Abstract
Introduction
Previous studies have identified common germline variants nominally associated with breast cancer survival. These associations have not been widely replicated in further studies. The purpose of this study was to evaluate the association of previously reported SNPs with breast cancer-specific survival using data from a pooled analysis of eight breast cancer survival genome-wide association studies (GWAS) from the Breast Cancer Association Consortium.
Methods
A literature review was conducted of all previously published associations between common germline variants and three survival outcomes: breast cancer-specific survival, overall survival and disease-free survival. All associations that reached the nominal significance level of P value <0.05 were included. Single nucleotide polymorphisms that had been previously reported as nominally associated with at least one survival outcome were evaluated in the pooled analysis of over 37,000 breast cancer cases for association with breast cancer-specific survival. Previous associations were evaluated using a one-sided test based on the reported direction of effect.
Results
Fifty-six variants from 45 previous publications were evaluated in the meta-analysis. Fifty-four of these were evaluated in the full set of 37,954 breast cancer cases with 2,900 events and the two additional variants were evaluated in a reduced sample size of 30,000 samples in order to ensure independence from the previously published studies. Five variants reached nominal significance (P <0.05) in the pooled GWAS data compared to 2.8 expected under the null hypothesis. Seven additional variants were associated (P <0.05) with ER-positive disease.
Conclusions
Although no variants reached genome-wide significance (P <5 x 10−8), these results suggest that there is some evidence of association between candidate common germline variants and breast cancer prognosis. Larger studies from multinational collaborations are necessary to increase the power to detect associations, between common variants and prognosis, at more stringent significance levels.
Electronic supplementary material
The online version of this article (doi:10.1186/s13058-015-0570-7) contains supplementary material, which is available to authorized users.
Introduction
Breast cancer is the most commonly diagnosed cancer in women, in the world, with an estimated 1.67 million new cancer cases diagnosed in 2012. Breast cancer mortality is the second most common cancer-related death in women in the more developed regions of the world and accounts for 15.4% of cancer-related deaths in women [1]. Breast cancer outcome is affected by several factors including: age, tumour size, tumour grade, extent of local and distal spread at diagnosis, oestrogen receptor (ER) status, human epidermal growth factor receptor 2 (HER2) status and treatment received. It is also likely that inherited host characteristics, such as genetic variants, are important [2].
The association between common germline genetic variation and breast cancer survival has been examined in many candidate gene studies investigating genes in pathways known to be involved in breast cancer [3]. These studies have identified numerous single nucleotide polymorphisms (SNPs) associated with outcome at nominal significance levels, but none have been widely replicated in further studies. The exceptions to this are three genome-wide association studies (GWAS) [4-6] and a study from the Breast Cancer Association Consortium, which had substantial power to detect associated variants with large effect sizes (hazard ratio (HR) >2) [7]. Two of those GWAS have reported significant associations for three polymorphisms (rs9934948, rs3784099, rs4778137) [4,6]. The aim of this study was to evaluate the association of previously reported SNPs with prognosis using data from a hypothesis-generating pooled analysis of eight breast cancer survival GWAS from ten studies including 37,954 breast cancer cases [8].
Methods
Literature review
Studies reporting common polymorphisms associated with breast cancer prognosis were identified by searching both Google Scholar and Pubmed. We searched Google Scholar using the search terms: ‘breast cancer’, ‘survival’, ‘prognosis’, ‘polymorphisms’ and ‘SNPs’. The search terms for Pubmed were ‘breast cancer’ AND (‘survival’ OR ‘prognosis’) AND (‘polymorphism’ OR ‘SNP’). The references of all identified studies were then individually interrogated for any additional studies. The search was last updated on 6 June 2014. We considered studies to be eligible for inclusion if they reported an association between a germline genetic variant and at least one of the following end points: overall survival, disease-free survival and breast cancer-specific survival (BCSS). Studies evaluating the prognostic importance of rare high-penetrance variants with minor allele frequency <2% in BRCA1, BRCA2 and CHEK2 were omitted from the review. Only one study conducted ER subtype-specific analyses.
For the purposes of comparison, all studies that used genetic models that grouped together two genotypes into a single category were defined as using ‘dominance models’. This category includes both dominant and recessive models as each study's definition of a dominant or recessive model is dependent on which allele is the major or minor allele, whether they consider the effect allele to be bi-directional, or whether they focus on only the risk allele.
Genome-wide association studies
We used data from a combined analysis of eight breast cancer GWAS, from ten studies [9-19], that had genotype data from a genome-wide SNP array and had collected follow-up time data for the 37,954 breast cancer cases [8]. Genotype and sample quality control were carried out separately for each study. In short, SNPs were excluded based on: low call rate, minor allele frequency <1% and significant deviation of genotype frequencies from the Hardy-Weinberg equilibrium. Samples were excluded for: low call rate, ambiguous gender, relatedness and extreme heterozygosity. We also excluded subjects of less than 90% European ancestry. Sample ancestry was determined separately for each GWAS included in the meta-analysis using either principal component analysis, multi-dimensional scaling or LAMP based on ethnicities from HapMap samples. Samples with less than 90% European ancestry were excluded. As different genotyping arrays had been used for the different studies, imputation had been performed using a reference panel from the 1000 Genomes Project [8,20]. We utilised the imputed data for the SNPs of interest in this study. Details of the pooled studies are shown in Additional files 1 and 2.
Cox proportional hazards models were fitted to assess the association of genotype with breast cancer-specific mortality under a co-dominant (log-additive) genetic model using the likelihood ratio test. The models were adjusted for principal components in order to minimise the effect of population substructure, and the Collaborative Oncological Gene-environment Study (COGS) [16] dataset was stratified by study. Each survival GWAS was analysed separately and the results were harmonised and combined using a standard inverse-variance weighted fixed-effects meta-analysis. In order to compare the results with the published associations we used a one-sided test based on the reported direction of effect. In the initial analysis all 56 SNPs' models were unadjusted for prognostic factors. However, we conducted multivariable analysis of the previously reported SNPs that were significantly associated with survival adjusting for age, stage and grade using 29,360 samples from the COGS study.
Results
Literature review
We identified 46 publications reporting nominally significant associations between 62 germline variants and survival after a breast cancer diagnosis. Details of each variant and the reported association with breast cancer prognosis are shown in Additional file 3. The median sample size was 890 cases; the smallest study had 85 cases and the largest 25,853. Fifty-nine variants were from 44 candidate gene studies and three variants were identified through GWAS. The candidate genes were involved in the following pathways: DNA repair, cell cycle control, matrix metalloproteinases, immune response, drug response, tumour progression, vitamin D receptors and miscellaneous other pathways (Table 1). Findings from the identified publications were infrequently replicated; only six variants out of the 62 were reported in at least one subsequent publication.
Table 1.
Previously identified breast cancer survival genes in cancer-related pathways
| Pathway | Nearest gene | References |
|---|---|---|
| DNA repair | XRCC1, XRCC2, XRCC3, RAD51B, LIG4, ERCC2 | [6,28-32] |
| Cell cycle control | CCND1, CCND3, PRKAG2, TP53, SIPA1, FGFR2, PPP2R2B | [28,33-38] |
| Matrix metalloproteinases | MMP7, MMP8, MMP2, SERPINE1, TIMP-3 | [23,40-44] |
| Immune and drug response, metabolism | Il-10, IL-6, IL-21, MPO, GSTP1, COMT, CYP19A1, CYP1A1, SULT1E1, NEF2L2, TLR4, SLC28A3, CD24, CD44, NQO1 | [14,22,24,45-57] |
| Tumour progression | NOS3, VEGF, NME1, SELE, GNAS1, ZFP36, TGF | [58-64] |
| Vitamin D receptors | RXRA, VDR | [65,66] |
| Miscellaneous | TOX3, MTHFR, COX11, OCA2, PLAUR. | [4,7,34,65,67] |
NB: the genes mentioned here are the candidate genes listed in the previous publications or are the nearest gene to the single nucleotide polymorphism (SNP) and are not necessarily the genes on which the SNPs have a functional effect.
