Abstract
Purpose
Estrogen receptor (ER) α predicts the natural history of breast cancer without intervening therapy. Here we have optimized the detection of a somatic mutation, an A908G transition of ERα, and examined its association with clinical and biological features of invasive breast cancer.
Experimental Design
We compared two methods of sequencing to detect the A908G ERα mutation. We then utilized primer extension sequencing with genomic DNA isolated from invasive breast tumors to determine whether the mutation was associated with clinical outcome in 267 axillary node-negative and positive breast tumors. The presence of the mutation and clinical variables were analyzed for association with recurrence-free survival (RFS) and overall survival (OS) by Cox proportional hazards regression models.
Results
We determined that dye-labeled terminator sequencing was not sensitive for detection of the A908G ERα mutation. The mutation was detected at a high frequency (50%) in invasive breast tumors using primer extension sequencing, and was found to be associated with clinical measures of poor outcome, including larger tumor size and axillary lymph node-positivity. Although the mutation was associated with RFS in univariate analysis, it was not predictive of outcomes in multivariate analysis.
Conclusions
Consistent with our previous finding of a somatic ERα mutation in breast ductal hyperplasias, we now present evidence that the A908G mutation is present in invasive breast tumors using an optimized sequencing method. We determined that the mutation is significantly associated with aggressive biological tumor features, and with an unfavorable prognosis, but was not an independent prognostic marker in untreated patients.
Background
Estrogens play a crucial role in regulating the growth and differentiation of normal breast epithelium and breast cancers, with many of these cancers dependent on these hormones for their growth. Estrogens affect cellular processes by binding to their cognate receptors, ERs α and β, which function as transcription factors mediating the mitogenic effects of estrogen. ERα expression in normal breast epithelium is generally low, but significantly higher expression has been reported in premalignant lesions1, with the majority of breast tumors expressing both receptors2,3. Since prolonged endogenous estrogen exposure is a potential risk factor for invasive breast cancer4, we originally hypothesized that overexpression of ERα or the emergence of mutated receptors could be early events in tumor progression5. Subsequently, using manual genomic Sanger dideoxysequence analysis, we identified an A to G somatic mutation at ERα nucleotide 908 (A908G) from several usual ductal hyperplasias, which are early premalignant lesions. This mutation results in a lysine to arginine transition at residue 303 (K303R ERα)6. To date no other ERα mutation has been identified in more than a few invasive breast cancers [for a review see7]
Dye-labeled terminator genomic automated fluorescent sequencing has been used to screen for the A908G ERα mutation in human breast specimens from women in the United States and Japan, but the mutation was not detected using this methodology8–10. In another recent study, the mutation was detected, but at a low frequency, using single strand conformation polymorphism (SSCP) analysis, confirming our preliminary data that it was indeed present in invasive tumors11. In the current study, we first compared two genomic sequencing approaches (dye-labeled terminator automated fluorescent sequencing12 and primer extension sequencing13), and then extended the study using primer extension sequencing.
Our first objective was to determine the optimum sequencing method for detection of the mutation, and then to determine its prognostic utility in untreated patients. Our overall purpose was to determine whether the A908G mutation was associated with clinical outcome and natural history in a cohort of breast cancers without intervening adjuvant therapies. We present showing that the presence of the mutation identifies a subgroup of women with a worse outcome.
Materials and Methods
Patient Population and Tumor Specimens
In this study, we utilized 267 invasive breast cancers obtained from women in the United States, and maintained in an archived tumor bank of the Breast Center at Baylor College of Medicine (Houston, TX). The patients in this study were derived from a prospectively assembled tumor bank. Tumor samples were archived in the form of formalin-fixed, paraffin-embedded medium density tissue microarrays as described14. All samples were originally stored as fresh frozen tissues, and were fixed and arrayed relatively recently (2001). At the time of DNA extraction, the microarrays were approximately 4 years old. Patients were diagnosed between 1973 and 1993 with primary breast cancer, treated with mastectomy or lumpectomy plus axillary dissection, with or without post-operative radiation therapy, but none of the women underwent adjuvant tamoxifen therapy. Tumor and clinical characteristics of the cohort are summarized in Table 1. The median follow-up time was 76 months, and the tumors have previously been described as part of our Program Project database15. This study was approved by the Baylor College of Medicine Institutional Review Board according to NIH guidelines.
Table 1.
Clinical characteristics of breast cancer cases and by A908G ERα mutation status
| All (n=267) |
Mut (n=133) |
WT (n=134) |
P* | |
|---|---|---|---|---|
| Age (y), No. (%) | ||||
| ≤50 | 72(27.0) | 27(20.3) | 45(33.6) | 0.015 |
| >50 | 195(73.0) | 106(79.7) | 89(66.4) | |
| Tumor Size (cm), No. (%) | ||||
| 0–2 | 89(33.6) | 32(24.2) | 57(42.9) | 0.002 |
| >2–5 | 149(56.2) | 81(61.4) | 68(51.1) | |
| >5 | 27(10.2) | 19(14.4) | 8(6.0) | |
| Missing | 2 | |||
| Nodes, No. (%) | ||||
| Node Negative | 161(60.3) | 56(42.1) | 105(78.4) | <0.0001 |
| Node Positive | ||||
| 1–3 | 60(22.5) | 42(31.6) | 18(13.4) | |
| >3 | 46(17.2) | 35(26.3) | 11(8.2) | |
| S phase, No. (%) | ||||
| Low (0 to <6%) | 61(27.2) | 25(22.3) | 36(32.1) | 0.256 |
| Intermediate (≥6 to ≤10%) | 62(27.7) | 33(29.5) | 29(25.9) | |
| High (>10%) | 101(45.1) | 54(48.2) | 47(50.0) | |
| Missing | 43 | |||
| Ploidy, No. (%) | ||||
| Diploid | 91(38.9) | 45(37.8) | 46(40.0) | 0.732 |
| Aneuploid | 143(61.1) | 74(62.2) | 69(60.0) | |
| Missing | 33 | |||
| ER (fmol/mg), No.(%) | ||||
| Negative (<3) | 51(19.1) | 25(18.8) | 26(19.4) | 0.900 |
| Positive (≥3) | 216(80.9) | 108(81.2) | 108(80.6) | |
| PR (fmol/mg), No.(%) | ||||
| Negative (<5) | 108(42.0) | 54(42.9) | 54(41.2) | 0.791 |
| Positive (≥5) | 149(58.0) | 72(57.1) | 77(58.8) | |
| Missing | 10 | |||
| Median follow-up time (mo) | 76 | 72 | 78 |
χ2 test
Other Biological Factors
Several biomarkers have been previously measured on the tumor samples used in this study. Total ER and PR protein levels were measured by ligand binding assay, and PR-A and B protein isoforms were measured by immunoblot analysis as described elsewhere16. For the ligand binding assay, tumors with an ER content of at least 3 fmol/mg protein, and with a PR content of at least 5 fmol/mg protein were considered positive. ERα protein status was also determined by immunohistochemistry (IHC) using the ER-6F11 antibody from Novocastra (Newcastle upon Tyne, UK) as described previously; Allred scores of 3–8 were considered positive for ERα expression17. AIB1 levels were previously determined by immunoblot, with high levels associated with a better outcome in untreated patients18. S-phase fraction was calculated by flow cytometry at the time of original tissue collection19; cases were classified as low (<6% S-phase), intermediate (6–10% S-phase), or high (>10% S-phase). Levels of HER2 were also determined by IHC as previously described using a semiquantitative estimate of the proportion of positive staining on the entire slide with estimates ranging from 0–4, and higher values correlated with shorter disease-free survival20.
