Abstract
Cell therapy of heart diseases is emerging as one of the most promising known treatments in recent years. Transplantation of cardiac stem cells (CSCs) may be one of the best strategies to cure adult or pediatric heart diseases. As these patient-derived stem cells need to be isolated from small heart biopsies, it is important to select the best isolation method and CSC subpopulation with the best cardiogenic functionality. We employed three different protocols including c-KIT+ cell sorting, clonogenic expansion, and explants culture to isolate c-KIT+ cells, clonogenic expansion-derived cells (CEDCs), and cardiosphere-derived cells (CDCs), respectively. Evaluation of isolated CSC characteristics in vitro and after rat myocardial infarction (MI) model transplantation revealed that although c-KIT+ and CDCs had higher MI regenerative potential, CEDCs had more commitment into cardiomyocytes and needed lower passages that were essential to reach a definite cell count. Furthermore, genome-wide expression analysis showed that subsequent passages caused changes in characteristics of cells, downregulation of cell cycle-related genes, and upregulation of differentiation and carcinogenic genes, which might lead to senescence, commitment, and possible tumorigenicity of the cells. Because of different properties of CSC subpopulations, we suggest that appropriate CSCs subpopulation should be chosen based on their experimental or clinical use.
Introduction
In 2010, one in four deaths worldwide was attributed to cardiovascular diseases, a prevalent cause of death [1,2]. Recently, cardiac cell therapy with stem cells has become a promising approach to repair heart diseases. The candidate stem cell for transplantation must improve heart function without inducing an immune response, arrhythmias, or carcinogenesis [3]. Results of animal model studies and clinical trials suggest that transplantation of cardiac stem cells (CSCs), resident cardiac precursor cells in the heart, may be one of the best strategies to cure adult or pediatric heart diseases [4–8]. All CSCs populations contain most properties of stem cells: self-renewal, multi-lineage differentiation, and clonogenic potentials. CSCs can improve heart function after transplantation [9].
The first identified subpopulation of CSCs in rats was the c-Kit+ cells [10]. The other explored fraction of CSCs was the stem cell antigen (Sca-1)-expressing population [11]. This was followed by isolation of CSCs from heart biopsies cultured as explants, which were named cardiospheres [12]. Several studies have been carried out to isolate and characterize cardiac precursor cells (mainly c-Kit+, Sca-1+, and cardiospheres) from normal and pathologic fetal and adult heart biopsies [12–15]. Different protocols have been developed to improve CSC isolation methods, resulting in the generation of a specific population of CSCs [16–19]. In addition to cell sorting and culture of explants, clonogenic expansion of cells has been described as another CSCs isolation method [20]. Although cardiomyogenic and regenerative potential of all human CSC subgroups isolated from different published methods have been confirmed, the relationship and probable differences between these groups remains to be determined [3,21]. The choice of isolation method and/or CSCs subpopulation as the proper one using the separate data obtained from different studies is difficult. Furthermore, in vitro cell culture may cause unknown effects on cell characteristics. Some studies report gene expression changes in different cell types after in vitro subculturing [22,23]. Therefore, the expression profile of CSCs after passages may be a prerequisite for their application in cell therapy. CSCs are optional cell sources to clinically treat heart diseases, however, it is important to compare different CSC isolation methods and different CSC populations to determine the best CSC subpopulation with the best cardiogenic functionality and lower essential cell manipulation.
Here, we show that CSCs are present in heart biopsies of patients with tetralogy of Fallot (ToF), a common congenital disease [24]. We then studied the proliferative and myogenic potentials of CSCs by comparing properties of CSCs isolated by three different protocols, c-KIT+ cell sorting, expansion of clonogenic cells, and explant culturing both in vitro and in vivo. Finally, genome-wide expression analysis has been performed to evaluate the changes in transcript levels of CSCs during subsequent passages.
Materials and Methods
Sample sources
This study was performed according to an Institutional Ethical Committee of Royan Institute protocol. Human heart biopsies were obtained from Iranian pediatric patients (n=35) diagnosed with a congenital heart disease, ToF. Patients' parents gave their informed consent for study participation. Patients with cardiac hypertrophic tissues removed by routine heart surgery were included in the study. Biopsies were ∼1 cm3, obtained from surgical waste and removed from the right atria and ventricular myocardial tissues. All patients were 1–10 years of age. Males comprised 71.4% of participants (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/scd).
Isolation of cardiac precursor cells
We used three protocols to isolate the CSCs. In the first protocol we cultured the samples as explants, as described by Messina et al. [12]. Briefly, samples were washed twice with PBS− and 10% penicillin/streptomycin (Gibco; 15070-063), then cut into 1–2 mm3 pieces and treated once with 2 mg/mL trypsin (Gibco; 27250-018) and 1 mg/mL collagenase IV (Gibco; 17104-019) for 5 min. Pieces were cultured as explants on plates coated with 1 mg/mL fibronectin (Sigma-Aldrich; F0635) in cardiac explant medium [IMDM (Sigma-Aldrich; 13390), 10% FBS (Gibco; 10270-106), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM l-glutamine (Gibco; 25030-024), and 0.1 mM 2-mercaptoethanol (Sigma-Aldrich; M7522)]. After 3 weeks small phase-bright cells that had migrated over a layer of fibroblast-like cells were collected by light enzymatic digestion using 0.025% trypsin/EDTA (Gibco; 15400) at room temperature. Cells were cultured to form cardiospheres on poly D-lysine- (Sigma-Aldrich; P0899)-coated plates in cardiosphere medium that contained 35% IMDM/65% DMEM-Ham's F12, 2% B27, 0.1 mM 2-mercaptoethanol, 10 ng/mL EGF, 20 ng/mL bFGF, 40 nM cardiotrophin 1 (R&D Systems; 612-CD), 40 nM thrombin (Sigma-Aldrich; T7572), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine. Cardiospheres were plated on fibronectin-coated plates in CEM medium after 3–5 days to attain adherent growth.
For the second and third protocols, tissue specimens were dissociated by treatment with collagenase I (2 mg/mL; Sigma-Aldrich, C0130), collagenase II (2 mg/mL; Gibco, 17101-015), collagenase IV (1 mg/mL), and trypsin (2 mg/mL; Gibco, 27250-018) for 0.5–1 h at 37°C. The treatment times were gradually increased based on the undigested specimens. Enzymes were neutralized with medium that contained serum. The cell suspension was filtered through a 40 μm filter (BD Falcon; 9340329) and centrifuged at 1,500 rpm for 5 min. Next, the cell pellets were suspended in culture medium and counted. After this step, we used two protocols: clonogenic expansion of cardiac precursor cells [20] and cell sorting by the MACS system. For clonogenic expansion, the cell suspension was cultured in tissue culture flasks to remove the fibroblasts and cardiomyocytes. After 75 min the medium was collected, centrifuged, and counted after suspending in culture medium. The cells were diluted to reach a concentration of 10 cells/mL. Each 100 μL of suspension was added to one well of a fibronectin-coated 96-well plate. After 7–10 days cells that had clonogenic potential grew in the wells.
In the third protocol, cells were sorted by their c-KIT surface antigen using anti-human c-KIT microbead-conjugated antibody (MiltenyiBiotec; 130-091-332) according to the manufacturer's protocol. To characterize isolated cells we analyzed their surface markers by flow cytometry, their doubling time, and mRNA expression (detailed descriptions are provided in Supplementary Methods and Materials section).
In vitro cardiomyogenic differentiation
All three isolated passage 4 CSCs groups were seeded in tissue culture plates for adherent growth at 104 cells in six-well low attached plates (Costar; 3471) for sphere formation. Culture medium was changed with differentiation medium [45% IMDM/55% DMEM, 2% horse serum, 1% ITS (Gibco; 41400-045), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM l-glutamine, and 0.1 mM 2-mercaptoethanol, and cytokines for differentiation] when adherent cells reached 80% confluency and spheres reached an adequate size, after 3 days. To induce differentiation initiation, we tested 5-azacytidine (Santa Cruz; sc221003) in addition to TGF-b (R&D Systems; 240-B) and ascorbic acid (Sigma-Aldrich; A8960) [20]. Briefly, cells were treated with 5 μM 5-azacytidine for 3 days and medium was removed at day 4. TGF-b was added twice weekly, whereas ascorbic acid was added every 2 days. We plated the spheres on gelatin-coated plates after the end of each treatment. The medium was refreshed every 3 days with differentiation medium that contained no cytokines. To characterize the efficiency of cardiomyogenic differentiation, expression of cardiomyocytes structural markers were analyzed in both mRNA and protein levels besides ultra-structural analysis of cell arrangements (detailed descriptions are provided in Supplementary Methods and Materials section).
In vitro multipotency evaluation
To initiate differentiation into osteoblasts, we cultured 104 cells from all isolated passage 4 CSCs groups in 35 cm plates. When cells reached 80% confluency, the culture medium was changed to osteoinductive medium (DMEM-LG, 10% FBS, 10 mM β-glycerophosphate, 100 nM dexamethasone, 50 μg/mL ascorbic acid-2-phosphate, 100 U/mL penicillin, and 100 μg/mL streptomycin) for 21 days [25]. The medium was refreshed every 3 days and calcification was evaluated by Alizarin red staining at the end of the incubation period. For differentiation into adipocytes, cells were cultured in adipogenic differentiating medium (DMEM supplemented with 50 μg/mL ascorbic acid 3-phosphate, 100 nM dexamethasone, and 50 μg/mL indomethacin). Oil red staining was used to show the presence of adipogenic differentiation.
