Abstract
Background
Tumor suppressor gene p53 plays an important role in the maintenance of the genomic integrity, and mutation in the gene may alter an individual’s susceptibility to various carcinomas. P53 Arg72Pro or codon 72 polymorphism has been indicated to increase the risk of developing certain cancers such as bladder cancer and cervical cancer. Human papillomavirus (HPV) infection has been shown as a risk factor for certain cancers such as cervical cancer and oral cancer as well, and the HPV oncoprotein E6 may induce the degradation of p53 function. However, the association between p53 Arg72Pro polymorphism and the risk of oral cancer with HPV infection remains inconclusive. Therefore, this meta-analysis involving 5,614 participants was performed to investigate the relations among the p53 Arg72Pro polymorphism, HPV infection, and the risk of developing oral cancer.
Results
A search of the literature by PubMed, Embase, Web of Science, and China National Knowledge Infrastructure databases was conducted to identify studies based on the inclusion and exclusion criteria. Odds ratios with 95 % confidence intervals were combined using a random-effect model or a fixed-effect model. The current study was conducted with 13 studies consisting of 2,413 cases and 3,201 controls. Neither overall analysis nor stratified analyses detected any obvious evidence of association between p53 Arg72Pro polymorphism and oral cancer susceptibility in all genetic models. However, a significant association between p53 Arg72Pro polymorphism and the risk of oral cancer with HPV infection was detected in the Arg/Arg vs. Arg/Pro + Pro/Pro model.
Conclusion
In the current meta-analysis which used the quantitative data synthesis for the first time, our study demonstrated that p53 Arg72Pro polymorphism together with HPV infection might jointly alter an individual’s susceptibility to the risk of oral cancer. Our results suggested that p53 Arg72Pro polymorphism may partly contribute to the pathogenesis of oral cancer development.
Keywords: P53 codon 72, Human papillomavirus, Oral cancer, Polymorphism, Susceptibility, Meta-analysis
Background
The incident rate for oral cancer has been increasing recently. Research studies have suggested that smoking, alcohol consumption, and betel quid chewing are risk factors that predispose individuals to oral cancer [1–3]. Nevertheless, only some smokers, alcohol users, and betel quid users develop oral cancer, which indicated that it can be a multifactorial process associated with various risk factors for oral cancer development. These exogenous carcinogens may induce a defective DNA damage response, which may alter the expression of tumor suppressor genes apoptosis or may result in genomic instability [4, 5]. Accumulative evidence indicates that individual susceptibility to oral cancer also depends on genetic predisposition and viral infection [6, 7]. Therefore, both environmental and genetic factors may play an important role in the process of oral cancer development.
Many published studies have reported that oral carcinoma susceptibility is associated with gene polymorphism. In recent years, much attention has been focused on the p53 codon 72 Arg/Pro polymorphism. The p53 tumor suppressor gene is located at human chromosome 17 and encoding a 53-kDa nuclear phosphoprotein which plays a crucial role in cell cycle regulation, maintenance of genomic integrity, apoptosis, and challenge of environmental insults [8, 9]. Mutant p53 codon 72 may allow cells with environment-associated damaged DNA to enter the cell cycle, leading to the development of tumors [10, 11]. In fact, there have been extensive research studies demonstrated that p53 Arg72Pro polymorphism played an important role in developing cervical cancer in HPV-positive patients. Odds of developing cervical cancer was significantly higher with the p53 Arg allele in HPV associated cervical cancer. This association was not detected in HPV-negative patients [12]. In addition, the association between p53 Arg72Pro polymorphism and oral cancer has been investigated, however, the results were inconsistent.
HPV infection have been proved as an independent risk factor for the development of oral cancer [13, 14]. The viral E6 protein, which encoded by two high risk HPV types named HPV-16 and HPV-18, was testified to bind and inactivate the human p53 gene product, and marking it for destruction by the ubiquitin proteasome pathway [15–17]. Storey et al. suggested that the p53 Arg72Pro polymorphism plays a part in the development of HPV-associated cancer in 1998 for the first time [18]. Since then, researchers have investigated the combined influence of the Arg72Pro polymorphism and HPV infection in the risk of developing oral cancer, but the results remained inconclusive [19–21].
Therefore, whether or not p53 Arg72Pro polymorphism can increase the risk of oral cancer with HPV infection remains unclear. Based on the above reasons, we conducted this evidence-based quantitative meta-analysis to investigate the relationship between p53 polymorphisms and the risk of HPV-related oral cancer.