Tumor DNA Isolation, PCR Amplification, Sequencing
Tumor DNA was isolated from 2 mm of a 0.6 µm core tissue microarray using Qiagen DNeasy Tissue kits according to the manufacturer (Valencia, CA). The primer sequences used for PCR amplification and sequencing are shown in Table 2. For PCR amplification of ERα, an initial amplification using primers ERα 1 and 2 was performed with a denaturation step at 95° C for 10 min, followed by 35 cycles of denaturation at 95 ° C for 1 min, primer annealing at 60° C for 30 s, and primer extension at 72° C for 30 s. Upon completion of the cycling steps, a final extension at 72° C for 5 min was performed before the reaction was stored at 4° C. To remove unincorporated PCR primers and dNTPs from the PCR amplification, the samples were treated by adding 2 units of Exonuclease I (USB, Cleveland, OH), and 5 units of shrimp alkaline phosphatase (Roche Applied Science, Indianapolis, IN), for 1 h at 37° C and 15 min at 80° C. These PCR products were used for both dye-labeled terminator and primer extension sequencing.
Table 2.
Primer sequences for PCR amplification, dye-labeled terminator sequencing, and primer extension sequencing (SNaPshot)
| Primer | Use | Sequence (5’ to 3’) |
|---|---|---|
| ERα 1 | Forward PCR | ACATGAGAGCTGCCAACCTT |
| ERα 2 | Reverse PCR | GGAATAGAGTATCGGGGGCT |
| ERα 3 | Forward Extension | TTCATGATCAAACGCTCTAAGA |
| ERα 4 | Reverse Extension | ACAAGGCCAGGCTGTTC |
A negative control consisting of a PCR reaction without genomic DNA to ensure that no contaminating DNA was present, and a positive control of wild-type (WT) ERα genomic DNA from MCF-7 human breast cancer cells (previously determined to be WT sequence6, were run in parallel with all tumor PCR reactions. Plasmids containing either WT or the mutant A908G ERα sequence6 were utilized for PCR amplification in DNA mixing experiments to compare the dye-labeled terminator and primer extension sequencing methods.
Dye-labeled terminator automated fluorescent sequencing was performed with an ABI PRISM® BigDye Terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase, FS (Perkin-Elmer/Applied Biosystems Division, Foster City, CA) according to manufacturer’s recommendations using either the ERα 1 primer (forward), or the ERα 2 (reverse) primers. Primer extension sequencing was performed using the ABI PRISM® SNaPshot™ sequencing method (Perkin-Elmer/Applied Biosystems Division), which involves the extension of a primer that ends one nucleotide 5’ of the ERα 908 nucleotide using fluorescently labeled ddNTPs. The ERα 3 (forward) and the ERα 4 (reverse) primers were utilized for the extension SNaPshot™ reactions. All fluorescent sequencing products were analyzed on an ABI PRISM® 310 Genetic Analyzer capillary sequencer (Perkin-Elmer/Applied Biosystems Division). Data were analyzed with the ABI Gene Scan™ software package. Manual verification of sequencing results was also performed. All tumor DNAs were first sequenced using SNaPshot™ in the reverse direction. To confirm this result, another aliquot of DNA from mutation-positive tumors was then PCR amplified again, and resequenced using SNaPshot™ in the forward direction. Only those tumor samples with the A908G mutation detected on both the reverse and forward strand with SNaPshot™ sequencing were considered mutation positive.
Statistical Methods
Descriptive statistics were used to summarize tumor and clinical characteristics by mutation status. Differences between WT and A908G ERα mutation were compared using the chi-square test. Associations between mutation status and biological factors, for which the sample size was smaller, were assessed using the Fisher’s exact test.
RFS was calculated from the time of diagnosis to the date of the first proved recurrence or censored at last follow-up or death not due to cancer. OS was calculated from the time of diagnosis to death from any cause or censored at last follow-up. Follow-up was truncated at 120 months for purposes of plotting. Survival curves were estimated by the Kaplan-Meier method and compared using the log-rank test.
Cox proportional hazards regression was used to assess the associations between clinical characteristics and RFS or OS. Factors found to be significant as single exploratory variables were entered into a multivariate Cox model21. Mutation status and clinical characteristics included in the model were categorized as indicated in Table 1. The assumption of proportional hazards was tested for each candidate exploratory variable by incorporating a time-dependent interaction into the Cox regression models. As has been reported previously by us22 and others, ER was found to violate the assumption of proportional hazards for RFS. To correct for this in the context of the Cox model, we therefore opted for a Cox regression model incorporating a time-dependent covariate for ER. As an alternative approach, we constructed accelerated failure time (AFT) models with various distributions (Weibull, lognormal, etc) assumed for the failure time. All AFT models yielded essentially identical results to the Cox regression model (data not shown).
