Abstract
Efavirenz resistance during HIV-1 treatment failure is usually associated with the reverse transcriptase mutation K103N. L100I, V108I, or P225H can emerge after K103N and increase its level of efavirenz resistance. K103N + L100I is the most drug-resistant of the double mutants but is the least common clinically. We hypothesized that differences in replication efficiency, or fitness, influence the relative frequencies of these secondary efavirenz resistance mutations in clinical isolates. We measured fitness of each secondary mutant introduced into HIVNL4-3, alone and in combination with K103N, using growth competition assays in H9 cells. In the absence of efavirenz, the fitness of V108I was indistinguishable from wild type. K103N, L100I, and P225H were minimally, but consistently, less fit than wild type. K103N + L100I had a greater reduction in fitness and was less fit than K103N + V108I and K103N + P225H. The fitness defect of K103N + L100I relative to K103N was completely compensated for by the addition of the nucleoside resistance mutation L74V. In the presence of efavirenz, L100I was less fit than K103N, and K103N + L100I was more fit than K103N + V108I. Our studies suggest the primary driving force behind the selection of secondary efavirenz resistance mutations is the acquisition of higher levels of drug resistance, but the specific secondary mutations to emerge are those with the least cost in terms of replication efficiency. In addition, nucleoside and NNRTI resistance mutations can interact to affect HIV replication efficiency; these interactions may influence which mutations emerge during treatment failure. These studies have important implications for the design of more durable NNRTI–nucleoside combination regimens.
Keywords: HIV-1, Replication fitness, K103N, L100I, V108I, P225H, Efavirenz
Introduction
Efavirenz is a non-nucleoside reverse transcriptase inhibitor (NNRTI) that is recommended, in combination with two nucleoside analogs, for the treatment of HIV-1 infected individuals who are antiretroviral-naive (DHHS, 2005; Yeni et al., 2004). Although efavirenz is highly effective when used in this setting, efavirenz-resistant mutants of HIV-1 commonly develop if virologic failure occurs. The reverse transcriptase mutation K103N is the most common resistance mutation that emerges in patients failing efavirenz-containing antiretroviral regimens and confers cross-resistance to the two other available NNRTIs, nevirapine and delavirdine (Bacheler et al., 2000, 2001).
After the emergence of K103N during efavirenz treatment failure, secondary mutations (L100I, V108I, and P225H) can develop that augment the efavirenz resistance of K103N (Bacheler et al., 2000). However, the degree of drug resistance conferred by these secondary resistance mutations does not uniformly predict their relative frequencies in clinical isolates during treatment failure. In early studies of efavirenz, K103N + P225H and K103N + V108I were the most common double mutants, each occurring in approximately 30–40% of resistant isolates, with K103N + L100I occurring in approximately 10% (Bacheler et al., 2000). When placed into the laboratory strain HIVNL4-3, secondary mutations increase the efavirenz IC90 of K103N. However, the relatively uncommon genotype K103N + L100I confers approximately 25- to 30-fold more efavirenz resistance than the more common genotypes K103N + V108I or K103N + P225H (Bacheler et al., 2001). Our previous work with primary resistance mutations to nevirapine, delavirdine, and efavirenz found that mutants which confer significant drug resistance but develop uncommonly during virologic failure in patients have significant reductions in replication fitness, as measured by growth competition assays in T cell lines (Archer et al., 2000; Gerondelis et al., 1999; Wang et al., 2006). These differences in relative replication efficiency would presumably affect the relative prevalence of these mutants before therapy is initiated (Coffin, 1995) and could reduce the chance that these mutants would emerge in patients during drug therapy. In addition, we found that less-fit mutants with modest (2- to 3-fold) increases in drug resistance still were outgrown by the more-fit, less drug-resistant K103N mutant in the presence of drug (Gerondelis et al., 1999; Wang et al., 2006). We therefore hypothesized that differences in relative replication efficiency could account for the lower than expected frequency of the K103N + L100I double mutant in clinical isolates.
To determine whether fitness differences could account for the relative prevalence of efavirenz resistance mutations in clinical isolates, we performed competition experiments in the absence of drug between a wild-type laboratory strain of HIV-1 and the site-directed mutants L100I, V108I and P225H, alone and in combination with K103N. Because of observations that K103N + L100I was frequently associated with the nucleoside resistance mutation L74V (Ait-Khaled et al., 2003; Demeter et al., 2004), we also determined the effect of L74V on the fitness of K103N + L100I. We then performed selected competition experiments in the presence of efavirenz to determine how the presence of drug affects these fitness differences.
Results
Relative fitness of L100I, K103N, V108I, and P225H in the absence of efavirenz
Before evaluating the impact of the secondary efavirenz resistance mutations L100I, V108I, and P225H on the relative fitness of K103N, we first tested the effects of each of these mutations individually on HIV-1 replication fitness. We introduced each mutation separately into pNL4-3, using PCR-mediated site-directed mutagenesis, and produced virus stocks by transfecting each plasmid separately into 293 cells. Using a CD4-negative cell line for transfection minimizes the likelihood that extraneous mutations would be introduced during reverse transcription, since progeny virus cannot infect the cells. For comparison, we also produced a virus stock from pNL4-3 containing the delavirdine-resistant P236L mutation, which we have shown has reduced replication efficiency (Gerondelis et al., 1999). We then evaluated the relative fitness or replication efficiency of these mutants by co-infecting the H9 lymphoid cell line with each mutant in combination with wild-type NL4-3. These infections were performed at a low ratio of virus to cells, to minimize the chances of cells being co-infected by both variants. Under these conditions, the relative replication efficiency or fitness of each mutant can then be directly compared to wild-type virus in the same culture, by monitoring the change in relative prevalence of the mutant variant over time, using direct sequence analysis of PCR-amplified proviral DNA. We have used this approach successfully in previous studies to evaluate the relative fitness of other NNRTI-resistant variants (Archer et al., 2000; Wang et al., 2006).