Methods
Search strategy
Relevant articles were searched using combinations of search terms “oral”, “oral cavity”, “buccal”, “oropharynx”, “oral cancer”, “oral carcinoma”, “oral squamous cell carcinoma”, “ameloblastoma”, “P53”, “TP53”, “Arg72Pro”, “HPV”, “human papillomavirus”, “polymorphism”, “susceptibility”, and “gene variants”, in PubMed, Embase, Web of Science, and China National Knowledge Infrastructure databases, focusing on articles which were published from their earliest entry points to April 2014.
Inclusion and exclusion criteria
The following inclusion criteria were used for the selection of literature for meta-analysis: (1) published in English; (2) examined case–control studies investigating the association between HPV infection, Arg72Pro polymorphism, and the risk of oral cancer; (3) definite histopathologic diagnosis; and (4) genotype distribution in controls must be in Hardy-Weinberg equilibrium (HWE). Major exclusion criteria included: (1) the unpublished reports and abstracts; (2) when duplicated studies published by the same author, only the most recent publication study was chosen.
Data extraction
All the eligible articles were independently reviewed and extracted by two reviewers (YL and WY) according to the selection criteria listed above. Disagreement was resolved by the third independent investigator (JH). The following data were extracted from the each study: the first author, year of publication, country, ethnicity, genotyping methods, source of the controls, and genotype numbers from the cases and controls.
Statistical analysis
The STATA version 11.0 software (Stata Corporation, College Station, TX) was used to conduct the statistical analyses. The combined odds ratio (OR) with a corresponding 95 % confidence interval (CI) was estimated to evaluate the relationship among p53 Arg72Pro polymorphisms, HPV infection, and the risk of oral cancer. For control groups, the goodness-of-fit test (Chi-square test or Fisher exact test) was used to test the deviations from HWE. The following statistical models were used in the meta-analysis: the allelic model (Arg72 allele vs. Pro72 allele), the codominant model (homozygote comparison: Arg/Arg vs. Pro/Pro), the dominant model (Arg/Arg + Arg/Pro vs. Pro/Pro), and the recessive model (Arg/Arg vs. Arg/Pro + Pro/Pro). Statistics Q and I2 statistic were evaluated to investigate the between-study heterogeneity [22, 23]. Either the random-effect model or the fixed-effect model was used to calculate the pooled effect estimate either in the presence or in the absence of heterogeneity, respectively [24, 25]. Additionally, the Begg’s funnel plot and the Egger’s test were used to estimate the publication bias (p < 0.05 was considered statistically significant) [26, 27].
Results
Studies characteristics
As shown in Fig. 1, 13 studies with a total of 5,614 participants met the inclusion and exclusion criteria [19–21, 28–37]. The characteristics of these included articles were summarized in Table 1. All the related distribution of p53 codon 72 polymorphism genotype frequencies in cases and controls were summarized in Table 2.
Fig. 1.
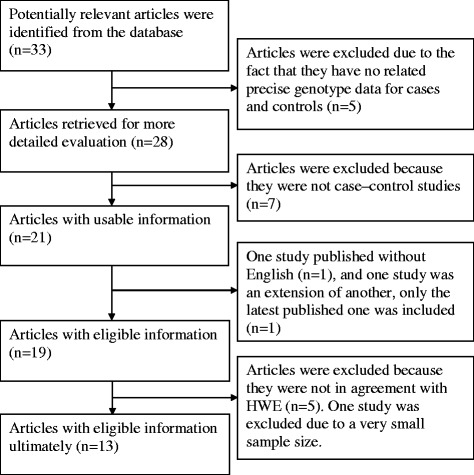
Flow diagram of the publication selection process
Table 1.