Meta-analysis findings
Results from the GWAS meta-analysis included 58 of the 62 previously identified variants discussed above. The SNP (rs2886162) was replaced by a perfectly correlated tagSNP (rs2364725, r2 = 1). Associations for four of the variants identified: rs4778137 in OCA2, rs3803662 in TOX3, rs1042522 in TP53 and rs2479717 in CCND1 were discovered in studies carried out by the Breast Cancer Association Consortium using sets of samples included in our GWAS meta-analysis. Therefore, we are unable to replicate these associations independently in the full dataset. The substantial sample overlap between the studies that identified associations with rs4778137 and rs3803662 means that there is little to be gained by attempting to replicate their associations in the additional samples included in the meta-analysis. However, the sample sizes in the studies identifying rs1042522 and rs2479717 were relatively small, so we evaluated their association with BCSS in the GWAS meta-analysis omitting the samples from studies used in the original publications. The two SNPs were evaluated in 29,224 and 31,434 samples respectively.
The results for the 56 SNPs evaluated in the meta-analysis are presented in Additional file 4. In the analysis of all cases, five SNPs (rs2981582, rs1800566, rs9934948, rs1800470 and rs3775775) were significant with one-sided P value <0.05, 51 SNPs were not significant at this nominal P value. The most significant association was for rs2981582 in FGFR2 (per G allele HR 1.09, 90% confidence interval (CI) 1.04 to 1.14, one-sided P value = 0.00085). All significantly associated SNPs had good imputation quality (r2 = 0.9 to 1). The imputation r2 for all 56 SNPs can be found in Additional file 4. No single SNP reached the stringent level of significance generally regarded as genome-wide significant (P value <5x10−8) but the number of moderately significant associations (5) was somewhat greater than that expected by chance (2.8). This is illustrated by the quantile-quantile plot shown in Figure 1. Seven SNPs not significantly associated with prognosis in all patients were significant in ER-positive disease. We found evidence of ER-positive specific associations with prognosis for seven out of the twelve SNPs nominally associated (P <0.05) with survival. These SNPs were not previously identified in patients with specifically ER-positive disease; however, our observations may agree with the previously reported results as most breast cancers are ER positive. We measured the level of heterogeneity between the studies included in the pooled analysis for the 12 SNPs associated with survival. There was moderate evidence of heterogeneity for the SNP rs2981582 (I2 = 41.1%, P value = 0.084). For all other SNPs there was low heterogeneity (I2 < 25%, P value >0.2). Details of the SNPs nominally associated with BCSS are shown in Table 2. The results for the nominally associated SNPs adjusted for age, stage and grade are shown in Additional file 5. The HRs for some of the SNPs were attenuated after adjustment. Also, the associations with BCSS of SNPs rs3775775 and rs2333227 were stronger in the multivariable analysis.
Figure 1.
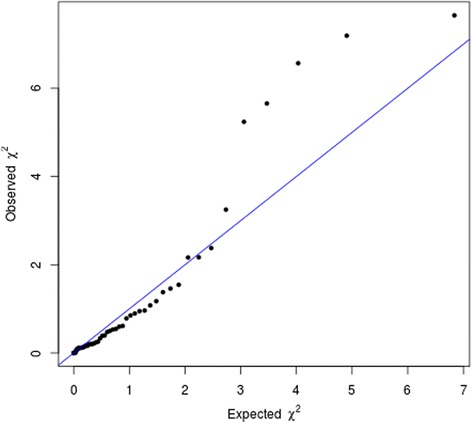
Quantile-quantile plot of results from look-up of previously reported associations in genome-wide association studies. Tests were one-sided with direction assumed from previous association.
Table 2.
Previously reported associations replicated in the meta-analysis
| All cases | ER-negative cases | ER-positive cases | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Published | Model | Effect allele | Effect allele freq | HR (90% CI) | One-sided | HR (90% CI) | One-sided | HR (90% CI) | One-sided |
| P value | P value | P value | |||||||||
| rs2981582 | FGFR2 | Bayraktar et al. [34] | Dominance | G | 0.57 | 1.09 (1.04-1.14) | 0.00085 | 1.08 (1.00-1.16) | 0.052 | 1.04 (0.98-1.10) | 0.15 |
| rs1800566 | NQO1 | Fagerholm et al. [14] | Dominance | A | 0.19 | 1.10 (1.03-1.17) | 0.0046 | 1.14 (1.03-1.25) | 0.015 | 1.04 (0.95-1.13) | 0.23 |
| rs9934948 | LOC100506172 | Shu et al. [6] (GWAS) | Co-dominance | T | 0.15 | 0.92 (0.86-0.98) | 0.011 | 0.90 (0.79-1.01) | 0.059 | 0.95 (0.86-1.04) | 0.18 |
| rs1800470 | TGF | Shu et al. [6] | Co-dominance | A | 0.61 | 0.95 (0.91-0.99) | 0.030 | 0.96 (0.88-1.04) | 0.20 | 0.95 (0.88-1.02) | 0.12 |
| rs3775775 | SULT1E1 | Choi et al. [47] | Dominance | G | 0.09 | 1.08 (1.00-1.16) | 0.046 | 1.17 (1.03-1.31) | 0.02 | 1.06 (0.95-1.17) | 0.18 |
| rs700519 | CYP19A1 | Long et al. [55] | Dominance | A | 0.03 | 1.10 (0.98-1.22) | 0.093 | 1.03 (0.83-1.23) | 0.40 | 1.30 (1.10-1.50) | 0.0050 |
| rs731236 | VDR | Perna et al. [66] | Co-dominance | G | 0.39 | 1.04 (1.00-1.08) | 0.056 | 1.03 (0.95-1.11) | 0.28 | 1.09 (1.02-1.16) | 0.017 |
| rs12900137 | CYP19A1 | Long et al. [55] | Dominance | C | 0.05 | 1.01 (0.91-1.11) | 0.47 | 0.94 (0.78-1.10) | 0.70 | 1.18 (1.02-1.34) | 0.032 |
| rs10477313 | PPP2R2B | Jamshidi et al. [35] | Dominance | T | 0.12 | 0.94 (0.87- 1.01) | 0.08 | 0.92 (0.79-1.05) | 0.15 | 0.88 (0.77-0.99) | 0.035 |
| rs2333227 | MPO | Ambrosone et al. [45] | Dominance | T | 0.21 | 1.03 (0.97-1.09) | 0.20 | 0.95 (0.87-1.03) | 0.78 | 1.09 (1.01-1.17) | 0.036 |
| rs1902586 | CYP19A1 | Long et al. [55] | Dominance | A | 0.05 | 1.01 (0.91-1.11) | 0.44 | 0.99 (0.83-1.15) | 0.54 | 1.16 (1.01-1.31) | 0.041 |
| rs28566535 | CYP19A1 | Long et al. [55] | Dominance | C | 0.05 | 1.00 (0.90-1.10) | 0.51 | 0.97 (0.81-1.13) | 0.60 | 1.15 (1.00-1.30) | 0.046 |
Hazard ratios are for breast cancer-specific survival using a Cox proportional hazards model corrected for principal components; hazard ratios, confidence intervals and P values are from a co-dominant model; P values refer to a one-sided test of association in the direction indicated in bold in the 90% CI of the HR; P values in bold indicate results that are nominally significant (P <0.05). HR, hazard ratio; CI, confidence interval; GWAS, genome-wide association study.