Cells isolated from three different patients were randomly collected and used to assess colony formation potential. Passage 3 cells were harvested and suspended in culture medium after washing with IMDM 2% FBS. A total of 104 cells from each sample were diluted in 200 μL of culture medium and added to 1.8 mL methylcellulose media (1:10). Thoroughly mixed samples were dispensed into one well of six-well culture plates. Spaces that surrounded the wells were filled with sterile water. Cells were allowed to incubate undisturbed at 37°C in a 5% CO2 humidified incubator. Formed colonies were counted after 14–16 days.
Illumina chip microarray analysis
We generated biotin-labeled cRNA from 200 ng total RNA using an Illumina® TotalPrep™ RNA Amplification kit (Applied Biosystems; AMIL1791). IVT reaction was performed overnight and cRNA was biotinylated. RNA and biotinylated cRNA concentrations were checked with a Nanodrop ND-1000 and quality was controlled by an Agilent's Bioanalyzer electrophoresis station. A total of 750 ng from each cRNA sample was hybridized into Illumina's Sentrix Human HT-12 v4 Expression Bead Chips at 58°C overnight according to the “Illumina Whole-Genome Gene Expression Direct Hybridization Assay Guide” (part 11322355, rev.A). Hybridizations were detected with 1 mg/mL cyanine3-streptavidine. Chips were scanned by an Illumina Bead Array Reader (factor=1.5, PMT=552, filter=100%) and the numerical results were extracted with Genome Studio v. 2010.2; Gene Expression Module v. 1.7.0. The Lumi software package was used for raw data background correction and normalization.
Microarray data analysis
Genespring software (version 12) was used for the hierarchical clustering of significantly expressed probes [analysis of variance (ANoVA) P-value <0.05 and fold change (FC)≥2]. For analysis, ANOVA and an arbitrary threshold (FC≥2) were used for the FC to select differentially expressed genes. The transcriptome data were published in the NCBI Gene Expression Omnibus (GEO) database (Acc. ID: GSE43447). The determination of correct number of clusters was based on both probes and replications. The cluster distance metrics were Euclidean and the linkage rule was Centroid. Gene ontology analysis was performed using genes that showed a significant expression change at passage 3 compared with passage 6 and we categorized statistically overrepresented GO-Terms by using BINGO (version 2.44) software with the Benjamini and Hochberg multiple tests and false discovery rate (FDR) multiple test correction. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to extract significant pathways via DAVID Bioinformatics Resources version 6.7 [26].
Myocardial infarction animal model study
An open-chest, ischemia protocol was used to generate an anterior myocardial infarction (MI). Male Wistar rats (250–300 g) were housed and handled in accordance with recommendations stated in the NIH guide for care and use of laboratory animals. To suppress the immune reaction, rats were treated with 210 mg/mL cyclosporine-A 2 days before the surgery, which was added to their drinking water. The immune suppression was continued throughout the experiment. Before surgery, animals were anesthetized with ketamine (100 mg/kg) and xylazine (5 mg/kg), then endotracheally intubated and ventilated (Harvard apparatus). A left-lateral thoracotomy was performed to expose the left anterior descending artery (LAD). The LAD was ligated with 6.0 vicryl sutures. ST-T changes were monitored during this period. Animals were divided into groups of treated or vehicle. Groups received either 1×106 of each of the isolated CSCs subpopulations or PBS, which were injected to the margins and center of the infracted area, respectively. All layers of the muscle and skin were sutured and rats were oxygenated for 2–3 h postsurgery. The animals' hearts were harvested for histological studies 8 weeks later.
Histological studies
Recipient rat hearts were fixed in 4% formaldehyde for 24 h at room temperature and then cut transversely and embedded in paraffin (Merck; 107150). Tissue sections (6 μm thick) were obtained by using a rotary microtome (Microm). We used Masson trichrome (MT) and hematoxylin and eosin (H & E) staining to evaluate the changes in thickness of the infarcted area with light microscopy. To visualize the blood vessels in infarcted area anti alpha smooth muscle actin (α-SMA, 1:100; Abcam, ab7817) antibody was used. Paraffinized sections of rat heart tissue were rinsed with dH2O and treated with antigen retrieval reagent (Dako; s1700). Sections were then stained with primary antibodies and visualized under fluorescence microscope (Olympus; IX71).
Infarction size and blood vessel density measurements
H&E and MT-stained sections that contained the infarct area were used to calculate the infarction middle length [27]. The middle length between the left ventricle (LV) epicardial and endocardial surfaces of five sections that contained infarction per animal was measured by ImageJ software. This length was also measured for their infarction area. The ratio percentage of the sum of all measured infarction middle lengths to the sum of all measured LV middle lengths was determined to be the infarction size. To measure blood vessel density, the number of α-SMA-positive blood vessels was counted for five fields per infarcted section [28]. We collected six slides for each animal and the average number of blood vessels per field for the groups was calculated as the blood vessel density. To normalize the data, we compared the blood vessel density of the transplanted groups to the vehicle group.
Statistical analysis
All data were showed as mean±standard deviation (SD). To analyze the significant differences between study groups, the Student's t-test and one-way ANOVA with Tukey's HSD and LSD post hoc methods were used by SPSS 16.0 software. As a nonparametric method, Kruskal–Wallis test was used. Data with P≤0.05 were determined to be statistically significant.
Results
Phenotype analysis of isolated cells by three different protocols showed cells were cardiac committed, but not fully differentiated with some differences
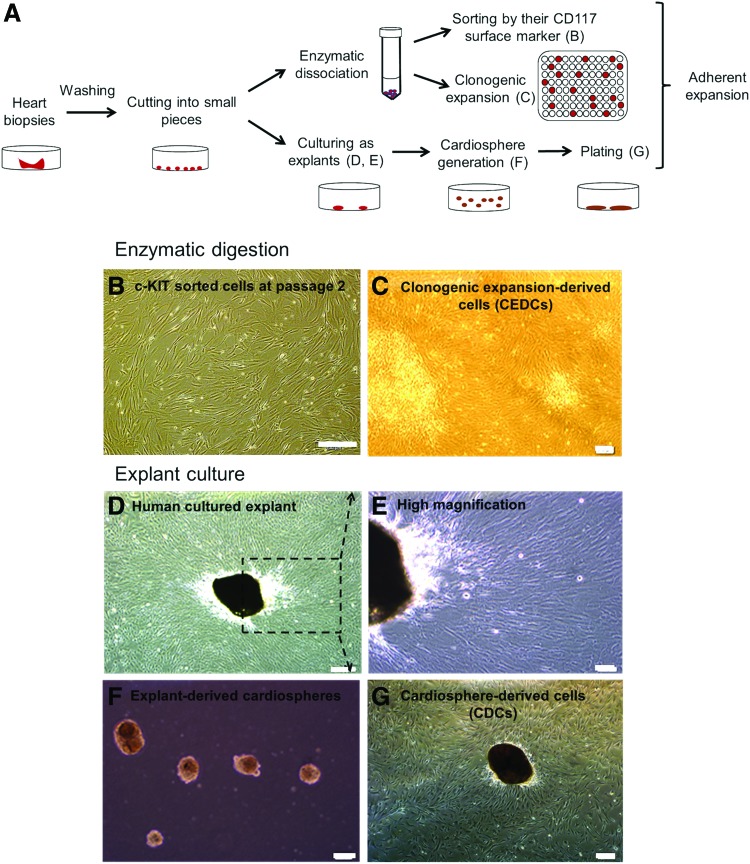
We used three different protocols including c-KIT positive cell sorting, clonogenic expansion, and explants culture to isolate c-KIT+ cells, clonogenic expansion-derived cells (CEDCs), and cardiosphere-derived cells (CDCs), respectively (Fig. 1A). Minced heart tissues were either cultured as explants or underwent enzymatic digestion. The resultant cell suspension was sorted according to the c-KIT surface antigen or cultured as one cell in each well of a 96-well plate (Fig. 1A). Cells in each of the three groups that grew after a 7–10-day lag phase showed fibroblast-like morphologies (Fig. 1B–G).
FIG. 1.
Isolation of cardiac stem cells (CSCs) using three different protocols. (A) Schematic diagram of our laboratory isolation protocol. After mincing, the transferred tissues were divided into two groups. One group was cultured as explants and the second group was enzymatically treated to generate a cell suspension. Digested tissues were either sorted by a MACS system or diluted and cultured at a 1 cell/well density to generate clones. Cultured cells that grew in the wells and generated colonies were pooled together and passaged into a larger plate after colony generation. (B) c-KIT+ cells were sorted by the MACS system at passage 2. (C) CSCs were generated from one cell at passage 2. (D) Human cultured explants on fibronectin coated plates after 10–14 days. A monolayer of fibroblast cells grew from the explants and a number of bright phase cells migrated over the fibroblast cells. (E) Human cultured explants at a higher magnification as indicated by inset in D. (F) Cardio-spheres derived from human explants on poly D-lysine coated plates. (G) Generated cardio-spheres from explants plated on fibronectin-coated plate at day 4. Scale bar=200 μm. Color images available online at www.liebertpub.com/scd
Population doubling time measurement
The measured population doubling time (PDT) for c-KIT+ cells (group 1) was 32.7±4.7 h, for CEDCs (group 2) it was 22.2±1.8 h, and for CDCs (group 3) it was 36.0±4.9 h. The PDT for CEDCs was significantly lower than c-KIT+ and CDCs (P≤0.05). In addition, cell morphology changed into elongated cells with vacuole-shaped cytoplasm in all three groups when the number of passages increased.