Clonal analysis is generally considered to be more accurate than direct sequencing of PCR-amplified products for the quantification of minority viral variants but is labor-intensive. We reasoned that direct sequence analysis of bulk PCR product would give similar results to clonal analysis in our growth competition assays, since we were using site-directed mutant viruses that differed only in the specific mutation being examined. Thus, variation in surrounding nucleotide sequence, which can influence efficiency of amplification during PCR or specificity of fluorescent-labeled terminator incorporation during the sequencing reaction, would not be present to complicate our assessment of the prevalence of a given mutant. In addition, we took the added precaution of averaging the relative peak heights for a mutant base from both sense and antisense primers that were within 300 nucleotides of the codon being assayed. To verify the concordance between bulk sequencing and clonal analysis, we compared the ratio of mutant virus for each mutant codon at selected time points using bulk sequence analysis with the ratio derived by clonal analysis. Analysis of 24 consecutive, unselected clonal sequences from at least one growth competition experiment per mutant verified that the measurement of peak heights at each mutant base correlates well with estimates of mutant proportions using clonal analysis (Fig. 1).
Fig. 1.
Correlation between quantification of mutant prevalence by direct sequencing of PCR product versus clonal analysis. Results from five independent growth competition assays using the L100I, K103N, V108I, P225H, or P236L mutants were pooled for use in this analysis. The same PCR amplified product was either directly sequenced or cloned into a bacterial plasmid and transformed into E. coli, with subsequent sequence analysis of DNA isolated from each clone. Twenty-four consecutive, unselected clonal sequences were utilized for each determination. The results of one assay were not known when analyzing the results of the other assay.
Using growth competition assays in H9 cells in the absence of efavirenz, we found that the K103N mutant has a modest but consistent fitness defect, compared to wild-type NL4-3 (Fig. 2A). Similar to K103N, the other secondary efavirenz resistance mutations had minor adverse effects on replication fitness, particularly when compared with P236L. Although wild-type virus clearly has an advantage over K103N, P225H, and L100I, these mutants still persist at moderate levels after 2 weeks in culture (Figs. 2A, C, D), whereas the P236L mutant is completely overgrown by day 11 (Fig. 2E). Under the conditions we used, we were unable to detect a reliable reduction in fitness of the V108I mutant relative to wild-type virus over a 2-week time period (Fig. 2B).
Fig. 2.
Growth competition assays between efavirenz-resistant and wild-type NL4-3, inoculated at a ratio of 1:1, based on p24 antigen content. The reference strain in each case is wild-type NL4-3. (A) K103N. (B) V108I. (C) P225H. (D) L100I. (E) P236L.
It appeared from the statistical analysis of relative fitness values that V108I and P225H were more fit than L100I or K103N (Table 1). In order to experimentally verify the existence of these small differences in relative fitness values, we directly competed these single mutants against each other in growth competition experiments. These direct comparisons established a hierarchy of relative fitness of V108I > P225H > K103N ~L100I (data not shown), although the magnitude of these fitness differences is modest, and takes 2–3 weeks to detect in culture.
Table 1.
Relative fitness values of NL4-3 virus containing site-directed single efavirenz resistance mutations compared to wild type NL4-3
| No. replicates | 1 + s a, b | SD | P value c | |
|---|---|---|---|---|
| NL4-3 wild type (reference) | 1 | |||
| K103N | 10 | 0.94 | 0.03 | <0.0001 |
| L100I | 7 | 0.93 | 0.03 | 0.0007 |
| V108I | 6 | 0.99 | 0.01 | NSd |
| P225H | 6 | 0.96 | 0.02 | 0.01 |
| P236L | 4 | 0.60 | 0.06 | 0.001 |
Results shown are means of replicate assays.
s = ln[(Mt/M0)/(Rt/R0)]/t; t is the time after co-infection, Rt is the proportion of reference strain at time t, Mt is the proportion of mutant at time t, R0 is the proportion of reference strain at time 0, M0 is the proportion of mutant at time 0 (Holland et al., 1991).
t Test for comparisons of (1 + s) value for each mutant and the reference strain.
NS, not significant, P > 0.05.
These findings are compatible with the observation that K103N is commonly selected for during clinical failure of NNRTIs, even though K103N confers only moderate levels of NNRTI resistance. We have proposed that larger deficits in fitness, such as those seen with the V106A, G190S and P236L mutants, can retard or prevent the emergence of an NNRTI-resistant variant during therapy (Archer et al., 2000; Gerondelis et al., 1999; Wang et al., 2006). V108I and P225H confer minimal levels of efavirenz resistance by themselves (fold changes in IC90 of 1.6- and 1.2-fold, respectively, compared to wild-type virus) (Bacheler et al., 2001), and this lack of resistance is presumably the reason why these highly fit mutants are rarely seen in the absence of K103N (Bacheler et al., 2000).