Discussion
There have been few studies focused on the replication of sub-genome-wide significant associations identified previously. Previous replication studies have focused on reporting the SNPs with the strongest evidence of association. We have found some evidence to support previously reported associations between common germline genetic variants and breast cancer prognosis. However, the moderate evidence for some variants provides a rationale for continued research efforts to identify such variants. Significant variants were for the most part candidates in cancer-related genes as is shown in Table 1. Despite the larger sample size and therefore increased power to detect true associations with prognosis in comparison to previous studies, a possible reason for associations failing to reach genome-wide significance may still be limited power. Figure 2a illustrates that for our analysis with 2,900 survival events from 37,954 cases, there is limited power to detect associations at stringent significance levels for modest effect sizes based on a variant with a 0.3 minor allele frequency. Figure 2b shows that almost five times as many events would be needed to detect with 80 per cent power at P value <10−8 an allele with a minor allele frequency of 0.3 that confers a HR of 1.1.
Figure 2.
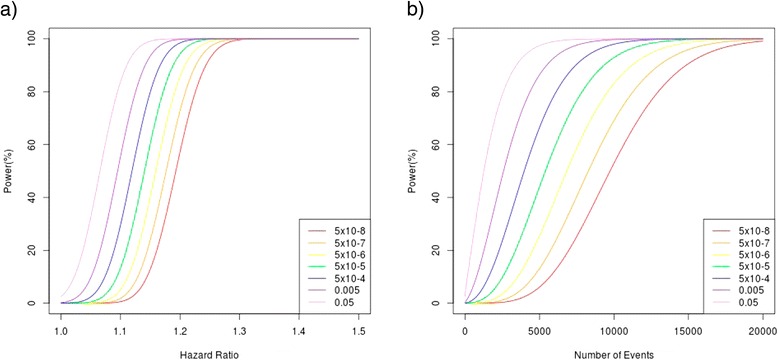
Power (%) to detect true associations with survival time across a range of minor allele frequencies and numbers of events. (a) Power (%) to detect true associations with survival time over a range of effect sizes at increasing orders of significance given a minor allele frequency of 0.3 and 2,900 events. We used an imputation r2 = 0.8 to account for suboptimal imputation. (b) Power (%) to detect true associations with survival time for increasing numbers of events, at increasing orders of significance, given a minor allele frequency of 0.3 and an effect size of 1.1. We used an imputation r2 = 0.8 to account for suboptimal imputation.
In a two-sided test, five of the previously reported associations with prognosis were significantly associated with BCSS in the GWAS meta-analysis but had discordant directions of effect to the original results. These discrepancies may be caused by differing ethnicity between the sample populations [21] as the meta-analysis is specific to patients with European ancestry whereas the five original studies consider non-European populations [6,22-24]. On the other hand, they may also represent false positive associations in both discovery and replication data.
Many previously published studies used a dominance model to evaluate associations. We only used a co-dominant model to detect association in the GWAS. This is justified because thousands of common variants [25] associated with a range of diseases have been identified using a co-dominant model with little or no evidence for dominance. It seems unlikely that breast cancer survival would differ substantially from other phenotypes in any true, underlying genetic model. Where the true underlying model is co-dominant this approach will maximise statistical power. While it is possible that some variants may be truly associated under a dominance model, for example through loss of heterozygosity of the specific germline variant in the tumour, we would still have reasonable power to detect such an association with the large sample size of the GWAS under a co-dominant model.
A further way to increase power to detect robust associations with prognosis is to reduce the level of heterogeneity in the phenotype. Studies focusing on identifying subtype-specific associations will have increased power to detect variants associated with a particular subtype than an analysis on all patients will have. In particular, studies considering disease subtypes, for example ER-negative disease, may provide valuable information into the reasons for known prognostic differences between subtypes. We identified seven SNPs associated with ER-positive disease. These SNPs were not previously identified in specifically ER-positive disease, however, our observations may agree with the previously reported results as most breast cancers are ER positive. In addition, studies looking at interactions with specific treatments, most notably adjuvant chemotherapy, hormonal therapy and adjuvant radiotherapy, may further inform targeted treatment of subgroups of patients according to their inherited genetic information. Some of the previously reported associations with prognosis were found in specific subgroups of patients; however, as yet the sizes of these studies are limited. Large subtype-specific studies are needed in order to investigate interactions with particular subgroups effectively. The generation of sufficiently large studies to deliver strongly significant results, as well as having good outcome and treatment data to enable powerful subtype-specific analyses, will only be possible by combining data resources through large-scale global collaborations. Case-control studies including approximately 100,000 cases are now being conducted to identify common variants associated with risk. It seems a realistic goal to carry out case-cohort studies of a similar size. Reliable identification of SNPs associated with breast cancer prognosis may help to understand the molecular mechanisms of tumour progression and metastasis. Ultimately, this may lead to the development of new therapeutic targets. Polygenic risk scores based on multiple risk alleles have been shown to have potentially useful discrimination [26]. Similar polygenic prognostic scores may improve discrimination of prognostic and treatment benefit tools such as PREDICT [27].
Conclusions
We have found limited evidence to support the assertion that germline genetic variation influences outcome after a diagnosis of breast cancer. Large studies with detailed clinical and follow-up information are needed in order to achieve sufficient statistical power to detect associations at stringent significance thresholds. In addition, power can also be increased by reducing the level of phenotype heterogeneity, which will also provide valuable insights into prognostic differences between subgroups.
Acknowledgements
This study would not have been possible without the contributions of the following: D. C. Tessier, F. Bacot, D. Vincent, S. LaBoissière and F. Robidoux and the staff of the genotyping unit, (Genome Quebec); J. Stone, S. McBean, J. Hadlington, A. Mustafa, K. Cook (Illumina); M. Angelakos, J. Maskiell, G. Dite, T. Selander, N. Weerasooriya (ABCFS); R. van Hien, L. Braaf, F. Hogervorst, S. Verhoef, E. Rutgers, F. Atsma (ABCS); N. McInerney, G. Colleran, A. Rowan, A. Jones (BIGGS); P. Bugert (BSUCH); D. U. Andersen, M. B. Arnadottir, A. Bank, D. K. Hansen (CGPS); G. Pita, C. Alonso, D. Herrero, N. Álvarez (CNIO-BCS); H. Ziegler, S. Wolf, V. Hermann (ESTHER); D. Greco, K. von Smitten, I. Erkkilä, T. Heikkinen, K. Aaltonen (HEBCS); E. Myöhänen, H. Kemiläinen (KBCP); G. Peuteman, D. Smeets, T. Van Brussel, K. Corthouts (LMBC); J. Heinz, N. Obi, A. Vrieling, S. Behrens, U. Eilber, M. Celik, T. Olchers (MARIE); B. Peissel, G. Scuvera, D. Zaffaroni, M. Barile, I. Feroce (MBCSG); K. Mononen, M. Otsukka (OBCS); T. Selander, N. Weerasooriya (OFBCR); E. Krol-Warmerdam, J. Blom. J. Molenaar (ORIGO); L. Brinton, M. Sherman, N. Szeszenia-Dabrowska, B. Peplonska, W. Zatonski, P. Chao, M. Stagner (PBCS); P. Bos, J. Blom, E. Crepin, A. Nieuwlaat, A. Heemskerk (RBCS); S. Higham, I. Brock, S. Balasubramanian, D. Connley, H. Cramp (SBCS); J. Young, C. Twelves, A. L. Vallier, S. Ingle, R. Hardy (PGSNPS); M. Rübner, S. Landrith, S. Oeser, L. Häberle (BBCC).
