Abstract
Human estrogen receptors (ERs) alpha and beta are crucially involved in the regulation of mammary growth and development. Normal breast tissues display a prevalently expression of ER beta than ER alpha, which drastically increases during breast tumorogenesis. So, it is reasonable to assume how a dysregulation of the two estrogen receptor subtypes may induce breast cancer development. However, the molecular mechanism underlying the opposite role played by the two estrogen receptors on tumor cell growth remains to be elucidated.
In the present study, we have demonstrated that ER beta overexpression in breast cancer cells decreases cell proliferation and down-regulates ER alpha mRNA and protein content along with a concomitant repression of estrogen-regulated genes. Transient transfection experiments, using a vector containing the human ER alpha promoter region, showed that elevated levels of the ER beta down-regulated basal ER alpha promoter activity. Furthermore, side-directed mutagenesis and deletion analysis have revealed that the proximal GC-rich motifs at −223 and −214 is crucial for the ER beta-induced ER alpha down-regulation in breast cancer cells. This occurred through ER beta-Sp1 protein-protein interaction within the ER alpha promoter region and the recruitment of a corepressor complex containing NCoR/SMRT (nuclear receptor corepressor/silencing mediator of retinoic acid and thyroid hormone receptor), accompanied by hypoacetylation of histone H4 and displacement of RNA polymerase II. Silencing of NCoR gene expression by RNA interference reversed the down-regulatory effect of ER beta on ER alpha gene expression and cell proliferation.
Our results provide evidence for a novel mechanism by which overexpression of ER beta through NCoR is able to down regulate ER alpha gene expression, thus inhibiting ER alpha’s driving role on breast cancer cell growth.
Keywords: Breast Cancer, Estrogen Receptor alpha and beta, NCoR/SMRT corepressors
Introduction
Estrogens play an important role in the mammary gland development, but they are also known to be involved in the mammary carcinogenesis (Heldring N et al. 2007; Hall JM et al. 2001). These steroid hormones exert their biological effects via interaction with two different isoforms of estrogen receptors, ER alpha and ER beta, each encoded by unique genes, but with a common structural and functional organization. Binding of the estrogen ligands to ERs in the nucleus results in receptor phosphorylation, dimerization, and recruitment of specific coregulator proteins, termed coactivators, which enhance binding of the receptor complex to promoter regions of target genes known as the estrogen response elements (EREs) and augment the receptor’s transcriptional activity (Osborne CK et al. 2001; Schiff R et al. 2005). In addition to the genomic action, ERs, located in the cell membrane or cytoplasm, may initiate rapid cellular signalings (Losel RM et al., 2003; Cato AC et al., 2002), involved in an intricate network of crosstalk with growth factor pathways (Ordonez-Moran P and Munoz A 2009; Bai Z and Gust R 2009; Nemere I et al., 2003). In addition, ERs may bind to DNA in a non classical way through its interaction with other transcription factors (Safe S and Kim K 2004; Safe S 2001) For instance, ERα/Sp1, ERα/AP-1, and ERβ/AP-1-mediated transactivation through binding GC-rich and AP-1 motifs have been extensively investigated at this regard (Safe S and Kim K 2004; Safe S 2001; Paech K et al. 1997; Webb P et al., 1999; Bjornstrom L and Sjoberg M 2005; Kushner PJ et al., 2000; Saville B et al. 2000; Catalano S et al., 2007; Mauro L et al., 2007; Panno ML et al., 2006). Indeed, the latter indirect mechanism may occur in absence of natural ligand, in a cell type and gene dependent context (De Amicis F et al. 2009), and may also involve recruitment of corepressors as NCoR and SMRT in order to inhibit basal cell transcription machinery (Li X et al., 2003).
Both ERs subtypes are expressed in human mammary tissue (Nilsson S et al., 2001; Heldring N et al., 2007) with only 7–10% of the epithelial cells expressing ER alpha and 80–85% expressing ER beta. In contrast, expression of ER alpha is increased in breast cancer cells, where it acts as a mediator of cell proliferation and has been shown to be an effective therapeutic target for decades. The role of ER beta in breast cancer is less clear and its prognostic value is still under debate. It is estimated that ER beta is expressed in approximately half of human primary breast cancers, but its expression is lost during breast cancer progression, most likely due to promoter hypermethylation (Zhao C et al., 2008) Moreover, ER beta protein levels have been linked to good prognosis, prolonged disease-free survival and response to antiestrogen treatment (Skliris et al., 2003; Hopp et al, 2004). Many cell-based studies suggest that ER beta acts as a negative modulator of ER alpha action and can negatively regulate breast cancer proliferation (Williams C et al., 2007; Chang EC et al., 2006; Paruthiyil S et al., 2004). Indeed, inducible expression of ER beta in ER alpha-positive breast cancer cells inhibited estrogen stimulated proliferation and tumor angiogenesis and growth in xenograft experiments.(Hodges-Gallagher L et al., 2008; Hartman J et al. 2006; Ström A et al., 2004) Overexpression of ER beta or ER beta cx isoforms also decreased ER alpha transcriptional activity concomitantly with a reduced expression of estrogen-regulated genes, such as vascular endothelial growth factor (VEGF) or progesterone receptor (PR) (Saji S et al., 2002; Buteau-Lozano H et al., 2002; Omoto Y et al., 2003). In ER-negative cells ectopically expressing the two ERs, ER beta reduced the sensitivity of the cells to estrogen treatment on growth (Nilsson S et al., 2001; Pettersson K et al., 2000) and inhibited cyclin D1 gene activation.
Given the markedly enhanced ratio ER alpha/ER beta in early breast cancers (Foley EF et al., 2000; Bardin A et al., 2004) and the opposite roles of the two ERs in regulating cell proliferation and differentiation, it is imperative to dissect the molecular mechanisms underlying the dysregulation of these processes in cancer cells. Therefore, aim of this study is to investigate if ER beta may play a direct inhibitory role on ER alpha expression and gene promoter activity. Here, we demonstrate that ER beta through its interaction with Sp1 protein recruits NCoR corepressor in the promoter region upstream the transcription start site of ER alpha gene, thus down-regulating its expression.
Results
ER beta overexpression down-regulates ER alpha expression in breast cancer cells
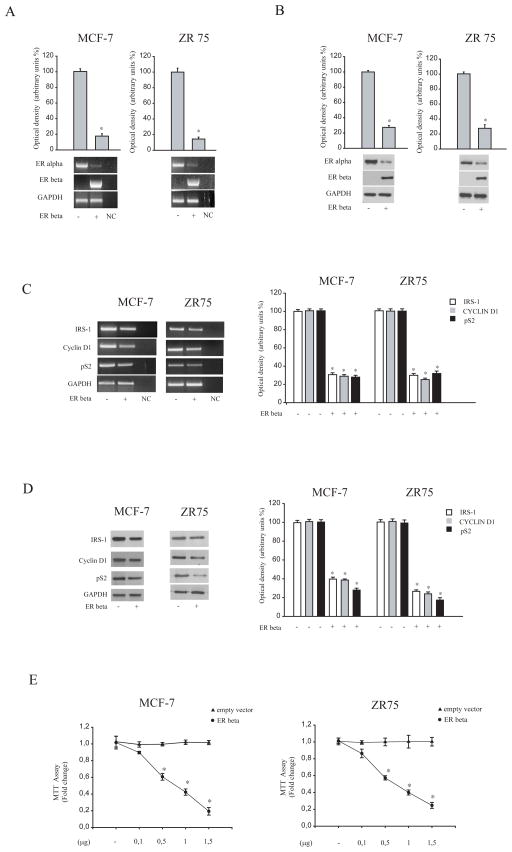
Although several data have demonstrated that ER beta negatively interferes with ER alpha signaling in breast cancer cells, it is still remained unexplored how ER beta may affect ER alpha gene expression. To this aim, ER alpha-positive MCF-7 and ZR75 breast cancer cells were transiently transfected with an ER beta expression vector and ER alpha expression was evaluated by RT-PCR and Western blotting analysis. As shown in Figure 1 A and B, ectopically expressed ER beta reduced ER alpha expression in terms of mRNA and protein content in both MCF-7 and ZR75 cells. Concomitantly, ER beta overexpression markedly decreased mRNA levels (Figure 1C) and protein (Figure 1 D) expression of classical estrogen-regulated genes, such as insulin receptor substrate 1(IRS-1), pS2 and cyclin D1 in both cell lines. These findings were well correlated with a dose-dependent inhibition mediated by ER beta on cell proliferation, as revealed by MTT assays (Figure 1F).
Figure 1. Overexpressed ER beta down-regulates ER alpha expression in breast cancer cells.
