Abstract
Spatial and temporal regulation of gene expression is achieved through instructions provided by the distal transcriptional regulatory elements known as enhancers. How enhancers transmit such information to their targets has been the subject of intense investigation. Recent advances in high throughput analysis of the mammalian transcriptome have revealed a surprising result indicating that a large number of enhancers are transcribed to noncoding RNAs. Although long noncoding RNAs were initially shown to confer epigenetic transcriptional repression, recent studies have uncovered a role for a class of such transcripts in gene-specific activation, often from distal genomic regions. In this review, we discuss recent findings on the role of long noncoding RNAs in transcriptional regulation, with an emphasis on new developments on the functional links between long noncoding RNAs and enhancers.
Where have all these noncoding RNAs been?
Recent large-scale genomics approaches have revealed the presence of a large number of non-protein-coding RNAs [1–8]. The RNA-dependent functions of tRNA, rRNA and small nucleolar RNA (snoRNA) have been appreciated for years, including important functions in several cellular processes [9,10]. More recently discovered miRNAs are being recognized as essential regulators of translational regulation and other processes [4]. Transcripts encoding proteins, mRNAs, are processed co-transcriptionally through a complex RNA splicing machinery to yield a mature mRNA [11]. Additionally, most mRNAs are not only spliced to remove intronic sequences, but also proceed on their 3′-end by means of intricate nuclear polyadenylation machinery [12]. These processes are vital components of the mRNA maturation cycle, which culminates in a final mRNA species that can be translated into a functional protein. Intriguingly, recent studies have identified thousands of transcripts that do not contain any recognizable protein-coding capacity, yet undergo processes such as splicing and polyadenylation [1–5]. Although not all such transcripts are spliced and polyadenylated, most display such mRNA-like characteristics. These transcripts are commonly referred to as long noncoding RNAs (ncRNAs), a large and heterogeneous class of RNA transcripts involved in many cellular processes [13]. The multiple functions of long ncRNAs are just starting to emerge, and the mechanisms through which they mediate their functions are subject to intense ongoing investigation. What makes these long ncRNA transcripts so interesting is that, although they do not encode for a protein, they have intimate regulatory roles in transcription modulation. Such regulatory roles suggest that ncRNAs contain extensive information stored in the form of specific structural conformation or nucleotide sequence that goes beyond the genetic code used for translation of protein-coding genes. Unraveling this new code for ncRNA function is the foremost challenge for future research on long ncRNAs.
In this review, we discuss the most recent progress in identifying and functionally characterizing long ncRNAs through various large-scale approaches. We present recent evidence of long ncRNAs functioning as enhancers to mediate activating functions across large genomic distances. Furthermore, recently proposed scenarios of long ncRNA mechanisms are discussed, aimed at understanding the impact of RNA in transcriptional regulation.
The latest estimates contend that approximately 1.5% of the human genome is transcribed into mRNA [6,7]. This leaves most of the transcribed human genome encoding RNA without any specified function [7,14]. The identification of thousands of long ncRNAs through large-scale approaches suggests that mRNAs could easily be outnumbered by ncRNAs. This raises the question as to why so little is known about their function and why they have been underappreciated. Perhaps a critical reason is the low expression level of long ncRNAs, which is, on average, 10–20 times less than the expression level of protein-coding genes [5]. Another important issue relates to the possibility that many of such low expressing long ncRNAs have been considered to result from spurious transcription by RNA polymerase II and might not represent bona fide functional RNAs. Moreover, although analysis of the evolutionary conservation of protein-coding RNAs is relatively simple owing to conservation of codon usage, ncRNAs pose a challenge for evolutionary biologists [10]. Indeed, it is difficult to ascertain the evolutionary conservation of human long ncRNAs beyond that of mammalian species. Although the advent of new sequencing technologies has, to a large extent, overcome the detection hurdles that hindered identification of RNAs expressed at such low levels, a detailed knowledge is still lacking of the specific RNA codes that confer the function of long ncRNAs and allow for evolutionary comparisons.
Are they really noncoding and does size matter?