Results
SNaPshot Primer Extension Sequencing is Sensitive for Detection of the A908G ERα Mutation
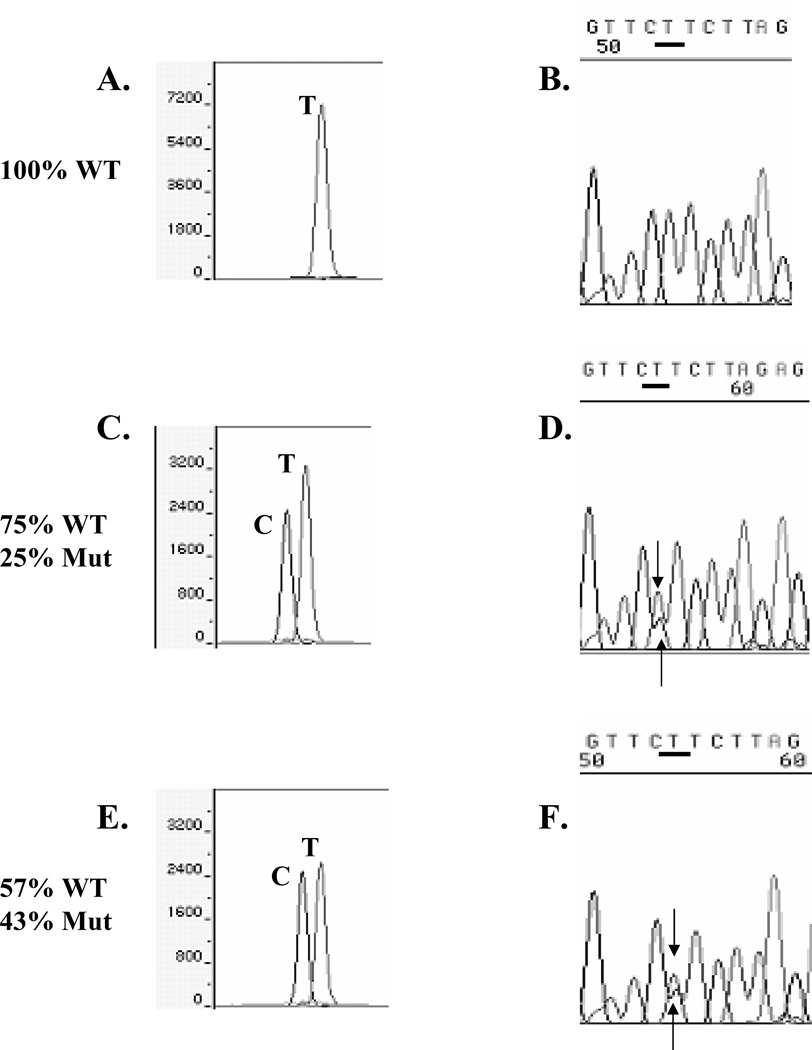
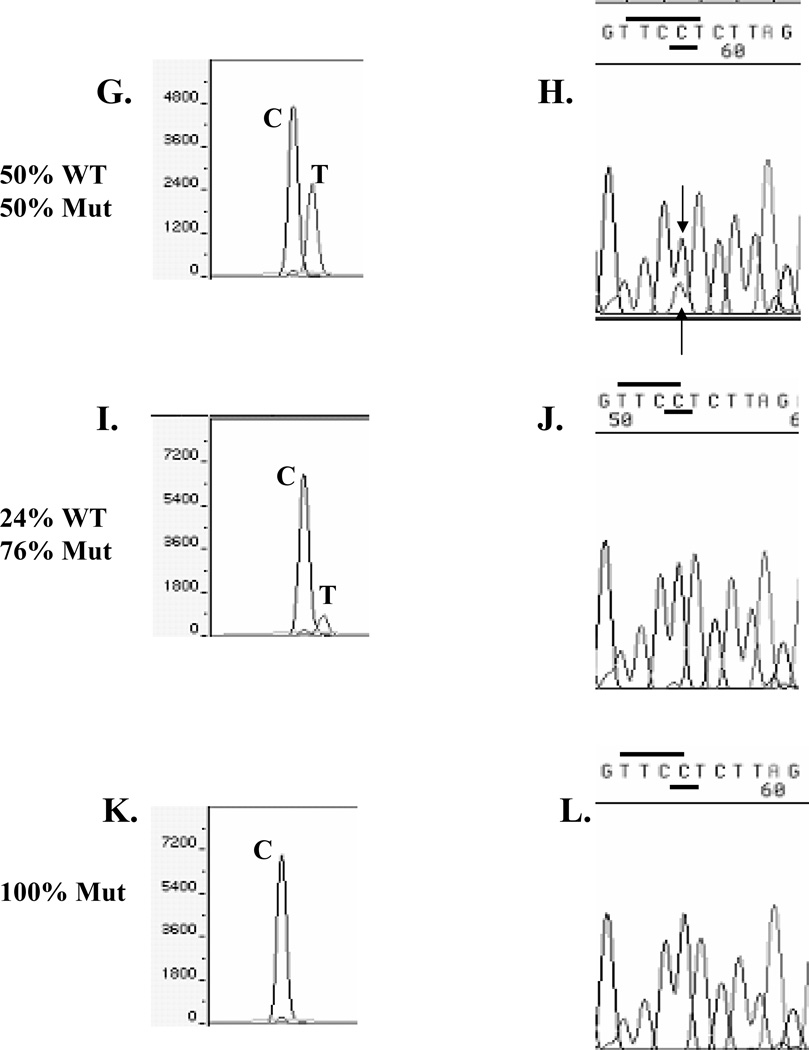
We compared two sequencing approaches, SNaPshot™ primer extension sequencing and dye-labeled terminator sequencing, using primers oriented in the reverse direction, because we had previously observed that reverse strand DNA sequencing was most robust (data not shown). As a control for our ability to detect the mutation, we performed mixing experiments of PCR-amplified DNA from either WT or mutant-containing plasmids (Fig. 1), simulating all the experimental conditions to be utilized for the clinical samples. In this experiment we sequenced DNA series by varying the ratio of mutant DNA to WT DNA. As expected, 100% WT DNA demonstrated a single SNaPshot™ T reverse sequence base peak (panel A), and the correct automated base call of T using dye-labeled terminator sequencing (panel B). With 25% or 43% mutant-containing DNAs, both WT and mutant bases were correctly genotyped using SNaPshot™ (panels C and E, respectively). However in these 25% and 43% mutant-containing DNA mixes, dye-labeled terminator sequencing called the samples homozygous WT A (upper arrows, panels D and F), although a mutant C peak was visible in the chromatograms (lower arrow); the peak height of the WT T base was visibly reduced due to the presence of two bases at this position. As expected, the SNapShot™ method was not a quantitative procedure, but rather a yes/no base call (panels C and E), compared to the automated fluorescent sequence algorithm which miscalled or missed the smaller mutant peak. When WT and mutant DNAs were equally present, dye-labeled terminator sequencing correctly called a mutant C base, but the WT T base was visible under the C peak (arrow, panel H). In all of the series, even with 76% mutant-containing DNA mixes (Panel I), SNapShot™ correctly identified both WT and mutant peaks. We conclude that the ERα A908G base peak heights were reduced in the dye-labeled terminator sequencing profiles. One complication with the dye-labeled terminator technology is that the pattern of termination can be non-uniform from differential incorporation of the dideoxyterminators, resulting in uneven peak heights12,23.
Fig. 1.
Experiment varying the amount of A908 WT and mutant A908G ERα plasmid DNAs which were sequenced using SNaPshot™ (panels A, C, E, G, I, K) and dye-labeled terminator (panels B, D, F, H, J, L) with reverse sequencing primers. The TTCT (WT) and TTCC (Mut) base pair combination is overlined, and the WT or mutant C 908 reverse strand bases are underlined.