(A) Bottom panel, Western Blot analysis of ER alpha and ER beta in total protein extracts from MCF-7 and ZR75 cells, transiently transfected with either empty vector (−) or ER beta expression plasmid. GAPDH was used as loading control. Upper panel, the histograms represent the mean ± S.D. of three independent experiments in which band intensities were evaluated in terms of optical density arbitrary units and expressed as the percentage of the control assumed to be100%. (B) Bottom panel, total RNA was isolated from MCF-7 and ZR75 cells transfected with either empty vector (−) or ER beta expression plasmid and reverse transcribed. cDNA was subjected to PCR using specific primers for ER alpha, ER beta and GAPDH. NC, negative control, RNA sample without the addition of transcriptase. Upper panel, the histograms represent the mean ± S.D. of three independent experiments in which band intensities were evaluated in terms of optical density arbitrary units and expressed as the percentage of the control assumed to be 100%. (C) Left panel, Total RNA was isolated from MCF-7 and ZR75 cells transfected with either empty vector (−) or ER beta expression plasmid and reverse transcribed. cDNA was subjected to PCR using specific primers for IRS-1, pS2, Cyclin D1 and GAPDH. NC, negative control, RNA sample without the addition of transcriptase. Right panel, the histograms represent the mean ± S.D. of three different experiments in which band intensities were evaluated in terms of optical density arbitrary units and expressed as the percentage of the control assumed to be 100%.
(D) Left panel, Western Blot analysis of IRS-1, pS2, Cyclin D1 in total protein extracts from MCF-7 and ZR75 cells transfected with either empty vector (−) or ER beta expression plasmid. GAPDH was used as loading control. Right panel, the histograms represent the mean ± S.D. of three different experiments in which band intensities were evaluated in terms of optical density arbitrary units and expressed as the percentage of the control assumed to be 100%. *, P<0.01 ER beta-transfected cells compared to empty vector-transfected cells.
(E) MTT growth assays in MCF-7 and ZR75(E) cells transfected with empty vector or ER beta expression vector (0,1/0,5/1/1,5 μg/well) for six days. Cell proliferation is expressed as fold change ± S.D. relative to empty vector-transfected cells and is representative of three different experiments each performed in triplicate. *, P< 0.05 ER beta-transfected cells compared to empty vector-transfected cells.
Overexpressed ER beta mediates down-regulation of ER alpha via a GC proximal region of its promoter
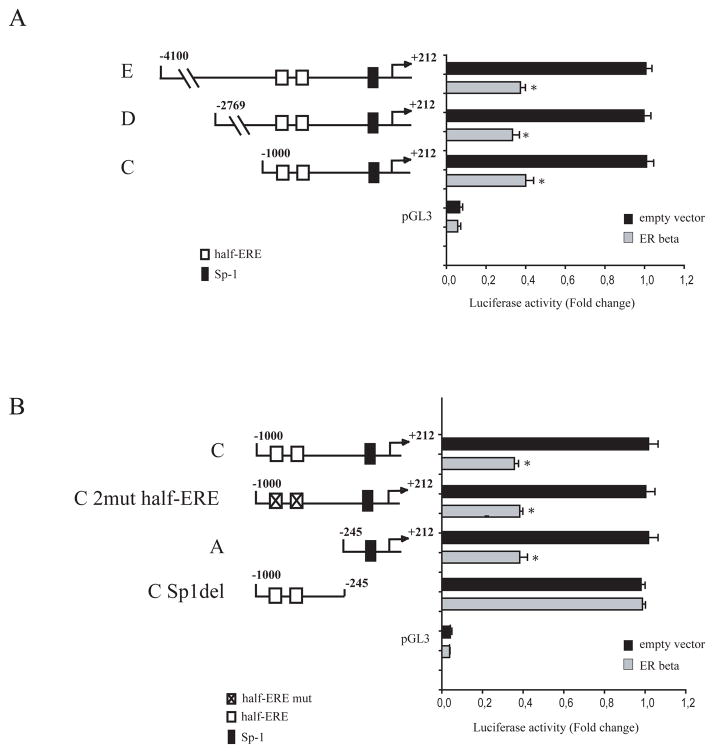
To analyze how ER beta interferes with ER alpha gene transcription, we transiently transfected breast cancer cell lines with a luciferase reporter plasmid containing the human ER alpha promoter region spanning from −4100 bp to +212 bp. As shown in Figure 2A, a significant decrease in ER alpha promoter activity was observed in MCF-7 cells when ER beta was overexpressed. The human ER alpha promoter contains multiple consensus sites for several transcription factors, including a CAT box, TFIID, AP2γ, and Sp-1 motifs (deGraffenried LA et al., 2004). To identify the region within the ER alpha promoter responsible for ER beta-mediated inhibitory effects, we transiently transfected MCF-7 cell lines with plasmids containing a series of 5′ deleted segments of human ER alpha promoter. Schematic representation of these constructs is shown in Figure 2. In transfection experiments performed using p-4100/+212 (E), p-2769/+212 (D), and p-1000/+212 (C) plasmids, the responsiveness to ER beta was still maintained, suggesting that the region from −1000 to +212 might be involved in the transrepression mechanisms exerted by ER beta (Figure 2A). Thus, we focused our attention on the latter construct p-1000/+212, and we evidenced, upstream to the initiation transcription site, two half ERE (−867/−861 and −894/−888) and one Sp-1 (−223/−214) sites, putative effectors of ER signaling (Salvatori L et al., 2003; Jennifer R et al., 2005). We observed that in MCF-7 cells transiently transfected with the ER alpha promoter plasmid bearing 2 half ERE-mutated sites (C 2mut half-ERE) or with a deleted construct of ER alpha promoter containing Sp-1 site (p-245/+212, A) ER beta-mediated down-regulation still persisted. In contrast, deletion of the Sp-1 site (C Sp-1 del) completely abrogated ER beta effects (Figure 2B). Similar results were obtained in ZR75 breast cancer cell line (data not shown). Taken together, our findings demonstrated that the down-regulatory effects of ER beta on ER alpha gene expression require Sp1 sequence motif. Taking into account that functional domains of Sp1 are involved in protein-protein interactions with other transcription regulatory molecules, such as the corepressors SMRT, NCoR and BCoR (BCL6 corepressor) (Huynh KD et al., 2000; Zamir I et al., 1997; Chen JD and Evans, RM 1995), to inhibit cell transcription machinery, we wondered whether the same corepressors were recruited by ER beta/Sp1 complex and then down-regulates ER alpha gene expression.
Figure 2. ER alpha promoter activity is down regulated by ER beta overexpression, and deletion of the GC-proximal promoter region abrogates this effect.
(A–B) Left panel, schematic representation of constructs of the ER alpha gene promoter used in this study.). Each fragment was subcloned into the pGL3 vector. (A–B) Right panel, plasmids containing ER alpha promoter fragments were transiently cotransfected in MCF-7 cells in presence or absence of ER beta expression plasmid. After 24 hours of transfection, luciferase activities were normalized to the Renilla Luciferase as internal transfection control and data where reported as fold change. The values represent the means ± S.D. of three different experiments each performed in triplicate. pGL3: basal activity measured in cells transfected with pGL3 basal vector. *, P< 0,05 ER beta-transfected cells compared to empty vector-transfected cells.