The average length of a primary transcript for most defined long ncRNAs is approximately 1 kilobase (kb) [5]. The conventional description for ncRNAs arbitrarily defines them as having a length of more than 200 nucleotides. However, there are known examples of long ncRNAs spanning genomic regions of more than 100 kb [such as antisense Igf2r RNA (Air)] [15]. The noncoding properties are often defined as the absence of a predicted open reading frame (ORF) [16,17]. A basic challenge is to define experimentally whether a transcript is coding or noncoding and, as such, an annotation of long ncRNAs inevitably relies on the absence of protein-coding potential. Although the presence of a predicted ORF of a certain length is a good indicator of a gene with protein-coding potential, the occurrence of predicted shorter ORFs are harder to interpret. Given that random occurrences of short ORFs are frequent, this is not a reliable indicator of whether a transcript is coding for a protein or is a noncoding transcript. Although coding potential can be addressed to some extent using in vitro translation assays, these are only suggestive at best, and are mostly reliable in case of positive results. The presence of a protein in vivo corresponding to an observed ORF is good evidence that the sequence is encoding a protein, whereas a confirmation of negative output is not possible. However, recent technological advances have provided a possible way to address coding potential both in vivo and genome wide [18]. Using a methodology called ribosome profiling for studying the translational versus mRNA stability effects of miRNAs, additional data are made available that can be used to study the extent of translation of almost any expressed RNA in a cell. In this approach, ribosomes are immobilized on the mRNAs and the ribosome–mRNA complexes are then subjected to RNase treatment. The RNA bound by a ribosome is therefore protected from the RNase enzyme and can be purified and sequenced. Comparing these ribosome-bound RNA fragments to total RNA sequencing reads, gives an estimate of the extent of translation of a given RNA transcript. Although the presence of ribosomes on a specific RNA does not necessarily mean that it is being actively translated, the absence of ribosome association with a potential long ncRNA supports its lack of protein-coding potential. Thus, this methodology could be used to address the occurrence of noncoding transcriptson a large scale without prior assumptions of coding potential.
As discussed above, long ncRNAs show several characteristics common to mRNAs. Both classes of polyadenylated and non-polyadenylated long ncRNAs have been characterized [1–5,8]. Several instances of multi-exonic long ncRNA spliced into mature long ncRNAs have been shown [1–3,5,19]. These processes occur by the same splicing apparatus that functions on multi-exonic mRNAs. It has been suggested that some RNA transcripts mediate dual functions [20]. Although the RNA is coding for a protein and is acting as an mRNA, it might also have other functions, which rely strictly on the RNA structure or sequence. This is an intriguing possibility that could apply to all mRNAs. The challenge of studying such complex dual functions of RNA transcripts requires a thorough understanding of the properties that are required for a long ncRNA to be functional.
How to find your noncoding RNA
Whereas the identification of functional mRNAs has relied on sequence conservation across species, long ncRNAs appear to be harder to define reliably. Many mRNAs show very high conservation between distantly related species, as conservation of the codon usage is essential for synthesis of the correct proteins [21]. Sequence conservation for RNA function seems to be not as important in long ncRNAs, which is suggestive of functions that are more dependent on structure or short nucleotide stretches in the RNA sequences [2,5,10]. Several studies have addressed the identification of long ncRNAs using large-scale approaches, which has resulted in the uncovering of thousands of putative long ncRNAs [1–5,8] (Figure 1). The different methods used for identifying long ncRNAs and the various criteria utilized, has led to a lack of consistency in nomenclature. Several reports refer to them as long intervening or intergenic ncRNAs (lincRNAs), whereas others simply refer to them as long ncRNAs [1,3–5,13,17,19]. We recently defined a set of long ncRNAs that activate transcription and, therefore, we referred to these as ncRNA-activating (ncRNA-a), indicating that they are a subgroup of long ncRNAs but with activating functions [5]. It is clear that more studies aimed at defining common functional modalities as well as structural features of such growing classes of RNAs will be needed to arrive at a more informative nomenclature for such a diverse group of RNAs.
Figure 1.