Funding
This work was supported by the following grants.
Higher level funding
The COGS project was funded through a European Commission's Seventh Framework Programme grant (agreement number 223175 - HEALTH-F2-2009-223175). The Breast Cancer Association Consortium (BCAC) is funded by Cancer Research-UK (C1287/A10118 and C1287/A12014). Meetings of the BCAC have been funded by the European Union COST programme (BM0606). ELAN Program of the University Hospital Erlangen (BBCC).
Personal support
AP is funded by a MRC studentship. DE is a Principal Research Fellow of Cancer Research UK. JH is a National Health and Medical Research Council (NHMRC) Australia Fellow. MS. is a NHMRC Senior Research Fellow. GT is an NHMRC Senior Principal Research Fellow. DL is supported by the FWO and the KULPFV/10/016-SymBioSysII. JL is a UNESCO-L'Oréal International Fellow. RB was a Cancer Institute NSW Fellow. KAP is a National Breast Cancer Foundation Fellow (Australia).
Funding of constituent studies
These are listed by funding agency, with each grant number in parentheses
Academy of Finland (266528);
Addenbrookes Charitable Trust;
Agency for Science, Technology and Research of Singapore;
Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923, PI08/1120);
Baden Württemberg Ministry of Science, Research and Arts;
Breast Cancer Campaign (2009PR42);
Breast Cancer Research Foundation;
Canadian Institutes of Health Research (CIHR Team in Familial Risks of Breast Cancer program);
Cancer Australia;
Cancer Councils of New South Wales, Victoria, Tasmania and South Australia;
Cancer Foundation of Western Australia;Cancer Fund of North Savo;
Cancer Research UK (C1287/A10118, C1287/A12014, A7572, A10124, A11699, A16561, C507/A6306, C10097/A7484,C1275/A11699);
Chief Physician Johan Boserup and Lise Boserup Fund;
Danish Breast Cancer Group;
Danish Medical Research Council;
Deutsche Krebshilfe (70-2892-BR I, PBZ_KBN_122/P05/2004);Dietmar-Hopp Foundation;
Dutch Cancer Society (1997-1505, 2004-3124, NKI2007-3839, 2009-4318, NKI2009-4363);
Dutch government (NWO 184.021.007);
Dutch National Genomics Initiative;
ELAN-Fond of the University Hospital of Erlangen;
European Community´s Seventh Framework Programme (HEALTH-F2-2009-223175) Federal Ministry of Education and Research, Germany (01KH0402);
Finnish Cancer Society;
Fondazione IRCCS Istituto Nazionale Tumori;
Genome Spain Foundation;
German Cancer Research Center (DKFZ);
Hamburg Cancer Society;
Helmholtz Society;
Helsinki University Central Hospital Research Fund;
Italian Association for Cancer Research(AIRC);
Kuopio University Hospital special Government Funding;
National Health and Medical Research Council of Australia (209057, 251553 and 504711);
National Breast Cancer Foundation (Australia);
NIHR Cambridge Biomedical Research Centre;
Nordic Cancer Union;
Märit and Hans Rausings Initiative Against Breast Cancer; Nordic Cancer Union;
Polish Foundation of Science (PBZ_KBN_122/P05/2004);
Queensland Cancer Fund;
Red Temática de Investigación Cooperativa en Cáncer;
Sigrid Juselius Foundation;
Susan G. Komen Breast Cancer Foundation;
Stichting tegen Kanker (232-2008 and 196-2010);
United States National Institutes of Health (BBMRI-NL-CP16, CA69638, CA69417, CA06503, CA116201, CA122340, CA128978, CA63464, CA54281, CA098758, CA132839, CA164920, CA98216, CA098233, CA148065, CA98710, CA98758, and Intramural Research Program of National Institutes of Health and National Cancer Institute);
UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Guy's & St. Thomas' NHS Foundation Trust in partnership with King's College London, and University of Oxford;
University of Eastern Finland strategic funding;
Victorian Health Promotion Foundation;
Victorian Breast Cancer Research Consortium; YORKSHIRE CANCER RESEARCH (S295, S299, S305PA).
Abbreviations
- BCSS
breast cancer-specific survival
- CI
confidence interval
- COGS
Collaborative Oncological Gene-environment Study
- ER
oestrogen receptor
- GWAS
genome-wide association study (studies)
- HER2
human epidermal growth factor receptor 2
- HR
hazard ratio
- SNP
single nucleotide polymorphism
Additional files
Study information for GWAS included in meta-analysis [ 8 ].
Samples included in meta-analysis by study [ 8 ].
Previously reported associations with breast cancer survival.
Look-up of previously reported associations in meta-analysis.
Multivariable analysis results adjusting for age, stage and grade in samples from the COGS dataset.
Footnotes
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
AP, PDP, MS and PAF conceived of the study. The data analysis was done by QG, AP, CC, SK, KM and JT. The data management was conducted by KM, ML, MKB, QW and JD. The manuscript was drafted by AP, PDP and MS and all authors contributed to the editing and critical review. The sample collection for the participating studies was conducted by: PK, BEH, CH, FS, LM, JF, RY, DC, RD, SG, MMG, SH, RH, AH, RK, MM, WW, MB, FC (BPC3); PDP, AD, MiS (SEARCH); KC, HD, ME (pKarma); DL, CW, KL, CO (LMBC); SEB, BN, SFN, HF (CGPS); JC, AR, PS, DF (MARIE); HN, CB, KA, RF, TM (HEBCS); FJC, JO, EH, CV (MCBCS); IA, JK, GG, AMM (OFBCR); CH, BEH, FS, LM (MEC); MS, SaC, AB, FH, StC (ABCS); JH, HT, CA, MCS (ABCFS); AC, SC, MR (SBCS); GGG, RM, CM (MCCS); RW, KP, AJV, MG (OBCS); MH, AnH, JM, AO (RBCS); MGC, SJC, JoL, JF (PBCS); FM, AS, RonY, BB (BSUCH); ES, IT, MK, NM (BIGGS); HB, KB, BH (ESTHER); AM, VeK, VmK, JMH (KBCP); PH, JiL, JSB, KH (SASBAC); PD, RT, CS (ORIGO); PR, PP, SM, FF (MBCSG); PF, MWB, AIH, AE (BBCC); GC, RB, KAP (kConFab/AOCS); JB, PZ, JIAP, PM (CNIO-BCS); AJ, JL, JG, KD (SZBCS); UH, MaK, HU, TR (SKKDKFZS); SaM (KARBAC); VK, SN (NBCS); DiE, NR, GE (UK2); JA, HE, CP, LH, JAD, SB (PGSNPs); SFC, CaC (METABRIC); DH, SL (NHS); DE (BCAC). All authors read and approved the final draft of the manuscript.
Contributor Information
Ailith Pirie, Email: ap736@medschl.cam.ac.uk.