The NCoR corepressor is recruited with Sp-1 to ER alpha promoter region
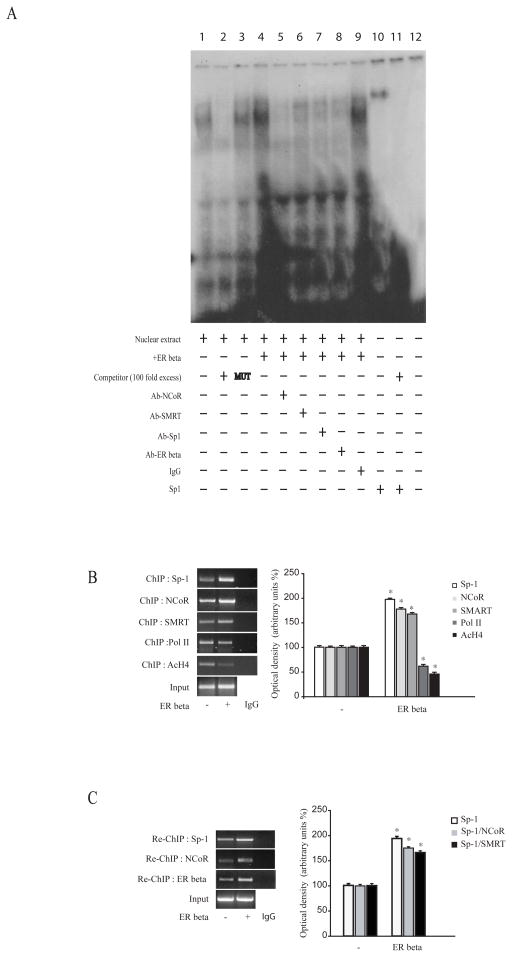
The specific role of the Sp-1 motif in mediating the inhibitory role of ER beta on ER alpha gene expression was investigated using electromobility shift assays (EMSA) and chromatin immune-precipitation (ChIP) assays. Using synthetic radiolabeled oligonucleotides bearing the Sp-1 motif present in the ER alpha promoter region (Figure 3A, lane 1), we observed in nuclear extracts from MCF-7 cells the formation of a protein complex, which was abrogated by incubation with 100 fold molar excess of unlabeled probe (Figure 3A, lane 2), demonstrating the specificity of the DNA-binding complex. This inhibition was no longer observed when mutated oligodeoxyribonucleotide probe was used as competitor (Figure 3A, lane 3). Interestingly, overexpression of ER beta strongly increased the DNA-binding protein complex compared with control samples (Figure 3A, lane 4). The inclusion of an anti-Sp-1, an anti-ER beta, an anti-NCoR and an anti-SMRT antibodies in the reaction immunodepleted the specific band, confirming the presence of these proteins in the complex (Figure 3A, lanes 5–8). However, immunodepletion occurred in a higher extend in the presence of NCoR antibody. Non specific IgG did not affect Sp-1 complex formation (Figure 3A, lane 9). Recombinant Sp-1 protein revealed a complex migrating at the same level as that of nuclear extracts from cells (Figure 3A, line 10).
Figure 3. ER beta recruits corepressors to the Sp-1 site in the ER alpha gene promoter.
(A) Nuclear extracts from MCF-7 cells transfected with either empty vector or ER beta expression plasmid were incubated with a double-stranded Sp-1 specific sequence probe labeled with [γ32P] ATP and subjected to elettrophoresis in a 6 % polyacrylamide gel (lanes 1 and 4). Competition experiments were performed adding as competitor a 100-fold molar excess of unlabeled probe (lane 2 and lane 11) or a 100-fold molar excess of unlabeled oligonucleotide containing a mutated Sp-1 motif (lane 3). Nuclear extracts from MCF-7 over-expressing ER beta were incubated with anti-NCoR (lane 5) or anti-SMRT (lane 6) or anti-Sp-1 (lane 7) or anti-ER beta (lane 8) or IgG (lane 9), incubated with probe. Lane 10, Sp-1 protein. Lane 12, probe alone. (B) Left panel, MCF-7 cells transfected with either empty vector (−) or ER beta expression plasmid were cross-linked with formaldehyde, and lysed. The pre-cleared chromatin was immune-precipitated with specific anti-Sp-1, anti-NCoR, anti-SMRT, anti-Polymerase II and anti-AcH4 antibodies, and with a normal mouse serum (IgG) as negative control. A 5 μl volume of each sample and input were analyzed by PCR with specific primers, as detailed in Materials and Methods Section, to amplify ER alpha promoter sequence containing Sp-1 site. Right panel, the histograms represent the mean ± S.D. of three separate experiments in which band intensities were evaluated in terms of optical density arbitrary units and expressed as percentage of the control, which was assumed to be 100%. (C) Left panel, Chromatin immunoprecipitated with anti-Sp-1 antibody was re-immunoprecipitated with anti-NCoR or anti-ER beta antibodies. A 5 μl volume of each sample and input were analyzed by PCR with specific primers, as detailed in Materials and Methods Section, to amplify ER alpha promoter sequence containing Sp-1 site. Right panel, the histograms represent the mean ± S.D. of three separate experiments in which band intensities were evaluated in terms of optical density arbitrary units and expressed as percentage of the control, which was assumed to be 100%. *, P <0.01 ER beta-transfected cells compared to empty vector-transfected cells.
Moreover, to better evaluate the involvement of Sp-1 and NCoR/SMRT corepressors in ER beta-mediated ER alpha-down-regulation at the promoter level, ChIP assays were performed. Using specific antibodies against Sp-1, NCoR, SMRT, RNA-polymerase II and acetyl histone H4 protein-chromatin complexes were immunoprecipitated from MCF-7 cells transfected either with an empty vector or an ER beta expression vector (Figure 3B). PCR using primers spanning the Sp-1 binding element in the ER alpha promoter region clearly showed an enhanced recruitment of Sp-1 and NCoR and slightly of SMRT upon ER beta overexpression to the ER alpha gene promoter, the corepressor DAX-1 not detected under the same experimental conditions (data not shown). These results were concomintant with a lower association of RNA-Polymerase II and acetyl histone H4 to ER alpha regulatory region, indicating that the chromatin in this region is probably in a less permissive environment for gene transcription. Re-ChIP assays confirmed the increased NCoR and ER beta occupancy of the Sp-1-containing region within the ER alpha promoter in cells overexpressing ER beta (Figure 3C).
NCoR knockdown reverses ER beta-mediated effects on ER alpha down-regulation and cell proliferation
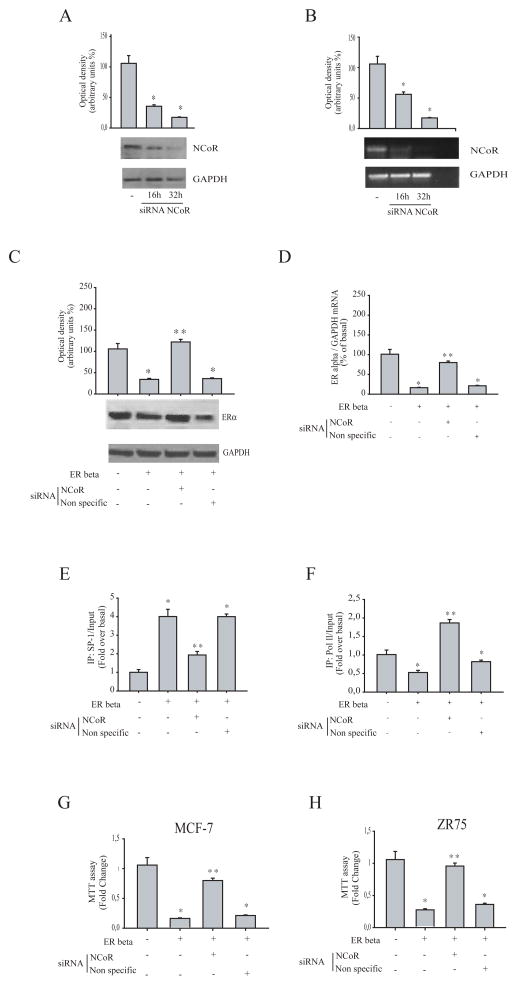
To ascertain the involvement of NCoR on ER beta-related downregulation of ER alpha, NCoR siRNA knockdown experiments were performed in MCF-7 cells transfected with an ER beta expression vector. Silencing of NCoR gene expression (evaluated by western blot and RT-PCR analysis, Figure 4A and B) restored both protein and mRNA of ER alpha expression, while no changes were observed after transfection of cells with a scrambled dsRNA (Figure 4 C and D). We also showed in MCF-7 cells overexpressing ER beta that the increased Sp-1 recruitment to ER alpha gene promoter was abrogated in precence of NCoR siRNA. (Figure 4E) Concomitantly, the recruitment of RNA polymerase II in the same region (Figure 4F).
Figure 4. Effects of NCoR silencing on ER beta-mediated down regulation of ER alpha expression, Sp-1 recruitment to ER alpha promoter and cell proliferation.
(A) Western Blot analysis for NCoR in MCF-7 cells transfected with non specific siRNA or targeted against human NCoR (100 nM) for 16 and 32 hours. GAPDH was used as a loading control. (B) RT-PCR for NCoR or GAPDH in MCF-7 cells transfected as above described. NC: negative control, RNA sample without the addition of reverse transcriptase. *,P <0.01 NCoR siRNA-transfected cells compared to non specific siRNA-transfected cells. (C) Western Blot analysis for ER alpha in MCF-7 cells transfected with either empty vector (−) or ER beta expression plasmid in presence of non-specific or NCoR siRNA. GAPDH was used as loading control. (D) RNA was extracted from MCF-7 cells transfected with either empty vector (−) or ER beta expression plasmid in presence of non-specific or NCoR siRNA, reverse transcribed and cDNA was subjected to Real-Time RT-PCR for analyzing ER alpha mRNA levels. Data represent the mean ± S.D. of values from three separate RNA samples expressed as percentage of control (−) assumed to be 100%. Each sample was normalized to GAPDH mRNA content. (E, F) MCF-7 cells transfected with either empty vector (−) or ER beta expression plasmid in presence of non-specific or NCoR siRNA, were crosslinked with formaldehyde, and lysed. The precleared chromatin was immunoprecipitated with anti-Sp-1 (E) and anti-RNA polymerase II (Pol II, F) antibodies. A 5 μl volume of each sample and input was analyzed by real-time PCR using specific primers to amplify ER alpha promoter sequence, including the Sp-1 site. Similar results were obtained in two independent experiments. MTT assays in MCF-7 (G) and ZR75 (H) cells transfected as indicated. Results are expressed as fold change ± S.D relative to empty vector-transfected cells and are representative of three different experiments each performed in triplicate. *,P <0.01 ER beta-transfected cells compared to empty vector-transfected cells. **,P <0.01 NCoR siRNA-transfected cells compared to ER beta-transfected cells.