Main characteristics of studies included in the meta-analysis
| Study | Country | Ethnicity | Control source | Genotyping Methods | Sample size (case/control) |
|---|---|---|---|---|---|
| Patel KR et al. [28] | India | Asian | healthy | PCR-RFLP | 79/110 |
| Wang Z et al. [18] | USA | Caucasian | healthy | PCR-RFLP | 320/321 |
| Ji X et al. [19] | USA | Caucasian | healthy | PCR-RFLP | 188/342 |
| Kuroda Y et al. [29] | Japanese | Asian | Hospital | PCR-RFLP | 100/271 |
| Kitkumthorn N et al. [30] | Thailand | Asian | healthy | PCR-RFLP | 78/94 |
| Chen X et al. [31] | USA | Caucasian | healthy | PCR-RFLP | 326/349 |
| Zemleduch T et al. [32] | Caucasian | Caucasian | healthy | PCR-RFLP | 123/300 |
| Ihsan R et al. [33] | India | Asian | healthy | PCR-RFLP | 116/278 |
| Tu HF et al. [34] | Taiwan | Asian | healthy | DNA sequence | 189/116 |
| Summersgill KF et al. [20] | USA | Caucasian | Hospital | PCR-CTPP | 190/308 |
| Misra C et al. [35] | India | Asian | Hospital | PCR-RFLP | 308/342 |
| Lin YC et al. [36] | Taiwan | Asian | unknown | PCR-RFLP | 297/280 |
| Saini R et al. [37] | Malaysia | Asian | healthy | PCR-CTPP | 99/90 |
Table 2.
Distribution of p53 codon 72 genotypes among oral cancer in cases and controls
| Cases (n) | Controls (n) | ||||||
|---|---|---|---|---|---|---|---|
| First author | Arg/Arg | Arg/Pro | Pro/Pro | Arg/Arg | Arg/Pro | Pro/Pro | P-value of HWE in controls |
| Patel KR et al. [28] | 32 | 29 | 18 | 30 | 58 | 22 | 0.528 |
| Wang Z et al. [18] | 43 | 41 | 15 | 24 | 15 | 2 | 0.860 |
| Ji X et al. [19] | 103 | 74 | 11 | 179 | 140 | 23 | 0.532 |
| Kuroda Y et al. [29] | 41 | 44 | 15 | 109 | 117 | 45 | 0.159 |
| Kitkumthorn N et al. [30] | 35 | 40 | 3 | 27 | 47 | 20 | 0.957 |
| Chen X et al. [31] | 183 | 121 | 22 | 181 | 144 | 24 | 0.518 |
| Zemleduch T et al. [32] | 55 | 52 | 16 | 176 | 104 | 20 | 0.389 |
| Ihsan R et al. [33] | 30 | 63 | 23 | 63 | 143 | 72 | 0.619 |
| Tu HF et al. [34] | 53 | 106 | 30 | 41 | 60 | 15 | 0.337 |
| Summersgill KF et al. [20] | 102 | 70 | 18 | 168 | 112 | 28 | 0.144 |
| Misra C et al. [35] | 87 | 155 | 66 | 85 | 159 | 98 | 0.203 |
| Lin YC et al. [36] | 96 | 155 | 46 | 72 | 152 | 56 | 0.135 |
| Saini R et al. [37] | 22 | 40 | 37 | 28 | 39 | 23 | 0.215 |
HWE: Hardy–Weinberg equilibrium
Meta-analysis results
The association between p53 Arg72Pro polymorphism and the risk of oral cancer in total population
A total of 13 studies were included in the meta-analysis to examine the association between p53 Arg72Pro polymorphism and the risk of oral cancer. There was no evidence of a significant association in any genetic model (Arg72 allele vs. Pro72 allele: OR = 1.05, 95 % CI: 0.90- 1.23; Arg/Arg vs. Pro/Pro: OR = 1.11, 95 % CI: 0.81- 1.52; Pro/Pro vs. Arg/Arg + Arg/Pro: OR = 0.94, 95 % CI: 0.72- 1.21; Arg/Arg vs. Arg/Pro + Pro/Pro: OR = 1.07, 95 % CI: 0.91- 1.26; all p values >0.05; Figs. 2, 3, 4 and 5, Table 3). However, significant heterogeneity across the studies was present in four genetic models (P = 0.000, 0.002, 0.013, 0.023 for the allelic genetic model, the homozygote comparison model, the dominant model and the recessive model, respectively Table 3).
Fig. 2.
