Abstract
In the past few years, technological advances in nucleotide sequencing have culminated in a greater understanding of the complexity of the human transcriptome. Notably, the discovery that distal regulatory elements known as enhancers are transcribed and such enhancer-derived transcripts (eRNAs) serve a critical function in transcriptional activation has added a new dimension to transcriptional regulation. Here we review recent insights into the tissue-specific and temporal-specific gene regulation brought about by the discovery of eRNAs.
Introduction
Genome-wide analysis of transcription factor binding sites has revealed a majority of their chromatin residence at distal intragenic and intergenic regions that exhibit features associated with enhancers [1–6]. There is a general consensus that enhancers govern tissue-specific and temporal-specific regulation of gene expression [7–9]. These findings are in line with the importance of enhancers in signal-dependent transcriptional responses and the evolutionary conservation of enhancer elements. Mapping co-activator binding sites and unique chromatin modification signatures at potential enhancer-like sequences provided further insight into the characterization of enhancers. It was found that most active enhancers are not only marked by p300/CBP binding but also contain histone H3 monomethyl lysine 4 and H3 lysine27 acetylation modifications [3,10,11]. The exact function of such histone modifications at enhancers is not known. However, recent biochemical and genetic studies have identified the MLL3/MLL4-containing complexes in deposition of monomethyl H3 lysine 4 at enhancers [12]. It is likely that such enhancer-associated histone marks are serving as a signal or a platform for important enhancer-binding factors or activities, as was shown for HP1 binding to H3 lysine 9 methylation marks at heterochromatic regions of the genome [13]. Importantly, since enhancers are critical regulatory regions during cellular differentiation, their modulation during disease progression could be of utmost relevance. Recent experiments have revealed that many silenced embryonic stem cell DNase I-hypersensitive sites (DHSs), which are a general feature of enhancers, are reactivated in cancer [14]. Surprisingly, many of the reactivated DHSs corresponded to sites distinct from those that were active in cell lineages from which the malignancy was derived [14]. Collectively, these results point to enhancers as critical elements whose reactivation may underlie disease-inducing gene expression programs.
Transcription at enhancers
While enhancers were known to bind sequence-specific transcription factors, the association of RNA polymerase II (RNAPII) and general transcription factors with enhancers came as a surprise [15••,16•]. Further experiments indicated that enhancers are transcribed as long noncoding RNAs and such transcripts displayed stimulus-dependent activation [15••,17••]. Importantly, a number of experiments pointed to a strong correlation between the transcription of enhancers and increased expression of their neighboring genes, suggesting a functional relevance for enhancer-mediated transcription [15••,17••]. The precise nature of enhancer-derived transcripts (named eRNAs) has not been defined. While transcriptional co-activator CBP and RNAPII are centered at sequence-specific transcription factor sites, eRNAs are transcribed bi-directionally emanating from the RNAPII peaks. Interestingly, while a large number of enhancers are transcribed bi-directionally, there is a group of enhancers that are induced uni-directionally [18,19•]. Currently, it remains unclear why some enhancers are transcribed from both strands and others are unidirectional. It is likely that the nature and number of transcription factor binding sites at a given enhancer determine the directionality of transcription. Moreover, additional regulatory co-factors may cooperate with RNAPII to govern the directionality of eRNAs.
Of critical importance is the mechanism by which the primary transcripts of eRNAs proceed to their mature forms. While the majority of eRNAs are reported to be monoexonic and not polyadenylated, there have been cases where eRNAs are spliced and polyadenylated [15••,16•,17••]. Moreover, it is unknown whether a single enhancer is transcribed as a mixture of polyadenylated and non-polyadenylated eRNAs or if different classes of enhancers express one or the other form of the transcript. However, there is a general agreement that the majority of the reported eRNAs correspond to transcripts of about 2–5 kilobases (kb) in length. While eRNAs were originally detected using high-throughput RNA sequencing of steady state levels of total RNAs, it is becoming clear that techniques such as Global nuclear run-on sequencing (Gro-Seq), which measures the pioneering rounds of transcription, are more suitable for the detection and quantitation of eRNA changes [18,19•,20]. This may be the result of the general instability of eRNAs compared to that of messenger RNAs, consistent with the observation that eRNAs are predominantly not polyadenylated.