Cell surface marker analysis
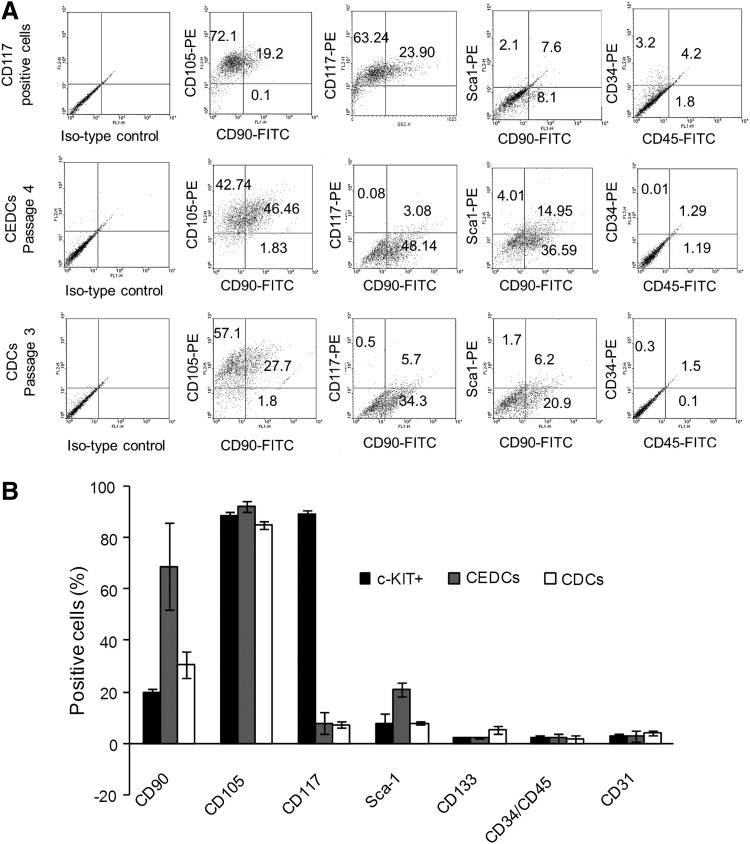
Cells from all three groups were dually stained with three different groups of antibodies: mesenchymal stem cell (MSCs) markers (CD90 and CD105), blood cell and endothelial markers (CD34, CD45, CD133, and CD31), and CSCs markers (CD117 and Sca-1) before flow cytometry analyses. Isolated cells stained positive for CD90 and CD105 and negative for CD34, CD45, CD133, and CD31 (less than 5% expression for each marker) in all three groups. CD117+ sorted cells were more than 89%±2% positive for c-KIT antigen and negative for Sca-1 surface marker, whereas colonies obtained from the clonogenic expansion protocol were 21.0%±2.7% positive for Sca-1. CDCs were positive for both CD117 (7.2%±1.2%) and Sca-1 (7.9%±0.7%) markers (Fig. 2A, B and Supplementary Table S2).
FIG. 2.
Flow cytometry results of isolated cells from all three protocols. (A) Chart of flow cytometry analyses. Expression of mesenchymal, endothelial, hematopoietic and cardiac surface markers is compared. The most important differences between these results are from expressions of c-KIT and Sca-1. (B) Surface marker distribution on congenital patients' derived CSCs isolated from the three different methods.
Gene expression analysis
Gene expression analysis of stemness markers (SOX2, c-KIT, and ABCG2), cardiac transcription factors (ISL-1, NKX2.5, GATA4, and MEF-2c), and cardiac structural genes (α-MHC, β-MHC, and MLC2v) showed that c-KIT+, CEDC, and CDC cells expressed c-KIT, SOX2, ABCG2, GATA4, and MEF-2c but not cardiac structure genes. NKX2.5 and ISL-1 expressions were not detected in any group (Supplementary Table S4).
In vitro differentiation potential assays indicated that all isolated cells were multipotent
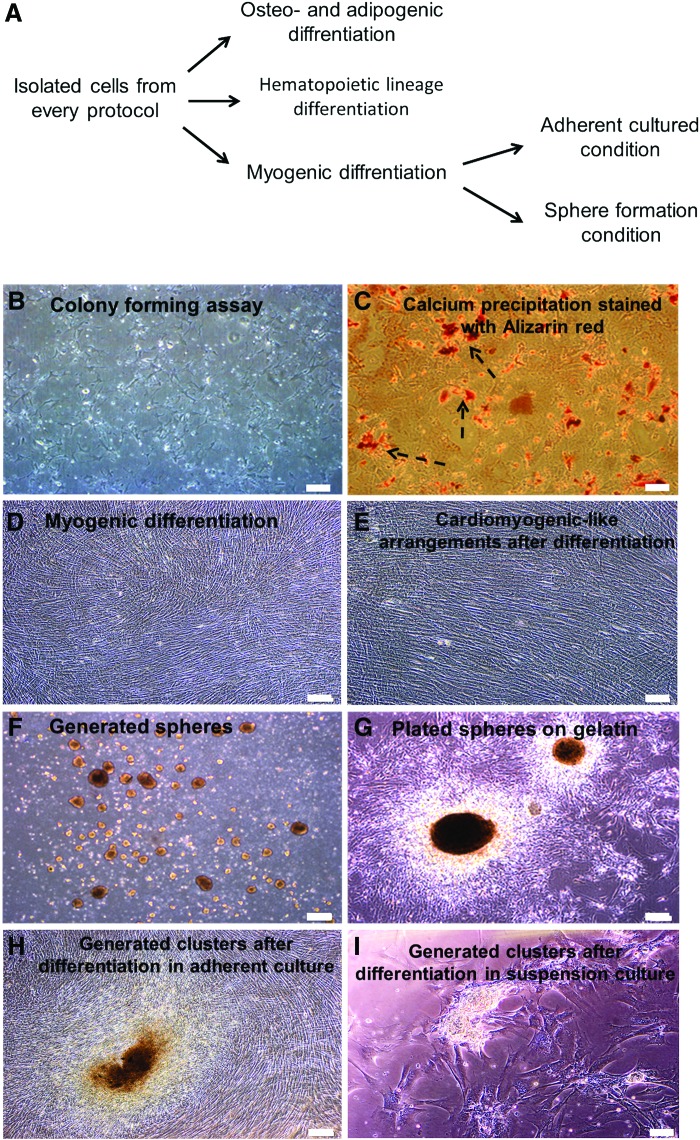
In vitro cardiomyogenic differentiation assay: We used Smits' protocol to assess the potential of isolated cells to differentiate into cardiomyocytes [20]. Isolated cells were cultured under adherent and suspension culture conditions followed by cardiomyogenic induction (Fig. 3A, D, F). At the end of the differentiation period spheres were plated on fibronectin-coated plates (Fig. 3G). Nonbeating cardiomyocyte-like clusters (Fig. 3H, I) and cardiomyocyte-like morphologies (Fig. 3E) were observed in differentiated cells from all three groups after the end of the differentiation period.
FIG. 3.
Differentiation potential assays. (A) A schematic diagram of our laboratory differentiation protocol. To investigate the differentiation potential of isolated cells we examined their potential for osteogenic, myogenic and hematopoietic differentiation. (B) Fibroblast-like colonies generated in methyl cellulose medium. There were no hematopoietic colonies in methyl cellulose medium that optimized for hematopoietic lineage proliferation. (C) Calcium precipitation in cells stained with Alizarin red after osteogenic differentiation (arrows). (D) Multilayer morphology of differentiated adherently cultured cells. (E) Arrangement of cells in the plate into cardiomyocyte-like cells 2 weeks after the end of differentiation. (F) Sphere formation of CSC on ultra-low attachment plates to evaluate the effect of cell-cell communication on cardiogenic differentiation. (G) Spheres plated on gelatin coated plates at the end of the differentiation procedure. (H) Some clusters generated in plates at the end of the differentiation procedure. (I) Generation of clusters after the end of differentiation procedure in plated spheres. Scale bar=200 μm. As the obtained phenotypic results in all three main cell groups (c-KIT+ cells, CEDCs and CDCs) was the same, figures of only c-KIT+ cells are presented. Color images available online at www.liebertpub.com/scd
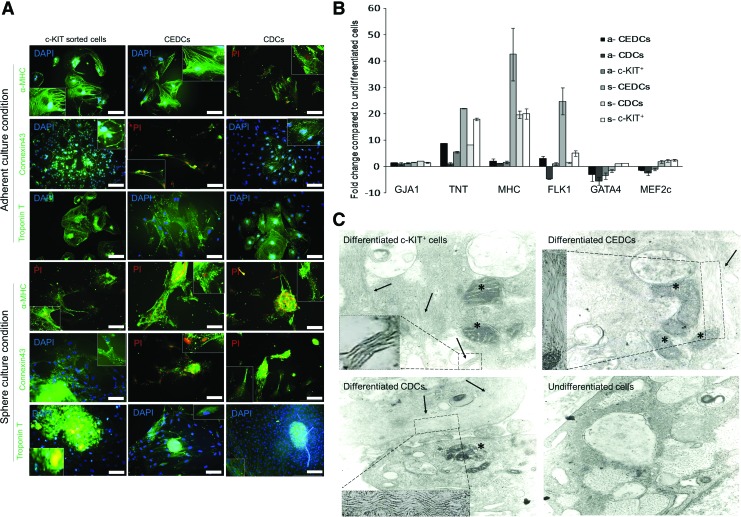
Reverse transcriptase PCR (RT-PCR) analysis of all groups showed that GATA4, NKX2.5, MEF-2c, α- and β-MHC, and MLC-2v expressed after differentiation. Immune-staining results showed production of MHC, CONNEXIN43 (GJA1), and cTnT after treatment with cytokines (Fig. 4A). According to quantitative real-time RT-PCR (qRT-PCR) analysis, there was significantly higher expression of cardiac structural genes under sphere conditions compared with adherent conditions (P≤0.01, Fig. 4B). We observed that cardiac structural genes such as cTnT, GJA1, and α-MHC upregulated through differentiation compared with the undifferentiated control group (P≤0.01). There were no significant differences between the three cell groups in expression of the end stage cardiac marker GJA1. However, α-MHC expressed more in sphere CEDCs (s-CEDCs) compared to the other groups (P≤0.01). The expression of cTnT significantly upregulated in CEDCs compared to c-KIT+ and CDCs (P≤0.01). GATA4 and MEF2c, two cardiac transcription factors, were downregulated after differentiation under adherent conditions and upregulated under sphere conditions with the exception of CEDCs, which showed downregulation for GATA4 (P≤0.01 sphere vs. adherent conditions). Flk-1, a marker for cardiovascular progenitor cells, upregulated in all of the groups except adherent cultured CDCs, which showed downregulation compared with the control group (P≤0.01, Fig. 4B).