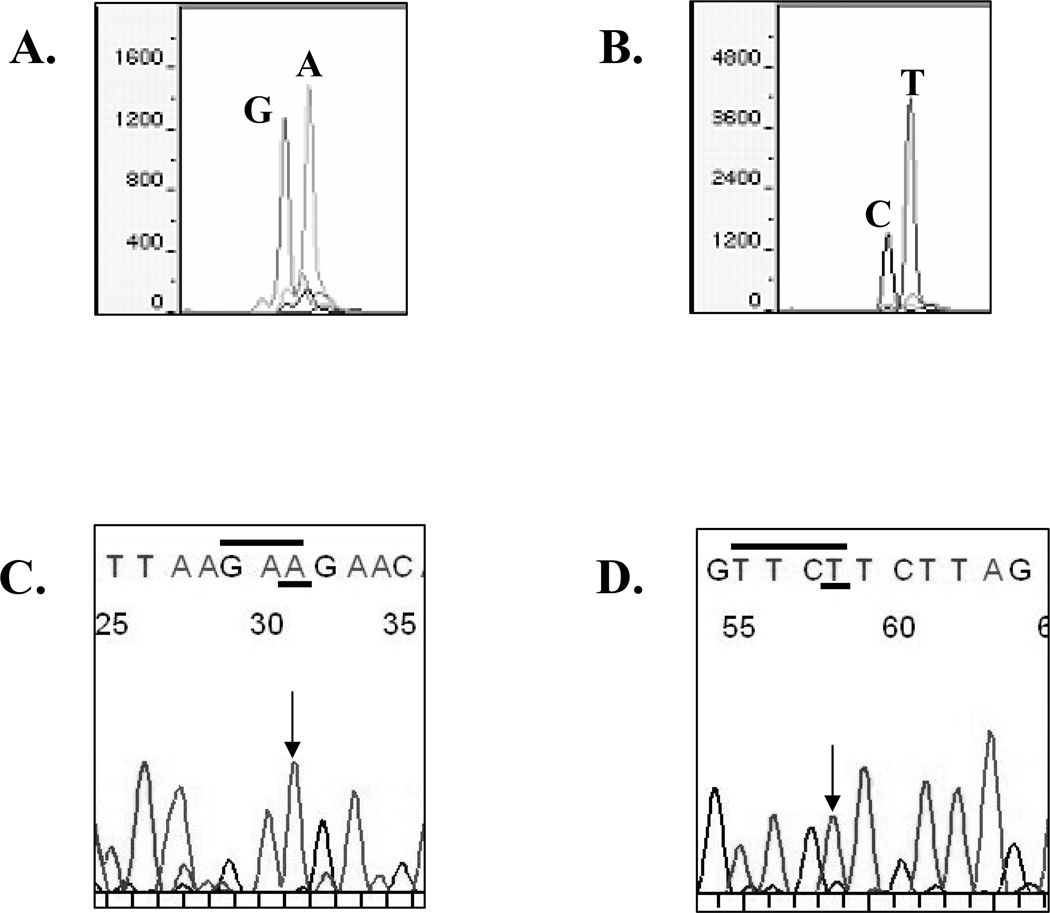
To further compare the two methods, genotyping data obtained from DNA of a representative invasive ductal tumor are shown in Fig. 2. We compared the two sequencing approaches using sequencing primers oriented in both the forward (panels A and C) and reverse directions (panels B and D) so as to sequence both DNA strands of the tumor. The tumor exhibited heterozygosity at ERα 908 with a WT sequence (A nucleotide) and the mutated base (G nucleotide) using the SNaPshot™ sequencing method on the forward strand (panel A). The reverse strand SNaPshot™ sequencing data confirmed heterozygosity at the 908 ERα residue (panel B). Interpretation of the SNaPshot™ sequencing data for this tumor was straight-forward because a positive/negative answer on heterzygotes was obtained using this method. The dye-labeled terminator sequencing of this heterozygote tumor revealed only the WT A nucleotide sequence on the forward strand (panel C, arrow and underlined), and a WT T nucleotide in the sequencing reaction of the reverse strand (panel D, arrow and underlined). We conclude that the ERα A908G mutation can occur in invasive breast tumors, and that the primer extension method is more sensitive for detection of the mutation at this location within ERα.
Fig. 2.
SNaPshot™ sequencing of a representative invasive breast tumor in the forward direction (panel A), and reverse direction (panel B). The same tumor DNA sequenced using dye-labeled terminator in the forward direction (panel C), and the reverse direction (panel D). The WT nucleotide at residue 908 in the forward direction is A, and the reverse direction it is T. The mutant residue is a G (forward direction) and a C (reverse direction). The 908 base call using the automated fluorescent technique is denoted with an arrow.
The A908G ERα Mutation and Correlations with Other Clinical and Biologic Variables
We next performed SNaPshot sequencing on primary breast tumors from a cohort of 267 untreated patients with known clinical outcomes with the goal of examining the role of the mutation in the natural history of these patients (Table 1). Most patients were > 50 years of age, and had tumors that were 2–5 cm in size with an intermediate to high S-phase fraction. Approximately 40% of the cases were node-positive at diagnosis, a frequency which is consistent with the SEER population-based database (www.seer.cancer.gov/seerstat). Most of the tumors were also aneuploid, and approximately 81% of tumors expressed ER and 58% expressed PR (as determined by ligand binding assays) at the time of diagnosis.
Tumor DNAs were first sequenced in the reverse direction using SNaPshot™, and then all of the mutation-positive cases were confirmed using sequencing of the forward DNA strand. Only tumors with the A908G mutation detected on both strands were considered positive. The mutation was detected in 133/267 (49.8%) of the breast tumor samples; the distribution of clinical variables of tumors containing either the WT sequence or the A908G mutation are also shown in Table 1. When we first reported the mutation in premalignant breast lesions, we hypothesized that the mutation might confer a selective advantage in postmenopausal women due to its ability to respond with increased proliferation to the low levels of hormone present in postmenopausal women6. In this cohort of patients with invasive lesions, we found the mutation to be significantly more frequent in women >50 years of age compared to WT sequence (79.7% vs. 66.4%, p=0.015). This result is consistent with our hypothesis that the mutation may play a role in postmenopausal women, but might be of less importance in the biology of tumors from premenopausal women.
Similarly, we predicted that if the A908G ERα mutation indeed conferred a selective advantage, it might be present in tumors from patients with worse outcomes24,25. In a small pilot study which we previously performed using manual Sanger dideoxysequencing of 50 invasive tumors, we found that the mutation was more frequent in lymph node-positive tumors. The A908G ER α mutation status was significantly associated with lymph node-positivity (57.9% vs. 21.6%, p<0.0001)26. The higher frequency of the mutation in axillary lymph node-positive tumors in the current prognostic study is consistent with our earlier results using laborious manual radioactive sequencing technique. Furthermore, in the current study we found that the presence of the mutation was significantly associated with larger tumor size (p=0.002). Both axillary lymph node-positivity and larger tumor size are established clinical variables associated with a poorer outcome27. The mutation was not associated with S-phase fraction, ploidy, or with the levels of ER or PR as measured by ligand binding assay. Thus, those tumors that contained the mutation were associated with worse clinical characteristics compared to WT ERα tumors.
The majority of cases were clinically ER-positive by ligand binding assay for both the mutant and WT, 81.2 vs. 80.6%, respectively (Table 1). As classified by IHC, fewer patients in the mutant-positive group were classified as being ER-positive as compared to WT (65.8 vs. 80.6%, respectively, Table 3). When comparing IHC to ligand binding assay, ER-positivity rates were similar by both methods in WT tumors (P=0.77, McNemar’s test), while the rate of ligand-binding assay ER-positivity was higher than IHC detected ER-positivity in mutant tumors (P>0.01, McNemar’s test). These results suggest several possibilities. Perhaps the IHC method is not be as sensitive for detection of mutant protein, or that the localization of the mutant protein is different from WT, since only nuclear protein is classified as ER-positive using IHC, whereas total ER is measured in ligand binding assays. These possibilities will be pursued in further studies. Regardless of the reason for this discordance in ER status, the mutation can occur in ER-negative cases. It is known that ERα can be lost during tumor progression; ~18% of recurrences present as ER-negative when the primary lesion was ER-positive28. Therefore the mutation may play a role early during tumor progression in some tumors, and ERα loss is a secondary event as has been demonstrated in HER2 and BRCA-1 models systems29,30
Table 3.