Fitness of L100I compared to K103N in the presence of efavirenz
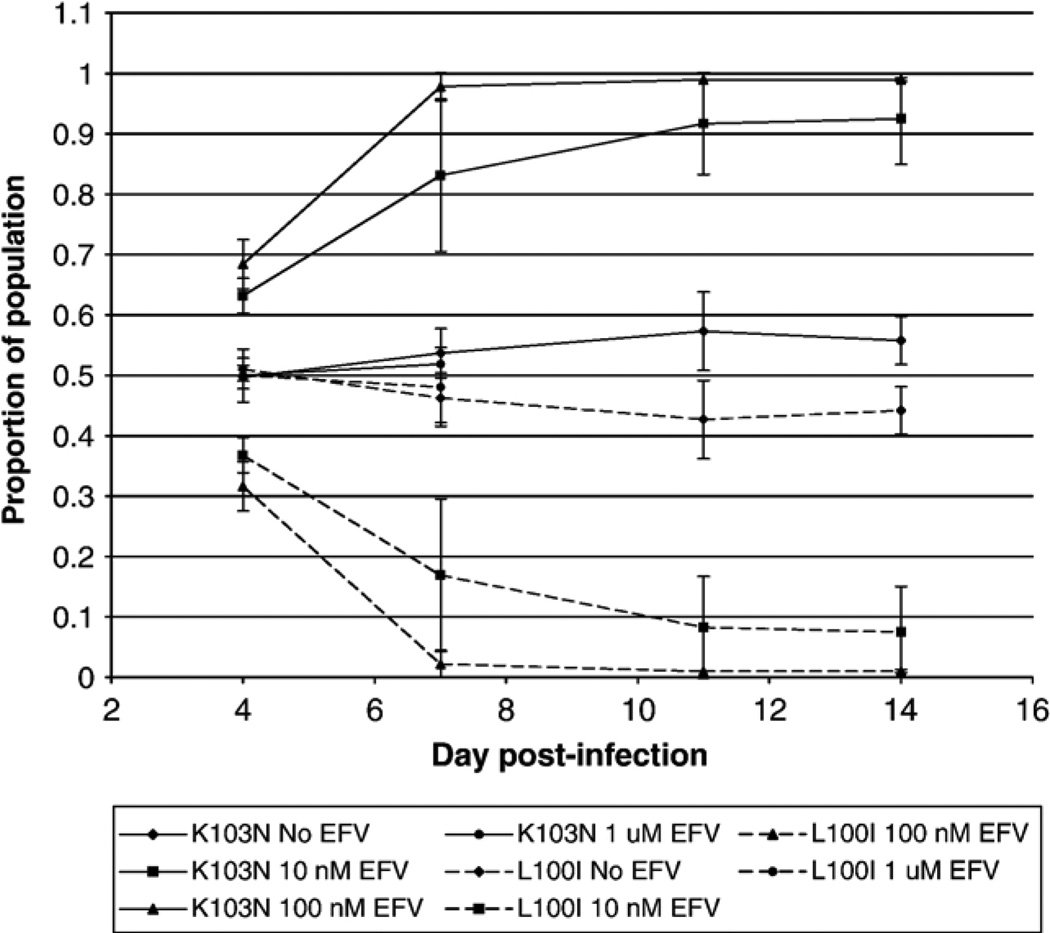
The assay we used to measure replication fitness did not detect a major reduction in fitness of the L100I mutant, which is uncommon clinically despite having a similar level of efavirenz resistance to K103N, as measured with traditional drug susceptibility assays (Bacheler et al., 2001; Petropoulos et al., 2000). Thus, relative fitness, as measured in this assay, does not explain the much more frequent occurrence clinically of K103N compared to L100I (Bacheler et al., 2000). In order to assess how the presence of efavirenz affected the relative fitness of these two mutants, we performed growth competition experiments in the presence of different concentrations of efavirenz (Fig. 3). We found that, although K103N has a similar fitness compared to L100I in the absence of efavirenz, the K103N mutant rapidly outgrows the L100I mutant in the presence of both 10 nM and 100 nM efavirenz (Fig. 3). In the presence of 1 µM efavirenz, there was little virus growth, and we were unable to amplify PCR product at timepoints later than 7 days after infection. In order to verify the published efavirenz susceptibilities of the L100I and K103N mutants, we performed a modification of a previously published drug susceptibility assay (Japour et al., 1993), using H9 cells instead of human peripheral blood mononuclear cells (PBMCs). These studies demonstrated similar efavirenz IC50 values for K103N (6.5 nM) and L100I (5.3 nM).
Fig. 3.
Growth competition assays between K103N and L100I, in the absence and presence of efavirenz. The L100I and K103N variants were inoculated at a ratio of 1:1, based on p24 antigen content. Solid lines, proportion of K103N, measured by direct sequence analysis at codon 103. Dotted lines, proportion of L100I, inferred from sequence analysis at codon 103. The same results were obtained when the prevalence of L100I was measured directly by sequence analysis at codon 100 (data not shown). ♦, No efavirenz;▪, 10 nM efavirenz;▲, 100 nM efavirenz; •, 1 µM efavirenz.
Fitness of K103N + L100I, K103N + V108I, and K103N + P225H compared to K103N in the absence of efavirenz
We next compared the effects of the secondary efavirenz resistance mutations L100I, V108I, and P225H on the relative replication fitness of K103N in the absence of efavirenz. We found that both V108I and P225H have minimal adverse effects on the replication fitness of K103N (Table 2). Therefore, these secondary mutations substantially augment the efavirenz resistance of K103N with little cost in terms of replication fitness. In contrast, a more substantial fitness impairment is seen with K103N + L100I relative to K103N (Table 2). This fitness impairment may explain why K103N + L100I is seen less commonly than K103N + P225H and K103N + V108I in some studies of resistance during efavirenz treatment failures (Bacheler et al., 2000).