Finally, the anti-proliferative effects exerted by ER beta were completely reversed in the presence of NCoR siRNA knockdown in MCF-7 and ZR75 breast cancer cells (Figure 4 G and H), addressing the crucial role of NCoR in mediating the ER beta-induced inhibitory effects on breast cancer cell proliferation.
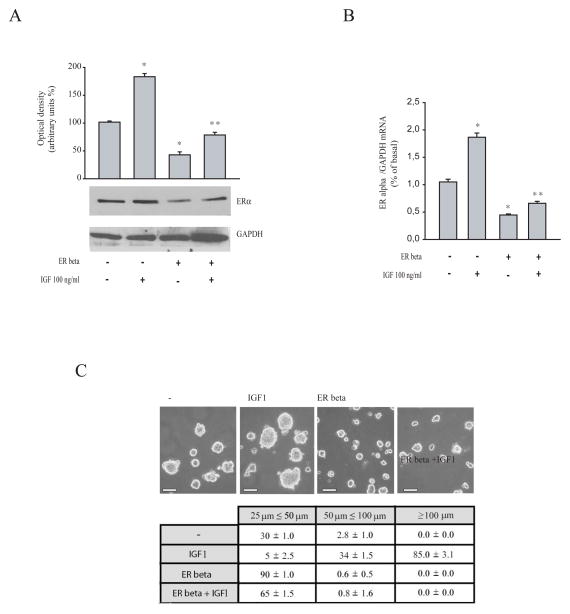
ER beta antagonizes IGF1-mediated up-regulatory effects on ER alpha expression and three-dimensional cell growth in MCF-7 cells
It has been previously demonstrated that IGF-1 and insulin can increase ER alpha expression and stimulate proliferation in breast cancer cells (Lannigan DA 2003; Andò S et al., 1998; Lee AV et al., 1997; Panno ML et al., 1996). Thus, we investigated the ability of ER beta to reverse the IGF-1 effects on ER alpha expression in MCF-7 cells, by western blotting and real time PCR analysis (Figure 5A and B). As expected, IGF-1 enhanced ER alpha protein and mRNA levels and ER beta overexpression significantly abrogated this increase. Then, the effects of ER beta an IGF1-induced growth were also assessed using three-dimensional MCF-7 cell culture, that simulate “in vivo” the biological features of tumors (Mauro L and Surmacz E 2004) Our results showed that ER beta overexpression inhibited the enhanced cell growth induced by IGF-1 exposure, as evidenced by the extent of aggregation scored by measuring the spheroid diameters (Figure 5C).
Figure 5. Overexpressed ER beta reverses IGF1-enhanced ER alpha expression and cell-cell adhesion.
(A) MCF-7 cells transiently transfected with either empty vector (−) or ER beta expression plasmid were treated with vehicle or IGF1 (100ng/ml) for 48 hours. Total proteins were extracted and Western Blot analysis was performed to evaluate the expression of ER alpha. GAPDH was used as loading control. (B) Real-Time RT-PCR analyzing ER alpha mRNA levels in cells transfected and treated as indicated. Data represent the mean + S.D. of values from three separate RNA samples expressed as percentage of control. Each sample was normalized to GAPDH RNA content. *,P<0.01 IGF-1 treatment compared to vehicle treated-cells. **,P<0.01 ER beta overexpressing cells compared to empty vector-transfected cells. (C) MCF-7 cells were transiently transfected with either empty vector (−) or ER beta expression plasmid and growth as three-dimensional cultures in the presence or absence of IGF (100 ng/ml, 48h). Scale bar=25 μm. The extent of aggregation was scored by measuring the spheroid diameters. The values represent the sum of spheroids in 10 optical fields under ×10 magnification.
Discussion
In this study, we show for the first time that ER beta overexpression down-regulates ER alpha gene expression in a ligand-independent manner in ER alpha-positive breast cancer cells. This occurs through ER beta interaction with Sp-1 and NCoR corepressor recruitment within the human ER alpha promoter region, up-stream the initiating transcription site.
ER alpha and ER beta have both overlapping and distinct expression patterns, and mammary gland development in animal models requires ER signaling. It has been hypothesized that dysregulated ER isoform expression may induce to an abnormal cell proliferation and survival, thus impacting mammary tumorogenesis. It is also well known how ER alpha expression is increased and ER beta expression is decreased in early breast cancers, whereas expression of both receptors declines in more invasive cancers (Roger P et al., 2001; Leygue E, et al., 1998; Jarvinen TA et al., 2000) Expression of ER beta is lost in other early tumor types respect from normal tissue, leading to the hypothesis that ER beta may function as a tumor suppressor (Chang EC et al., 2006; Williams C et al., 2007) Data coming from cell studies have suggested that ER beta negatively interferes with ER alpha signaling in breast cancer cells, and mediates anti-proliferative effects (Pettersson K et al., 2000; Williams C et al., 2007). ER beta over-expression inhibits tumor establishment and growth as well as E2-induced tumor formation “in vivo” in mouse xenografts of ER alpha-positive MCF-7 and T47D breast cancer cells (Paruthiyil S et al., 2004; Behrens D et al., 2007; Hartman J et al., 2006) Indeed, ER beta induces inhibition of classical estrogen-regulated genes, such as VEGF and PDGFβ (Hartman J et al., 2006). Recently, Song and Pan (Song K and Pan ZZ, 2012) demonstrated that ER alpha-mediated estrogenic activity in the mammary gland can be opposed by the ER beta and selective agonists such as DPN may be explored for the development of better hormone replacement therapy (HRT) regimens to reduce or eradicate the risk for breast cancer.
In the majority of clinical studies, ER beta expression indicates a favorable response to adjuvant tamoxifene (Tam) therapy, and patients with ER alpha+/ER beta+ tumors appear to respond at least as well as or better to endocrine therapy than patients with ER alpha+/ER beta-tumors. In addition, in Tam-treated patients, high ER beta expression well correlates with increased overall (Mann S et al., 2001; Hopp TA et al., 2004), and disease-free survival (Hopp TA et al., 2004), no disease progression (Murphy LC et al., 2002), or no relapse within five years (Fleming FJ et al., 2004; Esslimani-Sahla M et al., 2004). Thus, ER beta has emerged as an important potential marker for predicting response to endocrine therapy.
These findings led us to investigate the molecular mechanism through which ER subtypes are regulated in breast cancer cells. Here, we have demonstrated that ER beta overexpression in a ligand-independent manner resulted in inhibition of ER alpha in term of mRNA and protein content in breast cancer cells. Similar inhibitory effects were also obtained on the expression of estrogen-dependent genes such as IRS-1, pS2, cyclin D1. These data underline how ER beta-induced ER alpha down-regulation may arise via transcriptional mechanisms. Therefore, we focused on the molecular mechanisms by which ER beta mediates repression of ER alpha gene expression and on the biological consequences of ER beta overexpression on growth of breast cancer cells.