Functionality of enhancer RNAs
Initial reports correlating the transcription of eRNAs and their neighboring protein-coding genes were suggestive of the functional relevance of eRNAs in transcriptional activation. However, the exact requirements for eRNAs in activation of their target genes were not determined. Two reports provided evidence linking the long noncoding RNAs to transcriptional activation. Using knockdown approaches, it was shown that long noncoding RNAs positively regulate their neighboring protein-coding genes [21••]. Analysis of the genomic sites for noncoding RNA (termed ncRNA-a3) that regulated the TAL1 gene (also known as SCL) revealed its intimate association with enhancers as defined by enrichment in monomethyl H3K4 and acetylated H3K27 marks [21••,22]. This locus produced bi-directional transcripts that were polyadenylated and spliced. A similar approach was used to show that a noncoding RNA termed HOTTIP activates several 5′ HOXA genes in vivo [23]. HOTTIP also corresponded to a spliced and polyadenylated transcript. While these experiments revealed a role for long noncoding RNAs in activation of neighboring protein-coding genes, they did not assess the specific signaling pathway(s) that is regulated by long noncoding RNAs.
Subsequent studies analyzing the activation of transcription through a number of different signaling pathways extended the role of eRNAs in enhancer function. Genome-wide studies revealed p53 association with a large number of distal binding sites [24•]. Detailed analysis of two distinct extragenic p53 sites revealed the expression of eRNAs that were stimulated following treatment of cells with Nutlin-3, an inducer of p53. Importantly, depletion of eRNAs corresponding to each enhancer abrogated the Nutlin-3 induction of transcription of the targeted protein-coding gene [24•]. A similar scenario was observed following activation of estrogen-responsive genes in MCF7 cells [19•]. Treatment of MCF7 cells with estradiol (E2) induced the binding of estrogen receptor alpha (ER-alpha) to a large number of extragenic binding sites. Majority of the E2-upregulated protein-coding genes contained an E2/ER-alpha-binding enhancer within 200 kb from their transcription start sites [19•]. Most of these E2-induced enhancers produced bi-directional eRNAs as measured by Gro-Seq. Critically, depletion of eRNAs corresponding to a number of enhancers diminished the E2-induced activation of their target genes. It was further shown that at least in the case of one enhancer involved in activation of FOXC1 gene, only the sense strand of eRNA contained the activating function. This suggests that perhaps only one strand of eRNAs confers transcriptional activation [19•].
Further evidence to the functionality of eRNAs was provided following studies of nuclear receptor Rev-Erbs in mouse macrophages [20]. It was found that Rev-Erbs predominantly bound extragenic and intragenic enhancer-like regions to repress their neighboring protein-coding genes. This was highlighted for two Rev-Erbs-responsive genes, Mmp9 and Cx3cr1. Evidence was found of bidirectional transcription corresponding to eRNAs from a majority of Rev-Erbs-associated extragenic enhancers. Detailed examination of two distinct sites adjacent to Mmp9 and Cx3cr1 genes revealed that Rev-Erbs binding at these enhancers resulted in repression of eRNAs expression, leading to silencing of the targeted genes. Moreover, depletion of eRNAs corresponding to each enhancer led to a specific repression of neighboring protein-coding genes, demonstrating the functional importance of eRNAs. Significantly, they show that only one strand of the eRNA is involved in the activating function, which begs the question of the functional importance of the other strand. It is important to note that most protein-coding genes also contain bidirectional transcription of unknown function [25]. Taken together, it is clear that most activity-dependent enhancers express eRNAs and in nearly all cases examined, such eRNAs are endowed with functional information required for transcriptional activation.