FIG. 4.
Characterization of cardiomyogenic differentiation efficiency. (A) Immune-staining was performed for three different CSCs subpopulations, which were differentiated into cardiomyocytes under two different culture conditions. cTnT, α-MHC and Connexin 43 were stained with fluorescent-conjugated antibodies (Scale bar=200 μm). (B) Real time reverse transcriptase PCR analyses for all six differentiation groups. Cardiac structural genes up-regulated after differentiation in all groups, whereas cardiac transcription factors downregulated in the adherent groups and upregulated in the sphere groups after myogenic differentiation. Flk-1 expression was upregulated in all groups except for adherently cultured CDCs. Expression of cardiac structural genes under the sphere condition was significantly higher than the adherent condition (P≤0.05). (C) Transmission electron microscopy results for isolated cells after differentiation. Myofibril polymers are shown with arrows (higher magnification of myofibril polymers are presented in dashes). Their arrangement is not obvious in the pictures. Of note, we observed large mitochondria (stars) in the cytoplasm of differentiated cells, which might be a good sign of cardiomyocyte differentiation of the cells. These changes are compared with undifferentiated control cells. Insets show higher magnifications of endoplasmic reticulum. Color images available online at www.liebertpub.com/scd
The differentiated cells underwent ultra-structure analysis to investigate their cardiac structural arrangements. Electronic microscopy showed myofibril polymerization 4 weeks after differentiation induction but no appropriate arrangement was observed in the cell. In addition, myofibril polymerization was not observed in the control groups (Fig. 4C). Numerous sarcoplasmic reticulum, big mitochondria with densely packed cristae, large accumulations of glycogen, and atrial natriuretic factor granules were observed in the differentiated cells (Fig. 4C).
Multipotency assay for isolated cells
We evaluated the potential for isolated cells to differentiate into adipo- and osteogenic lineages. Culture media were replaced with osteoinductive medium and adipogenic differentiating medium, which were refreshed every 3 days. Calcium precipitation was observed by Alizarin red staining, which suggested the capacity of isolated cells to differentiate into osteoblasts (Fig. 3C). However, no oil vacuoles were observed after adipogenic differentiation (data not shown).
Hematopoietic lineage differentiation assay
Flow cytometric analysis of isolated cells showed that they did not express hematopoietic markers. Their culture in methylcellulose also confirmed that these cells did not belong to a hematopoietic lineage. After 14 days in culture medium, only mesenchymal-like cells grew in all of the plates; we observed no hematopoietic line colonies (Fig. 3B).
Genome-wide expression analysis revealed upregulation of developmental, metastatic, and cancer-related genes and downregulation of cell cycle-related genes over passaging
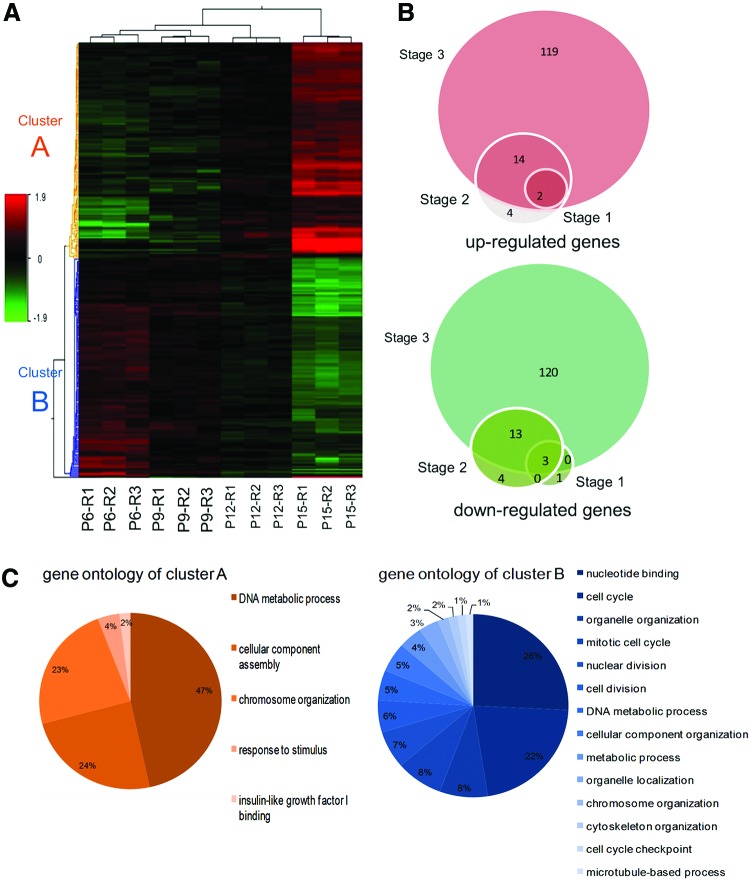
We investigated the effects of passaging on expression profiles of CSCs by analyzing CEDCs' transcriptome at passages 6, 9, 12, and 15, before the cells' morphological changes were visible. Three biological replicates were used for each of the four passages to determine gene expression analysis with microarray. Our results indicated that 280 distinct transcripts showed statistically significant differential expression (P<0.05 and FC≥2) in at least one passage compared with passage 6. Based on their expression pattern, the differentially expressed transcripts were categorized into two distinct groups: (1) 92 unique transcripts, which showed no expression in passage 6 and consequently were upregulated through the passages and (2) the expression level of 142 unique transcripts that decreased after subsequent passages compared to passage 6 (Fig. 5A). Overall, we observed upregulation in 2, 20, and 135 transcripts and downregulation in 4, 20, and 136 transcripts in passages 9, 12, and 15 compared to passage 6 (Fig. 5B).
FIG. 5.
Whole genome expression analysis using microarray technology. (A) Hierarchical clustering of 280 differentially expressed genes was performed using the median signal intensity for each replicate. Three biological replicates of cells in passages 6, 9, 12 and 15 were compared and showed high intraclass correlations compared with interclass correlations. Two distinct clusters were distinguishable based on the expression patterns of cells, including transcripts that were up-regulated through passages that were highly expressed in passage 15 (cluster A) and transcripts downregulated by increasing the passages (cluster B). (B) Venn diagram showing the number of up- and downregulated genes in passages 9 (stage 1), 12 (stage 2) and 15 (stage 3) compared with passage 6. The number of significantly changed transcripts (P≤0.05, FC≥2) through subsequent passages is increased. (C) Functional classification of differentially expressed transcript was performed using BINGO software. It was shown that the main significant gene ontology of up-regulated transcripts (cluster A) was the DNA metabolic process while they were nucleotide binding and cell cycle for downregulated transcripts (cluster B). Color images available online at www.liebertpub.com/scd
Verification of differentially expressed mRNA
To corroborate the microarray results, qRT-PCR analysis was performed on ten differentially expressed genes. We included six upregulated (IGFBP2, MYH11, ECM1, ANGPTL4, PLAU, and CCND2) and four downregulated genes (UBE2C, ID1, CDC20, and PTTG1). In 9 out of 10 reactions, the microarray-derived differential expression was confirmed at a 90% confidence level. In one case, PTTG1, downregulation of mRNA at passage 15 that was detected by microarray could not be confirmed by qRT-PCR (Supplementary Fig. S1).
Functional analysis of differentially expressed genes
We utilized BINGO (version 2.44), a plugin of Cytoscape (using multiple test correction FDR and a P-value of 0.05), to analyze gene ontology of the differentially expressed genes. Upregulated genes (cluster A) were mainly enriched for genes involved in the DNA metabolic process and cellular component assembly ontology while downregulated transcripts (cluster B) enriched for genes involved in cell cycle phases (cell cycle, mitotic cell cycle, nuclear and cell division, and cell cycle checkpoints) (Fig. 5C and Supplementary Fig. S2). Total significantly changed gene ontology for both A and B clusters are brought in Supplementary Fig. S3, which are classified as biological process, cellular component, and molecular function.
Involved pathways and gene regulatory network
KEGG analysis also showed six significant pathways that included the cell cycle, PPAR and p53 signaling pathways, complement and coagulation cascades, systemic lupus erythematosus, and biosynthesis of unsaturated fatty acids (Supplementary Figs S4 and S5).