ER alpha and ER beta are transcriptional factors that can regulate gene expression through several different modes including direct DNA binding (acting as homodimers or as heterodimers) or through tethering to other transcription factors such as activating protein-1 (AP-1) and stimulating protein 1(Sp1) (Paech K et al., 1997; Saville B et al., 2000). This has been most extensively investigated in relationship to protein complexes involving Sp1 and ER alpha at GC boxes, which are classic binding sites for members of the Sp1 family of transcription factors (Duan R et al., 1998; Vyhlidal C et al., 2000; Krishnan V et al., 1994; Porter W et al., 1997) Many studies have observed that ER alpha is able to enhance binding of Sp1 to its site in several promoter regions (Catalano S et al., 2007; Mauro L et al., 2007; Panno ML et al., 2006) The analysis of different functional motifs present within the ER alpha proximal promoter (deGraffenried LA et al., 2004) has identified two half-ERE and one Sp-1 responsive elements, as potential targets of ER beta. Functional experiments using ER alpha promoter-deleted or mutated constructs have shown that Sp-1 sequence is an important prerequisite for the down-regulatory effects of ER beta on ER alpha promoter activity. These results were well supported by electrophoretic mobility shift assays, which revealed a marked increase in a specific DNA-binding complex in nuclear extracts from MCF-7 overexpressing ER beta. This complex was immune-depleted by anti-Sp-1 and anti-ER beta antibodies, suggesting the presence of these proteins in the complex. Furthermore, we observed an enhanced recruitment of Sp-1 and ER-beta to the ER alpha promoter, that was concomitant with a decrease in RNA polymerase II and acetyl histone H4 recruitment, further supporting a negative role for ER beta inmodulating ER alpha gene transcriptional machinery.
A recent study reported that the ZFDBD (Zinc Finger DNA Binding Domain) and ID (Inhibitory Domain) domains of Sp-1 are involved in protein-protein interactions with other transcription regulatory molecules, such as the corepressors SMRT, NCoR and BCoR (BCL6 corepressor) (Huynh KD et al., 2000; Zamir I et al., 1997; Chen JD and Evans, RM 1995) These corepressors interact with unliganded nuclear receptors, through an elongated helix of sequence LXXI/HIXXXI/L, alternatively referred to as the CoRNR-box (Nagy L et al., 1997; Hu X and Lazar MA, 1999; Webb P et al., 2000). It has been recently documented that NCoR and SMRT are also recruited by both ER and PR in the presence of ligand antagonist to regulate transcription of different genes (Jiang S et al., 2006; Townson SM et al., 2006).. Our results demonstrate that NCoR was the corepressor crucially recruited on the Sp-1 site of the ER alpha gene promoter together with Sp-1 and ER beta. Generally, NCoR and SMRT share the same molecular architecture, interact with many of the same transcription factors, and assemble into similar corepressor complexes (Ghisletti S et al., 2009). We also detected a slight recruitment of SMRT in the same experimental conditions. Finally, the contribution of NCoR corepressor factor in ER beta-mediated effects emerges from experiments showing that silencing of NCoR gene expression is able to reverse the inhibitory effects of ER beta on ER alpha mRNA and protein, Sp-1 recruitment to the ER alpha promoter gene and cell growth proliferation.
Previous “in vitro” studies have shown that insulin and IGF1 up-regulate the ER alpha expression as well as its DNA binding capacity. (Lannigan DA 2003; Andò S et al., 1998; Lee AV et al., 1997; Panno ML et al., 1996). We demonstrate how ER beta reduces the stimulatory effects induced by IGF1 on ER alpha expression and on the three-dimensional cell growth, and became a negative modulator of the well known cross-talk between ER alpha and IGF1-R signaling pathways.
In conclusion, we suggest that inhibition of ER alpha by ER beta is a critical regulatory pathway of ER-positive cells, addressing prospectively how therapeutic tools able to potentiate ER beta action and thereby deplete intratumoral ER alpha content may inhibit breast cancer cell growth and progression
Materials and Methods
Reagents and antibodies
The following components were obtained from the given respective companies, with their addresses in brackets. Dulbecco’s Modified Eagle’s Medium/Nutrient Mixture F-12 Ham, DMEM, 100 bp DNA ladder, l-Glutamine, penicillin, streptomycin, bovine serum albumine, phosphate-buffered saline were purchased from by Invitrogen (Carlsbad, CA, USA), Sephadex G50 spin columns and poly (dI-dC) by Roche (Indianapolis, IN, USA). GoTaq DNA polymerase, T4 polynucleotide Kinase, Dual luciferase kit, FuGENE 6, Sp-1 human recombinant protein and CMV renilla luciferase plasmid were provided by Promega (Madison, WI, USA). The RETROscript kit and DNase I were purchased from Ambion (Austin, TX, USA). Aprotinin, leupeptin, phenylmethylsulfonyl fluoride, sodium orthovanadate, formaldehyde, NP-40, MTT, dimethyl sulphoxide, proteinase K, tRNA, by Sigma (Milan, Italy). Antibodies against ER alpha and beta, IRS-1, cyclin D1, pS2, GAPDH, NCoR, Sp-1, AcH4 and polymerase II (N20) were provided by Santa Cruz Biotechnology (Santa Cruz, CA, USA). ECL System and [ 32P]ATP were purchased by PerkinElmer (Wellesley, MA, USA), Salmon sperm DNA/protein A agarose by UBI (Chicago, IL, USA), Triazol, SYBR Green Universal PCR Master Mix by Biosystems (Forster City, CA, USA).
Cell cultures
MCF-7 and ZR 75 breast cancer cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). MCF-7 cells were cultured in DMEM-F12 containing 5 % fetal bovine serum, 1% l-Glutamine, 1 mg/ml penicillin–streptomycin. ZR75 cells were maintained in complete DMEM supplemented with 10% fetal bovine serum, 1% l-Glutamine and 1 mg/ml penicillin–streptomycin.
Plasmids
The plasmids containing the human ER alpha promoter region or its deletions (E: p–4100/+212; D: p–2769/+212; C:p–1000/+212, A: p–245/+212) were kindly provided by Prof Fuqua (Baylor College of Medicine, Houston, TX, USA). These fragments were inserted into the luciferase vector pGL3. Deletion of Sp-1 sequence in ER alpha gene promoter was generated by PCR using as template p1000 construct with following primers: forward 5′-GCGGTACCCGAAAGATCGAGT TGTAGGAC-3′ and reverse 5′-CGCTCGAGTTATATAGGGAAGACTGGGCTTAAAATA-3′. The amplified DNA fragment was digested with Kpn I and Xho I and ligated into pGL3-basic vector. The resulting plasmid encoding the human ER alpha gene promoter containing the desired deletion was designed as C Sp-1 del and the sequence was confirmed by nucleotide sequence analysis. The plasmid encoding the human ERβ gene was a gift from JA Gustafsson (Department of Biosciences and Nutrition, Karolinska Institute, Sweden).
Site-directed mutagenesis
The mutation of the two half-ERE sites in the ER alpha gene promoter plasmid was created by site-directed mutagenesis using QuickChange kit (Stratagene, La Jolla, CA), according to manufactures’s method. We used as template the human ER alpha promoter C (p–1000/+212) with the following mutagenic primers (mutation are shown as lowercase letters): forward 5′-CATAAT TGCCTTTGCTTTGGTTCGTGgttTGAGGTTATGTTTGGTATGAAAAG-3′, 5′-CGTGACCTG AGGTTATGTTTGGTATGAAAAGactACATTTTATATTCAGTTTTCTGAAG-3′, and reverse 5′ CTTTTCATACCAAACATAACCTCAaacCACGAACCAAAGCTTTGGCAATTATG-3′, 5′ CTT CAGAAAACTGAATATAAAATGTagtCTTTTCATACCAAACATAACCTCAGGTCACG 3′. Mutation was confirmed by DNA sequencing and the resulting plasmid was designed as C 2mut half-ERE.
Western blot analysis
Cells were grown in 10 cm dishes to 70–80% confluence, transfected and treated as indicated and then lysed in 500 μl of 50 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 2 mM sodium fluoride, 0,5 % sodium deoycholate, 0,1 % SDS and a mixture of protease inhibitors (Aprotinin, PMSF and Na-orthovanadate). Equal amounts of total proteins were resolved on a 11% SDS-polyacrylamide gel and then electroblotted onto a nitrocellulose membrane. Blots were incubated overnight at 4°C with specific primary antibodies. The antigen-antibody complex was detected by incubation of membranes for 1 hour at room temperature with peroxidase-coupled goat anti-rabbit or anti-mouse IgG and revealed using the ECL System. Blots were then exposed to film and bands of interest were quantified by densitometer (Mod 620 BioRad, USA). Blots are representative of at least three independent experiments.