Correlation of the A908G ERα Mutation with Other Biologic Variables*
| Mut | WT | P* | |
|---|---|---|---|
| ERα1, No.(%) | |||
| Negative (0,2) | 39 (34.2) | 14 (19.4) | 0.032 |
| Positive (3–8) | 75 (65.8) | 58 (80.6) | |
| ER-β2, No.(%) | |||
| Negative (<3) | 34 (53.1) | 10 (55.6) | 1.000 |
| Positive (≥3) | 30 (46.9) | 8 (44.4) | |
| PR-A2, No.(%) | |||
| Negative (0 –<1) | 39 (60.9) | 8 (44.4) | 0.282 |
| Positive (≥1) | 25 (39.1) | 10 (55.6) | |
| PR-B2, No.(%) | |||
| Negative (0 –<1) | 42 (65.6) | 7 (38.9) | 0.057 |
| Positive (≥1) | 22 (34.4) | 11 (61.1) | |
| HER23, No.(%) | |||
| Negative (<2) | 57 (75.0) | 26 (92.9) | 0.055 |
| Positive (≥2) | 19 (25.0) | 2 (7.1) | |
| AIB14, No. (%) | |||
| High (>1.61) | 19 (29.7) | 5 (27.8) | 1.000 |
| Low (≤ 1.61) | 45 (70.3) | 13 (72.2) | |
| Ki675, No.(%) | |||
| Negative (≤5%) | 10(33.3) | 11 (35.5) | 1.000 |
| Positive (>5%) | 20(66.7) | 20 (64.5) |
Fisher’s exact test. A number of biomarkers have been previously studied on many of the cases included in this study; not all cases were assessed for all markers.
ERα was previously determined by immunohistochemistry and contains both node-negative and positive tumors.
ERβ and PR-A, B isoforms were previously determined by immunoblot analysis and contain only node-positive tumors.
HER2 was previously determined by immunohistochemcistry and contains only node-positive tumors.
AIB1 was previously determined by immunoblot analysis and contains only node-positive tumors.
Ki67 was previously determined by immunohistochemistry and contains only node-negative tumors.
The mutation was not correlated with ERβ levels, the PR-A isoform, or the proliferation marker Ki67 (Table 3). However, there was a weak but nonsignificant correlation between the mutation and the absence of the PR-B isoform (p=0.057). This result is interesting in that we have previously reported that low PR-B levels were associated with a poorer outcome in tamoxifen-treated breast cancer patients16. The presence of the mutation was also more frequent in HER2-positive tumors, but this association did not quite reach statistical significance (25.0% vs. 7.1%, p=0.055). The significance of the potential associations with PR-B and HER2 are currently under study.
A908G ERα Mutation Status and Prognosis
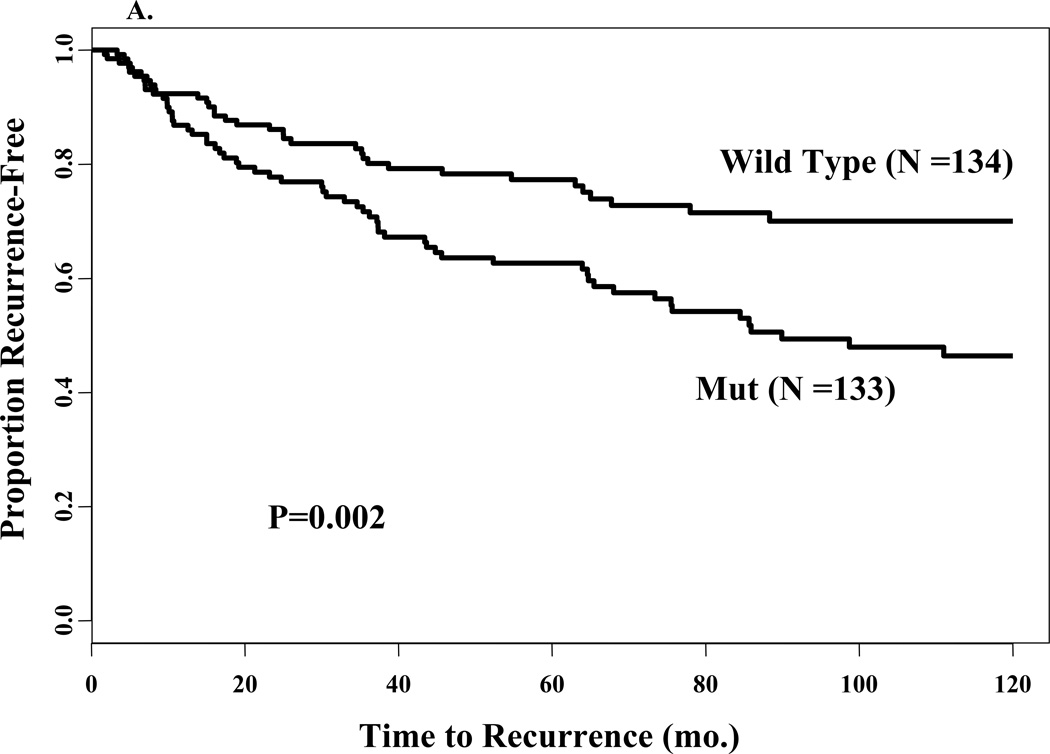
To examine the relationship between the mutation and prognosis, or natural progression of the disease, we included in our study only those patients who did not receive adjuvant therapy. The mutation was significantly associated with shorter time to recurrence (Fig. 3A, log rank test, P = 0.002); 10-year recurrence-free survival (RFS) was 46% (95% CI = 36% to 56%) compared to a RFS of 70% (95% CI = 60% to 78%) for WT ERα patients. Age and PR status by ligand binding assay were not prognostic in this group of patients (data not shown). However, several of the other clinical variables investigated in this study had statistically significant associations with a worse RFS including tumor size (P<0.0001), lymph nodes (P<0.0001), S-phase fraction (P=0.029), ploidy (P=0.024), and ER status (P =0.014). ER was found to violate the assumption, and to adjust for this we also added a time-dependent term. When the A908G mutation status and these other variables were included in a Cox multivariate analysis, only tumor size, lymph nodes, and ER status were significantly associated with RFS and remained in the model (Table 4). The model revealed that the mutation was not associated with poor RFS after adjusting for tumor size, lymph nodes, ER status and a time-dependent covariate of ER status (Table 4, P=0.526). In this final model, positive nodes >3 [HR, 3.28; 95% CI, 1.97–5.44], tumor size > 5cm [HR, 2.65; 95% CI, 1.40–5.03] and ER-negative [HR, 4.37; 95% CI, 2.16–8.85] were significantly associated with RFS. These results are consistent with previous observations that axillary lymph node status is a powerful prognostic factor in breast cancer27.