String data base was used to create interaction regulatory network for genes, which expressed differentially in three stages. The string data base detected 230 genes from 280 transcripts and made a network with 50 unique genes and 182 interactions for the beginning. Next, the network was filtered based on experiments, databases, and texts. This resulted in a network with 34 genes and 43 interactions. We categorized 16 out of 34 genes as cell cycle-related genes at different stages such as interphase and mitosis. A total of 14 genes out of these 16 transcripts were downregulated through passages. Overall, nine genes of the network were upregulated through passages (Supplementary Fig. S6).
In vivo functionality assay unveiled more regenerative potential of c-KIT+ and CDCs compared with CEDCs
The influence of cell transplantation on fibrosis attenuation
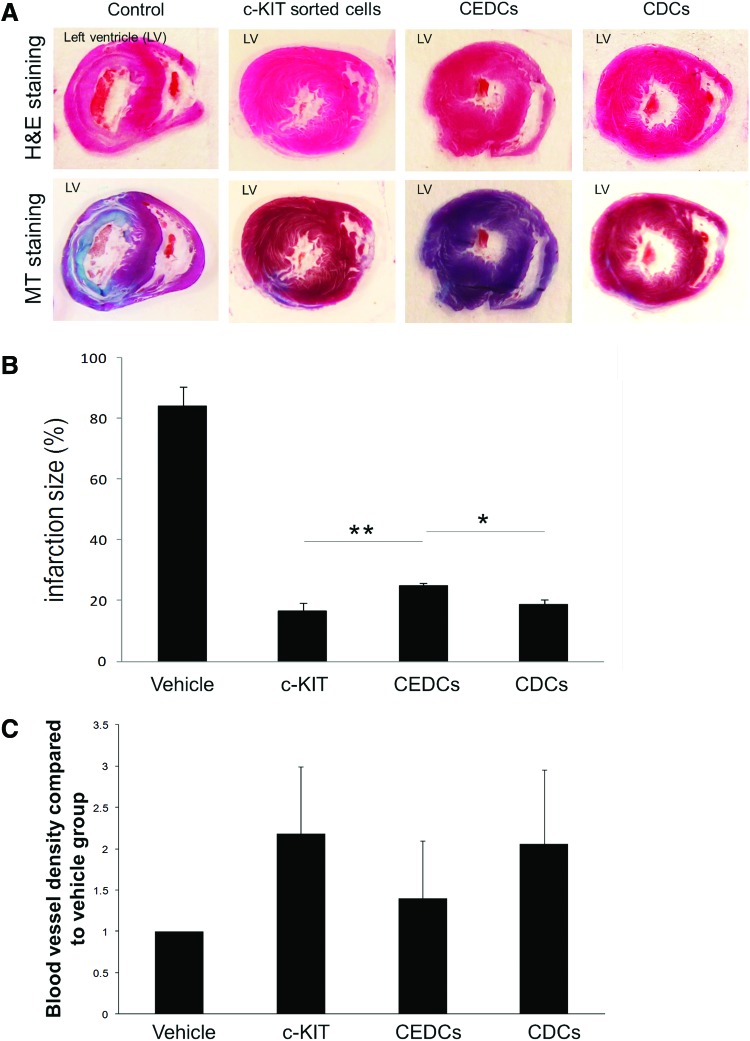
To determine whether isolated cells have cardiac regeneration abilities, the isolated cell groups (c-KIT+, CEDCs, and CDCs) were transplanted into infarcted area of rat MI model. Eight weeks after cell transplantation, histological studies of the rats' hearts showed attenuation of the damaged area (fibrosis) and LV wall thickening compared with the vehicle groups (Fig. 6A and Supplementary Fig. S7). Quantitative analysis of the infarction size in transplanted and vehicle groups using infarction middle length measurement showed significant attenuation of fibrosis up to five-fold in the treated groups compared with the vehicle group (P≤0.01, Fig. 6B). The obtained infarct size in vehicle group was 84.3%±6.4%. This size was calculated as 16.6%±2.7% for the c-KIT+ transplanted group, 24.8%±1.1% for CEDCs transplanted group, and 18.6%±1.7% for the CDCs transplanted group. There was a significantly higher decrease in infarction size in the c-KIT+ and CDCs groups compared with the CEDCs-treated group (P≤0.05).
FIG. 6.
Evaluation of the effect of CSCs transplantation on infarct size. (A) Hematoxylin and eosin (H&E) and Masson's trichrome (MT) staining of infracted rat hearts 8 weeks after transplantation showed attenuation of infarction size in the left ventricle (LV) of all three transplanted groups compared to the control groups. (B) Quantification of infarction size using H&E and MT stained sections showed a significant decrease in infarction length 8 weeks after transplantation in the treated groups compared to the vehicle group (P≤0.01 transplanted groups vs. vehicle group). Attenuation in c-KIT+ cells and CDC groups were significantly higher than the clonogenic expansion-derived cell (CEDC) group (P≤0.05). (C) Quantification of the blood vessel density and normalization of the transplanted group compared to the vehicle group (average of αSMA-positive blood vessels counted in five fields of six sections with infarction for each transplanted animal compared to the vehicle group; P≤0.05). As the chart shows the blood vessel density of c-KIT+ and CDCs transplanted groups were significantly higher than the CEDCs transplanted group (P≤0.05). The data were analyzed using Kruskal–Wallis test a nonparametric statistical method. *: P<0.05, **: P<0.01. Color images available online at www.liebertpub.com/scd
Blood vessel density measurement
To estimate the regenerative potential of transplanted cells in the infarcted area, the measured blood vessel densities of each transplanted group that were positive for αSMA were normalized to the vehicle group (P≤0.05) (Supplementary Fig. S8). Figure 6C shows that blood vessel densities similarly increased in transplanted c-KIT+ (2.2±0.8-fold) and CDCs (2.1±0.8-fold) compared with the vehicle group, whereas the blood vessel density in the CEDCs group increased 1.4±0.7-fold compared with the vehicle group (P≤0.05 c-KIT+ and CDCs vs. CEDCs).
Discussion
Exploration of CSCs in adult mammalian hearts has challenged opinion that the heart was considered a postmitotic organ without any internal regeneration potential [29]. In the last decade numerous studies have characterized different CSC subpopulations and evaluated their molecular and functional properties. The first isolated CSCs subpopulation in the rat heart was c-KIT+ whose regenerative potential has been proven [30]. Recently, the safety and efficiency of c-KIT+ administration has been studied in SCIPIO, a clinical trial [6]. Another CSCs population with high cardiogenic potential are cardiospheres. Their safety and efficiency has been confirmed in the CADUCEUS study [5]. Sca-1-expressing cells are another population of CSCs initially discovered in mice. Cells that express the Sca-1-like epitope exist in the human heart.
Various methods have been described to isolate and characterize CSCs. Each focuses on one CSC population, confirming its regenerative potential as the best cell therapy source. To the best of our knowledge this study, for the first time, has compared three subpopulations of human CSCs isolated from three common protocols and evaluated the effects of subsequent passages on whole genome expression. We showed that CSCs exist in ToF patients. These CSCs had cardiomyogenic potential. Our findings demonstrated that although CEDCs needed lower subsequent passages to reach a definite cell count they were more committed into cardiomyocytes. However, c-KIT+ and CDCs showed higher cardiogenic potential than CEDCs in vivo. We determined that subsequent passages caused changes in cellular characteristics. Hence, we have proposed that the cell culture period should be decreased to have safe cell populations.
Myocardial tissues were removed from children during surgeries and used as cardiac precursor cell sources. Expression of cardiac stemness surface markers in freshly obtained cell suspension from whole enzymatic digestion of cardiac tissue samples showed the presence of CSCs in the right atrium and ventricle biopsies of patients. In most studies, existence and characteristics of human CSCs have been evaluated in normal and diseased heart tissues. It is shown that the number of CSCs increases immediately after MI compared with healthy tissues and falls down in chronic stage. However, ToF is a congenital heart defect with anatomical abnormalities and the results of ischemic heart diseases cannot be generalized to it; so, identification of CSCs in congenital heart patients is needed. Previously, isolation of CSCs from the right atrium of human neonates, infants, and young children's heart biopsies with congenital diseases was performed by an explant culture. It reported that the number of CSCs changed during postnatal maturation with the most abundance in neonatal period. Isolated cells had regenerative potential and they could be used as autologous cell sources for cell therapy of heart patients [8].
To find the best subpopulation of CSCs suitable for clinical uses, we have attempted to research the three common protocols, which are based on different properties of cardiac precursor cells. CSCs have the potential to colonize a stem cell colony and the capability to migrate from a small tissue sample and be sorted according to their surface stemness markers. Expression of cardiac transcription factors and stemness markers has shown that all isolated cells were committed CSCs that did not express cardiac structural genes. Cell surface antigen analysis showed that most CEDCs were Sca-1+, whereas CDCs were included in both Sca-1+ and c-KIT+ CSCs groups. Our findings confirmed previous studies, which reported that Sca-1+ cells had clonogenic potential [31,32] and CDCs were included in both subgroups of CSCs [12,16]. Until now the most important difference of isolated cells was based on expression of surface markers that included CD117 and Sca-1. PDT measurement showed that CEDCs divided significantly faster than the other two populations.