RT-PCR and Real-time RT-PCR
Total cellular RNA was extracted from cells using “TRIAZOL Reagent” as suggested by the manufacturer. All RNA was treated with DNase I and purity and integrity of RNA were confirmed spectroscopically and by gel electrophoresis prior to use. Two micrograms of total RNA was reverse transcribed in a final volume of 50 μl using a RETROscript kit as suggested by the manufacturer. cDNA was diluted 1:5 in nuclease free water, aliquoted and stored at 20°C. The cDNAs obtained were further amplified for ER alpha, ER beta, IRS-1, PS2, Cyclin D1, NCoR and GAPDH genes using the following primers: forward 5′-AGATCCAAGGGAACGAGCT-3′ and reverse 5′-TTCTCCAGGTAGTAGGGCA-3′ (ER alpha); forward 5′-CCTTCCTCCTATGTAGAC AGC-3′ and reverse 5′-TCTCTCTGTTTACAGGTAAGG T-3′ (ER beta); forward 5′-CTCAAC ATCTCCCCCTTC TC-3′ and reverse 5′-CAAATCCCATAT CCT CGT CC-3′ (36B4); forward 5′-AGGATATTTAATTTGCCTCGGG-3′ and reverse 5′-AAGCGTTTGTGCATGCTCTTG-3′ (IRS-1); forward 5′-TTCTATCCTAATACCATCGACG-3′ and reverse 5′-TTTGAGTAGTCAAAG TCAGAGC-3′ (PS2); forward 5′-TCTAAGATGAAGGAGACCATC-3′ and reverse 5′-GCGGTA GTAGGACAGGAAGTTGTT-3′ (Cyclin D1); forward 5′-GCCACTGTATAACCAGCCAT-3′ and reverse 5′-CCTCCATAAGCCCATTCATG-3′ (NCoR); forward 5′-GACAACTTT GGTATCGTG GA-3′ and reverse 5′-TACCAG GAAATGAGCTTGAC-3′ (GAPDH). ER alpha gene expression was also evaluated by real-time RT-PCR. Briefly, 5 μl of diluited (1:3) cDNA was analyzed in triplicate by real-time PCR in an iCycler iQ Detection System (Bio-Rad) using SYBR Green Universal PCR Master Mix, following the manufacturer’s recommendations. Each sample was normalized on its GAPDH mRNA content. Primers used for the amplification were: forward 5′-CAC CAT TGA TAA AAA CAG GAG GAA-3′ and reverse 5′-CTCCCTCCTCTTCGGTCTTTT C-3′ (ER alpha); forward 5′-CCCACT CCTCCACCTTTGAC3′ and reverse 5′-TGTTGCTGT AGCCAAATTCGTT-3′ (GAPDH). The relative gene expression levels were calculated as described (Catalano et al., 2007)
Transient transfection and luciferase assays
MCF-7 cells were transfected using the FuGENE 6 reagent with ER alpha gene promoter, different deleted segments, C 2mut half-ERE or C Sp-1 del in the presence or absence of pCMV5-hERβ, for 16 hours. Empty vectors were used to ensure that DNA concentrations were constant in each transfection. CMV renilla luciferase plasmid was used to normalize the efficiency of the transfection. The firefly and renilla luciferase activities were measured using Dual Luciferase Kit. The firefly luciferase data for each sample were normalized on the basis of transfection efficiency measured by renilla luciferase activity.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from MCF-7 and MCF-7 overexpressing ER beta as previously described (Andrews and Faller 1991). The probe was generated by annealing single stranded oligonucleotides and labeled with [γ32P] ATP and T4 polynucleotide kinase, and then purified using Sephadex G50 spin columns. The DNA sequences used as probe or as cold competitor are the following (mutations are shown as lowercase letters): 5′-TCG TGC GCC CCC GCC CCC TGG CCG TG-3′, 5′ CAC GGC CAG GGG GCG GGG GCG CAC GA-3′ (Sp-1); 5′-TCG TGC GCC CC ata C CCC TGG CCG TG-3′, 5′-CAC GGC CAG GGG tat GGG GCG CAC GA-3 mutated Sp-1. As positive control we used Sp-1 human recombinant protein (1 μl). The protein binding reactions were carried out in 20 μl of buffer (20 mM HEPES pH 8, 1 mM EDTA, 50 mM KCl, 10 mM DTT, 10% glicerol, 1 mg/ml BSA, 50 μg/ml poli dI/dC) with 50,000 cpm of labeled probe, 10 μg of MCF-7 nuclear protein and 5 μg of poly (dI-dC). The above-mentioned mixture was incubated at room temperature for 20 min in the presence or absence of unlabeled competitor oligonucleotide. For experiments involving Sp-1, NCoR, SMRT, IgG and ERβ antibodies, the reaction mixture was incubated with these antibodies at 4°C for 12 h. The entire reaction mixture was electrophoresed through a 6% polyacrylamide gel in 0.25 X Tris borate-EDTA for 3 h at 150 V. Gel was dried and subjected to autoradiography at −70°C.
RNA interference (RNAi)
MCF-7 cells were cotransfected with an empty vector or an ER beta expression vector and RNA duplex of stealth RNAi targeted human NCoR mRNA sequence 5′-UUG UUU GGC UCU GGA GAC CUC UUG C-3′ (Invitrogen) or with a stealth RNAi control (Invitrogen) to a final concentration of 100 nM using Lipofectamine 2000 as recommended by the manufacturer. After 5 h the transfection medium was changed with SFM in order to avoid Lipofectamine 2000 toxicity. The effects of NCoR gene silencing were evaluated on ER alpha expression (36 hours), Sp-1 recruitment to ER alpha promoter (16 hours) and cell proliferation by MTT assay (96 hours).
Chromatin immunoprecipitation assays and Re-ChIP assay
According to the ChIP assay procedure previously described (Catalano et al.,.2010) cells were grown in 100 mm dishes to 50–60% confluence for 16 hours. The cells were then crosslinked with 1% formaldehyde at 37°C for 10 min and sonicated. The supernatants were immunocleared with salmon sperm DNA/protein A agarose for 1 hour at 4°C and immunoprecipitated with anti-Sp-1, anti-NCoR, anti-SMRT, anti-AcH4 and anti-poymerase II antibodies. The anti-Sp-1 immunoprecipitated samples were re-immunoprecipitated with an anti-NCoR or anti-ER beta antibodies. A normal mouse serum IgG as negative control. Pellets were washed, eluated with elution buffer (1% SDS, 0.1 M NaHCO3) and digested with proteinase K (0.5 mg/ml) at 45°C for 1 hour. DNA was obtained by phenol/chloroform/isoamyl alcohol extractions, precipitated with 70% EtOH and resuspended in 20 μl of TE buffer. 5 μl of each sample was used for PCR amplification with the following primers flanking Sp-1 sequence present in the ER alpha promoter region: 5′-GCACATAAGGCAGCACATTA-3′ (forward) and 5′-TGGCTTAAACATCACTCCAG-3′ (reverse). The PCR conditions were 1 min at 94°C, 1 min at 65°C, and 2 min at 72°C. The amplification products obtained in 35 cycles were analyzed in a 2% agarose gel and visualized by ethidium bromide staining. In another set of experiments, 5μl volume of each sample and input DNA was used for real-time PCR using the primers flanking Sp-1 sequence in the human ER alpha promoter region: 5′-TCGTGCGCCCCCGCCCCCTGCCCGTG-3′ and 5′-CCAAAGAGCAGC TTCCCTGA-3′. Real-Time PCR was performed as described above. Final results were calculated using ΔΔCt method, using input Ct values instead of the GAPDH mRNA. The basal sample was used as calibrator.
Cell proliferation assay
Cell viability was determined by using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenylformazan (MTT) assay. Cells (3×104 cell/ml) were transfected as indicated. The MTT assay was performed as the following: 100 μl MTT stock solution in PBS (2mg/ml) was added into each well and incubated at 37 °C for 2 hours followed by media removal and solubilisation in 500 μl dimethyl sulphoxide. After shaking the plates for 15 min, the absorbance in each well, including the blanks, was measured at 570 nm. The data are representative of three independent experiments, each performed in triplicate.
Three-dimensional spheroid culture
MCF-7 cells plated in single-cell suspension in 2% agar-coated plates were transfected with empty vector or plasmid encoding the human ER beta and untreated or treated with IGF 100 nM for 48 h. To generate three-dimensional spheroids, the plates were rotated for 4 h at 37 °C. The three-dimensional cultures were photographed using a phase-contrast microscope (Olympus, Milan, Italy). The extend of aggregation was scored by measuring the spheroids with an ocular micrometer. The spheroids between 25 and 50, 50 and 100 and >100 μm were counted in 10 different fields under ×10 magnification.
Statistical analysis
Data were analyzed for statistical significance using a two-tailed student’s Test, performed by Graph Pad Prism 4. Standard deviations (S.D.) are shown.
Acknowledgments
This work was supported by Reintegration AIRC/Marie Curie International Fellowship in Cancer Research to IB.
This work was supported by………
Footnotes
Disclosure of Potential Conflict of Interest: The authors declare they have no conflict of interest.