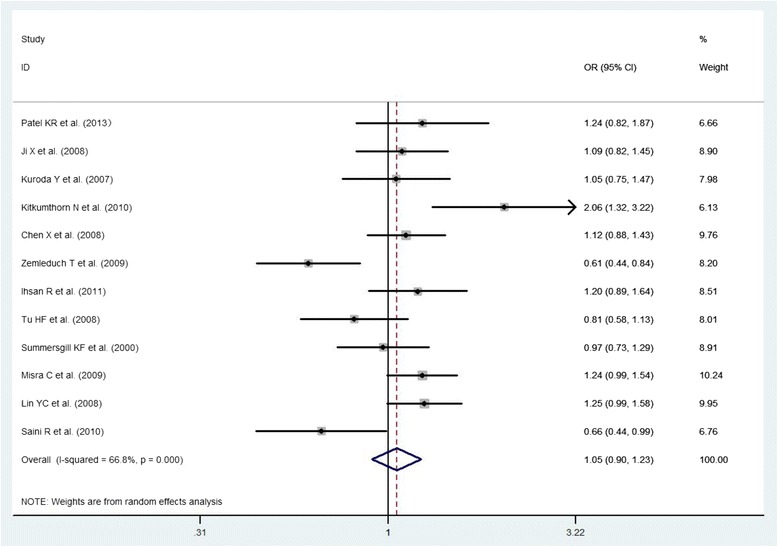
The association between p53 Arg72Pro polymorphism and the risk of oral cancer in total population (Arg72 allele vs. Pro72 allele)
Fig. 3.
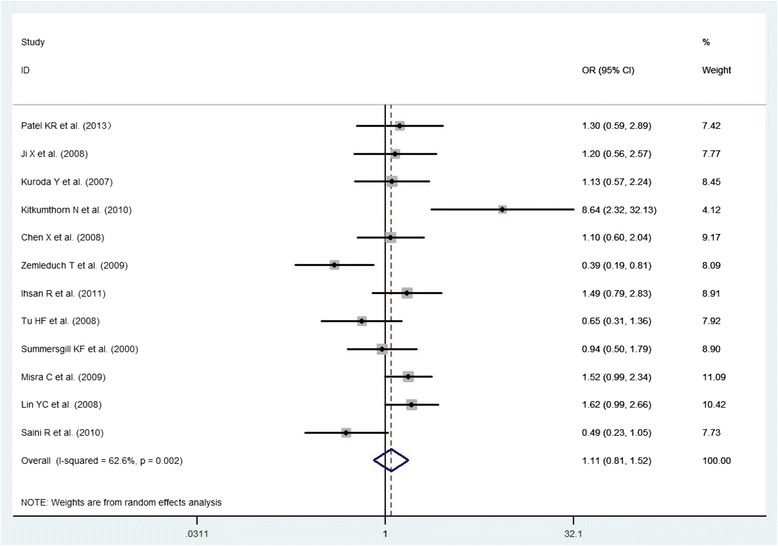
The association between p53 Arg72Pro polymorphism and the risk of oral cancer in total population (Arg/Arg vs. Pro/Pro)
Fig. 4.
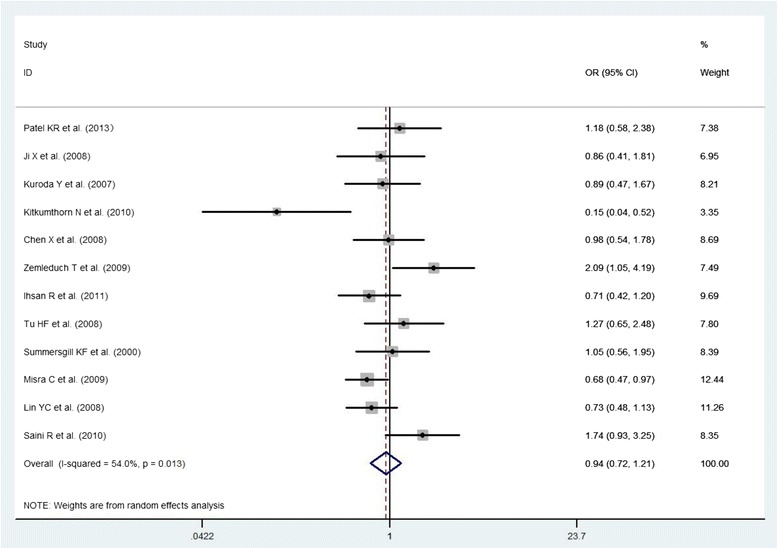
The association between p53 Arg72Pro polymorphism and the risk of oral cancer in total population (Arg/Arg + Arg/Pro)
Fig. 5.
