SUMMARY
Paramutation is an epigenetic phenomenon that has been observed in a number of multicellular organisms. The epigenetically silenced state of paramutated alleles is not only meiotically stable but also “infectious” to active homologous alleles. The molecular mechanism of paramutation remains unclear, but components involved in RNA-directed DNA methylation (RdDM) are required. Here, we report a multi-copy pRD29A-LUC transgene in Arabidopsis thaliana that behaves like a paramutation locus. The silent state of LUC is induced by mutations in the DNA glycosylase gene ROS1. The silent alleles of LUC are not only meiotically stable but also able to transform active LUC alleles into silent ones, in the absence of ros1 mutations. Maintaining silencing at the LUC gene requires action of multiple pathways besides RdDM. Our study identified specific factors that are involved in the paramutation-like phenomenon and established a model system for the study of paramutation in Arabidopsis.
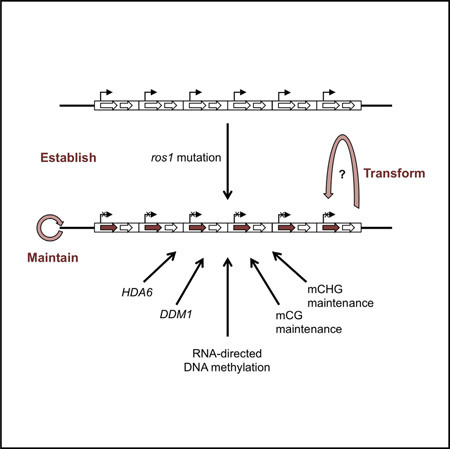
Graphical abstract
INTRODUCTION
Paramutation is an epigenetic phenomenon that involves trans interactions between two homologous sequences that usually exhibit different transcriptional activities (Chandler and Stam, 2004). One of the two homologous alleles (termed “paramutagenic”) is able to transform the other homologous allele (termed “paramutable”) into a new paramutagenic allele. The first reported example of paramutation is the maize red1 (r1) gene, which encodes a transcription factor that regulates anthocyanin synthesis and confers red color to corn kernels when strongly expressed (Brink, 1956). When the weakly expressed r1’ and the strongly expressed r1 alleles are combined by crossing, the F1 and F2 progenies all exhibit the phenotype of the r1’ plants because the r1’ allele transforms r1 into r1’. The newly transformed allele is also meiotically heritable and is able to transform active r1 alleles. Paramutation represents a special case where the epigenetic state of a gene is not only stable through meiosis but also changes the epigenetic state of its homologous sequences.
Almost every case of paramutation identified so far is associated with DNA repeats. Paramutation of the booster (b1) locus in maize, for example, is regulated by seven tandem repeats of an 853-bp sequence that are located ~100 kb upstream of the b1 gene. Moreover, the seven tandem repeats are both necessary and sufficient for the paramutation of the b1 gene (Stam et al., 2002). In plants, silencing of repetitive sequences including transposons is important for maintaining genome integrity and for plant development. Stable silencing typically requires removal of epigenetic modifications associated with transcriptional activation, such as histone acetylation and trimethylation at histone H3 lysine 4 (H3K4me3), and with deposition of repressive epigenetic modifications, such as DNA methylation and/or methylation at histone H3 lysine 9 (H3K9me1/2). DNA methylation in plants can occur at both symmetric sequence contexts (CG and CHG, where H = A, C, T) and asymmetric sequence contexts (CHH). Maintenance of the three types of DNA methylation (CG, CHG, and CHH) involves different processes associated, respectively, with DNA replication, histone modifications (H3K9me1/2), and small interfering RNAs (siRNAs). The siRNA-guided DNA methylation process, called RNA-directed DNA methylation (RdDM), is also required for de novo DNA methylation of all three types.
Genetic screens in maize have identified six genes that are required for paramutation (reviewed in Hollick, 2012). Five of those have homologs in Arabidopsis that are involved in siRNA generation. Mutation of the sixth gene also leads to a decrease in the siRNAs generated from the paramutation locus, suggesting that siRNAs likely play an important role in the trans interaction between paramutagenic and paramutable alleles. In Drosophila, Piwi-interacting RNA (piRNA) is required for paramutation of P-transposable element-derived transgenes (de Vanssay et al., 2012).
The exact role of siRNA in paramutation, however, is still unclear. The siRNA is not sufficient to confer paramutation. In Arabidopsis, 24-nt siRNAs are generated from thousands of loci, most of which are DNA repeats, but paramutation has not been reported for any of them (Lee et al., 2012; Zhang et al., 2007). In addition, siRNAs can be detected from both the paramutagenic and paramutable alleles (Arteaga-Vazquez et al., 2010). In maize, direct evidences showing that RNA polymerase IV transcribes the loci that undergo paramutation are lacking (Erhard et al., 2009). However, overexpression of a hairpin RNA that can be processed into the same 24-nt siRNAs can induce the paramutable B-I allele into the paramutagenic B’ (Arteaga-Vazquez et al., 2010), suggesting that the effect of siRNAs on paramutation may depend on siRNA level.