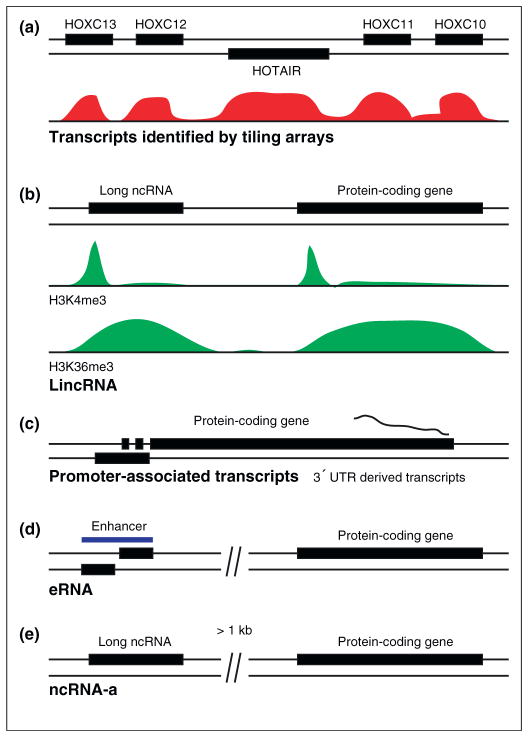
Different ways to identify long noncoding RNAs (ncRNAs). Thousands of long ncRNAs have been recently identified using various approaches. This figure summarizes the approaches used for the identification of large sets of long ncRNAs. (a) Using tiling arrays, regions that are being transcribed can be identified (intensity from detected transcripts are depicted in red). In the HOX cluster, hundreds of long ncRNAs have been identified as transcribed regions outside of protein-coding genes using tiling arrays. Among the better studied ones are HOTAIR [1] and HOTTIP [19]. (b) A large class of long ncRNAs in both mouse [2] and human [3] has been identified based on histone marks associated with active transcription. The presence of a H3K4me3 mark is indicative of the start site of an actively transcribed gene, and H3K36me3 often marks the body of the transcribed gene. These long ncRNAs have been called lincRNAs, for long intervening ncRNAs. (c) Several long ncRNAs, and shorter derivatives, are transcribed from 3′ untranslated regions (UTRs) of protein-coding genes and from around their promoters [7,24,25]. The function of these ncRNAs is unknown, but speculated to be involved in the regulation of transcription of the genes they are coexpressed with. (d) Enhancer-associated RNA (eRNA) is another class of long ncRNAs observed for thousands of enhancers in mouse [4]. Long ncRNAs are transcribed bidirectionally from the enhancer region, and speculated to have active roles in the regulation of nearby genes. (e) A class of long ncRNAs called ncRNA-a (activating ncRNA) can mediate the induction of a protein-coding gene at a distance, resembling classically defined enhancers [5]. ncRNA-a transcripts are defined from the GENCODE annotation of the human genome, as those ncRNAs residing at least 1 kb away from any known protein-coding gene.
One of the recent large-scale studies to identify long ncRNAs in human fibroblasts used tiling-arrays covering the sequence of the HOX cluster [1] (Figure 1a). This analysis revealed the presence of 231 long ncRNAs across a wide panel of human tissues analyzed. One of these, termed ‘HOX antisense intergenic RNA’ (HOTAIR), was further shown to act as a trans-repressive factor, as described below. More recently, a similar approach has identified a long ncRNA, termed ‘HOXA transcript at the distal tip’ (HOTTIP), which mediates transcriptional activation in cis [19]. An alternative approach for identifying long ncRNAs has utilized the chromatin signatures associated with actively transcribed genes as a measure of ongoing RNA polymerase II transcription at putative ncRNA loci [2,3] (Figure 1b). Chromatin modifications, such as histone 3 lysine 4 trimethylation (H3K4me3) at the promoter of a gene and histone 3 lysine 36 trimethylation (H3K36me3) along the body of the transcribed genes, occur at many genes transcribed at an intermediate to high level [22]. By using tiling arrays to define H3K4me3 and H3K36me3 profiles (defined as K4-K36 domains) across four mouse cell types, 1600 long ncRNAs have been identified in mouse [2]. These experiments were extended to an analysis of chromatin signatures in human cells by using K4-K36 domains in various human cell lines to further delineate 3300 long ncRNAs [3]. These large groups of transcripts were shown to be regulated by transcription factors, and suggested to associate with chromatin-modifying complexes to mediate their functions [1–3,23]. However, this approach suffers from the fact that a large number of ncRNAs might be silenced in a given cell type and, therefore, might not be identified using active chromatin marks. Moreover, although K4-K36 trimethylation domains might in large part delineate active genes, it is formally possible that such domains also represent sites for other important biological events, such as recombination hot spots.