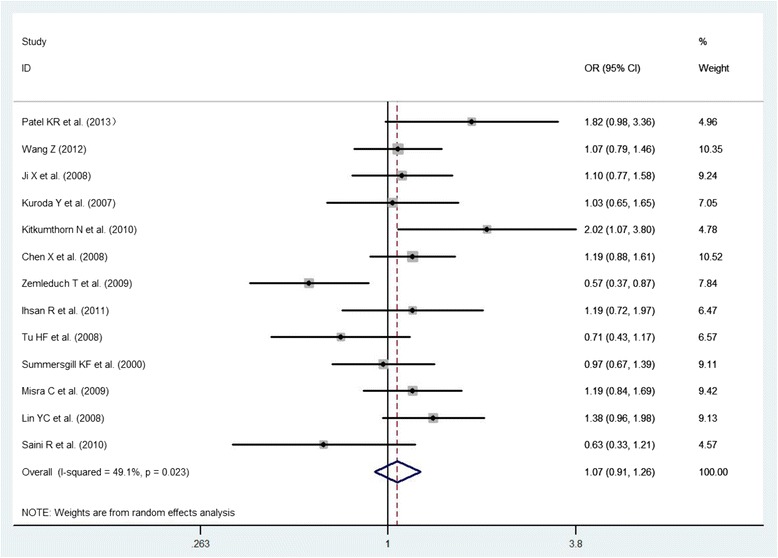
The association between p53 Arg72Pro polymorphism and the risk of oral cancer in total population (Arg/Arg vs. Arg/Pro + Pro/Pro)
Table 3.
Association, test heterogeneity and publication bias for p53 Arg72Pro polymorphism and the risk of oral cancer
| Comparison | Number of studies | Sample size (case/control) | Test of association | Test of heterogeneity | Publication bias | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95 %CI | P value | Model | Q | P value | I2 | P value (Begg’s) | P value (Egger’s) | |||
| Arg72 allele vs. Pro72 allele | |||||||||||
| Total | 12 | 2,093/2,880 | 1.054 | 0.905-1.228 | 0.500 | R | 33.16 | 0.000 | 66.8 % | 0.300 | 0.202 |
| Caucasian | 4 | 827/1,299 | 0.933 | 0.722-1.206 | 0.597 | R | 9.92 | 0.019 | 69.7 % | 0.221 | 0.175 |
| Asian | 8 | 1,266/1,581 | 1.128 | 0.934-1.362 | 0.210 | R | 19.40 | 0.007 | 63.9 % | 0.902 | 0.717 |
| Arg/Arg vs. Pro/Pro | |||||||||||
| Total | 12 | 2,093/2,880 | 1.109 | 0.807-1.524 | 0.523 | R | 29.39 | 0.002 | 62.6 % | 0.360 | 0.415 |
| Caucasian | 4 | 827/1,299 | 0.870 | 0.621-1.218 | 0.416 | F | 6.04 | 0.110 | 50.3 % | 0.462 | 0.312 |
| Asian | 8 | 1,266/1,581 | 1.277 | 0.860-1.898 | 0.226 | R | 19.36 | 0.007 | 63.8 % | 0.536 | 0.914 |
| Arg/Arg + Arg/Pro vs. Pro/Pro | |||||||||||
| Total | 12 | 2,093/2,880 | 0.936 | 0.723-1.211 | 0.613 | R | 23.90 | 0.013 | 54.0 % | 0.161 | 0.423 |
| Caucasian | 4 | 827/1,299 | 1.142 | 0.823-1.583 | 0.427 | F | 3.81 | 0.283 | 21.2 % | 0.806 | 0.451 |
| Asian | 8 | 1,266/1,581 | 0.846 | 0.613-1.169 | 0.312 | R | 17.10 | 0.017 | 59.1 % | 0.711 | 0.990 |
| Arg/Arg vs. Arg/Pro + Pro/Pro | |||||||||||
| Total | 13 | 2,413/3,201 | 1.069 | 0.907-1.259 | 0.426 | R | 23.56 | 0.023 | 49.1 % | 0.511 | 0.302 |
| Caucasian | 5 | 1,147/1,620 | 0.975 | 0.777-1.224 | 0.828 | R | 8.54 | 0.074 | 53.2 % | 0.060 | 0.054 |
| Asian | 8 | 1,266/1,581 | 1.161 | 0.917-1.471 | 0.215 | R | 13.19 | 0.068 | 46.9 % | 0.902 | 0.883 |
| HPV infection | 5 | 396/213 | 0.677 | 0.478-0.959 | 0.027 | F | 0.93 | 0.920 | 0.0 % | 0.462 | 0.400 |
Model Abbreviations: R = random-effect; F = fixed-effect
The association between p53 Arg72Pro polymorphism and the risk of oral cancer in a specific population
In order to determine the major cause for the heterogeneity, a stratified analysis of the specific populations was performed. Eight studies were conducted in Asian populations and five studies were conducted in Caucasian populations. No significant association between the risk of oral cancer and p53 codon 72 polymorphism was detected in the Asian and the Caucasian groups in any genetic model (Table 3). Significant heterogeneity was detected in both groups in all genetic models, except for Pro/Pro vs. Arg/Arg + Arg/Pro in the Caucasian group (Table 3).