Table 2.
Relative fitness values of NL4-3 with site-directed double efavirenz resistance mutations compared to NL4-3 virus containing K103N alone
| No. replicates | 1 + s a, b | SD | P value c | |
|---|---|---|---|---|
| K103N (reference) | 1 | |||
| K103N + L100I | 8 | 0.84 | 0.01 | <0.0001 |
| K103N + V108I | 8 | 0.92 | 0.02 | 0.03 |
| K103N + P225H | 8 | 0.99 | 0.01 | NSd |
Results shown are means of replicate assays.
s = ln[(Mt/M0)/(Rt/R0)]/t, t is the time after co-infection, Rt is the proportion of reference strain at time t, Mt is the proportion of mutant at time t, R0 is the proportion of reference strain at time 0, M0 is the proportion of mutant at time 0 (Holland et al., 1991).
t Test for comparisons of (1 + s) value for each mutant and the reference strain.
NS, not significant, P > 0.05.
To directly verify the fitness differences seen when these mutants were compared to K103N, we performed pairwise growth competitions between each of these double mutants and found that K103N + V108I is substantially more fit than K103N + L100I (1 + s = 1.322 [SD 0.16]) and somewhat less fit than K103N + P225H (1 + s = 0.94 [SD 0.031]). These data support a relative fitness hierarchy of K103N ≃ K103N + P225H > K103N + V108I > K103N + L100I.
Fitness of K103N + L100I compared to K103N + V108I and K103N in the presence of efavirenz
We also evaluated the relative fitness of K103N + L100I and K103N + V108I in the presence of efavirenz, in order to determine whether escalating concentrations of efavirenz could overcome the replication defect of the more drug-resistant K103N + L100I mutant. Growth competition assays between K103N + V108I and K103N + L100I demonstrated that K103N + L100I was more fit than the K103N + V108I mutant at 100 nM efavirenz, and that this improved fitness persisted at higher drug concentrations (Fig. 4). We also found similar results when comparing the relative fitness of K103N + L100I to K103N in the presence and absence οf efavirenz (data not shown).
Fig. 4.
Growth competition assay between K103N + L100I and K103N + V108I in the absence and presence of efavirenz. Mutant viruses were inoculated at a ratio of 1:1, based on p24 antigen content. Results represent the average ± standard error of the mean of at least three independent experiments. Solid lines, prevalence of K103N + V108I, based on sequence analysis at codon 108. Dotted lines, inferred prevalence of K103N + L100I, based on sequence analysis at codon 108. Sequence analysis at codon 100 gave the same results (data not shown). ◊, No efavirenz; Δ, 100 nM efavirenz; ○, 1 µM efavirenz, ×, 10 µM efavirenz.
Effect of the nucleoside resistance mutation L74V on the relative fitness of K103N + L100I
We had noted an apparent association of L74V with K103N + L100I in two studies of efavirenz combination therapy in highly treatment experienced patients (Ait-Khaled et al., 2003; Demeter et al., 2004).We tested the hypothesis that L74V might improve the replication capacity of K103N + L100I, by performing growth competition assays using the double and triple mutants. We found that in the absence of drug, K103N + L100I + L74V was substantially more fit than K103N + L100I, 1 + s = 1.21 (SD 0.03) (Fig. 5). Direct competitions of the K103N + L100I + L74V triple mutant with K103N demonstrated no detectable differences in replication capacity (1 + s = 1.01 [SD 0.05]), suggesting that L74V fully compensates for the fitness reduction conferred by L100I when combined with K103N. Growth competition assays in the presence of efavirenz (100 nM–10 µM) demonstrated no significant increase in efavirenz resistance of the triple mutant relative to K103N + L100I (data not shown).
Fig. 5.
Growth competition assay between K103N + L100I and K103N + L100I + L74V (inoculated at a 75:25 ratio, respectively, based on p24 antigen content) in the absence of efavirenz. Results represent the average ± standard error of the mean of at least three independent experiments.
Discussion
Our studies support the hypothesis that differences in replication capacity influence the frequency with which specific NNRTI resistance mutations occur in clinical isolates. K103N, the most commonly reported mutant, has only a minor reduction in replication efficiency, that is substantially less than those we have measured for other, less common NNRTI-resistant variants, such as V106A, P236L, and G190S (Archer et al., 2000; Gerondelis et al., 1999; Wang et al., 2006). Reductions in replication efficiency also appear to explain the less frequent occurrence in most studies of the highly resistant K103N + L100I double mutant, relative to K103N + V108I and K103N + P225H.
Our studies do have limitations, and our data should be interpreted with some caution. Our data were generated using site-directed mutants of NL4-3, a laboratory strain of HIV-1. Since mutations arise clinically in the context of polymorphisms and background mutations in reverse transcriptase, we cannot be certain whether the fitness effects of mutations in NL4-3 can fully explain their frequency in patient isolates. Studies that describe fitness effects of these mutations within the genetic backbone of clinical isolates may be more clinically relevant.
Additionally, we used H9 cells rather than PBMCs in our competition experiments. It is possible that fitness differences might be affected by cellular factors involved in viral replication such as dNTP concentration and that the PBMCs may better reflect the effect of mutations on viral replication in patients. However, our studies in which fitness of NNRTI-resistant variants was measured both in cell lines and in PBMCs did not show substantial differences in results (Gerondelis et al., 1999). One would expect the most differences in relative fitness between PBMCs and cell lines for mutants that affect affinity for nucleotides, such as the M184V mutant (Back et al., 1996). Our biochemical studies have not to date identified such abnormalities in NNRTI-resistant variants (Domaoal et al., in press; Wang et al., 2006).