Enhancer RNAs mechanism of action
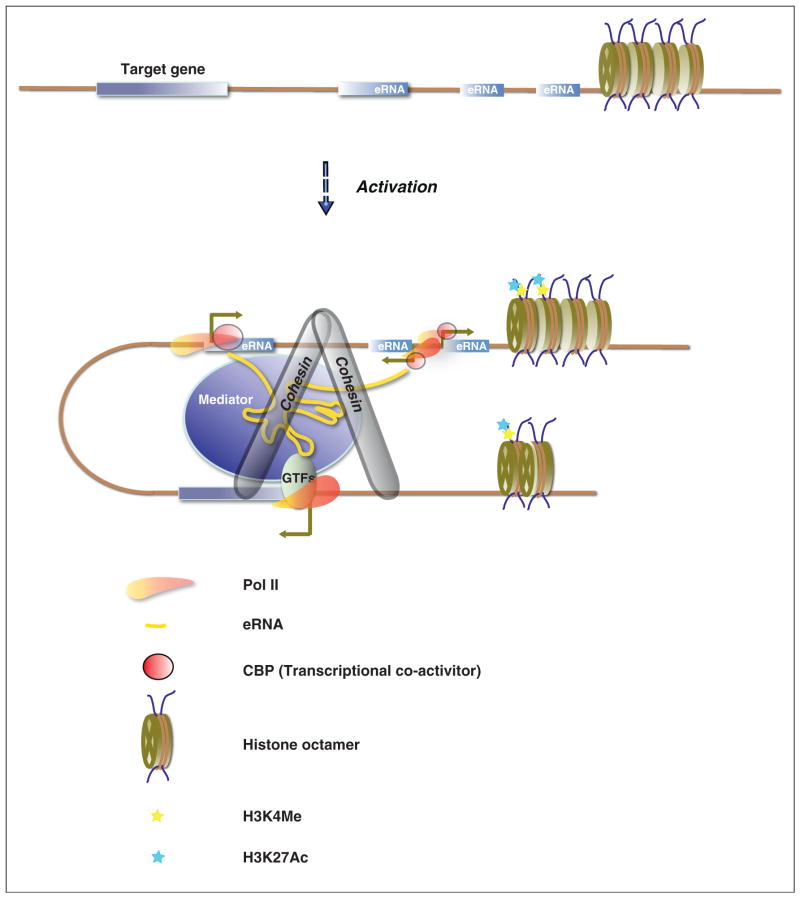
How distal regulatory elements confer transcriptional activation of their target genes has remained a puzzle in transcription. The prevailing idea put forward from recent advances in understanding the three-dimensional structure of the genome stipulates the formation of a DNA loop connecting the distal regulatory sites and the core promoter elements of the protein-coding gene. In such a scenario sequence-specific DNA binding factors and their associated co-factors mediate the association of the enhancer and their targeted promoter sequences. Recent experiments have suggested that two multi-protein complexes, Mediator and Cohesin, play an important role in such stimulus-dependent chromatin looping [26,27]. More importantly, these experiments have suggested a role for eRNAs in either the establishment or the maintenance of enhancer–promoter contacts. Depletion of long noncoding RNAs involved in activation of SNAI1 or TAL1 resulted in decreased chromatin residence of Mediator at the promoter of these genes concomitant with diminished DNA looping as measured by chromosome conformation capture (3C) [22]. Similarly, knockdown of estrogen-induced eRNAs reduced the enhancer–promoter looping at a number of estrogen-responsive genes. The decrease in chromatin looping was accompanied with reduced levels of Cohesin occupancy at estrogen-induced enhancers [19•]. Collectively, these results paint a framework by which the Mediator and Cohesin complexes cooperate with the eRNAs to promote enhancer–promoter interactions (Figure 1).
Figure 1.
eRNAs induce chromatin looping and transcriptional activation. Stimulus-induced expression of eRNAs cooperate with transcriptional cofactors such as Mediator and Cohesin to promote chromatin looping and transcriptional activation.
While the augmentation of DNA looping serves as one possible mechanism by which eRNAs confer their responsiveness, there are other proposed mechanisms of action. The overall hypothesis envisions a role for eRNAs as a guide to recruit sequence-specific factors, chromatin remodeling or chromatin modifying complexes to the targeted promoters. Two recent studies proposed the association of activating long noncoding RNAs to transcription factors SOX2 or androgen receptor, resulting in increased chromatin residence of the transcription factors and activation of their specific gene expression programs [28,29]. In a different study, the long noncoding RNA HOTTIP was shown to recruit the MLL complex to the HOXA cluster resulting in its activation [23]. More recently, the eRNAs expressed from the core enhancer (CE) element of the MYOD gene were shown to increase RNAPII occupancy and chromatin accessibility at the MYOD promoter by the proposed recruitment of chromatin remodeling complexes [30]. Therefore, the overarching theme for these studies suggests the physical association of the eRNAs with a component of transcription regulatory machinery, resulting in increased chromatin association of a specific factor and enhancement of transcription. While it is likely that there may be common themes by which eRNAs expressed from different regulatory regions mediate their responsiveness, it would not be surprising to find specific eRNAs that utilize novel strategies to activate transcription.