Several studies based on large-scale sequencing efforts have also revealed an extensive class of promoter-associated transcripts [7,24–26] (Figure 1c). Although most of these are shown to be short transcripts, there is a large class of promoter-associated RNAs that could encode longer RNAs. Additionally, a class of long ncRNAs is expressed from genomic regions that were previously defined as 3′ untranslated regions [27]. Sequence conservation in regions outside of protein-coding genes, and corresponding evidence that such sequences are transcribed, has also been used to identify long ncRNAs in the brain [8]. This study points out distinct coexpression patterns, where protein-coding genes and long ncRNAs are coexpressed in the brain in the adjacent genomic regions, suggestive of a regulatory interplay of the two classes of RNAs [8].
One of the most extensive annotations of the human genome (GENCODE) has been performed under the framework of the ENCODE project [28]. The GENCODE human genome database has been compiled to represent not only protein-coding genes, but also ncRNAs. GENCODE annotation is based on experimental evidence, including sequencing studies of spliced RNAs, cDNAs and ESTs, and comprises an extensive resource for long ncRNAs. In a recent study, the GENCODE annotation was used to define a set of long ncRNAs in multiple cell lines. Importantly, additional filtering criteria were used to arrive at a class of potential long ncRNAs that do not overlap protein-coding genes [5]. This approach provided a set of 3019 long ncRNAs from the annotation of approximately 30% of the human genome. The current estimates based on GENCODE annotation, puts the number of ncRNAs close to 10 000, once the annotation has been completed.
Although, in theory, all such approaches should result in a similar collection of long noncoding transcripts, the identified sets differ significantly both in number, characteristics and sequence of proposed long ncRNAs. This is probably a measure of the incomplete characterization of the ncRNA species and their complexity of expression patterns, reflecting both their low endogenous levels and tissue-specific expression patterns. Furthermore, an important concern with some long ncRNAs is whether they are independent transcripts or merely rare extensions or alternative splice forms of protein-coding genes. A recent study found evidence for the existence of ncRNAs in close proximity to protein-coding genes that could be deemed as extensions of primary mRNA sequences [29]. However, there is now increasing experimental evidence for a large number of independent long ncRNA transcripts that are distal to protein-coding genes and are supported by large-scale chromatin immunoprecipitation (ChIP) experiments for RNA polymerase II and transcription factor binding [30].
NcRNAs could be a transcriptional turn off
The best examples of an inhibitory role for long ncRNAs are described in processes such as X-inactivation and imprinting, where they silence gene expression in cis [31]. X-inactive specific transcript (Xist), an ncRNA that is expressed from the inactive X chromosome, was the first long ncRNA found to be involved in transcriptional silencing of the X chromosome. It was initially reported to be ‘a candidate for a gene either involved in or uniquely influenced by the process of X inactivation’ [16]. Xist is unique among ncRNAs described thus far, as it silences an entire X chromosome through a phenomenon that has been described as ‘coating’ or ‘painting’ the chromosome [32]. Although the precise molecular events that lead to transcriptional silencing by Xist have not yet been elucidated, the structural changes seen at the inactive X chromosome implicates the formation of a transcriptionally non-permissive chromatin structure [32]. These include the presence of histone modifications [23] as well as histone variants [33] that are often associated with transcriptional inactivation [33,34].