Finally, the clinical relevance of competitions in the presence of efavirenz should be interpreted with caution. Efavirenz is highly protein bound in patients, and it is difficult to extrapolate specific concentrations in cell culture (in the presence of only 20% serum) to the clinical setting. Thus, our observations concerning the changes in relative fitness of mutants in the presence of efavirenz indicate general trends but do not necessarily indicate that these specific efavirenz concentrations will be associated with the same mutation prevalence in treated patients.
We speculate that there is a significant fitness cost associated with very high levels of NNRTI resistance, and that it is difficult to select a single mutant that is highly fit and highly NNRTI-resistant. It seems plausible that a mutation with large effects on drug binding may sufficiently distort the structure of the surrounding region, leading to greater dysfunction of the reverse transcriptase, with concomitantly greater reductions in replication efficiency. This correlation between increased NNRTI resistance and decreased replication capacity was observed in a recent study of mutations at codon 190, using a single cycle replication assay (Huang et al., 2003). Similar observations have also been made for mutations conferring resistance to the fusion inhibitor, enfuvirtide (Lu et al., 2004). Thus, the frequent initial selection for K103N in clinical samples may reflect the fact that it has an optimal balance between replication efficiency and drug resistance, at least to currently available NNRTIs.
Higher levels of efavirenz resistance are instead achieved through the selection of secondary resistance mutations, usually those at V108I and P225H. These mutations by themselves confer little, if any efavirenz resistance, and minimal reductions in replication efficiency. Interestingly, when combined with K103N, they substantially augment the level of efavirenz resistance with little or no adverse effects on replication efficiency. The mechanism(s) of how V108I and P225H augment the efavirenz resistance of K103N without directly affecting resistance themselves has not been studied. However, crystallographic studies support a role for K103N in stabilizing the closed form of the NNRTI binding pocket (Hsiou et al., 2001). Thus, V108I and P225H may further augment K103N's ability to stabilize the closed form of the NNRTI binding pocket. In contrast to V108I and P225H, the augmented resistance conferred by the addition of L100I to K103N comes with the cost of a substantial reduction in the replication efficiency.
Our studies suggest that high levels of drug can overcome the reduced fitness of K103N + L100I. This finding contrasts with our previous studies of G190S, in which we were unable to identify a concentration of efavirenz at which the less-fit, more drug-resistant G190S mutant could overgrow the K103N mutant (Wang et al., 2006). Taken together, these studies suggest that minor increases in drug resistance are not sufficient to overcome moderate reductions in replication efficiency, but that larger increases in drug resistance, such as the 30-fold difference in resistance between K103N + L100I and K103N + V108I, may allow selection for a mutant with reduced replication efficiency in the presence of drug. Thus, it is possible that high efavirenz concentrations could account for the selection of K103N + L100I in some patients.
Since the degree of drug resistance as measured by standard drug susceptibility assays is similar for K103N and L100I, we had originally expected that L100I would be substantially less fit than K103N. Growth competition assays in the absence of efavirenz did not support this hypothesis, but those performed in the presence of efavirenz demonstrated that L100I is more susceptible to efavirenz than K103N. This finding suggests that competition experiments can provide supplemental information on the susceptibilities of drug-resistant variants to that provided by standard drug susceptibility assays and may thus lead to a more complete identification of mutations that contribute to drug resistance. An alternative explanation for the low frequency of L100I in clinical isolates is that it has a deficit in replication fitness not detected by our studies of replication in a T cell line.
We found that the L74V mutation compensates for the fitness impairment of K103N + L100I. It is interesting that an association between L74V and K103N + L100I was observed in two clinical studies (Ait-Khaled et al., 2003; Demeter et al., 2004), suggesting that this combination may be favored because of the improved replication efficiency of the triple mutant. This hypothesis is further supported by the observation in one study that L74V could be selected for in the absence of concomitant nucleoside therapy (Demeter et al., 2004). Other investigators have observed that L74V can be selected for in vitro by an NNRTI alone (Kleim et al., 1996) and have shown that L74V improves the replication efficiency of a number of mutants at codon 190 (Huang et al., 2003). Our studies indicate that the beneficial effect of L74V on the replication efficiency of NNRTI-resistant mutants is not limited to those at codon 190.
The interaction between L74V and these NNRTI resistance mutations is just one example of the interplay that occurs between nucleoside analogs and NNRTIs. For example, the NNRTI-resistant mutants L100I and Y181C each sensitize HIV-1 to the nucleoside analog zidovudine (AZT) (Byrnes et al., 1994; Larder, 1992, 1994). The Y181C mutation sensitizes to AZT by reducing nucleoside excision (Selmi et al., 2003). In addition, several nucleoside analog mutations can result in efavirenz hypersusceptibility (Shulman et al., 2001, 2004). One implication of these findings is that the choice of which nucleoside analogs are given with an NNRTI may influence the development of NNRTI resistance. These mutation interactions are the most likely explanation for the observation that concomitant therapy with AZT prevents the emergence of Y181C during therapy with the NNRTI nevirapine (Richman et al., 1994). These findings suggest that other nucleoside resistance mutations, particularly thymidine analog mutations that confer resistance to AZT, may also affect the replication fitness of NNRTI-resistant mutants. Further studies of potential interactions between NNRTI and nucleoside analog resistance mutations are underway in our laboratory.