Outlook and biological implications of enhancer RNAs
It is becoming clear that achieving a detailed molecular understanding of enhancers will require a greater insight into eRNA biogenesis, mechanism of action and regulation. Since enhancers control tissue-specific and temporal-specific gene expression, understanding the dynamic regulation of eRNAs during development and in disease progression will be of utmost importance. Recent studies have started to shed some light on the biological relevance of eRNAs. An enhancer-like RNA was shown to regulate neurogenin 1 expression during mouse cortical development [31]. The eRNA expressed from the CE element upstream of the MYOD1 locus was shown to regulate the MyoD expression during the differentiation of C2C12 cells, underscoring the importance of eRNAs in myogenic gene expression programs [30]. In a different study, it was shown that an eRNA expressed from p53-bound enhancer region 2 (p53BER2) activated the protein-coding gene PAPPA in MCF7 cells [24•]. As expected, while treatment of MCF7 cells with infrared (IR) irradiation led to a G1 arrest, depletion of p53 or p21 could rescue such cell cycle arrest, confirming that the arrest is indeed p53 dependent. Importantly, depletion of PAPPA or the eRNA (derived from the p53BER2) regulating the PAPPA also had a significant inhibitory effect on the p53-induced cell-cycle arrest. The critical importance of these studies is the idea that targeting of specific eRNAs may be the key methodology that would allow for locus-specific, tissue-specific, and temporal-specific regulation of gene expression. It was possible to deplete the eRNAs controlling the Mmp9 gene leading to diminished Mmp9 expression in vivo [20] using lipofectamine-siRNA delivery following induction of sterile peritonitis in mice. These experiments provided the proof of principle that targeting of eRNAs to reduce mRNA expression could be a viable methodology to control locus-specific gene expression. It is clear that in the coming years improved technologies for targeting oligonucleotides will provide us with an opportunity to modulate aberrantly expressed eRNAs in vivo, which could bring forth a new era for enhancer therapy.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
•of special interest
•• of outstanding interest
- 1.Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 3.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol. 2010;30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 9.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 11.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by trithorax-related, the drosophila homolog of mammalian MLL3/MLL4. Genes Dev. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 14.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. In this work the authors show that neuronal activity-dependent enhancers produced bidirectional transcription and therefore highlighted the importance of transcription at active enhancers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C, Eick D, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol. 2011;18:956–963. doi: 10.1038/nsmb.2085. The authors present compelling evidence for binding of general transcription factors and RNA polymerase II at enhancers leading to transcription. Moreover, they delineate regions where transcription factors occupy large regions of the genome identifying regulatory landscapes. [DOI] [PubMed] [Google Scholar]
- 17••.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. The authors report transcription at macrophage-induced enhancers and show that such enhancer-derived transcripts correlate with activation of neighboring genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. The authors show that depletion of eRNAs expressed from estrogen-binding sites diminishes the activation of the target genes. Moreover, they present evidence that eRNAs are invovled in DNA looping and they invoke the recruitment of Cohesin as a mechanism in transcriptional activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, et al. RevErbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, et al. Long noncoding rnas with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. Using knockdown approaches the authors show that long noncoding RNAs display activating function and many of them overlap previously defined enhancer sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating rnas associate with mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, de Laat W, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. Using Nutlin to induce p53 responsiveness, the authors show that eRNAs expressed from distal p53-binding sites can activate the neighboring p53 responsive genes. [DOI] [PubMed] [Google Scholar]
- 25.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, Young RA. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, Bland MJ, et al. Architectural protein subclasses shape 3d organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, Evans CP, et al. IncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onoguchi M, Hirabayashi Y, Koseki H, Gotoh Y. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc Natl Acad Sci U S A. 2012;109:16939–16944. doi: 10.1073/pnas.1202956109. [DOI] [PMC free article] [PubMed] [Google Scholar]