References
- Andò S, Panno ML, Salerno M, Sisci D, Mauro L, Lanzino M, Surmacz E. Role of IRS-1 signaling in insulin-induced modulation of estrogen receptors in breast cancer cells. Biochem Biophys Res Commun. 1998;253:315–9. doi: 10.1006/bbrc.1998.9330. [DOI] [PubMed] [Google Scholar]
- Andrews NC, Faller DV. A rapid microperation technique foe extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Z, Gust R. Breast cancer, estrogen receptor and ligands. Arch Pharm (Weinheim) 2009;342:133–149. doi: 10.1002/ardp.200800174. [DOI] [PubMed] [Google Scholar]
- Bardin A, Boulle N, Lazennec G, Vignon F, Pujol P. Loss of ERβ expression as a common step in estrogen-dependent tumor progression. Endocr Relat Cancer. 2004;11:537–551. doi: 10.1677/erc.1.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens D, Gill JH, Fichtner I. Loss of tumourigenicity of stably ERβ-transfected MCF-7 breast cancer cells. Mol Cell Endocrinol. 2007;274:19–29. doi: 10.1016/j.mce.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19:833–42. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- Buteau-Lozano H, Ancelin M, Lardeux B, Milanini J, Perrot-Applanat M. Transcriptional regulation of vascular endothelial growth factor by estradiol and tamoxifen in breast cancer cells: a complex interplay between estrogen receptors α and β. Cancer Res. 2002;62:4977–4984. [PubMed] [Google Scholar]
- Catalano S, Malivindi R, Giordano C, Gu G, Panza S, Bonofiglio D, Lanzino M, Sisci D, Panno ML, Andò S. Farnesoid X Receptor through its binding to Steroidogenic Factor 1 responsive element inhibits aromatase expression in tumor Leydig cells. J Biol Chem. 2010;285:5581–5593. doi: 10.1074/jbc.M109.052670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano S, Rizza P, Gu G, Barone I, Giordano C, Marsico S, Casaburi I, Middea E, Lanzino M, Pellegrino M, Andò S. Fas ligand expression in TM4 Sertoli cells is enhanced by estradiol in situ production. J Cell Physiol. 2007;211(2):448–56. doi: 10.1002/jcp.20952. [DOI] [PubMed] [Google Scholar]
- Cato AC, Nestl A, Mink S. Rapid actions of steroid receptors in cellular signaling pathways. Science STKE. 2002 doi: 10.1126/stke.2002.138.re9. [DOI] [PubMed] [Google Scholar]
- Chang EC, Frasor J, Komm B, Katzenellenbogen BS. Impact of estrogen receptor β on gene networks regulated by estrogen receptor α in breast cancer cells. Endocrinology. 2006;147:4831–4842. doi: 10.1210/en.2006-0563. [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- De Amicis F, Zupo S, Panno ML, Malivindi R, Giordano F, Barone I, Mauro L, Fuqua SA, Andò S. Progesterone receptor B recruits a repressor complex to a half-TRE site of the estrogen receptor α gene promoter. Mol Endocrinol. 2009;23(4):454–65. doi: 10.1210/me.2008-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deGraffenried LA, Hopp TA, Valente AJ, Clark RA, Fuqua SAW. Regulation of the estrogen receptor α minimal promoter by Sp1, USF-1 and ERα. Breast Cancer Res and Treat. 2004;85:111–120. doi: 10.1023/B:BREA.0000025398.93829.78. [DOI] [PubMed] [Google Scholar]
- Duan R, Porter W, Safe S. Estrogen-induced c-fos protooncogene expression in MCF-7 human breast cancer cells: role of estrogen receptor Sp1 complex formation. Endocrinology. 139: 1981–1990. doi: 10.1210/endo.139.4.5870. [DOI] [PubMed] [Google Scholar]
- Esslimani-Sahla M, Simony-Lafontaine J, Kramar A, Lavaill R, Mollevi C, Warner M, et al. Estrogen receptor beta (ER beta) level but not its ER beta cx variant helps to predict tamoxifen resistance in breast cancer. Clin Cancer Res. 2004;10:5769–5776. doi: 10.1158/1078-0432.CCR-04-0389. [DOI] [PubMed] [Google Scholar]
- Fleming FJ, Hill AD, McDermott EW, O’Higgins NJ, Young LS. Differential recruitment of coregulator proteins steroid receptor co-activator-1 (SRC-1) and silencing mediator for retinoid and thyroid receptors to the estrogen receptor-estrogen response element by β-estradiol and 4-hydroxytamoxifen in human breast cancer. J Clin Endocrinol Metab. 2004;89:375–383. doi: 10.1210/jc.2003-031048. [DOI] [PubMed] [Google Scholar]
- Foley EF, Jazaeri AA, Shupnik MA, Jazaeri O, Rice LW. Selective loss of estrogen receptor β in malignant human colon. Cancer Res. 2000;60:245–248. [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of infiammatory and anti-infiammatory signaling pathways. Gene Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- Hartman J, Lindberg K, Morani A, Inzunza J, Strom A, Gustafsson JA. Estrogen receptor beta inhibits angiogenesis and growth of T47D breast cancer xenografts. Cancer Res. 2006;66 :11207–11213S. doi: 10.1158/0008-5472.CAN-06-0017. [DOI] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, Gustafsson JA. Estrogen receptors: How do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Hodges-Gallagher L, Valentine C, Bader S, Kushner P. Estrogen receptor beta increases the efficacy of antiestrogens by effects on apoptosis and cell cycling in breast cancer cells. Breast Cancer Res Treat. 2008;109:241–250. doi: 10.1007/s10549-007-9640-6. [DOI] [PubMed] [Google Scholar]
- Hopp TA, Weiss HL, Parra IS, Cui Y, Osborne CK, Fuqua SA. Low levels of estrogen receptor beta protein predict resistence to tamoxifen therapy in breast cancer. Clinical Cancer Research. 2004;10:7490–7499. doi: 10.1158/1078-0432.CCR-04-1114. [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- Jarvinen TA, Pelto-Huikko M, Holli K, Isola J. Estrogen receptor β is coexpressed with ERα and PR and associated with nodal status, grade, and proliferation rate in breast cancer. Am J Pathol. 2000;156:29–35. doi: 10.1016/s0002-9440(10)64702-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz Jennifer R, Petz Larry N, Nardulli Ann M. Cell- and Ligand-specific Regulation of Promoters Containing Activator Protein-1 and Sp1 Sites by Estrogen Receptors α and β. J Biol Chem. 2005;280(1):347–54. doi: 10.1074/jbc.M407879200. [DOI] [PubMed] [Google Scholar]
- Jiang S, Meyer R, Kang K, Osborne CK, Wong J, Oesterreich S. Scaffold attachment factor SAFB1 suppresses estrogen receptor α-mediated transcription in part via interaction with nuclear receptor corepressor. Mol Endocrinol. 2006;20:311–320. doi: 10.1210/me.2005-0100. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Wang X, Safe S. Estrogen receptor–Sp1 complexes mediate estrogen-induced cathepsin D gene expression in MCF-7 human breast cancer cells. J Biol Chem. 269 [PubMed] [Google Scholar]
- Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–7. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- Lannigan DA. Estrogen receptor phosphorylation Steroids. 2003;68:1–9. doi: 10.1016/s0039-128x(02)00110-1. [DOI] [PubMed] [Google Scholar]
- Lee AV, Weng C-N, Jackson JG, Yee D. Activation of estrogen receptor-mediated gene transcription by IGF-I in human breast cancer cells. J Endocrinol. 1997;152(1):39–47. doi: 10.1677/joe.0.1520039. [DOI] [PubMed] [Google Scholar]
- Leygue E, Dotzlaw H, Watson PH, Murphy LC. Altered estrogen receptor α and β messenger RNA expression during human breast tumorigenesis. Cancer Res. 1998;58:3197–3201. [PubMed] [Google Scholar]
- Li X, Wong J, Tsai SY, Tsai MJ, O’Malley BW. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol Cell Biol. 2003;23:3763–3773. doi: 10.1128/MCB.23.11.3763-3773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losel RM, Falkenstein E, Feuring M, Schultz A, Tillmann HC, Rossol-Haseroth K, Wehling M. Nongenomic steroid action: controversies, questions, and answers. 2003. [DOI] [PubMed] [Google Scholar]
- Mann S, Laucirica R, Carlson N, Younes PS, Ali N, Younes A, et al. Estrogen receptor beta expression in invasive breast cancer. Hum Pathol. 2001;32:113–118. doi: 10.1053/hupa.2001.21506. [DOI] [PubMed] [Google Scholar]