Qi Guo, Email: qg209@medschl.cam.ac.uk.
Peter Kraft, Email: pkraft@hsph.harvard.edu.
Sander Canisius, Email: s.canisius@nki.nl.
Diana M Eccles, Email: d.m.eccles@soton.ac.uk.
Nazneen Rahman, Email: Nazneen.Rahman@icr.ac.uk.
Heli Nevanlinna, Email: Heli.Nevanlinna@hus.fi.
Constance Chen, Email: cjchen@hsph.harvard.edu.
Sofia Khan, Email: sofia.khan@helsinki.fi.
Jonathan Tyrer, Email: jpt34@medschl.cam.ac.uk.
Manjeet K Bolla, Email: mkh39@medschl.cam.ac.uk.
Qin Wang, Email: qw232@medschl.cam.ac.uk.
Joe Dennis, Email: jgd29@cam.ac.uk.
Kyriaki Michailidou, Email: km533@medschl.cam.ac.uk.
Michael Lush, Email: mjl81@medschl.cam.ac.uk.
Alison M Dunning, Email: amd24@medschl.cam.ac.uk.
Mitul Shah, Email: ms483@medschl.cam.ac.uk.
Kamila Czene, Email: kamila.czene@ki.se.
Hatef Darabi, Email: hatef.darabi@ki.se.
Mikael Eriksson, Email: mikael.eriksson@ki.se.
Dieter Lambrechts, Email: diether.lambrechts@med.kuleuven.be.
Caroline Weltens, Email: caroline.weltens@uzleuven.be.
Karin Leunen, Email: karin.leunen@uzleuven.be.
Chantal van Ongeval, Email: Chantal.VanOngeval@uzleuven.be.
Børge G Nordestgaard, Email: boerge.nordestgaard@regionh.dk.
Sune F Nielsen, Email: sune.fallgaard.nielsen@regionh.dk.
Henrik Flyger, Email: henrik.flyger@regionh.dk.
Anja Rudolph, Email: a.rudolph@dkfz.de.
Petra Seibold, Email: p.seibold@dkfz.de.
Dieter Flesch-Janys, Email: flesch@uke.uni-hamburg.de.
Carl Blomqvist, Email: carl.blomqvist@helsinki.fi.
Kristiina Aittomäki, Email: kristiina.aittomaki@hus.fi.
Rainer Fagerholm, Email: Heli.Nevanlinna@hus.fi.
Taru A Muranen, Email: taru.a.muranen@helsinki.fi.
Janet E Olsen, Email: olsonj@mayo.edu.
Emily Hallberg, Email: Hallberg.emily@mayo.edu.
Celine Vachon, Email: vachon.celine@mayo.edu.
Julia A Knight, Email: knight@lunenfeld.ca.
Gord Glendon, Email: gglendon@uhnresearch.ca.
Anna Marie Mulligan, Email: AnnaMarie.Mulligan@uhn.ca.
Annegien Broeks, Email: a.broeks@nki.nl.
Sten Cornelissen, Email: s.cornelissen@nki.nl.
Christopher A Haiman, Email: Christopher.Haiman@med.usc.edu.
Brian E Henderson, Email: Behender@usc.edu.
Frederick Schumacher, Email: fschumac@usc.edu.
Loic Le Marchand, Email: loic@crch.hawaii.edu.
John L Hopper, Email: j.hopper@unimelb.edu.au.
Helen Tsimiklis, Email: htsi@unimelb.edu.au.
Carmel Apicella, Email: capic@unimelb.edu.au.
Melissa C Southey, Email: msouthey@unimelb.edu.au.
Simon S Cross, Email: s.s.cross@sheffield.ac.uk.
Malcolm WR Reed, Email: m.w.reed@sheffield.ac.uk.
Graham G Giles, Email: Graham.Giles@cancervic.org.au.
Roger L Milne, Email: Roger.Milne@cancervic.org.au.
Catriona McLean, Email: C.McLean@alfred.org.au.
Robert Winqvist, Email: robert.winqvist@oulu.fi.
Katri Pylkäs, Email: katri.pylkas@oulu.fi.
Arja Jukkola-Vuorinen, Email: arja.jukkola-vuorinen@ppshp.fi.
Mervi Grip, Email: mervi.grip@ppshp.fi.
Maartje J Hooning, Email: m.hooning@erasmusmc.nl.
Antoinette Hollestelle, Email: a.hollestelle@erasmusmc.nl.
John WM Martens, Email: j.martens@erasmusmc.nl.
Ans MW van den Ouweland, Email: a.vandenouweland@erasmusmc.nl.
Federick Marme, Email: frederikmarme@gmail.com.
Andreas Schneeweiss, Email: Andreas.Schneeweiss@med.uni-heidelberg.de.
Rongxi Yang, Email: R.Yang@dkfz.de.
Barbara Burwinkel, Email: Barbara.Burwinkel@med.uni-heidelberg.de.
Jonine Figueroa, Email: figueroaj@mail.nih.gov.
Stephen J Chanock, Email: chanocks@mail.nih.gov.
Jolanta Lissowska, Email: lissowsj@coi.waw.pl.
Elinor J Sawyer, Email: chanocks@mail.nih.gov.
Ian Tomlinson, Email: iant@well.ox.ac.uk.
Michael J Kerin, Email: michael.kerin@nuigalway.ie.
Nicola Miller, Email: nicola.miller@nuigalway.ie.
Hermann Brenner, Email: h.brenner@dkfz.de.
Katja Butterbach, Email: k.butterbach@dkfz.de.
Bernd Holleczek, Email: b.holleczek@gbe-ekr.saarland.de.
Vesa Kataja, Email: vesa.kataja@kuh.fi.
Veli-Matti Kosma, Email: veli-matti.kosma@uef.fi.
Jaana M Hartikainen, Email: jaana.hartikainen@uef.fi.
Jingmei Li, Email: jingmei@gmail.com.
Judith S Brand, Email: judith.brand@ki.se.
Keith Humphreys, Email: Keith.Humphreys@ki.se.
Peter Devilee, Email: p.devilee@lumc.nl.
Robert AEM Tollenaar, Email: r.a.e.m.tollenaar@lumc.nl.
Caroline Seynaeve, Email: c.seynaeve@erasmusmc.nl.
Paolo Radice, Email: paolo.radice@istitutotumori.mi.it.
Paolo Peterlongo, Email: paolo.peterlongo@ifom.eu.
Siranoush Manoukian, Email: siranoush.manoukian@istitutotumori.mi.it.
Filomena Ficarazzi, Email: filomena.ficarazzi@ifom.eu.
Matthias W Beckmann, Email: fk-direktion@uk-erlangen.de.
Alexander Hein, Email: alexander.hein@uk-erlangen.de.
Arif B Ekici, Email: arif.ekici@uk-erlangen.de.
Rosemary Balleine, Email: rosemary.balleine@sydney.edu.au.
Kelly-Anne Phillips, Email: Kelly.Phillips@petermac.org.
kConFab Investigators, Email: Georgia.Trench@qimrberghofer.edu.au.
Javier Benitez, Email: jbenitez@cnio.es.
M Pilar Zamora, Email: zamorapilar@gmail.com.
Jose Ignacio Arias Perez, Email: jiarias@msn.com.
Primitiva Menéndez, Email: tiva@hca.es.
Anna Jakubowska, Email: aniaj@sci.pam.szczecin.pl.
Jan Lubinski, Email: lubinski@sci.pam.szczecin.pl.