We evaluated the myogenic differentiation potential of the isolated cells in two different culture manners, adherent, and sphere. Since the generation of spheres imitates the heart's three-dimensional niche condition [33], we have attempted to evaluate the effect of sphere generation on myogenic differentiation. We hypothesized that cell–cell communication might be an advantage for myogenic differentiation, so we cultured isolated cells in suspension media and allowed the cells to grow under sphere conditions. There were no beating areas in the adherent or sphere conditions. Analysis of cells after differentiation at the molecular, protein and ultra-structure levels suggested that their differentiation into cardiomyocytes was done with functionality. Observation of beating colonies in differentiated human CSCs using cytokines has not been reported. Cardiac structural genes, proteins and polymerization of myofibrils, all signs of cardiomyocytes, were obvious in the differentiated cells and confirmed isolated cells' myogenic potential (Fig. 4). Significantly higher expression of cardiac structural genes in sphere conditions than adherent conditions conferred that the sphere condition could improve cardiomyogenic differentiation of cardiac precursor cells as we have previously hypothesized. Downregulation of cardiac transcription factors under adherent conditions showed transition of isolated cells from their precursor state to a differentiated mode, but higher expression of these genes under sphere conditions could be the existence of different cell types in different developmental stages. Thus, due to the presence of cells in other differentiation levels, cardiomyocyte purity of sphere condition was lower than under adherent conditions. We could consider that cells under sphere conditions received different cell signals than the cells under adherent conditions. By considering the advantages and disadvantages of our sphere culture condition, this new protocol could be recommended as an optional myogenic differentiation method.
It was important to know whether isolated cells were committed and differentiated only into cardiomyocytes or if they were multi-potent. It has been frequently reported that CSCs are multi-potent [32]; our differentiation findings also confirmed this potential (Fig. 3). We did not detect any hematopoietic line colonies after the isolated cells were cultured in methyl cellulose medium; the grown colonies only included fibroblast-like cells (Fig. 3). This observation rejected the relation of isolated cells to blood cells. In addition it has been reported that cell colonies with mesenchymal morphologies would be generated when mouse CSCs were cultured in colony forming unit-fibroblasts. Analysis of the generated colonies revealed that they expressed surface mesenchymal markers and had a broad range of differentiation potential into cardiomyocytes, smooth muscle cells, endothelial cells, adipocytes and bone and cartilage cells [34]. Our results supported previous findings.
To evaluate and compare the regenerative abilities of the three isolated CSC subpopulations, we transplanted them into an infarct area of a rat MI model. Infarct size measurement showed that all cell groups caused attenuation in fibrosis size, but c-KIT+ and CDCs had significantly higher scar improvement than CEDCs. Measured blood vessel density in the infarction area also confirmed this finding (Fig. 6). It should be noted that several reports have considered that the beneficial action of transplanted CSCs (c-KIT+, Sca-1+ and CDCs) is sometimes because of secretion of paracrine factors and activation of endogenous pool of CSCs. Also, CSCs subpopulations express receptors of paracrine ligands that explain activation of endogenous CSCs compartment in response to stem cell transplantation including MSCs [35–41]. It has also been shown that in short-term follow-up after transplantation, the observed improvement in heart function is most because of paracrine effects and is independent of cardiomyogenic differentiation [42]. So, it will be important to further evaluate and compare the paracrine effects of these patients-derived CSCs groups. A newly published article determined the effective events (except transplanted cells differentiation) that improve infarcted heart function including reduction in cell apoptosis [43]. These phenomenons besides to engraftment and differentiation of transplanted CSCs make them more suitable for improvement of MI. Overall, regarding in vitro cardiomyogenic and in vivo cell transplantation results, an important point was noted. CEDCs expressed cardiac structural genes significantly higher than c-KIT+ and CDCs after differentiation, but their infarct area regeneration was significantly lower. A newly published article in which the molecular differences of three mouse CSCs subpopulations was studied by microarray technology demonstrated that c-KIT+ cells were more primitive than Sca-1+ and side population cells [44]. It can be explained that besides to cardiomyogenic potential, other cells properties including paracrine activity, incorporation into blood vessels and differentiation potential into all cardiac lineages are as effective factors.
CEDCs were more cardiomyogenic and they needed fewer passages to reach definite cell counts. c-KIT+ and CDCs had higher regeneration potential, yet they needed additional subsequent passages. Therefore, we used microarray technology to evaluate the effect of passages on genome-wide expression of the cells in four different passages (6, 9, 12 and 15). As we aimed to evaluate only the effects of passages on cells' transcription pattern, it was sufficient to study only one cell group; so, CEDCs were analyzed by microarray technology. Our results showed that the number of differentially expressed transcripts increased by passaging. Based on gene ontology analysis results, cell cycle, differentiation, and specification were major groups of down- and upregulated transcripts, respectively (Supplementary Figs. S2 and S3 part A). An overall view of Supplementary Tables S7 and S9 showed that the transcripts with the highest expression in passage 15 that were categorized in developmental process ontology were mostly for muscle and vasculature development. However, highly downregulated transcripts were mainly associated to cell cycle (Supplementary Tables S8 and S10 and Supplementary Fig. S2). Passaging mostly affected the cell cycle process and induced senescence in the cells. Downregulation of cell cycle checkpoint genes after subsequent passages caused no regulation of the cell cycle phases. In a similar experiment, microarray analysis of mouse cultured bone marrow MSCs at three different ages showed that aging caused reductions in the cell cycle, growth factor and also apoptotic gene expression and differentiation potential. This might suggest that MSCs lost some of their properties due to aging. It was reported that these changes were stable even after subsequent passages [22]. In addition, another study investigated and analyzed the mechanism of porcine coronary endothelial cell dysfunction after passages 1 and 4. Whole genome expression analysis revealed that passages caused senescence in the cells by generating oxidative stress and reduction in active oxygen metabolites [23].
We observed remarkable increase in the expression of several transcripts including myosin, heavy chain 11 smooth muscle (MYH11), insulin-like growth factor binding protein family (IGFBP2 and IGFBP5) and cyclin D2 (CCND2) through passaging. Among highly downregulated transcripts, there were inhibitors of DNA binding 1 (ID1), cell division cycle 20 (CDC20) and ubiquitin-conjugating enzyme E2C (UBE2C). MYH11 and its isoforms are differentially expressed during muscle cell maturation [45]. It has frequently been reported that CSCs are committed to all cardiac lineages (cardiomyocytes, endothelial, and smooth muscle cells) [29,32,46,47]. The upregulation of MYH11 might suggest spontaneous differentiation of cells into smooth muscle cells (one of the cell lines that CSCs are committed to) through passages. It was shown that members of IGFBP family such as IGFBP4 and IGFBP3, which are independent of binding to insulin growth factor, are involved in the processes of cardiogenesis [48–50]. A new subset of cardiac precursor cells that expressed insulin-like growth factor 1 receptor (IGF-1R) was found to have higher proliferation and myogenic differentiation potential compared with other CSCs subgroups [51]. Overexpression of IGFBP2, which was highly expressed in our microarray results, has been reported in common brain tumors. [52]. Higher expression of IGFBP family members after cell passages might suggest the induction of differentiation, commitment, and tumorigenecity of the cells during subsequent passaging. The ID family (ID1 to ID4) has been shown to regulate cell proliferation, differentiation, and control of the cell cycle progression in numerous cell types. This family showed high expression in proliferative cells and a decline in expression in cellular differentiation and senescence. Loss of ID family function, especially ID1, has been associated with replicative senescence [53,54]. The most important signaling pathways in this regard are p16 INK4A/pRB and p53; their inactivation is required for overriding senescence [53,54]. Depending on the cell condition, ID1 has different functions. It has been considered that ID1 has a negative effect on cell differentiation. Recently, it has been shown that its upregulation can regulate cell differentiation including cardiogenesis, which plays an important role in cardiac development [55,56]. Although downregulation of ID1 in addition to other cell cycle-related genes such as UBE2C and CDC20 after passages causes cell senescence some undesired changes in cardiogenic properties of the cells may be induced by downregulation of the ID1 transcript.
We observed the upregulation of cyclinD2 and its cyclin-dependent kinase (CDK6) in passage 15 compared with passage 6. Inhibition of cyclinD2/CDK6 complex causes G1 cell cycle arrest due to stress signals involved in both cell cycle and P53 signaling pathways (Supplementary Fig. S5). Another involved altered gene in the P53 signaling pathway was PERP, which was upregulated through passages. Expression of PERP causes apoptosis in cells. On the other hand, the gene regulatory network study showed that most of the involved downregulated transcripts were related to the cell cycle, whereas a number of important transcripts were upregulated (Supplementary Fig. S6). According to reports, oxidized low density lipoprotein (lectin-like) receptor 1 (OLR1) is involved in the regulation of Fas-induced apoptosis [57]. It has been shown that matrix metalloproteinase1 (MMP1), which is involved in the breakdown of extracellular matrix proteins, plays a role in tissue remodeling and metastasis [58–60] through collagen destruction, which causes progression in cardiac dysfunction [61]. The role of plasminogen activator urokinase (PLAU) and plasminogen activator tissue (PLAT) in extracellular matrix degradation and possible tumor cell migration and proliferation has also been highlighted [62–65]. ell cycle-related genes were downregulated through subsequent passages. It could be proposed that the cells entered into the senescence phase, the upregulation of some metastatic and cancer-related genes led us to hypothesis whether cell characteristics might change with increased passaging. In addition, downregulation of other genes such as secreted frizzled-related protein 1 (SFRP1), survivin (BIRC5), and BUB1, which play a role in regulation of the Wnt signaling pathway [66,67], inhibition of apoptosis [68–70], and regulation of spindle check point proteins [71–73], respectively, have supported this hypothesis. Upregulation of some developmental, metastatic, and cancer-related genes in addition to downregulation of cell cycle-related genes, which was observed in our microarray results, indicated the distinct properties of the cells in higher passages compared to those in low passages.