The mechanisms underlying the fitness impairment of K103N + L100I and compensation by L74V are unknown. The NNRTI resistance mutations V106A, G190A, G190S, and P236L have no effects on DNA polymerization from a DNA primer but do reduce rates of RNase H cleavage (Archer et al., 2000; Gerondelis et al., 1999; Wang et al., 2006). The G190S and G190A mutants also reduce priming of DNA synthesis from tRNALys, 3 (Wang et al., 2006). The G190E mutant, which has a much greater impairment in replication, reduces both DNA polymerization and RNase H cleavage, and the addition of L74V improves both these biochemical functions (Boyer et al., 1998). Studies of reverse transcriptase polymerase and RNase H function for the K103N + L100I and K103N + L100I +L74V mutants should add to our understanding of the biochemical mechanisms underlying fitness impairments and the virus' ability to correct them.
In summary, our data, using site-directed mutants of a laboratory strain, suggest that fitness differences in addition to relative drug resistance can explain the reported frequencies of resistance mutations during clinical failure of efavirenz-based therapies, and that growth competition experiments may be better able to detect relative drug resistance than standard susceptibility assays. In our studies, L74V compensates for the K103N + L100I fitness impairment, and this may account for its preferential selection in combination with this NNRTI-resistant variant in clinical samples. Further studies are needed to delineate the clinical relevance of compensatory mutations on the evolution of drug resistance mutations in patients and to describe the biochemical mechanisms underlying this compensation.
Materials and methods
Reagents and cell lines
The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases: the infectious HIV-1 molecular clone pNL4-3 from Malcolm Martin. Oligonucleotides were purchased from Oligos Etc (Wilsonville, OR). Efavirenz was obtained from Dupont Pharmaceuticals (50 mg/10 ml of DMSO, stored at −20 °C). The 293 and H9 cell lines were obtained from ATCC (Manassa, VA). 293 cells were grown in DMEM (Life Technologies, Gaithersburg, MD) with 10% fetal bovine serum, 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM glutamine. H9 cells were grown in RPMI 1640 (ATCC) with 20% fetal bovine serum, 100 U/ml penicillin, 100 U/ml streptomycin. Both cell lines were incubated in 5% CO2 at 37 °C.
Site-directed mutagenesis
The L100I, K103N, V108I, P225H, K103N + L100I, K103N + V108I, K103N + P225H, K103N + L100I +L74V mutations were each introduced into the vector pRHA1 (Gerondelis et al., 1999), which contains a 4.1-kb region of pNL4-3 flanked by the SphI and EcoRI restriction sites, using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The pol region of HIV-1 containing the relevant mutations was subcloned from pRHA-1 into pNL4-3 using ApaI and AgeI. Individual clones of each mutant were isolated and sequenced to verify the integrity of the cloning sites, the presence of the appropriate reverse transcriptase resistance mutation(s), and the absence of spurious mutations. We also utilized a previously characterized clone of pNL4-3 containing the delavirdine resistance mutation P236L for comparison (Gerondelis et al., 1999). 293 cells were transiently transfected with each pNL4-3 mutant construct using lipofection (SuperFect; Qiagen, Santa Clarita, CA). Infections were performed in triplicate. Cell-free supernatants were harvested and assayed for p24 antigen content on day 3. At least two independently generated virus stocks were used in separate experiments with similar results.
Efavirenz susceptibility assays
We measured the efavirenz susceptibilities of selected mutants using a modification of the ACTG-DoD consensus assay (Japour et al., 1993). Virus stocks were titered in H9 cells. 4 × 106 H9 cells were infected with 4000 TCID50 of virus for one hour at 37 °C, 5% CO2. 2 × 105 cells were then incubated in a volume of 200 µl medium alone or with escalating concentrations of efavirenz (range 0.5–100 nM, final concentration). Virus growth at day 7 was determined by p24 ELISA, and IC50 was calculated using the median effect equation (Chou and Talalay, 1984) (KaleidaGraph 3.51, Synergy Software).
Growth competition assays for relative fitness of wild-type and efavirenz-resistant mutants of HIV-1
The relative replication fitness of efavirenz-resistant mutants of NL4-3 was measured using growth competition experiments in H9 cells. H9 cells (2.5 × 106) were infected with a pair of virus stocks at a total of 50–200 ng p24 per competition (estimated MOI < 0.005, data not shown), in a final volume of 2 ml at 37 °C in 5% CO2 for 1 h. Cells were then washed and resuspended in 5 ml H9 medium (or in 5 ml of H9 medium containing the specified concentration of efavirenz). After 7 days (1 passage), 1 ml of culture supernatant was used to infect an additional 2.5 × 106 H9 cells. Cultured cells (2.5 ml) were harvested and pelleted at days 3 or 4, 7, 11, and 14. For a subset of experiments, a third passage was performed, and cultured cells were harvested at day 21. Genomic DNA was harvested from cell pellets using the QIAamp DNA Blood Mini kit (Qiagen). A region of the pol gene encompassing reverse transcriptase codons 74, 100, 103, 108, and 225 was amplified using PCR (Platinum Taq High Fidelity; Invitrogen) and the following primers: RT18 (5′-GGA AAC CAA AAA TGA TAG GGG GAA TTG GAG G-3′; and RT21 (5′-CTG TAT TTC TGC TAT TAA GTC TTT TGATGG G-3′). Cycling conditions were those recommended by the manufacturer, using an annealing temperature of 55 °C. Direct sequencing of PCR products was performed using BigDye version 3.1 fluorescent-labeled terminators and AmpliTaq (Applied Biosystems). The relative prevalence of mutant variants at each passage was quantitated by averaging the relative peak heights using sense and antisense sequencing primers. Each growth competition experiment between a given pair of mutants was performed in duplicate or triplicate.
