40 mM MOPS, pH 7.5, 1 mM MgCl2, 2 mM DTT, 2U/µL RNase inhibitor and 1 mM ATP.
Supplementary Material
Acknowledgements
We thank R. K. Sigel and O. Fedorova for many helpful and stimulating discussions and for commenting on the manuscript, and A. Feig, K. Musier-Forsyth and Rajan Lamichhane for protein gifts. This work was supported by the NIH (R01GM085116 to DR, R01GM050313 to AMP) and the NSF (MCB-0747285 to DR). AMP is an HHMI investigator.
Footnotes
Author Contributions DR and AMP conceived and designed the experiments. KSK performed and analyzed the experiments with help from AS. All authors wrote the manuscript.
REFERENCES
- 1.Linder P. Quick guide: DEAD-box proteins. Curr Biol. 2000;10:R887. doi: 10.1016/s0960-9822(00)00857-5. [DOI] [PubMed] [Google Scholar]
- 2.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang HR, et al. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci U S A. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 5.Halls C, et al. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Campo M, et al. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- 8.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 9.Toor N, Hausner G, Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin PZ, Pyle AM. The architectural organization and mechanistic function of group II intron structural elements. Curr Opin Struct Biol. 1998;8:301–308. doi: 10.1016/s0959-440x(98)80062-6. [DOI] [PubMed] [Google Scholar]
- 12.Pyle AM, Lambowitz AM. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor Laboratory Press; 2006. pp. 469–505. [Google Scholar]
- 13.Koch JL, Boulanger SC, Dib-Hajj SD, Hebbar SK, Perlman PS. Group II introns deleted for multiple substructures retain self-splicing activity. Mol Cell Biol. 1992;12:1950–1958. doi: 10.1128/mcb.12.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michels WJ, Jr, Pyle AM. Conversion of a group II intron into a new multiple-turnover ribozyme that selectively cleaves oligonucleotides: elucidation of reaction mechanism and structure/function relationships. Biochemistry. 1995;34:2965–2977. doi: 10.1021/bi00009a028. [DOI] [PubMed] [Google Scholar]
- 15.Qin PZ, Pyle AM. Stopped-flow fluorescence spectroscopy of a group II intron ribozyme reveals that domain 1 is an independent folding unit with a requirement for specific Mg2+ ions in the tertiary structure. Biochemistry. 1997;36:4718–4730. doi: 10.1021/bi962665c. [DOI] [PubMed] [Google Scholar]
- 16.Su LJ, Waldsich C, Pyle AM. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res. 2005;33:6674–6687. doi: 10.1093/nar/gki973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steiner M, Karunatilaka KS, Sigel RK, Rueda D. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci U S A. 2008;105:13853–13858. doi: 10.1073/pnas.0804034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steiner M, Rueda D, Sigel RK. Impact of Ca2+ on Single Molecule Folding of a Group II Intron Ribozyme. Angewandte Chem. Int. Ed. 2009;48:9739–9742. doi: 10.1002/anie.200903809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noah JW, Lambowitz AM. Effects of maturase binding and Mg2+ concentration on group II intron RNA folding investigated by UV cross-linking. Biochemistry. 2003;42:12466–12480. doi: 10.1021/bi035339n. [DOI] [PubMed] [Google Scholar]
- 20.Zhang A, Derbyshire V, Salvo JL, Belfort M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- 21.Clodi E, Semrad K, Schroeder R. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J. 1999;18:3776–3782. doi: 10.1093/emboj/18.13.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fedorova O, Solem A, Pyle AM. Protein-facilitated folding of group II intron ribozymes. J Mol Biol. 2010;397:799–813. doi: 10.1016/j.jmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waldsich C, Pyle AM. A folding control element for tertiary collapse of a group II intron ribozyme. Nat Struct Mol Biol. 2007;14:37–44. doi: 10.1038/nsmb1181. [DOI] [PubMed] [Google Scholar]
- 24.Fedorova O, Waldsich C, Pyle AM. Group II intron folding under near-physiological conditions: collapsing to the near-native state. J Mol Biol. 2007;366:1099–1114. doi: 10.1016/j.jmb.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 26.Lamichhane R, et al. RNA looping by PTB: evidence using FRET and NMR spectroscopy and for a role in splicing repression. Proc Natl Acad Sci U S A. 2010;107:4105–4110. doi: 10.1073/pnas.0907072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hopkins JF, Panja S, McNeil SA, Woodson SA. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 2009;37:6205–6213. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bifano AL, Turk EM, Caprara MG. Structure-guided mutational analysis of a yeast DEAD-box protein involved in mitochondrial RNA splicing. J Mol Biol. 2010;398:429–443. doi: 10.1016/j.jmb.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zingler N, Solem A, Pyle AM. Dual roles for the Mss116 cofactor during splicing of the ai5γ group II intron. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq530. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao R, Rueda D. RNA folding dynamics by single-molecule fluorescence resonance energy transfer. Methods. 2009;49:112–117. doi: 10.1016/j.ymeth.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 33.Ha T, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 34.Christian TD, Romano LJ, Rueda D. Single Molecule Measurements of Synthesis by DNA Polymerase with Base-Pair Resolution. Proc Natl Acad Sci U S A. 2009;106:21109–21114. doi: 10.1073/pnas.0908640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shuman S, Spencer E, Furneaux H, Hurwitz J. The role of ATP in in vitro vaccinia virus RNA synthesis effects of AMP-PNP and ATP gamma S. J Biol Chem. 1980;255:5396–5403. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.