Jacek Gronwald, Email: jgron@sci.pum.edu.pl.
Katarzyna Durda, Email: k.durda@onet.pl.
Ute Hamann, Email: u.hamann@dkfz-heidelberg.de.
Maria Kabisch, Email: m.kabisch@dkfz-heidelberg.de.
Hans Ulrich Ulmer, Email: hu.ulmer@klinikum-mittelbaden.de.
Thomas Rüdiger, Email: Thomas.Ruediger@klinikum-karlsruhe.de.
Sara Margolin, Email: Sara.Margolin@karolinska.se.
Vessela Kristensen, Email: Vessela.N.Kristensen@rr-research.no.
Siljie Nord, Email: silje.nordgard@gmail.com.
NBCS Investigators, Email: Vessela.N.Kristensen@rr-research.no.
D Gareth Evans, Email: Gareth.Evans@cmft.nhs.uk.
Jean Abraham, Email: ja344@medschl.cam.ac.uk.
Helena Earl, Email: hme22@cam.ac.uk.
Christopher J Poole, Email: christopher.poole@warwick.ac.uk.
Louise Hiller, Email: l.hiller@warwick.ac.uk.
Janet A Dunn, Email: j.a.dunn@warwick.ac.uk.
Sarah Bowden, Email: s.j.bowden@bham.ac.uk.
Rose Yang, Email: royang@mail.nih.gov.
Daniele Campa, Email: d.campa@dkfz-heidelberg.de.
W Ryan Diver, Email: Ryan.diver@cancer.org.
Susan M Gapstur, Email: Susan.gapstur@cancer.org.
Mia M Gaudet, Email: Mia.gaudet@cancer.org.
Susan Hankinson, Email: shankinson@schoolph.umass.edu.
Robert N Hoover, Email: hooverr@exchange.nih.gov.
Anika Hüsing, Email: a.huesing@dkfz.de.
Rudolf Kaaks, Email: r.kaaks@dkfz.de.
Mitchell J Machiela, Email: Mitchell.machiela@nih.gov.
Walter Willett, Email: wwillett@hsph.harvard.edu.
Myrto Barrdahl, Email: m.barrdahl@Dkfz-Heidelberg.de.
Federico Canzian, Email: f.canzian@dkfz-heidelberg.de.
Suet-Feung Chin, Email: Suet-Feung.Chin@cruk.cam.ac.uk.
Carlos Caldas, Email: carlos.caldas@cruk.cam.ac.uk.
David J Hunter, Email: dhunter@hsph.harvard.edu.
Sara Lindstrom, Email: slindstr@hsph.harvard.edu.
Montserrat Garcia-Closas, Email: Montse.GarciaClosas@icr.ac.uk.
Fergus J Couch, Email: couch.fergus@mayo.edu.
Georgia Chenevix-Trench, Email: Georgia.Trench@qimrberghofer.edu.au.
Arto Mannermaa, Email: arto.mannermaa@uef.fi.
Irene L Andrulis, Email: andrulis@lunenfeld.ca.
Per Hall, Email: Per.Hall@ki.se.
Jenny Chang-Claude, Email: j.chang-claude@dkfz-heidelberg.de.
Douglas F Easton, Email: dfe20@medschl.cam.ac.uk.
Stig E Bojesen, Email: stig.egil.bojesen@regionh.dk.
Angela Cox, Email: a.cox@shef.ac.uk.
Peter A Fasching, Email: peter.fasching@uk-erlangen.de.
Paul DP Pharoah, Email: pp10001@medschl.cam.ac.uk.
Marjanka K Schmidt, Email: mk.schmidt@nki.nl.
References
- 1.IARC. http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx.
- 2.Hartman M, Lindström L, Dickman PW, Adami H-O, Hall P, Czene K. Is breast cancer prognosis inherited? Breast Cancer Res. 2007;9:R39. doi: 10.1186/bcr1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ribelles N, Santonja A, Pajares B, Llácer C, Alba E. The seed and soil hypothesis revisited: current state of knowledge of inherited genes on prognosis in breast cancer. Cancer Treat Rev. 2014;40:293–299. doi: 10.1016/j.ctrv.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Azzato EM, Tyrer J, Fasching PA, Beckmann MW, Ekici AB, Schulz-Wendtland R, et al. Association between a germline OCA2 polymorphism at chromosome 15q13.1 and estrogen receptor-negative breast cancer survival. J Natl Cancer Inst. 2010;102:650–662. doi: 10.1093/jnci/djq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azzato EM, Pharoah PDP, Harrington P, Easton DF, Greenberg D, Caporaso NE, et al. A genome-wide association study of prognosis in breast cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:1140–1143. doi: 10.1158/1055-9965.EPI-10-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shu XO, Long J, Lu W, Li C, Chen WY, Delahanty R, et al. Novel genetic markers of breast cancer survival identified by a genome-wide association study. Cancer Res. 2012;72:1182–1189. doi: 10.1158/0008-5472.CAN-11-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fasching P, Pharoah PDP, Cox A, Nevanlinna H, Bojesen SE, Karn T, et al. The role of genetic breast cancer susceptibility variants as prognostic factors. Hum Mol Genet. 2012;21:3926–3939. doi: 10.1093/hmg/dds159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Q, Schmidt MK, Kraft P, Canisius S, Chen C, Khan S, et al. JNCI J Natl Cancer Inst 2015;107(5):djv081 doi:10.1093/jnci/djv081.
- 9.Abraham JE, Guo Q, Dorling L, Tyrer J, Ingle S, Hardy R, et al. Replication of genetic polymorphisms reported to be associated with taxane-related sensory neuropathy in patients with early breast cancer treated with paclitaxel. Clin Cancer Res. 2014;20:2466–2475. doi: 10.1158/1078-0432.CCR-13-3232. [DOI] [PubMed] [Google Scholar]
- 10.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Earl HM, Vallier AL, Hiller L, Fenwick N, Young J, Iddawela M, et al. Effects of the addition of gemcitabine, and paclitaxel-first sequencing, in neoadjuvant sequential epirubicin, cyclophosphamide, and paclitaxel for women with high-risk early breast cancer (Neo-tAnGo): an open-label, 2 × 2 factorial randomised phase 3 trial. Lancet Oncol. 2014;15:201–212. doi: 10.1016/S1470-2045(13)70554-0. [DOI] [PubMed] [Google Scholar]
- 12.Eccles D, Gerty S, Simmonds P, Hammond V, Ennis S, Altman DG. Prospective study of Outcomes in Sporadic versus Hereditary breast cancer (POSH): study protocol. BMC Cancer. 2007;7:160. doi: 10.1186/1471-2407-7-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Einarsdóttir K, Darabi H, Li Y, Low YL, Li YQ, Bonnard C, et al. ESR1 and EGF genetic variation in relation to breast cancer risk and survival. Breast Cancer Res. 2008;10:R15. doi: 10.1186/bcr1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fagerholm R, Hofstetter B, Tommiska J, Aaltonen K, Vrtel R, Syrjäkoski K, et al. NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat Genet. 2008;40:844–853. doi: 10.1038/ng.155. [DOI] [PubMed] [Google Scholar]
- 15.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poole CJ, Hiller L, Howard HC, Dunn JA, Canney P, Wardley AM, et al. tAnGo: a randomized phase III trial of gemcitabine (gem) in paclitaxel-containing, epirubicin/cyclophosphamide-based, adjuvant chemotherapy (CT) for women with early-stage breast cancer (EBC) ASCO Meet Abstr. 2008;26:506. [Google Scholar]
- 18.Poole CJ, Earl HM, Hiller L, Dunn JA, Bathers S, Grieve RJ, et al. Epirubicin and cyclophosphamide, methotrexate, and fluorouracil as adjuvant therapy for early breast cancer. N Engl J Med. 2006;355:1851–1862. doi: 10.1056/NEJMoa052084. [DOI] [PubMed] [Google Scholar]
- 19.Siddiq A, Couch FJ, Chen GK, Lindström S, Eccles D, Millikan RC, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5373–5384. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012; 491:56–65. [DOI] [PMC free article] [PubMed]
- 21.Bryant EK, Dressen AS, Bunker CH, Hokanson JE, Hamman RF, Kamboh MI, et al. A multiethnic replication study of plasma lipoprotein levels-associated SNPs identified in recent GWAS. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long JR, Cai Q, Shu XO, Cai H, Gao YT, Zheng W. Genetic polymorphisms in estrogen-metabolizing genes and breast cancer survival. Pharmacogenet Genomics. 2007;17:331–338. doi: 10.1097/FPC.0b013e32801a3bfe. [DOI] [PubMed] [Google Scholar]
- 23.Slattery ML, John E, Torres-Mejia G, Stern M, Lundgreen A, Hines L, et al. Matrix metalloproteinase genes are associated with breast cancer risk and survival: the Breast Cancer Health Disparities Study. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You Y, Deng J, Zheng J, Hu M, Li N, Wu H, et al. IL-21 gene polymorphism is associated with the prognosis of breast cancer in Chinese populations. Breast Cancer Res Treat. 2013;137:893–901. doi: 10.1007/s10549-012-2401-1. [DOI] [PubMed] [Google Scholar]
- 25.Hindorff LA, MacArthur J, Morales J, Junkins HA, Hall PN, Klemm AK et al: A catalog of published genome-wide association studies. Available at: www.genome.gov/gwastudies. Accessed 22 Aug 2013.