Whereas Xist function to silence large regions of the X chromosome, the ncRNAs associated with the phenomenon of imprinting silence a small number of genes, often forming gene clusters of 3–15 genes [15]. One such ncRNA that has been studied in some detail is Air, a polyadenylated, unspliced transcript that is transcribed from a genomic region of more than 100 kb [15]. Insertion of a polyadenylation cassette leading to truncation of the Air transcript resulted in the derepression of imprinted genes in the locus, which is suggestive of a direct role for long ncRNAs in imprinting of insulin-like growth factor 2 receptor (Igf2r) [15]. The reported effects of Air on imprinting expand to genes located several hundreds of kilobases away, demonstrating the long-ranging effects of long ncRNAs [35]. Although it is clear that ncRNAs involved in imprinting could mediate their responsiveness in cis at long distances from their locus of expression, the mechanism by which such interactions are established and epigenetically maintained remain an enigma.
Unlike the action of long ncRNAs in imprinting and X inactivation, HOTAIR, a long ncRNA which is expressed from the HOXC locus, was recently described to mediate transcriptional silencing through a trans-acting mechanism [1]. A tiling array-based study was used to identify hundreds of long ncRNAs encoded by the human Hox cluster, including the HOTAIR ncRNA. The expression of HOTAIR negatively regulates the expression of several genes transcribed from the HOXD locus, situated on a different chromosome from the HOXC cluster of genes. Knockdown of HOTAIR using small interfering RNAs led to the increased expression of genes encoded in the HOXD locus, suggesting an RNA-dependent trans-acting mechanism. HOTAIR was shown to interact with the polycomb complex PRC2 and the depletion of HOTAIR led to increased levels of histone 3 lysine 27 trimethylation at the promoters of genes in the HOXD locus [1]. Therefore, the mechanism through which HOTAIR mediates repression of gene expression in trans has been suggested to involve its binding to the PRC2 complex, which it then directs to promoters of genes in the HOXD locus [1]. HOTAIR was later identified as a factor with a potential to reprogram chromatin states, with a possible role in metastasis [36]. More recent studies pointed to the enhancer of zeste [23] subunit of PRC2 as the subunit with RNA-binding potential, which could be regulated via phosphorylation during the cell cycle [23,37]. ANRIL (i.e. antisense ncRNA in INK4 locus) has also been linked to transcriptional repression via interaction with both PRC1 and PRC2 [38,39]. However, what is not clear from these studies is the mode by which either ANRIL or HOTAIR mediate the recruitment of either of these repressive complexes to a specific target site. Because both PRC1 and PRC2 regulate a large number of protein-coding genes, binding to a specific ncRNA is expected to provide further specificity for their target recognition. However, it is currently not clear whether critical RNA sequences or a specific RNA structural motif provide the additional determinants that might be required for the recruitment of either of the repressive complexes to a particular target.
NcRNAs could enhance transcription
A host of recent reports suggests that ncRNAs also mediate transcriptional activation and that such transcriptional potentiation is a common function of a large class of ncRNAs [40,41]. In both mouse and human cells, long ncRNAs are reported to be associated with transcriptional enhancers [4,5,19,30] (Figure 1d). Although previous reports have suggested transcriptional activity at enhancers resulting in enhancer-associated transcripts, whether the resulting RNA mediated biologically relevant functions was unclear [42–46]. Recent advances in the analysis of enhancers based on specific chromatin signatures combined with high throughput sequencing of the transcriptome have revealed the prevalence of ncRNA at active enhancers [4,30]. Predominantly, through a correlative analysis of ncRNAs and the nearby protein-coding genes, these studies have pointed to an important role for long ncRNAs in the positive regulation of nearby genes [4,8,30]. Although initial studies suggested that such positive regulation of transcription by a neighboring genes could be attributed to ripples of transcription (i.e. transcriptional activity of the neighboring gene resulting in waves of transcription throughout the locus) [47], recent evidence has pointed to the importance of long ncRNAs and not the act of transcription per se in mediating such transcriptional activation [4,5,19,48].