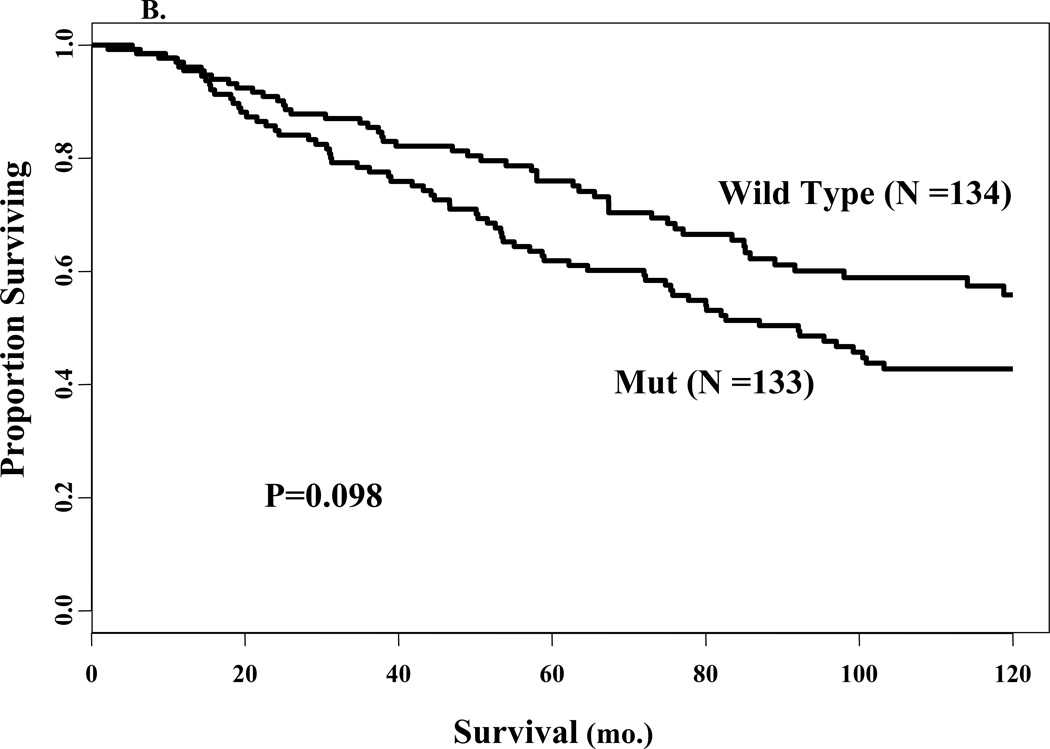
Fig. 3.
Univariate survival analyses according to mutation status. Kaplan-Meier survival curves for proportion recurrence free (panel A) and proportion surviving (panel B) for patients stratified by wild type or mut sequence in ERα. The number of events in each group and the P values are also shown.
Table 4.
Cox regression model of A908G ERα mutation with time-dependent covariate for ER on recurrence-free survival
| Variable | HR (95% CI) | P |
|---|---|---|
| Mutation Status | 0.5258 | |
| WT | 1.00 | |
| Mut | 1.14 (0.76–1.71) | |
| Tumor Size | 0.0119 | |
| 0–2 cm | 1.00 | |
| >2–5 cm | 1.42 (0.92–2.18) | |
| >5 cm | 2.65 (1.40–5.03) | |
| Node No. | <0.0001 | |
| Node Negative | 1.00 | |
| Node Positive (1–3) | 1.31 (0.81–2.11) | |
| Node Positive (>3) | 3.28 (1.97–5.44) | |
| ER | <0.0001 | |
| Positive (≥3) | 1.00 | |
| Negative (<3) | 4.37 (2.16–8.85) | |
| Time-dependent ER* | 0.0058 | |
| Positive (≥3) | 1.00 | |
| Negative (<3) | 0.97 (0.94–0.99) |
Indicates decreasing detrimental effect of ER-negativity over time (months).
Differences in OS between the patient groups (Fig. 3B, P=0.098) did not achieve significance, although disease-specific survival was significant when patients were censored at death from other causes or last follow-up (P=0.0028, data not shown). This is because early deaths tend to be disease-related while later deaths are not. Tumor size (P<0.0001), lymph nodes (P<0.0001) and S-phase fraction (P=0.003) were significantly associated with OS in the univariate Cox regression analyses. Thus, the A908G ERα mutation was not an independent prognostic factor for RFS or OS, most probably due to its clear association with tumor size and positive nodal status.
Discussion
This is the first study to evaluate the clinical utility of the A908G ERα mutation in human breast cancer. Our data help to reconcile our earlier finding in hyperplasias6, and these current results demonstrating the mutation in invasive breast cancers helps to reconcile results reported by others who utilized dye-labeled terminator sequencing methods for detection of the A908G ERα mutation8–10. The mutation was clearly visualized in our current study using SNapShot™ sequencing. It has been demonstrated that there are reproducible peak height patterns using the dye-labeled terminator sequencing method31,32, and that specific three or four-base pair combinations can affect base pair heights of the 3’ base. For instance, the three-base combination GAA results in a small peak height for the 3’ base (A)31,33, and this three-base combination is the same sequence in the ERα 908 WT forward direction (sequence overlined in Fig. 2, panel C). Similarly, sequence-dependent incorporation may be complicating the discrimination of the mutant C base on the reverse strand as well. The sequence TTCC is problematic in that the 3’ C peak can be smaller using dye terminator chemistry32, and this is the same sequence preceding the mutation in the ERα reverse strand (sequence overlined in Fig. 2, panel D). Conway et al. have also reported the problem of using the dye-labeled terminator sequencing and restriction digestion methods for detection of this mutation34. Thus, the identification of ERα 908 heterozygote individuals may be difficult because of uneven peak heights, especially in heterogeneous tumor samples where contaminating normal cells could further dilute the mutant signal as was reported by one group of investigators9. Therefore, the dye-labeled terminator sequencing method may not be the most suitable method for discriminating the ERα A908G mutation in mixed tissue types.
We have previously reported that the A908G ERα mutation was present in ~30% of typical breast hyperplasias, a type of early but non-obligate premalignant breast lesion6. Our demonstration herein that the mutation was present at a high frequency (~50%) in invasive breast cancers is suggestive that the mutation might play a role in cancer progression. Of course, a direct test of this hypothesis will require large epidemiologic studies of patients with premalignant lesions and long-term follow-up. It is known that at diagnosis, about 40% of patients with breast cancer will have histological evidence of axillary lymph node involvement, and that lymph node involvement is highly correlated with patient prognosis. Our data showing a statistically significant increase in the A908G ERα mutation in node-positive cancers provides supportive evidence that the mutation is correlated with poor prognosis, and in our univariate analyses, the mutation was significantly correlated with a worse outcome. The mutation may be a biomarker of increased risk in some tumors for tumor progression and metastatic dissemination, especially in postmenopausal women where cells which express the mutation might be at a growth advantage during the natural course of the disease. Tumor size and nodal status are independent and additive prognostic factors, with nodal status being our most reliable prognostic factor27. The mutation did not remain a significant independent variable in our multivariate models, which weakens the mutation’s impact as a single breast cancer diagnostic biomarker. Clearly the mutation does not independently predict RFS or OS in untreated patients. The failure of the mutation status to remain an independent prognostic variable in our multivariate models might also reflect its strong relationship to nodal status and tumor size. Although the study by Conway et al.34 did not have follow-up available on their patients, they did find a significant correlation between the A908G ERα mutation and higher grade breast tumors, suggestive that the mutation is associated with a more aggressive tumor type. Clearly additional larger studies are needed to clarify the role of this mutation in breast cancer biology. The occurrence of the mutation in ERα-positive premalignant hyperplastic lesions6, but its presence in ER-negative invasive tumors is reminiscent of that reported in HER2 and BRCA1 mutant transgenic mouse models, where hormones can influence early tumorigenesis, but ER loss is a secondary event in the progression of these tumors29,30.