The association between combined effect of p53 Arg72Pro polymorphism with HPV infection and the risk of oral cancer in total population
A total of five studies, including 396 cases and 213 controls, were included to evaluate the relations among HPV, p53 Arg72Pro polymorphism, and oral cancer susceptibility. The result showed that the association of HPV with p53 Arg72Pro variant genotypes displayed a statistical significance on oral cancer risk in the Arg/Arg vs. Pro carriers (Arg/Pro + Pro/Pro) model (OR: 0.68, 95 % CI: 0.48- 0.96, p = 0.028) (Fig. 2, Table 3). There was no significant heterogeneity among these studies (Q = 0.93, I2 = 0.0 %, P = 0.92; Table 3).
Publication bias
Begg’s funnel plots seemed to be approximately symmetrical in all meta-analyses (data not shown). Additionally, Egger’s tests did not reveal any obvious evidence of publication bias either (Table 3).
Discussion
Since the identification of the p53 codon 72 polymorphism, many studies have been devoted to explore the genetic effect of p53 Arg72Pro polymorphism on susceptibility of oral cancer. However, the evidence regarding the role of single nucleotide polymorphism of p53 Arg72Pro gene as a genetic marker for the risk of oral cancer is inconsistent. This prompted us to undertake the present meta-analysis to explore a more robust estimate of the relationship between p53 Arg72Pro genetic variant and the oral cancer susceptibility. In this study, we found that individuals who have genetic variants (Arg/Pro genotype or Pro/Pro genotype) may not have induced modification of oral cancer risk compared with those who carry wild-type genotype (Arg/Arg genotype). Same SNP may play different roles in the development of cancer in different ethnic populations. Therefore, the relation of p53 Arg72Pro polymorphism with oral cancer susceptibility might be affected by the different ethnic groups. Nevertheless, neither Arg/Arg genotype individuals nor Pro carriers have a significant association with oral carcinoma in the Asian group or the Caucasian group.
HPV belongs to a large virus family, the PAPOVA virus family. There are nearly a hundred types of HPV discovered in human [38]. In this family, some of the members are known to be high-risk oncogenic HPV type, such as HPV-16, HPV-18, HPV-33, and HPV-58. Through encoding oncogenic protein E6, high-risk HPV types inhibit p53 cell cycle tumor suppressor. The viral E6 protein has a powerful binding affinity for p53 protein resulting in its ubiquitination and destruction, thereby inducing degradation of p53 function and cell cycle out of control [15]. Therefore, p53 gene may have some interaction with HPV infection in susceptibility to HPV-associated oral cancer. Some investigators have found that joint action of the p53 codon 72 polymorphism with HPV is associated with the risk of oral cancer [19, 20, 30], but different conclusions were obtained by other investigators [21, 36]. Considering the above mentioned conflicting conclusions, a subgroup analysis of interaction of p53 gene polymorphism with HPV infection on oral cancer susceptibility was performed. Our study demonstrated a significant interaction between HPV infection and p53 Arg72Pro polymorphism on the risk of developing oral cancer in p53 Arg/Arg genotype carriers compared with p53 72Pro carriers.
The small sample size is a major limitation in this study. There were only five research articles investigating the interaction between the infection with HPV and p53 codon 72 polymorphism on the risk of oral carcinoma. Thus, additional studies with larger sample size are needed to further evaluate the impact of HPV infection and p53 Arg72Pro polymorphism on HPV-associated oral cancer susceptibility.
Conclusion
For the first time, the current study provided the quantitatively synthesized estimates for the effect of interaction between HPV infection and p53 Arg72Pro polymorphism on the risk of developing oral cancer. This combined effect might together alter an individual’s susceptibility to oral cancer. Our results suggested that p53 Arg72Pro polymorphism may partly contribute to the pathogenesis of oral cancer development. Further well-designed studies with reference to the interactions of gene-gene and gene-environment on p53 codon 72 polymorphism to oral carcinoma susceptibility are required.
Acknowledgement
The authors thank two anonymous reviewers for the careful reading of the manuscript and valuable comments.