Some of the earliest evidence for transcription at enhancers comes from studies performed on the β-globin locus, where the hypersensitive site 2 (HS2) enhancer was shown to be transcribed in a strand-specific manner [41]. The transcription of the HS2 enhancer could be observed in a reporter assay irrespective of its location or direction compared with the reporter gene [41,42]. These studies suggested that the HS2 enhancer was transcribed from an independent promoter, and led to the hypothesis that such enhancer-derived transcripts could have biologically significant functions. Interestingly, a later study in which a transcriptional terminator was inserted in the DNA sequences intervening the HS2 enhancer and β-globin promoter resulted in a concomitant abrogation of HS2-derived transcript as well as the HS2-mediated enhancement [42]. Although these experiments were both interpreted to indicate the importance of transcription through such intervening sequences, recent findings on the roles of ncRNAs in transcription indicate that the HS2 activation could putatively be mediated through an HS2 enhancer-derived ncRNA.
To further analyze the role of enhancer-derived RNAs, a genome-wide study in the mouse [4] addressed the identification of putative enhancers by chromatin modifications and binding of enhancer binding proteins p300/CBP. Although this study indicated the presence of nearly 12 000 enhancers, only a subset of these were shown to contain RNA polymerase II. Importantly, approximately 2000 of these sites were shown to be transcriptionally active and were differentially expressed upon membrane depolarization. However, the transcripts were reported to not be polyadenylated and bidirectional. In one specific example, an ncRNA distal to activity-regulated cytoskeleton-associated protein (Arc) was suggested to regulate expression of the Arc gene. Using a knockout model for the Arc gene, the expression of the ncRNA was shown to be dependent on the presence of the Arc promoter. These data suggested that transcription at the enhancer of Arc is somehow influenced by the activity of the Arc promoter. What is becoming clear from such analyses is that the enhancer and the promoter of the gene it regulates are in dynamic communication and, therefore, the activity of one might effect the functioning of the other. Although the above example paints a scenario in which there is a close association between transcription at the enhancer and its corresponding targeted promoter [4], earlier studies with the HS2 enhancer suggested that transcription at the enhancer was independent of the associated promoter [42]. Collectively, these studies point to different modes by which enhancers and their corresponding promoters might be communicating.
A recent study utilized the GENCODE annotation of the human genome, performed by the Human and Vertebrate Analysis consortium, to define long ncRNAs. The GENCODE annotation has the added advantage of also containing a catalog of long ncRNAs [5]. This study reported a set of transcripts annotated as noncoding and transcribed from unique loci of the human genome without overlap with protein-coding genes. Depletion of a set of these long ncRNAs in several human cell lines resulted in a concomitant decrease in their neighboring protein-coding genes. Detailed analysis of a long ncRNA residing in proximity to Snail homolog 1 (SNAI1) indicated the absolute requirement for the ncRNA sequences in mediating transcriptional activation. These results supported the notion that long ncRNAs can mediate their effects on transcription in cis. However, genome-wide analysis using microarrays following depletion of the long ncRNA proximal to SNAI1 showed regulation of several genes located on other chromosomes, suggesting a broader mechanism of action, possibly reflecting secondary or trans-mediated effects. More recently, chromosome conformation capture carbon copy (5C) was used to identify interactions in the HOXA locus involved in transcriptional regulation [19]. As enhancers are thought to mediate their function by looping DNA to bring them into proximity of the promoter of the regulated genes, 5C and similar approaches are promising for revealing the function of long ncRNAs. This study identified HOTTIP in association with the promoter regions of downstream 5′ HOXA genes. HOTTIP was shown to mediate enhancer-like effects on the adjacent genes through a mechanism involving direct interaction with the adaptor protein WDR5. This is a component of mixed-lineage leukemia (MLL)-containing complexes [49], and this association was shown to affect H3K4me3 through MLL recruitment to the HOXA locus.
Further complexity of long ncRNA regulation of gene expression is reflected by a study assessing the role of an ncRNA in transcriptional regulation of the Xist locus. Upstream of Xist lies a long ncRNA called Jpx, which was shown to regulate positively the expression of Xist RNA from the inactive X chromosome [48]. Such positive regulation of one ncRNA (Xist) by another ncRNA [48] suggests that the regulatory effects of long ncRNAs are not limited to protein-coding genes and represent a much broader reach of long ncRNA regulatory networks in gene expression in mammals. Indeed, there has been increasing evidence of the role for ncRNAs in dosage compensation. Besides the role for Xist RNA in mammalian dosage compensation, the Drosophila melanogaster dosage compensation complex contains two ncRNAs, the roX1 and roX2 RNAs, which have a critical role in dosage compensation of the male X chromosome [50,51]. Interestingly, although the functions of roX1 and roX2 ncRNAs are apparently redundant, there is very little sequence similarity between the two RNAs. It is likely that the secondary or tertiary structure of these long ncRNAs could be of primary importance for their function. Although most studies are aimed at identifying protein factors that mediate the function of long ncRNAs, it is possible that some long ncRNAs could be working independently of associated proteins through mechanisms resembling those of ribozymes.