- 26.Pharoah PDP, Antoniou AC, Easton DF, Ponder BAJ. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358:2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 27.Wishart GC, Azzato EM, Greenberg DC, Rashbass J, Kearins O, Lawrence G, et al. PREDICT: a new UK prognostic model that predicts survival following surgery for invasive breast cancer. Breast Cancer Res. 2010;12:R1. doi: 10.1186/bcr2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bewick MA, Conlon MSC, Lafrenie RM. Polymorphisms in XRCC1, XRCC3, and CCND1 and survival after treatment for metastatic breast cancer. J Clin Oncol. 2006;24:5645–5651. doi: 10.1200/JCO.2006.05.9923. [DOI] [PubMed] [Google Scholar]
- 29.Bewick MA, Lafrenie RM, Conlon MSC. Nucleotide excision repair polymorphisms and survival outcome for patients with metastatic breast cancer. J Cancer Res Clin Oncol. 2011;137:543–550. doi: 10.1007/s00432-010-0915-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castro E, Olmos D, Garcia A, Cruz JJ, González-Sarmiento R. Role of XRCC3, XRCC1 and XPD single-nucleotide polymorphisms in survival outcomes following adjuvant chemotherapy in early stage breast cancer patients. Clin Transl Oncol. 2014;16:158–165. doi: 10.1007/s12094-013-1055-8. [DOI] [PubMed] [Google Scholar]
- 31.Goode EL, Dunning AM, Kuschel B, Healey CS, Day NE, Ponder BAJ, et al. Effect of germ-line genetic variation on breast cancer survival in a population-based study. Cancer Res. 2002;62:3052–3057. [PubMed] [Google Scholar]
- 32.Lin WY, Camp NJ, Cannon-Albright LA, Allen-Brady K, Balasubramanian S, Reed MWR, et al. A role for XRCC2 gene polymorphisms in breast cancer risk and survival. J Med Genet. 2011;48:477–484. doi: 10.1136/jmedgenet-2011-100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azzato EM, Driver KE, Lesueur F, Shah M, Greenberg D, Easton DF, et al. Effects of common germline genetic variation in cell cycle control genes on breast cancer survival: results from a population-based cohort. Breast Cancer Res. 2008;10:R47. doi: 10.1186/bcr2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayraktar S, Thompson PA, Yoo SY, Do K, Sahin AA, Arun BK, et al. The relationship between eight GWAS-identified single-nucleotide polymorphisms and primary breast cancer outcomes. Oncologist. 2013;18:493–500. doi: 10.1634/theoncologist.2012-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jamshidi M, Schmidt MK, Dörk T, Garcia-Closas M, Heikkinen T, Cornelissen S, et al. Germline variation in TP53 regulatory network genes associates with breast cancer survival and treatment outcome. Int J Cancer. 2013;132:2044–2055. doi: 10.1002/ijc.27884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei R, Xu Y, Wei Y, Ouyang T, Li J, Wang T, et al. Association of SIPA1 545 C > T polymorphism with survival in Chinese women with metastatic breast cancer. Front Med. 2013;7:138–142. doi: 10.1007/s11684-013-0247-5. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt MK, Tommiska J, Broeks A, van Leeuwen FE, Van’t Veer LJ, Pharoah PDP, et al. Combined effects of single nucleotide polymorphisms TP53 R72P and MDM2 SNP309, and p53 expression on survival of breast cancer patients. Breast Cancer Res. 2009;11:R89. doi: 10.1186/bcr2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tommiska J, Eerola H, Heinonen M, Salonen L, Kaare M, Tallila J, et al. Breast cancer patients with p53 Pro72 homozygous genotype have a poorer survival. Clin Cancer Res. 2005;11:5098–5103. doi: 10.1158/1078-0432.CCR-05-0173. [DOI] [PubMed] [Google Scholar]
- 39.Toyama T, Zhang Z, Nishio M, Hamaguchi M, Kondo N, Iwase H, et al. Association of TP53 codon 72 polymorphism and the outcome of adjuvant therapy in breast cancer patients. Breast Cancer Res. 2007;9:R34. doi: 10.1186/bcr1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beeghly-Fadiel A, Shu XO, Long J, Li C, Cai Q, Cai H, et al. Genetic polymorphisms in the MMP-7 gene and breast cancer survival. Int J Cancer. 2009;124:208–214. doi: 10.1002/ijc.23859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beeghly-Fadiel A, Zheng W, Lu W, Long J, Zheng Y, Cai H, et al. Replication study for reported SNP associations with breast cancer survival. J Cancer Res Clin Oncol. 2012;138:1019–1026. doi: 10.1007/s00432-012-1174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Decock J, Long J-R, Laxton RC, Shu X-O, Hodgkinson C, Hendrickx W, et al. Association of matrix metalloproteinase-8 gene variation with breast cancer prognosis. Cancer Res. 2007;67:10214–10221. doi: 10.1158/0008-5472.CAN-07-1683. [DOI] [PubMed] [Google Scholar]
- 43.Peterson NB, Beeghly-Fadiel A, Gao YT, Long J, Cai Q, Shu X, et al. Polymorphisms in tissue inhibitors of metalloproteinases −2 and −3 and breast cancer susceptibility and survival. Int J Cancer. 2010;125:844–850. doi: 10.1002/ijc.24405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ambrosone CB, Ahn J, Singh KK, Furberg H, Sweeney C, Coles B, et al: 804 Polymorphisms in genes related to oxidative stress (MPO, MnSOD, CAT) and 805 survival after treatment for breast cancer. Cancer Res. 2005: 1105–11. [PubMed]
- 45.Ambrosone CB, Ahn J, Singh KK, Furberg H, Sweeney C, Coles B, et al: Polymorphisms in genes related to oxidative stress (MPO, MnSOD, CAT) and survival after treatment for breast cancer. 2005: 1105–11. [PubMed]
- 46.Buck K, Hug S, Seibold P, Ferschke I, Altevogt P, Sohn C, et al. CD24 polymorphisms in breast cancer: impact on prognosis and risk. Breast Cancer Res Treat. 2013;137:927–937. doi: 10.1007/s10549-012-2325-9. [DOI] [PubMed] [Google Scholar]