Conclusion
This study attempted to compare three different subpopulations of human CSCs (c-KIT+, CEDCs, and CDCs) based on their functional properties. Evaluation of isolated CSC characteristics in vitro and after MI model transplantation revealed that although c-KIT+ and CDCs had higher MI regenerative potential, CEDCs had more commitment into cardiomyocytes and needed lower passages that were essential to reach a definite cell count. On the other hand our genome-wide expression analysis showed that subsequent passages caused changes in characteristics of the cells, downregulation of the cell cycle-related genes, and upregulation of differentiation and carcinogenic genes, which led to senescence, commitment, and possible tumorigenicity of the cells. Our data suggested that using CSCs before passage 9 might be safe for clinical applications. We also noticed that a small myocardial biopsy (3–5 mm3) was sufficient to isolate CSCs by the explants culture method without any need for enzymatic treatments. However, for whole tissue enzymatic digestion, a greater amount of cardiac tissue is needed. Our results showed that each group has its own properties; therefore, the appropriate CSCs subpopulation should be selected based on their intended use, either experimental or clinical.
Supplementary Material
Acknowledgment
This study was supported by a grant provided from Royan Institute. The authors would like to appreciate Mina Abrari, Vahid Hajhosseini, Fahimeh Kashfi, Dr. Niloofar Sodeify, Razieh Karamzedeh, and Ehsan Janzamin for their technical assistance.
Author Disclosure Statement
The authors have declared there is no competing financial interest.
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, et al. , (2012). Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finegold JA, Asaria P. and Francis DP. (2013). Mortality from ischaemic heart disease by country, region, and age: statistics from World Health Organisation and United Nations. Int J Cardiol 168:934–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith RR, Barile L, Messina E. and Marban E. (2008). Stem cells in the heart: what's the buzz all about?. Part 2: arrhythmic risks and clinical studies. Heart Rhythm 5:880–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Files MD. and Boucek RJ. (2012). ‘Shovel-Ready’ applications of stem cell advances for pediatric heart disease. Curr Opin Pediatr 24:577–583 [DOI] [PubMed] [Google Scholar]
- 5.Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, Berman D, Czer LS, Marban L, Mendizabal A, et al. , (2012). Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet 379:895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolli R, Chugh AR, D'Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, et al. , (2011). Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet 378:1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Zhao X. and Huang L. (2013). Cardiac stem cells: a promising treatment option for heart failure. Exp Ther Med 5:379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mishra R, Vijayan K, Colletti EJ, Harrington DA, Matthiesen TS, Simpson D, Goh SK, Walker BL, Almeida-Porada G, et al. , (2011). Characterization and functionality of cardiac progenitor cells in congenital heart patients. Circulation 123:364–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith RR, Barile L, Messina E. and Marban E. (2008). Stem cells in the heart: what's the buzz all about?.—Part 1: preclinical considerations. Heart Rhythm 5:749–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, et al. , (2003). Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114:763–776 [DOI] [PubMed] [Google Scholar]
- 11.Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, Pocius J, Michael LH, Behringer RR, et al. , (2003). Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci U S A 100:12313–12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, Fiordaliso F, Salio M, Battaglia M, Latronico MV, et al. , (2004). Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res 95:911–921 [DOI] [PubMed] [Google Scholar]
- 13.Bearzi C, Rota M, Hosoda T, Tillmanns J, Nascimbene A, De Angelis A, Yasuzawa-Amano S, Trofimova I, Siggins RW, et al. , (2007). Human cardiac stem cells. Proc Natl Acad Sci U S A 104:14068–14073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaruba MM, Soonpaa M, Reuter S. and Field LJ. (2010). Cardiomyogenic potential of C-kit(+)-expressing cells derived from neonatal and adult mouse hearts. Circulation 121:1992–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Amario D, Fiorini C, Campbell PM, Goichberg P, Sanada F, Zheng H, Hosoda T, Rota M, Connell JM, et al. , (2011). Functionally competent cardiac stem cells can be isolated from endomyocardial biopsies of patients with advanced cardiomyopathies. Circ Res 108:857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang YL, Shen L, Qian K. and Phillips MI. (2007). A novel two-step procedure to expand cardiac Sca-1+ cells clonally. Biochem Biophys Res Commun 359:877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis DR, Kizana E, Terrovitis J, Barth AS, Zhang Y, Smith RR, Miake J. and Marban E. (2010). Isolation and expansion of functionally-competent cardiac progenitor cells directly from heart biopsies. J Mol Cell Cardiol 49:312–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carr CA, Stuckey DJ, Tan JJ, Tan SC, Gomes RS, Camelliti P, Messina E, Giacomello A, Ellison GM. and Clarke K. (2011). Cardiosphere-derived cells improve function in the infarcted rat heart for at least 16 weeks—an MRI study. PLoS One 6:e25669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye J, Boyle A, Shih H, Sievers RE, Zhang Y, Prasad M, Su H, Zhou Y, Grossman W, Bernstein HS. and Yeghiazarians Y. (2012). Sca-1+ cardiosphere-derived cells are enriched for Isl1-expressing cardiac precursors and improve cardiac function after myocardial injury. PLoS One 7:e30329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smits AM, van Vliet P, Metz CH, Korfage T, Sluijter JP, Doevendans PA. and Goumans MJ. (2009). Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and pathophysiology. Nat Protoc 4:232–243 [DOI] [PubMed] [Google Scholar]
- 21.Ellison GM, Nadal-Ginard B. and Torella D. (2012). Optimizing cardiac repair and regeneration through activation of the endogenous cardiac stem cell compartment. J Cardiovasc Transl Res 5:667–677 [DOI] [PubMed] [Google Scholar]
- 22.Wilson A, Shehadeh LA, Yu H. and Webster KA. (2010). Age-related molecular genetic changes of murine bone marrow mesenchymal stem cells. BMC Genomics 11:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee MY, Wang Y. and Vanhoutte PM. (2010). Senescence of cultured porcine coronary arterial endothelial cells is associated with accelerated oxidative stress and activation of NFkB. J Vasc Res 47:287–298 [DOI] [PubMed] [Google Scholar]
- 24.Apitz C, Webb GD. and Redington AN. (2009). Tetralogy of Fallot. Lancet 374:1462–1471 [DOI] [PubMed] [Google Scholar]
- 25.Matsuura K, Nagai T, Nishigaki N, Oyama T, Nishi J, Wada H, Sano M, Toko H, Akazawa H, et al. , (2004). Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. J Biol Chem 279:11384–11391 [DOI] [PubMed] [Google Scholar]
- 26.Fathi A, Hatami M, Hajihosseini V, Fattahi F, Kiani S, Baharvand H. and Salekdeh GH. (2011). Comprehensive gene expression analysis of human embryonic stem cells during differentiation into neural cells. PLoS One 6:e22856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takagawa J, Zhang Y, Wong ML, Sievers RE, Kapasi NK, Wang Y, Yeghiazarians Y, Lee RJ, Grossman W. and Springer ML. (2007). Myocardial infarct size measurement in the mouse chronic infarction model: comparison of area- and length-based approaches. J Appl Physiol 102:2104–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai W, Hale SL, Martin BJ, Kuang JQ, Dow JS, Wold LE. and Kloner RA. (2005). Allogeneic mesenchymal stem cell transplantation in postinfarcted rat myocardium: short- and long-term effects. Circulation 112:214–223 [DOI] [PubMed] [Google Scholar]
- 29.Barile L, Messina E, Giacomello A. and Marban E. (2007). Endogenous cardiac stem cells. Prog Cardiovasc Dis 50:31–48 [DOI] [PubMed] [Google Scholar]
- 30.Leri A, Kajstura J. and Anversa P. (2005). Cardiac stem cells and mechanisms of myocardial regeneration. Physiol Rev 85:1373–1416 [DOI] [PubMed] [Google Scholar]
- 31.Takamiya M, Haider KH. and Ashraf M. (2011). Identification and characterization of a novel multipotent sub-population of Sca-1(+) cardiac progenitor cells for myocardial regeneration. PLoS One 6:e25265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Nardo P, Forte G, Ahluwalia A. and Minieri M. (2010). Cardiac progenitor cells: potency and control. J Cell Physiol 224:590–600 [DOI] [PubMed] [Google Scholar]