Concluding remarks
It is clear that recent advances in genomic and proteomic technologies have revealed a wealth of additional information regarding the composition and the informational content of the human genome. A combination of high throughput sequencing of the transcriptome allowing for analysis of a variety of transcripts and the definition of the chromatin signatures that could be linked to such transcriptional outcomes, have begun to shed light on the general principles of genomic organization. A large number of noncoding regions are transcribed and the number of ncRNAs could turn out to surpass the number of protein-coding genes. Importantly, genome-wide analyses have revealed that only a small part of such transcripts are produced by RNA polymerase III, and the bulk of ncRNAs in mammalian cells are products of RNA polymerase II, thus resembling the transcripts of protein-coding genes to a large degree.
Although the past decade has resulted in great insights into understanding new cellular mechanisms mediated through small RNAs, such as miRNAs, knowledge of long ncRNAs has lagged behind. Important questions regarding the biogenesis of long ncRNAs, their mechanism of action and their scope of function remain. The possibility that a class of long ncRNAs (ncRNA-a, Figure 1e) could act to enhance transcription is an additional regulatory layer to the repertoire of functions of ncRNAs. We envision that, similar to miRNAs, long ncRNAs might also be processed to smaller mature transcripts and that such transcripts, by virtue of sequence homology to the DNA or the nascent RNA of the target genes, could bring about enhanced transcription (Figure 2). Given that ncRNAs are composed of large domains of repetitive sequences, such repetitive RNA sequences could in principle provide the sequence complementarity with similar repeats embedded in the RNA of the protein-coding targets. Indeed, such RNA repeats interactions might explain the chromosomal conformation changes that have been observed between the distal enhancer elements and their respective targets. Once such interactions are established, similar to transcription factors, ncRNAs could have diverse roles in biology, depending on the specific targets they regulate. Functional roles in chromosome segregation, DNA repair and cellular reprogramming are just a few possible processes that could be fined tuned through the action of long ncRNAs. Indeed, a recent report suggests a role for a long ncRNA, RNA-RoR, in regulation of reprogramming of human-induced pluripotent stem cells [52].
Figure 2.
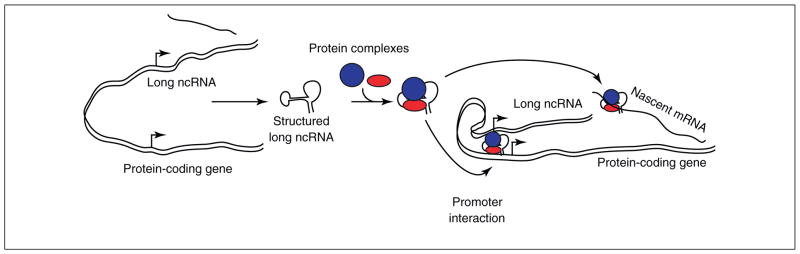
Possible mechanisms of long noncoding RNA (ncRNA) functions. The mechanisms of long ncRNA regulation of gene expression are still not well understood. From recent studies, the opinion that they work in complexes with proteins is emerging [1–4,19,26,40]. Summarized here is an overview of how long ncRNAs are currently speculated to function. The long ncRNA is expressed from an independent promoter and, in many cases, is spliced and polyadenylated. The structured, processed long ncRNA then associates to specific protein complexes. As both repressive and activating functions of long ncRNAs have been reported, it is likely that several different protein complexes can constitute these factors. The RNA–protein complex is then thought to target the promoter of the regulated gene, causing a conformational change and leading to altered gene expression. Alternatively, the long ncRNA–protein complex could target the nascent mRNA, making RNA–RNA hybrids and, thus, mediating immediately post-transcriptional control of gene expression.