- Mauro L, Catalano S, Bossi G, Pellegrino M, Barone I, Morales S, Giordano C, Bartella V, Casaburi I, Andò S. Evidences that leptin upregulates E-cadherin expression in breast cancer effects on tumor growth and progression. Cancer Res. 2007;67(7):3412–21. doi: 10.1158/0008-5472.CAN-06-2890. [DOI] [PubMed] [Google Scholar]
- Mauro L, Surmacz E. IGF-I receptor, cell-cell adhesion, tumor development and progression. J Mol Histol. 2004;35:247–253. doi: 10.1023/b:hijo.0000032356.98363.a2. [DOI] [PubMed] [Google Scholar]
- Murphy LC, Leygue E, Niu Y, Snell L, Ho SM, Watson PH. Relationship of coregulator and oestrogen receptor isoform expression to de novo tamoxifen resistance in human breast cancer. Br J Cancer. 2002;87:1411–1416. doi: 10.1038/sj.bjc.6600654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Nemere I, Pietras RJ, Blackmore PF. Membrane receptors for steroid hormones: signal transduction and physiological significance. Journal of Cellular Biochemistry. 2003;88:438–445. doi: 10.1002/jcb.10409. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, et al. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA. Mechanisms of estrogen action. Physiol Rev. 2001;81 :1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- Omoto Y, Eguchi H, Yamamoto-Yamaguchi Y, Hayashi S. Estrogen receptor (ER) β1 and ERβcx/β ERα function differently in breast cancer cell line MCF7. Oncogene. 2003;22:5011–5020. doi: 10.1038/sj.onc.1206787. [DOI] [PubMed] [Google Scholar]
- Ordonez-Moran P, Munoz A. Nuclear receptors: genomic and nongenomic effects converge. Cell Cycle. 2009;8:1675–1680. doi: 10.4161/cc.8.11.8579. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Schiff R, Fuqua SA, Shou J. Estrogen receptor: current understanding of its activation and modulation. Clinical Cancer Research. 2001;7:4338s–4342s. (discussion 4411s–4412s) [PubMed] [Google Scholar]
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS. Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- Panno ML, Mauro L, Marsico S, Bellizzi D, Rizza P, Morelli C, Salerno M, Giordano F, Andò S. Evidence that mouse IRS-1 belongs to the family gene which promoter is activated by ERα through its interaction with Sp-1. J Mol Endocrinol. 2006;36:91–105. doi: 10.1677/jme.1.01848. [DOI] [PubMed] [Google Scholar]
- Panno ML, Salerno M, Pezzi V, Sisci D, Maggiolini M, Mauro L, Morrone L, Andò S. Effect of estradiol and insulin on proliferative pattern and on estrogen and progesteron receptor contents in MCF-7 cells. J Cancer Res Clin Oncol. 1996;122:745–9. doi: 10.1007/BF01209122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paruthiyil S, Parmar H, Kerekatte V, Cunha GR, Firestone GL, Leitman DC. Estrogen receptor β inhibits human breast cancer cell proliferation and tumor formation by causing a G2 Cell Cycle Arrest. Cancer Res. 2004;64:423–428. doi: 10.1158/0008-5472.can-03-2446. [DOI] [PubMed] [Google Scholar]
- Pettersson K, Delaunay F, Gustafsson JA. Estrogen receptor β acts as a dominant regulator of estrogen signaling. Oncogene. 2000;19:4970–4978. doi: 10.1038/sj.onc.1203828. [DOI] [PubMed] [Google Scholar]
- Porter W, Saville B, Hoivik D, Safe S. Functional synergy between the transcription factor Sp1 and the estrogen receptor. Mol Endocrinol. 1997;11: 1569–1580. doi: 10.1210/mend.11.11.9916. [DOI] [PubMed] [Google Scholar]
- Roger P, Sahla ME, Makela S, Gustafsson JA, Baldet P, Rochefort H. Decreased expression of estrogen receptor β protein in proliferative preinvasive mammary tumors. Cancer Res. 2001;61:2537–2541. [PubMed] [Google Scholar]
- Safe S. Transcriptional activation of genes by 17β-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm. 2001;62:231–252. doi: 10.1016/s0083-6729(01)62006-5. [DOI] [PubMed] [Google Scholar]
- Safe S, Kim K. Nuclear receptor-mediated transactivation through interaction with Sp proteins. Prog Nucleic Acid Res Mol Biol. 2004;77:1–36. doi: 10.1016/S0079-6603(04)77001-4. [DOI] [PubMed] [Google Scholar]
- Saji S, Omoto Y, Shimizu C, Warner M, Hayashi Y, Horiguchi S, et al. Expression of estrogen receptor (ER) (beta)cx protein in ER(alpha)-positive breast cancer: specific correlation with progesterone receptor. Cancer Res. 2002;62:4849–4853. [PubMed] [Google Scholar]
- Salvatori L, Pallante P, Ravenna L, Chinzari P, Frati L, Russo MA, Petrangeli E. Oestrogens and selective oestrogen receptor (ER) modulators regulate EGF receptor gene expression through human ER α and β subtypes via an Sp1 site. Oncogene. 2003;22:4875–4881. doi: 10.1038/sj.onc.1206784. [DOI] [PubMed] [Google Scholar]
- Saville B, Wormke M, Wang F, Nguyen T, Enmark E, Kuiper G, Gustafsson JÅ, Safe S. Ligand-, cell-, and estrogen receptor subtype (_/_)-dependent activation at GC-rich. 2000. [DOI] [PubMed] [Google Scholar]
- Schiff R, Massarweh S, Shou J, Bharwani L, Arpino G, Rimawi M, Osborne C. Advanced concepts in estrogen receptor biology and breast cancer endocrine resistance: implicated role of growth factor signaling and estrogen receptor coregulators. Cancer Chemotherapy and Pharmacology. 2005;56(Suppl 7):10–20. doi: 10.1007/s00280-005-0108-2. [DOI] [PubMed] [Google Scholar]
- Skliris GP, Munot K, Bell SM, Carder PI, Lane S, Horgan K, Landsdown MR, Parkes AT, Hanby AM, Markham AF, et al. Reduced expression of estrogen receptor beta in invasive breast cancer and its re-expression using DNA methyl transferase inhibitors in a cell line model. Journal of Pathology. 2003;201:213–220. doi: 10.1002/path.1436. [DOI] [PubMed] [Google Scholar]
- Song K, Pan ZZ. Estrogen receptor beta agonist diaryalpropionitrile counteracts the estrogenic activity of estrogen receptor alpha agonist propylpyrazole-triol in the mammary glan of ovariectomized Sprague Dawley rats. Journal of steroid Biochemistry and molecular Biology. 2012;130:26–35. doi: 10.1016/j.jsbmb.2011.12.018. [DOI] [PubMed] [Google Scholar]
- Ström A, Hartman J, Foster JS, Kietz S, Wimalasena J, Gustafsson JA. Estrogen receptor β inhibits 17β-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proc Natl Acad Sci U S A. 2004;101 :1566–1571. doi: 10.1073/pnas.0308319100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson SM, Kang K, Lee AV, Oesterreich S. Novel role of the RET finger protein in estrogen receptor-mediated transcription in MCF-7 cells. Biochem Biophys Res Commun. 2006;349:540–548. doi: 10.1016/j.bbrc.2006.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyhlidal C, Samudio I, Kladde MP, Safe S. Transcriptional activation of transforming growth factor alpha by estradiol: requirement for both a GC-rich site and an estrogen response. doi: 10.1677/jme.0.0240329. [DOI] [PubMed] [Google Scholar]
- Webb P, Anderson CM, Valentine C, Nguyen P, Marimuthu A, West BL, Baxter JD, Kushner PJ. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs) Mol Endocrinol. 2000;14:1976–1985. doi: 10.1210/mend.14.12.0566. [DOI] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Valentine C, Lopez GN, Kwok GR, McInerney E, Katzenellenbogen BS, Enmark E, Gustafsson J-Å, Nilsson S, Kushner PJ. The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol Endocrinol. 1999;13:1672–1685. doi: 10.1210/mend.13.10.0357. [DOI] [PubMed] [Google Scholar]
- Williams C, Edvardsson K, Lewandowski SA, Strom A, Gustafsson JA. A genome-wide study of repressive effects of estrogen receptor beta on estrogen receptor alpha signaling in breast cancer cells. Oncogene. 2007 doi: 10.1038/sj.onc.1210712. [DOI] [PubMed] [Google Scholar]
- Zamir I, Dawson J, Lavinsky RM, Glass CK, Rosenfeld MG, Lazar MA. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci U S A. 1997;94: 14400–14405. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Dahlman-Wright K, Gustafsson JA. Estrogen receptor beta: an overview and update. Nucl Recept Signal. 2008;6 :e003. doi: 10.1621/nrs.06003. [DOI] [PMC free article] [PubMed] [Google Scholar]