Another obvious difference between our data and two other reports is the racial background of the population studied; the A908G ERα mutation has not been detected in Japanese breast cancers using genomic sequencing or restriction fragment length polymorphism analysis9,10. This disparity could also be related to the lower incidence of premalignant ductal hyperplasia, and tumor ERα-positivity in Japanese women35, or other ethnic and hormonal differences in etiology between the two countries36,37. This interesting possibility certainly warrants further study.
It has been reported that the A908G ERα mutation can be detected using SSCP followed by either manual radioactive sequence confirmation of tumors with abnormal band patterns or SNapShot™ sequencing, but at a much lower frequency (5.7%)34. These authors also reported that the A908G ERα mutation may be more frequent in mixed lobular/ductal breast tumors, and they confirmed our earlier finding that the mutation represents a somatic change in the breast. We had insufficient lobular cancers in this dataset to examine for histological correlations with the A908G ERα mutation. We have similarly utilized SSCP, and a number of other screening techniques, such as oligonucleotide array hybridization and mismatch cleavage, to detect the A908G ERα mutation in clinical breast samples (data not shown). However, we did not find that these alternative genotyping methods38–40 were preferable to SNaPshot™ because of the laborious optimization required, and decreased mutation detection sensitivity with some of these, such as SSCP41. The true population-based estimate of the frequency of the A908G will await further validation studies using optimized sequencing methods.
There are few examples of genes involved in breast cancer, which are mutated and confer a gain-of-function phenotype (hypersensitivity and enhanced substrate for phosphorylation) as we have demonstrated for this mutation6,24. The high frequency of the A to G transition in ERα is intriguing. Since the majority of spontaneous mutations are single base pair changes42, it will be important to determine whether the A908G ERα transition is at a spontaneous mutation “hot-spot”, or alternatively is driven by exogenous carcinogen exposures or endogenous DNA damage processes.
It is known that ERα is posttranslationally modified by protein acetylation and phosphorylation by a number of secondary messenger signaling cascades. We and others have shown that the K303R ERα mutation resides adjacent to a protein kinase A (PKA) and p21-activated kinase 1 (PAK-1) phosphorylation site at ERα S30524,43. It has been demonstrated that S305 phosphorylation promotes ligand hypersensitivity and ligand-independent activity of ERα, as well as up-regulation of the cell cycle regulatory protein cyclin D144. The K303R ERα mutation exhibits enhanced substrate efficiency for PKA signaling to and phosphorylation of the ERα S305 site. Further evidence of the importance of the ERα S305 site is data from Michalides et al.45, who have shown that PKA signaling to this site confers resistance to the antiestrogen tamoxifen. We do not yet know the role of the A908G ERα mutation as a predictive marker in breast cancer since this study focused on a prognostic evaluation of the mutation. It remains to be determined whether the mutation is a predictive marker in clinical trials using either tamoxifen or aromatase inhibitors. Hopefully, with a resolution of the technical differences between detection methods, and the use of sensitive and optimized methods for A908G ERα mutation detection as described here, larger studies to determine the potential clinical relevance of this mutation in different races, histological types, and treated tumor populations are justified.
ACKNOWLEDGMENTS
This work was supported by NCI RO1 CA58183, and Department of Defense Fellowship DAMD17-02-1-0278 to YC and DAMD17-03-1-0417 to MHH. The authors would like to thank Ms. Robin Brown for excellent administrative assistance.
REFERENCES
- 1.Allred DC, Mohsin SK, Fuqua SA. Histological and biological evolution of human premalignant breast disease. Endocr Relat Cancer. 2001;8:47–61. doi: 10.1677/erc.0.0080047. [DOI] [PubMed] [Google Scholar]
- 2.Elledge RM, Fuqua SAW. Estrogen and Progesterone Receptors. In: Harris JR, Lippman ME, Morrow M, editors. Diseases of the Breast. Philadelphia, Lippincott: Williams & Wilkins; 2000. pp. 471–488. [Google Scholar]
- 3.Hopp TA, Weiss HL, Parra IS, et al. Low levels of estrogen receptor beta protein predict resistance to tamoxifen therapy in breast cancer. Clin Cancer Res. 2004;10:7490–7499. doi: 10.1158/1078-0432.CCR-04-1114. [DOI] [PubMed] [Google Scholar]
- 4.Pike MC, Spicer DV, Dahmoush L, et al. Estrogens, progestins, normal breast cell proliferation, and breast cancer risk. Epidemiological Reviews. 1993;15:17–35. doi: 10.1093/oxfordjournals.epirev.a036102. [DOI] [PubMed] [Google Scholar]
- 5.McGuire WL, Chamness GC, Fuqua SAW. Estrogen receptor variants in clinical breast cancer. Molecular Endocrinology. 1991;5:1571–1577. doi: 10.1210/mend-5-11-1571. [DOI] [PubMed] [Google Scholar]
- 6.Fuqua SAW, Wiltschke C, Zhang QX, et al. A hypersensitive estrogen receptor-α mutation in premalignant breast lesions. Cancer Research. 2000;60:4026–4029. [PubMed] [Google Scholar]
- 7.Herynk MH, Fuqua SA. Estrogen receptor mutations in human disease. Endocr Rev. 2004;25:869–898. doi: 10.1210/er.2003-0010. [DOI] [PubMed] [Google Scholar]
- 8.Tebbit CL, Bentley RC, Olson JA, Jr, et al. Estrogen receptor alpha (ESR1) mutant A908G is not a common feature in benign and malignant proliferations of the breast. Genes Chromosomes Cancer. 2004;40:51–54. doi: 10.1002/gcc.20017. [DOI] [PubMed] [Google Scholar]
- 9.Tokunaga E, Kimura Y, Maehara Y. No hypersensitive estrogen receptor-alpha mutation (K303R) in Japanese breast carcinomas. Breast Cancer Res Treat. 2004;84:289–292. doi: 10.1023/B:BREA.0000019963.67754.93. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Yamashita H, Toyama T, et al. Estrogen Receptor alpha Mutation (A-to-G Transition at Nucleotide 908) Is Not Found in Different Types of Breast Lesions from Japanese Women. Breast Cancer. 2003;10:70–73. doi: 10.1007/BF02967628. [DOI] [PubMed] [Google Scholar]
- 11.Fuqua SAW, Cui Y, Mohsin SK, et al. The estrogen receptor alpha A908G mutation is present in invasive breast cancer. Proceedings of the American Association of Cancer Research. 2005;87 doi: 10.1158/1078-0432.CCR-06-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenblum BB, Lee LG, Spurgeon SL, et al. New dye-labeled terminators for improved DNA sequencing patterns. Nucleic Acids Res. 1997;25:4500–4504. doi: 10.1093/nar/25.22.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sokolov BP. Primer extension technique for the detection of single nucleotide in genomic DNA. Nucleic Acids Res. 1990;18:3671. doi: 10.1093/nar/18.12.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin MD, Hilsenbeck SG, Mohsin SK, et al. Breast tumors that overexpress nuclear metastasis-associated 1 (MTA1) protein have high recurrence risks but enhanced responses to systemic therapies. Breast Cancer Res Treat. 2006;95:7–12. doi: 10.1007/s10549-005-9016-8. [DOI] [PubMed] [Google Scholar]