Clonal analysis
To determine the accuracy of peak heights in quantifying the relative proportions of mutants present at each time point, PCR products generated at each time point were cloned into a vector (TOPO-TA, Invitrogen), transformed into MAX Efficiency DH5a cells (Invitrogen) and grown. Twenty-four sequential clonal sequences were obtained from each selected time point, and the relative proportion of each mutant on clonal analysis was compared to the peak heights reported from direct sequence analysis of the same PCR product. Results were obtained from each method without knowledge of results obtained using the other method.
Calculations of relative fitness
To quantify relative fitness, the proportions of each mutant at the relevant time points and the fitness difference (1 + the selection coefficient, s) were determined, according to the method of Holland et al., where s = ln[(Mt/M0) /(Rt/R0)] /t, t is the time after co-infection, Rt is the proportion of reference strain at time t, Mt is the proportion of mutant at time t, R0 is the proportion of reference strain at time 0, M0 is the proportion of mutant at time 0 (Holland et al., 1991). Relative fitness values are reported as (1 + s), compared to the reference strain's designated value of 1. The value (1 + s) is also analogous to the log relative fitness, described in a recent evaluation of different methods to quantify relative fitness (Wu et al., 2006). For selected competitions fitness was also calculated using the method of Maree and coworkers, which incorporates the fold expansion of virus over time, such that s = ln[(Mt/M0) /(Rt/R0)] /[ln(Wt/W0) + dt], where Wt is the p24 content of reference (usually wild type) virus at time t, W0 is the p24 content of reference (usually wild type) virus at time 0, d is the half-life of productively infected cells and is estimated to be 0.5 (Maree et al., 2000). This parameter has also been recently referred to as the production rate ratio (Wu et al., 2006). Since relative fitness values did not differ substantially between the two methods, we report our fitness values using the method of Holland.
Acknowledgments
This work was supported by NIH R01-AI-41387 to LMD and NIH K08-51154 to CEK.
References
- Ait-Khaled M, Rakik A, Griffin P, Stone C, Richards N, Thomas D, Falloon J, Tisdale M. HIV-1 reverse transcriptase and protease resistance mutations selected during 16–72 weeks of therapy in isolates from antiretroviral therapy-experienced patients receiving abacavir/efavirenz/amprenavir in the CNA2007 study. Antivir. Ther. 2003;8(2):111–120. [PubMed] [Google Scholar]
- Archer RH, Dykes C, Gerondelis P, Lloyd A, Fay P, Reichman RC, Bambara RA, Demeter LM. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 2000;74(18):8390–8401. doi: 10.1128/jvi.74.18.8390-8401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacheler LT, Anton ED, Kudish P, Baker D, Bunville J, Krakowski K, Bolling L, Aujay M, Wang XV, Ellis D, Becker MF, Lasut AL, George HJ, Spalding DR, Hollis G, Abremski K. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 2000;44(9):2475–2484. doi: 10.1128/aac.44.9.2475-2484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacheler L, Jeffrey S, Hanna G, D’Aquila R, Wallace L, Logue K, Cordova B, Hertogs K, Larder B, Buckery R, Baker D, Gallagher K, Scarnati H, Tritch R, Rizzo C. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 2001;75(11):4999–5008. doi: 10.1128/JVI.75.11.4999-5008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back NK, Nijhuis M, Keulen W, Boucher CA, Oude Essink BO, van Kuilenburg AB, van Gennip AH, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15(15):4040–4049. [PMC free article] [PubMed] [Google Scholar]
- Boyer PL, Gao HQ, Hughes SH. A mutation at position 190 of human immunodeficiency virus type 1 reverse transcriptase interacts with mutations at positions 74 and 75 via the template primer. Antimicrob. Agents Chemother. 1998;42(2):447–452. doi: 10.1128/aac.42.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes VW, Emini EA, Schleif WA, Condra JH, Schneider CL, Long WJ, Wolfgang JA, Graham DJ, Gotlib L, Schlabach AJ, et al. Susceptibilities of human immunodeficiency virus type 1 enzyme and viral variants expressing multiple resistance-engendering amino acid substitutions to reserve transcriptase inhibitors. Antimicrob. Agents Chemother. 1994;38(6):1404–1407. doi: 10.1128/aac.38.6.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Demeter L, DeGruttola V, Lustgarten S, Eshleman SH, Hammer S, Fischl M, Squires K. A genotypic score for efavirenz hypersusceptibility is associated with virologic response to efavirenz + indinavir +/− abacavir in nucleoside-experienced patients (ACTG 368). 11th Conference on Retroviruses and Opportunistic Infections; San Francisco, CA. 2004. Abstract #669. [Google Scholar]
- DHHS. Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents. 2005 [Google Scholar]
- Domaoal RA, Bambara RA, Demeter LM. The HIV-1 reverse transcriptase mutants resistant to non-nucleoside reverse transcriptase inhibitors do not adversely affect DNA synthesis: pre-steady state and steady state kinetic studies. J. Acquired Immune Defic. Syndr. doi: 10.1097/01.qai.0000222288.90201.33. in press. [DOI] [PubMed] [Google Scholar]