Abbreviations
- HPV
Human papillomavirus
- OR
Odds ratio
- CI
Confidence interval
- HWE
Hardy-Weinberg equilibrium
Footnotes
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the study: JH and HX. Searched Literature: YL, WY and JH. Analyzed the data: LY and WY. Determined the genotyping methods: SW. Wrote and revised the paper: LY, WY, YG, WH, LZ, YH and MS. All authors provided critical input in manuscript completion and approved the final version of the manuscript.
Contributor Information
Jun Hou, Email: houjun@ahmu.edu.cn.
Ying Gu, Email: ying.gu@stonybrook.edu.
Wei Hou, Email: wei.hou@stonybrookmedicine.edu.
Song Wu, Email: song.wu@stonybrook.edu.
Yin Lou, Email: dscao1966@126.com.
Wenyu Yang, Email: ywyu8001@163.com.
Ling Zhu, Email: 18756987937@163.com.
Yukun Hu, Email: ykhu992@yahoo.com.
Ming Sun, Email: sunming1124@163.com.
Haowei Xue, Email: xuehaowei@ahmu.edu.cn.
References
- 1.Tanaka T, Tanaka M, Tanaka T. Oral carcinogenesis and oral cancer chemoprevention: a review. Patholog Res Int. 2011;2011:431246. doi: 10.4061/2011/431246. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Petersen PE. Oral cancer prevention and control—the approach of the World Health Organization. Oral Oncol. 2009;45:454–460. doi: 10.1016/j.oraloncology.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 3.Zain RB, Ikeda N, Gupta PC, Warnakulasuriya S, van Wyk CW, Shrestha P, Axéll T. Oral mucosal lesions associated with betel quid, areca nut and tobacco chewing habits: consensus from a workshop held in Kuala Lumpur, Malaysia, November 25–27, 1996. J Oral Pathol Med. 1999;28:1–4. doi: 10.1111/j.1600-0714.1999.tb01985.x. [DOI] [PubMed] [Google Scholar]
- 4.Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 5.Zhong Y, Carmella SG, Upadhyaya P, Hochalter JB, Rauch D, Oliver A, Jensen J, Hatsukami D, Wang J, Zimmerman C, Hecht SS. Immediate consequences of cigarette smoking: rapid formation of polycyclic aromatic hydrocarbon diol epoxides. Chem Res Toxicol. 2011;24:246–252. doi: 10.1021/tx100345x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enwonwu CO, Meeks VI. Bionutrition and oral cancer in humans. Crit Rev Oral Biol Med. 1995;6:5–17. doi: 10.1177/10454411950060010401. [DOI] [PubMed] [Google Scholar]
- 7.Scully C, Bedi R. Ethnicity and oral cancer. Lancet Oncol. 2000;1:37–42. doi: 10.1016/S1470-2045(00)00008-5. [DOI] [PubMed] [Google Scholar]
- 8.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/S0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 9.Donehower LA, Bradley A. The tumor suppressor p53. Biochim Biophys Acta. 1993;1155:181–205. doi: 10.1016/0304-419x(93)90004-v. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki K, Matsubara H. Recent advances in p53 research and cancer treatment. J Biomed Biotechnol. 2011;2011:978312. doi: 10.1155/2011/978312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 12.Habbous S, Pang V, Eng L, Xu W, Kurtz G, Liu FF, Mackay H, Amir E, Liu G. p53 Arg72Pro polymorphism, HPV status and initiation, progression, and development of cervical cancer: a systematic review and meta-analysis. Clin Cancer Res. 2012;18:6407–15. doi: 10.1158/1078-0432.CCR-12-1983. [DOI] [PubMed] [Google Scholar]
- 13.Smith EM, Hoffman HT, Summersgill KS, Kirchner HL, Turek LP, Haugen TH. Human papillomavirus and risk of oral cancer. Laryngoscope. 1998;108:1098–1103. doi: 10.1097/00005537-199807000-00027. [DOI] [PubMed] [Google Scholar]
- 14.Portugal LG, Goldenberg JD, Wenig BL, Ferrer KT, Nodzenski E, Sabnani JB, Javier C, Weichselbaum RR, Vokes EE. Human papillomavirus expression and p53 gene mutations in squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 1997;123:1230–1234. doi: 10.1001/archotol.1997.01900110084011. [DOI] [PubMed] [Google Scholar]