- 47.Choi JY, Lee KM, Park SK, Noh DY, Ahn SH, Chung HW, et al. Genetic polymorphisms of SULT1A1 and SULT1E1 and the risk and survival of breast cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:1090–1095. doi: 10.1158/1055-9965.EPI-04-0688. [DOI] [PubMed] [Google Scholar]
- 48.Demichele A, Martin A, Mick R, Gor P, Wray L, Klein-Cabral M, et al. Interleukin-6 174G3 C polymorphism is associated with improved outcome in high-risk breast cancer. Cancer Res. 2003: 8051–56. [PubMed]
- 49.Duggan C, Ballard-Barbash R, Baumgartner RN, Baumgartner KB, Bernstein L, McTiernan A. Associations between null mutations in GSTT1 and GSTM1, the GSTP1 Ile(105)Val polymorphism, and mortality in breast cancer survivors. Springerplus. 2013;2:450. doi: 10.1186/2193-1801-2-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fasching PA, Loehberg CR, Strissel PL, Lux MP, Bani MR, Schrauder M, et al. Single nucleotide polymorphisms of the aromatase gene (CYP19A1), HER2/neu status, and prognosis in breast cancer patients. Breast Cancer Res Treat. 2008;112:89–98. doi: 10.1007/s10549-007-9822-2. [DOI] [PubMed] [Google Scholar]
- 51.Gerger A, Renner W, Langsenlehner T, Hofmann G, Knechtel G, Szkandera J, et al. Association of interleukin-10 gene variation with breast cancer prognosis. Breast Cancer Res Treat. 2010;119:701–705. doi: 10.1007/s10549-009-0417-y. [DOI] [PubMed] [Google Scholar]
- 52.Hartikainen JM, Tengström M, Kosma VM, Kinnula VL, Mannermaa A, Soini Y. Genetic polymorphisms and protein expression of NRF2 and Sulfiredoxin predict survival outcomes in breast cancer. Cancer Res. 2012;72:5537–5546. doi: 10.1158/0008-5472.CAN-12-1474. [DOI] [PubMed] [Google Scholar]
- 53.Jiang L, Deng J, Zhu X, Zheng J, You Y, Li N, et al. CD44 rs13347 C > T polymorphism predicts breast cancer risk and prognosis in Chinese populations. Breast Cancer Res. 2012;14:R105. doi: 10.1186/bcr3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JH, Kim Y, Choi JW, Kim YS. Clinicopathological significance of plasminogen activator inhibitor-1 promoter 4G/5G polymorphism in breast cancer: a meta-analysis. Arch Med Res. 2013;44:39–45. doi: 10.1016/j.arcmed.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Long JR, Kataoka N, Shu XO, Wen W, Gao YT, Cai Q, et al. Genetic polymorphisms of the CYP19A1 gene and breast cancer survival. Cancer Epidemiol Biomarkers Prev. 2006;15:2115–2122. doi: 10.1158/1055-9965.EPI-06-0464. [DOI] [PubMed] [Google Scholar]
- 56.Sweeney C, Mcclure GY, Fares MY, Stone A, Coles BF, Thompson PA, et al. Association between survival after treatment for breast cancer and glutathione S-transferase P1 Ile105Val polymorphism advances in brief association between survival after treatment for breast cancer and glutathione. Cancer Res. 2000: 5621–24. [PubMed]
- 57.Yang CX, Li CY, Feng W. Toll-like receptor 4 genetic variants and prognosis of breast cancer. Tissue Antigens. 2013;81:221–226. doi: 10.1111/tan.12096. [DOI] [PubMed] [Google Scholar]
- 58.Choi JY, Lee KM, Noh DY, Ahn SH, Lee JE, Han W, et al. Genetic polymorphisms of eNOS, hormone receptor status, and survival of breast cancer. Breast Cancer Res Treat. 2006;100:213–218. doi: 10.1007/s10549-006-9245-5. [DOI] [PubMed] [Google Scholar]
- 59.Kontogianni P, Zambirinis CP, Theodoropoulos G, Gazouli M, Michalopoulos NV, Flessas J, et al. The impact of the stromal cell-derived factor-1-3′A and E-selectin S128R polymorphisms on breast cancer. Mol Biol Rep. 2013;40:43–50. doi: 10.1007/s11033-012-1989-x. [DOI] [PubMed] [Google Scholar]
- 60.Lu H, Shu XO, Cui Y, Kataoka N, Wen W, Cai Q, et al. Association of genetic polymorphisms in the VEGF gene with breast cancer survival. Cancer Res. 2005;65:5015–5019. doi: 10.1158/0008-5472.CAN-04-2786. [DOI] [PubMed] [Google Scholar]
- 61.Otterbach F, Callies R, Frey UH, Schmitz KJ, Wreczycki C, Kimmig R, et al. The T393C polymorphism in the gene GNAS1 of G protein is associated with survival of patients with invasive breast carcinoma. Breast Cancer Res Treat. 2007;105:311–317. doi: 10.1007/s10549-006-9462-y. [DOI] [PubMed] [Google Scholar]
- 62.Qu S, Long J, Cai Q, Shu XO, Cai H, Gao YT, et al. Genetic polymorphisms of metastasis suppressor gene NME1 and breast cancer survival. Clin Cancer Res. 2008;14:4787–4793. doi: 10.1158/1078-0432.CCR-08-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shu XO. Genetic polymorphisms in the TGF-1 gene and breast cancer survival: a report from the Shanghai Breast Cancer Study. Cancer Res. 2004;64:836–839. doi: 10.1158/0008-5472.CAN-03-3492. [DOI] [PubMed] [Google Scholar]
- 64.Upadhyay R, Sanduja S, Kaza V, Dixon DA. Genetic polymorphisms in RNA binding proteins contribute to breast cancer survival. Int J Cancer. 2013;132:E128–E138. doi: 10.1002/ijc.27789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pande M, Thompson PA, Do KA, Sahin AA, Amos CI, Frazier ML, et al. Genetic variants in the vitamin D pathway and breast cancer disease-free survival. Carcinogenesis. 2013;34:587–594. doi: 10.1093/carcin/bgs369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perna L, Butterbach K, Haug U, Schöttker B, Müller H, Arndt V, et al. Vitamin D receptor genotype rs731236 (Taq1) and breast cancer prognosis. Cancer Epidemiol Biomarkers Prev. 2013;22:437–442. doi: 10.1158/1055-9965.EPI-12-0970-T. [DOI] [PubMed] [Google Scholar]
- 67.Martin DN, Boersma BJ, Howe TM, Goodman JE, Mechanic LE, Chanock SJ, et al. Association of MTHFR gene polymorphisms with breast cancer survival. BMC Cancer. 2006;6:257. doi: 10.1186/1471-2407-6-257. [DOI] [PMC free article] [PubMed] [Google Scholar]