- Gerondelis P, Archer RH, Palaniappan C, Reichman RC, Fay PJ, Bambara RA, Demeter LM. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5′-end- and DNA 3′-end-directed RNase H activities. J. Virol. 1999;73(7):5803–5813. doi: 10.1128/jvi.73.7.5803-5813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JJ, de la Torre JC, Clarke DK, Duarte E. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 1991;65(6):2960–2967. doi: 10.1128/jvi.65.6.2960-2967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiou Y, Ding J, Das K, Clark AD, Jr, Boyer PL, Lewi P, Janssen PA, Kleim JP, Rosner M, Hughes SH, Arnold E. The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance. J. Mol. Biol. 2001;309(2):437–445. doi: 10.1006/jmbi.2001.4648. [DOI] [PubMed] [Google Scholar]
- Huang W, Gamarnik A, Limoli K, Petropoulos CJ, Whitcomb JM. Amino acid substitutions at position 190 of human immunodeficiency virus type 1 reverse transcriptase increase susceptibility to delavirdine and impair virus replication. J. Virol. 2003;77(2):1512–1523. doi: 10.1128/JVI.77.2.1512-1523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japour AJ, Mayers DL, Johnson VA, Kuritzkes DR, Beckett LA, Arduino JM, Lane J, Black RJ, Reichelderfer PS, D’Aquila RT, et al. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. The RV-43 Study Group, the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Antimicrob. Agents Chemother. 1993;37(5):1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleim JP, Rosner M, Winkler I, Paessens A, Kirsch R, Hsiou Y, Arnold E, Riess G. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor-specific (RT Leu-74->Val or Ile and Val-75->Leu or Ile) HIV-1 mutants. Proc. Natl. Acad. Sci. U. S. A. 1996;93(1):34–38. doi: 10.1073/pnas.93.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder BA. 3′-Azido-3′-deoxythymidine resistance suppressed by a mutation conferring human immunodeficiency virus type 1 resistance to nonnucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 1992;36(12):2664–2669. doi: 10.1128/aac.36.12.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder BA. Interactions between drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. J. Gen. Virol. 1994;75(Pt. 5):951–957. doi: 10.1099/0022-1317-75-5-951. [DOI] [PubMed] [Google Scholar]
- Lu J, Sista P, Giguel F, Greenberg M, Kuritzkes DR. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20) J. Virol. 2004;78(9):4628–4637. doi: 10.1128/JVI.78.9.4628-4637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maree AF, Keulen W, Boucher CA, De Boer RJ. Estimating relative fitness in viral competition experiments. J. Virol. 2000;74(23):11067–11072. doi: 10.1128/jvi.74.23.11067-11072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, Tian H, Smith D, Winslow GA, Capon DJ, Whitcomb JM. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 2000;44(4):920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Havlir D, Corbeil J, Looney D, Ignacio C, Spector SA, Sullivan J, Cheeseman S, Barringer K, Pauletti D, et al. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 1994;68(3):1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmi B, Deval J, Alvarez K, Boretto J, Sarfati S, Guerreiro C, Canard B. The Y181C substitution in 3′-azido-3′-deoxythymidine-resistant human immunodeficiency virus, type 1, reverse transcriptase suppresses the ATP-mediated repair of the 3′-azido-3′-deoxythymidine 5′-monophosphate-terminated primer. J. Biol. Chem. 2003;278(42):40464–40472. doi: 10.1074/jbc.M302928200. [DOI] [PubMed] [Google Scholar]
- Shulman N, Zolopa AR, Passaro D, Shafer RW, Huang W, Katzenstein D, Israelski DM, Hellmann N, Petropoulos C, Whitcomb J. Phenotypic hypersusceptibility to non-nucleoside reverse transcriptase inhibitors in treatment-experienced HIV-infected patients: impact on virological response to efavirenz-based therapy. AIDS. 2001;15(9):1125–1132. doi: 10.1097/00002030-200106150-00007. [DOI] [PubMed] [Google Scholar]
- Shulman NS, Bosch RJ, Mellors JW, Albrecht MA, Katzenstein DA. Genetic correlates of efavirenz hypersusceptibility. AIDS. 2004;18(13):1781–1785. doi: 10.1097/00002030-200409030-00006. [DOI] [PubMed] [Google Scholar]
- Wang J, Dykes C, Domaoal RA, Koval CE, Bambara RA, Demeter LM. The HIV-1 reverse transcriptase mutants G190S and G190A, which confer resistance to non-nucleoside reverse transcriptase inhibitors, demonstrate reductions in RNase H activity and DNA synthesis from tRNA(Lys, 3) that correlate with reductions in replication efficiency. Virology. 2006;348:462–474. doi: 10.1016/j.virol.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Huang Y, Dykes C, Liu D, Ma J, Perelson AS, Demeter LM. Modeling and estimation of replication fitness of human immunodeficiency virus type 1 in vitro experiments by using a growth competition assay. J. Virol. 2006;80(5):2380–2389. doi: 10.1128/JVI.80.5.2380-2389.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeni PG, Hammer SM, Hirsch MS, Saag MS, Schechter M, Carpenter CC, Fischl MA, Gatell JM, Gazzard BG, Jacobsen DM, Katzenstein DA, Montaner JS, Richman DD, Schooley RT, Thompson MA, Vella S, Volberding PA. Treatment for adult HIV infection: 2004 recommendations of the International AIDS Society-USA Panel. JAMA. 2004;292(2):251–265. doi: 10.1001/jama.292.2.251. [DOI] [PubMed] [Google Scholar]