- 33.Li TS, Cheng K, Lee ST, Matsushita S, Davis D, Malliaras K, Zhang Y, Matsushita N, Smith RR. and Marban E. (2010). Cardiospheres recapitulate a niche-like microenvironment rich in stemness and cell-matrix interactions, rationalizing their enhanced functional potency for myocardial repair. Stem Cells 28:2088–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chong JJ, Chandrakanthan V, Xaymardan M, Asli NS, Li J, Ahmed I, Heffernan C, Menon MK, Scarlett CJ, et al. , (2011). Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell 9:527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chimenti I, Smith RR, Li TS, Gerstenblith G, Messina E, Giacomello A. and Marban E. (2010). Relative roles of direct regeneration versus paracrine effects of human cardiosphere-derived cells transplanted into infarcted mice. Circ Res 106:971–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang XL, Rokosh G, Sanganalmath SK, Yuan F, Sato H, Mu J, Dai S, Li C, Chen N, et al. , (2010). Intracoronary administration of cardiac progenitor cells alleviates left ventricular dysfunction in rats with a 30-day-old infarction. Circulation 121:293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li TS, Cheng K, Malliaras K, Smith RR, Zhang Y, Sun B, Matsushita N, Blusztajn A, Terrovitis J, et al. , (2012). Direct comparison of different stem cell types and subpopulations reveals superior paracrine potency and myocardial repair efficacy with cardiosphere-derived cells. J Am Coll Cardiol 59:942–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith RR, Barile L, Cho HC, Leppo MK, Hare JM, Messina E, Giacomello A, Abraham MR. and Marban E. (2007). Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation 115:896–908 [DOI] [PubMed] [Google Scholar]
- 39.Huang C, Gu H, Yu Q, Manukyan MC, Poynter JA. and Wang M. (2011). Sca-1+ cardiac stem cells mediate acute cardioprotection via paracrine factor SDF-1 following myocardial ischemia/reperfusion. PLoS One 6:e29246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuentes T. and Kearns-Jonker M. (2013). Endogenous cardiac stem cells for the treatment of heart failure. Stem Cells Cloning 6:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nadal-Ginard B, Ellison GM. and Torella D. (2014). The cardiac stem cell compartment is indispensable for myocardial cell homeostasis, repair and regeneration in the adult. Stem Cell Res 13:615–630 [DOI] [PubMed] [Google Scholar]
- 42.den Haan MC, Grauss RW, Smits AM, Winter EM, van Tuyn J, Pijnappels DA, Steendijk P, Gittenberger-De Groot AC, van der Laarse A, et al. , (2012). Cardiomyogenic differentiation-independent improvement of cardiac function by human cardiomyocyte progenitor cell injection in ischaemic mouse hearts. J Cell Mol Med 16:1508–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye L, Chang YH, Xiong Q, Zhang P, Zhang L, Somasundaram P, Lepley M, Swingen C, Su L, et al. , (2014). Cardiac repair in a porcine model of acute myocardial infarction with human induced pluripotent stem cell-derived cardiovascular cells. Cell Stem Cell 15:750–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dey D, Han L, Bauer M, Sanada F, Oikonomopoulos A, Hosoda T, Unno K, De Almeida P, Leri A. and Wu JC. (2013). Dissecting the molecular relationship among various cardiogenic progenitor cells. Circ Res 112:1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Babu GJ, Warshaw DM. and Periasamy M. (2000). Smooth muscle myosin heavy chain isoforms and their role in muscle physiology. Microsc Res Tech 50:532–540 [DOI] [PubMed] [Google Scholar]
- 46.Barile L, Chimenti I, Gaetani R, Forte E, Miraldi F, Frati G, Messina E. and Giacomello A. (2007). Cardiac stem cells: isolation, expansion and experimental use for myocardial regeneration. Nat Clin Pract Cardiovasc Med 4 Suppl 1:S9–S14 [DOI] [PubMed] [Google Scholar]
- 47.Partovian C. and Simons M. (2008). Stem cell therapies in cardiovascular disease A “realistic”. appraisal. Drug Discov Today Ther Strateg 5:73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu W, Shiojima I, Ito Y, Li Z, Ikeda H, Yoshida M, Naito AT, Nishi J, Ueno H, et al. , (2008). IGFBP-4 is an inhibitor of canonical Wnt signalling required for cardiogenesis. Nature 454:345–349 [DOI] [PubMed] [Google Scholar]
- 49.Li H, Zuo S, Pasha Z, Yu B, He Z, Wang Y, Yang X, Ashraf M. and Xu M. (2011). GATA-4 promotes myocardial transdifferentiation of mesenchymal stromal cells via up-regulating IGFBP-4. Cytotherapy 13:1057–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oikonomopoulos A, Sereti KI, Conyers F, Bauer M, Liao A, Guan J, Crapps D, Han JK, Dong H, et al. , (2011). Wnt signaling exerts an antiproliferative effect on adult cardiac progenitor cells through IGFBP3. Circ Res 109:1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D'Amario D, Cabral-Da-Silva MC, Zheng H, Fiorini C, Goichberg P, Steadman E, Ferreira-Martins J, Sanada F, Piccoli M, et al. , (2011). Insulin-like growth factor-1 receptor identifies a pool of human cardiac stem cells with superior therapeutic potential for myocardial regeneration. Circ Res 108:1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Hsieh D, Hsieh A, Stea B. and Ellsworth R. (2010). IGFBP2 promotes glioma tumor stem cell expansion and survival. Biochem Biophys Res Commun 397:367–372 [DOI] [PubMed] [Google Scholar]
- 53.Zebedee Z. and Hara E. (2001). Id proteins in cell cycle control and cellular senescence. Oncogene 20:8317–8325 [DOI] [PubMed] [Google Scholar]
- 54.Yokota Y. and Mori S. (2002). Role of Id family proteins in growth control. J Cell Physiol 190:21–28 [DOI] [PubMed] [Google Scholar]
- 55.Meng Q, Jia Z, Wang W, Li B, Ma K. and Zhou C. (2011). Inhibitor of DNA binding 1 (Id1) induces differentiation and proliferation of mouse embryonic carcinoma P19CL6 cells. Biochem Biophys Res Commun 412:253–259 [DOI] [PubMed] [Google Scholar]
- 56.Zhao Q, Beck AJ, Vitale JM, Schneider JS, Gao S, Chang C, Elson G, Leibovich SJ, Park JY, et al. , (2011). Developmental ablation of Id1 and Id3 genes in the vasculature leads to postnatal cardiac phenotypes. Dev Biol 349:53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imanishi T, Hano T, Sawamura T, Takarada S. and Nishio I. (2002). Oxidized low density lipoprotein potentiation of Fas-induced apoptosis through lectin-like oxidized-low density lipoprotein receptor-1 in human umbilical vascular endothelial cells. Circ J 66:1060–1064 [DOI] [PubMed] [Google Scholar]
- 58.Korem S, Resnick MB. and Kraiem Z. (1999). Similar and divergent patterns in the regulation of matrix metalloproteinase-1 (MMP-1) and tissue inhibitor of MMP-1 gene expression in benign and malignant human thyroid cells. J Clin Endocrinol Metab 84:3322–3327 [DOI] [PubMed] [Google Scholar]
- 59.McCawley LJ. and Matrisian LM. (2000). Matrix metalloproteinases: multifunctional contributors to tumor progression. Mol Med Today 6:149–156 [DOI] [PubMed] [Google Scholar]
- 60.Stamenkovic I. (2003). Extracellular matrix remodelling: the role of matrix metalloproteinases. J Pathol 200:448–464 [DOI] [PubMed] [Google Scholar]
- 61.Kim HE, Dalal SS, Young E, Legato MJ, Weisfeldt ML. and D'Armiento J. (2000). Disruption of the myocardial extracellular matrix leads to cardiac dysfunction. J Clin Invest 106:857–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sappino AP, Huarte J, Belin D. and Vassalli JD. (1989). Plasminogen activators in tissue remodeling and invasion: mRNA localization in mouse ovaries and implanting embryos. J Cell Biol 109:2471–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McMahon B. and Kwaan HC. (2008). The plasminogen activator system and cancer. Pathophysiol Haemost Thromb 36:184–194 [DOI] [PubMed] [Google Scholar]
- 64.Yoshizawa K, Nozaki S, Kitahara H, Kato K, Noguchi N, Kawashiri S. and Yamamoto E. (2011). Expression of urokinase-type plasminogen activator/urokinase-type plasminogen activator receptor and maspin in oral squamous cell carcinoma: Association with mode of invasion and clinicopathological factors. Oncol Rep 26:1555–1560 [DOI] [PubMed] [Google Scholar]
- 65.Thummarati P, Wijitburaphat S, Prasopthum A, Menakongka A, Sripa B, Tohtong R. and Suthiphongchai T. (2012). High level of urokinase plasminogen activator contributes to cholangiocarcinoma invasion and metastasis. World J Gastroenterol 18:244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dufourcq P, Leroux L, Ezan J, Descamps B, Lamaziere JM, Costet P, Basoni C, Moreau C, Deutsch U, Couffinhal T. and Duplaa C. (2008). Regulation of endothelial cell cytoskeletal reorganization by a secreted frizzled-related protein-1 and frizzled 4- and frizzled 7-dependent pathway: role in neovessel formation. Am J Pathol 172:37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bergmann MW. (2010). WNT signaling in adult cardiac hypertrophy and remodeling: lessons learned from cardiac development. Circ Res 107:1198–1208 [DOI] [PubMed] [Google Scholar]
- 68.Silke J. and Vaux DL. (2001). Two kinds of BIR-containing protein—inhibitors of apoptosis, or required for mitosis. J Cell Sci 114:1821–1827 [DOI] [PubMed] [Google Scholar]
- 69.Lamers F, van der Ploeg I, Schild L, Ebus ME, Koster J, Hansen BR, Koch T, Versteeg R, Caron HN. and Molenaar JJ. (2011). Knockdown of survivin (BIRC5) causes apoptosis in neuroblastoma via mitotic catastrophe. Endocr Relat Cancer 18:657–668 [DOI] [PubMed] [Google Scholar]
- 70.Mita AC, Mita MM, Nawrocki ST. and Giles FJ. (2008). Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res 14:5000–5005 [DOI] [PubMed] [Google Scholar]
- 71.Logarinho E. and Bousbaa H. (2008). Kinetochore-microtubule interactions “in check”. by Bub1, Bub3 and BubR1: The dual task of attaching and signalling. Cell Cycle 7:1763–1768 [DOI] [PubMed] [Google Scholar]
- 72.Bolanos-Garcia VM. and Blundell TL. (2011). BUB1 and BUBR1: multifaceted kinases of the cell cycle. Trends Biochem Sci 36:141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yao Y. and Dai W. (2012). Mitotic checkpoint control and chromatin remodeling. Front Biosci 17:976–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.