- 15.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 16.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 17.Huibregtse JM, Scheffner M, Howley PM. Acellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh IM, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirusassociated cancer. Nature. 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Sturgis EM, Zhang Y, Huang Z, Zhou Q, Wei Q, Li G. Combined p53-related genetic variants together with HPV infection increase oral cancer risk. Int J Cancer. 2012;131:E251–8. doi: 10.1002/ijc.27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji X, Neumann AS, Sturgis EM, Adler-Storthz K, Dahlstrom KR, Schiller JT, Wei Q, Li G. p53 codon 72 polymorphism associated with risk of human papillomavirus-associated squamous cell carcinoma of the oropharynx in never-smokers. Carcinogenesis. 2008;29:875–879. doi: 10.1093/carcin/bgn039. [DOI] [PubMed] [Google Scholar]
- 21.Summersgill KF, Smith EM, Kirchner HL, Haugen TH, Turek LP. p53 polymorphism, human papillomavirus infection in the oral cavity, and oral cancer. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2000;90:334–339. doi: 10.1067/moe.2000.107359. [DOI] [PubMed] [Google Scholar]
- 22.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–102. doi: 10.2307/3001666. [DOI] [Google Scholar]
- 23.Higgins JP, Thompson SE. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 24.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 25.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 26.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 27.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel KR, Vajaria BN, Begum R, Shah FD, Patel JB, Shukla SN, Patel PS. Association between p53 gene variants and oral cancer susceptibility in population from Gujarat, West India. Asian Pac J Cancer Prev. 2013;14:1093–1100. doi: 10.7314/APJCP.2013.14.2.1093. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda Y, Nakao H, Ikemura K, Katoh T. Association between the TP53 codon72 polymorphism and oral cancer risk and prognosis. Oral Oncol. 2007;43:1043–1048. doi: 10.1016/j.oraloncology.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Kitkumthorn N, Yanatatsaneejit P, Rabalert J, Dhammawipark C, Mutirangura A. Association of P53 codon 72 polymorphism and ameloblastoma. Oral Dis. 2010;16:631–635. doi: 10.1111/j.1601-0825.2010.01664.x. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Sturgis EM, El-Naggar AK, Wei Q, Li G. Combined effects of the p53 codon 72 and p73 G4C14-to-A4T14 polymorphisms on the risk of HPV16-associated oral cancer in never-smokers. Carcinogenesis. 2008;29:2120–2125. doi: 10.1093/carcin/bgn191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zemleduch T, Lianeri M, Rydzanicz M, Gajecka M, Szyfter K, Jagodziński PP. Contribution of polymorphism in codon 72 of TP53 gene to laryngeal cancer in Polish patients. Oral Oncol. 2009;45:683–686. doi: 10.1016/j.oraloncology.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 33.Ihsan R, Devi TR, Yadav DS, Mishra AK, Sharma J, Zomawia E, Verma Y, Phukan R, Mahanta J, Kataki AC, Kapur S, Saxena S. Investigation on the role of p53 codon 72 polymorphism and interactions with tobacco, betel quid, and alcohol in susceptibility to cancers in a high-risk population from North East India. DNA Cell Biol. 2011;30:163–171. doi: 10.1089/dna.2010.1119. [DOI] [PubMed] [Google Scholar]
- 34.Tu HF, Chen HW, Kao SY, Lin SC, Liu CJ, Chang KW. MDM2 SNP 309 and p53 codon 72 polymorphisms are associated with the outcome of oral carcinoma patients receiving postoperative irradiation. Radiother Oncol. 2008;87:243–252. doi: 10.1016/j.radonc.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 35.Misra C, Majumder M, Bajaj S, Ghosh S, Roy B, Roychoudhury S. Polymorphisms at p53, p73, and MDM2 loci modulate the risk of tobacco associated leukoplakia and oral cancer. Mol Carcinog. 2009;48:790–800. doi: 10.1002/mc.20523. [DOI] [PubMed] [Google Scholar]
- 36.Lin YC, Huang HI, Wang LH, Tsai CC, Lung O, Dai CY, Yu ML, Ho CK, Chen CH. Polymorphisms of COX-2 -765G > C and p53 codon 72 and risks of oral squamous cell carcinoma in a Taiwan population. Oral Oncol. 2008;44:798–804. doi: 10.1016/j.oraloncology.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Saini R, Tang TH, Zain RB, Cheong SC, Musa KI, Saini D, Ismail AR, Abraham MT, Mustafa WM, Santhanam J. Significant association of high-risk human papillomavirus (HPV) but not of p53 polymorphisms with oral squamous cell carcinomas in Malaysia. J Cancer Res Clin Oncol. 2011;137:311–320. doi: 10.1007/s00432-010-0886-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]