It is also likely that activating long ncRNAs interact with their target genes or their promoters through secondary or tertiary structural motifs. Such a mechanism could involve the recognition of specific structural motifs by a protein complex involved in transcriptional regulation, leading to formation of a protein–RNA or a protein–RNA–DNA hybrid structure. The mechanisms that have been explored thus far favor the association of long ncRNAs with transcriptional regulatory complexes, leading to their recruitment to specific targets [1,19,23,50,51]. Several long ncRNAs, including HOTAIR, have been suggested to interact with repressive chromatin-modifying complexes to carry out their regulatory functions. A similar scenario might be operating in cases where long ncRNA functions to enhance gene expression, as shown for the long ncRNA located in the HOXA cluster, HOTTIP [19]. Future technological advances that will enable rigorous biochemical isolation of ncRNAs and their associated protein complexes, combined with better immunological reagents for analyses of such ncRNAs, are needed to begin deciphering the mysteries of this brave new world of ncRNAs.
Acknowledgments
We wish to thank the members of the Shiekhattar Laboratory for helpful discussions. UAO is supported by the Danish Research Council.
References
- 1.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orom U, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 8.Ponjavic J, et al. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattick JS, et al. A global view of genomic information; moving beyond the gene and the master regulator. Trends Genet. 2010;26:21–28. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Ponting CP, et al. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Wahl MC, et al. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinger ME, et al. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct Genomic Proteomic. 2009;8:407–423. doi: 10.1093/bfgp/elp038. [DOI] [PubMed] [Google Scholar]
- 15.Sleutels F, et al. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 16.Brockdorff N, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 17.Jia H, et al. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo H, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulveling D, et al. When one is better than two: RNA with dual functions. Biochimie. 2011;93:633–644. doi: 10.1016/j.biochi.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Bernardi G, et al. Compositional patterns in vertebrate genomes: conservation and change in evolution. J Mol Evol. 1988;28:7–18. doi: 10.1007/BF02143493. [DOI] [PubMed] [Google Scholar]
- 22.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko S, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 26.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer TR, et al. Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res. 2011;39:2393–2403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrow J, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):S4.1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Bakel H, et al. Most ‘dark matter’ transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pauler FM, et al. Silencing by imprinted noncoding RNAs: is transcription the answer? Trends Genet. 2007;23:284–292. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 33.Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 34.Mermoud JE, et al. Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J Cell Biol. 1999;147:1399–1408. doi: 10.1083/jcb.147.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koerner MV, et al. The function of non-coding RNAs in genomic imprinting. Development. 2009;136:1771–1783. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotake Y, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Curr Opin Genet Dev. 2011 doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling J, et al. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J Biol Chem. 2004;279:51704–51713. doi: 10.1074/jbc.M404039200. [DOI] [PubMed] [Google Scholar]
- 43.Faedo A, et al. Identification and characterization of a novel transcript down-regulated in Dlx1/Dlx2 and up-regulated in Pax6 mutant telencephalon. Dev Dyn. 2004;231:614–620. doi: 10.1002/dvdy.20152. [DOI] [PubMed] [Google Scholar]
- 44.Kohtz JD, Fishell G. Developmental regulation of EVF-1, a novel non-coding RNA transcribed upstream of the mouse Dlx6 gene. Gene Expr Patterns. 2004;4:407–412. doi: 10.1016/j.modgep.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Feng J, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefevre P, et al. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell. 2008;32:129–139. doi: 10.1016/j.molcel.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ebisuya M, et al. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10:1106–1113. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- 48.Tian D, et al. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trievel RC, Shilatifard A. WDR5, a complexed protein. Nat Struct Mol Biol. 2009;16:678–680. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- 50.Franke A, Baker BS. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell. 1999;4:117–122. doi: 10.1016/s1097-2765(00)80193-8. [DOI] [PubMed] [Google Scholar]
- 51.Kelley RL, et al. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- 52.Loewer S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]