- 15.Bardou VJ, Arpino G, Elledge RM, et al. Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases. J Clin Oncol. 2003;21:1973–1979. doi: 10.1200/JCO.2003.09.099. [DOI] [PubMed] [Google Scholar]
- 16.Hopp TA, Weiss HL, Hilsenbeck SG, et al. Breast cancer patients with progesterone receptor PR-A-Rich tumors have poorer disease-free survival rates. Clin Cancer Res. 2004;10:2751–2760. doi: 10.1158/1078-0432.ccr-03-0141. [DOI] [PubMed] [Google Scholar]
- 17.Harvey JM. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. Journal of Clinical Oncology. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 18.Osborne CK, Bardou V, Hopp TA, et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95:353–361. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- 19.Brown R, Allred D, Clark G, et al. Prognostic value of Ki-67 compared to S-phase fraction in axillary node- negative breast cancer. Clinical Cancer Research. 1996;2:585–592. [PubMed] [Google Scholar]
- 20.Molina R, Ciocca DR, Tandon AK, et al. Expression of HER-2/neu oncoprotein in human breast cancer: a comparison of immunohistochemical and western blot techniques. Anticancer Res. 1992;12:1965–1971. [PubMed] [Google Scholar]
- 21.Collett D. Modeling survival data in medical research. New York: Chapman & Hall; 1994. [Google Scholar]
- 22.Hilsenbeck SG, Ravdin PM, de Moor CA, et al. Time-dependence of hazard ratios for prognostic factors in primary breast cancer. Breast Cancer Res. Treatment. 1998;52:227–237. doi: 10.1023/a:1006133418245. [DOI] [PubMed] [Google Scholar]
- 23.Kronick MN. Heterozygote determination using automated DNA sequencing technology. In: Taylor GR, editor. Laboratory methods for the detection of mutations and polymorphisms in DNA. New York: CRC Press; 1997. pp. 175–189. [Google Scholar]
- 24.Cui Y, Zhang M, Pestell R, et al. Phosphorylation of estrogen receptor α blocks its acetylation and regulates estrogen sensitivity. Cancer Research. 2004;64:9199–9208. doi: 10.1158/0008-5472.CAN-04-2126. [DOI] [PubMed] [Google Scholar]
- 25.Fuqua SAW. The role of estrogen receptors in breast cancer metastasis. J Mam Gland Bio Neoplasia. 2002;6:407–417. doi: 10.1023/a:1014782813943. [DOI] [PubMed] [Google Scholar]
- 26.Fuqua SAW, Hopp T, Van M, et al. An estrogen receptor α mutation that predicts metastatic breast cancer clinical behavior. 9th SPORE Investigators' Workshop. 2001:174. [Google Scholar]
- 27.Carter C, Allen C, Henson D. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer. 1989;63:181. doi: 10.1002/1097-0142(19890101)63:1<181::aid-cncr2820630129>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 28.Hull DF, Clark GM, Osborne CK, et al. Multiple estrogen receptor assays in human breast cancer. Cancer Research. 1983;43:413–416. [PubMed] [Google Scholar]
- 29.Bershtein LM, Alimova IN, Tsyrlina EV, et al. Mammary tumors in HER-2/NEU mice are characterized by low content of estrogen receptors-alpha and absence of progesterone receptors. Bull Exp Biol Med. 2003;135:580–581. doi: 10.1023/a:1025437620749. [DOI] [PubMed] [Google Scholar]
- 30.Katiyar P, Ma Y, Fan S, et al. Regulation of progesterone receptor signaling by BRCA1 in mammary cancer. Nucl Recept Signal. 2006;4:e006. doi: 10.1621/nrs.04006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker LT, Zakeri H, Deng Q, et al. AmpliTaq DNA polymerase, FS dye-terminator sequencing: analysis of peak height patterns. Biotechniques. 1996;21:694–699. doi: 10.2144/96214rr02. [DOI] [PubMed] [Google Scholar]
- 32.Parker LT, Deng Q, Zakeri H, et al. Peak height variations in automated sequencing of PCR products using Taq dye-terminator chemistry. Biotechniques. 1995;19:116–121. [PubMed] [Google Scholar]
- 33.Lee LG, Connell CR, Woo SL, et al. DNA sequencing with dye-labeled terminators and T7 DNA polymerase: effect of dyes and dNTPs on incorporation of dye-terminators and probability analysis of termination fragments. Nucleic Acids Res. 1992;20:2471–2483. doi: 10.1093/nar/20.10.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conway K, Parrish E, Edmiston SN, et al. The estrogen receptor-alpha A908G (K303R) mutation occurs at a low frequency in invasive breast tumors: results from a population-based study. Breast Cancer Res. 2005;7:R871–R880. doi: 10.1186/bcr1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stemmermann GN. The pathology of breast cancer in Japanese women compared to other ethnic groups: a review. Breast Cancer Res Treat. 1991;18(Suppl 1):S67–S72. doi: 10.1007/BF02633531. [DOI] [PubMed] [Google Scholar]
- 36.Deapen D, Liu L, Perkins C, et al. Rapidly rising breast cancer incidence rates among Asian-American women. Int J Cancer. 2002;99:747–750. doi: 10.1002/ijc.10415. [DOI] [PubMed] [Google Scholar]
- 37.Maskarinec G. Breast cancer--interaction between ethnicity and environment. In Vivo. 2000;14:115–123. [PubMed] [Google Scholar]
- 38.Elledge RM, Fuqua SAW, Clark GM, et al. Prognostic signifigance of p53 gene alterations in node-negative breast cancer. Breast Cancer Research and Treatment. 1993;26:225–235. doi: 10.1007/BF00665800. [DOI] [PubMed] [Google Scholar]
- 39.Moul JW, Theune SM, Chang EH. Detection of RAS mutations in archival testicular germ cell tumors by polymerase chain reaction and oligonucleotide hybridization. Genes Chromosomes Cancer. 1992;5:109–118. doi: 10.1002/gcc.2870050204. [DOI] [PubMed] [Google Scholar]
- 40.Cotton RG, Rodrigues NR, Campbell RD. Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci U S A. 1988;85:4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tolbert DM, Noffsinger AE, Miller MA, et al. p53 immunoreactivity and single-strand conformational polymorphism analysis often fail to predict p53 mutational status. Mod Pathol. 1999;12:54–60. [PubMed] [Google Scholar]
- 42.Loeb LA. A mutator phenotype in cancer. Cancer Research. 2001;61:3230–3239. [PubMed] [Google Scholar]
- 43.Wang RA, Mazumdar A, Vadlamudi RK, et al. P21-activated kinase-1 phosphorylates and transactivates estrogen receptor-alpha and promotes hyperplasia in mammary epithelium. Embo J. 2002;21:5437–5447. doi: 10.1093/emboj/cdf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balasenthil S, Barnes CJ, Rayala SK, et al. Estrogen receptor activation at serine 305 is sufficient to upregulate cyclin D1 in breast cancer cells. FEBS Lett. 2004;567:243–247. doi: 10.1016/j.febslet.2004.04.071. [DOI] [PubMed] [Google Scholar]
- 45.Michalides R, Griekspoor A, Balkenende A, et al. Tamoxifen resistance by a conformational arrest of the estrogen receptor alpha after PKA activation in breast cancer. Cancer Cell. 2004;5:597–605. doi: 10.1016/j.ccr.2004.05.016. [DOI] [PubMed] [Google Scholar]