Other factors in addition to RdDM are also involved in paramutation. It is proposed that paramutation may involve physical interaction between the two alleles, which can be mediated by the homologous DNA sequence itself or other proteins. A protein called CBBP (CXC-domain b1 repeat binding protein) was found to interact with the seven tandem repeats upstream of the b1 gene, and overexpression of the CBBP gene induces paramutation (Brzeska et al., 2010). The Arabidopsis genome does not contain a gene homologous to CBBP but does encode three proteins that also have the CXC domain. Functions of those proteins have not been reported.
In this study, we report a pRD29A-LUC transgene system in Arabidopsis whose behavior resembles that of classical paramutation. Silencing of the transgene is induced by ros1 mutations but can be maintained independent of ros1. The silenced allele acts as a paramutagenic allele and converts active pRD29A-LUC into a silenced one. Extensive genetic experiments found that not only genes involved in RdDM function but also genes involved in CG/CHG methylation and specific histone modifications are required to maintain the silenced state of the transgene. This system provides an excellent model for studying paramutation in the reference plant Arabidopsis, which will be facilitated by the abundant genetic and epigenetic resources in the community.
RESULTS
The ros1 Mutation Induces TGS of pRD29A-LUC
Transcription of the RD29A gene is activated by cold or salt stresses. We showed previously that the promoter of a pRD29A-LUC transgene was under dynamic regulation by two antagonizing processes: RdDM and active DNA demethylation. Through forward genetic screens, we identified many genes that function in RdDM and DNA demethylation (He et al., 2009). The original genetic screens were performed in the C24 ecotype. To utilize the abundant genetic resources in the Col ecotype, we introduced a similar vector that contains the pRD29A-LUC transgene into Col-0 plants (Figure S1A). As expected, the Col transgenic plants exhibit stable and strong luciferase signals upon salt or cold treatment (Figure 1A).
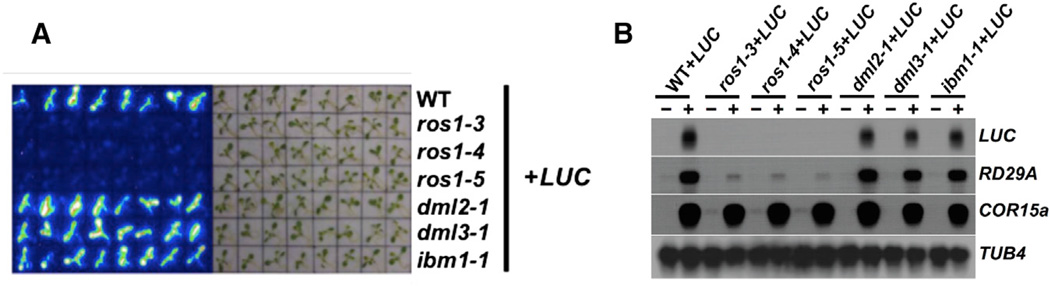
Figure 1. Silencing of the pRD29A-LUC Reporter Gene Specifically Requires the ros1 Mutation.
(A) Luminescence imaging of 2-week-old wild-type and mutant plants with the pRD29A-LUC transgene.
(B) Northern blotting analyses of the LUC transgene and endoRD29A. Two-week-old seedlings with indicated genotype before (indicated by “−”) and after (indicated by “+”) stress treatment were analyzed. TUB8 and COR15A each serve as the loading control and the control for normal cold response.
We crossed the Col-pRD29A-LUC line with several anti-silencing mutants. Consistent with previous findings, pRD29A-LUC is silenced by all three independent alleles of ros1: ros1–3, ros1–4, and ros1–5 (Figures 1A and S1B). pRD29A-LUC was not silenced, however, in other mutants that are also involved in anti-silencing, including dml2, dml3, and ibm1 (Figure 1A). DML2 and DML3 are homologs of ROS1. IBM1 encodes the histone demethylase specific for the lysine 9 of histone H3 (Miura et al., 2009). Similar levels of LUC transcripts can be detected in wild-type (WT), dml2, dml3, and ibm1 plants, but not in any of the ros1 mutants that are stress treated (Figure 1B), indicating that proper transcription of the pRD29A-LUC transgene specifically requires the action of ROS1.
When the transgenic RD29A promoter is methylated in the ros1-1 mutant, the endogenous RD29A promoter (endoRD29A) is also methylated and silenced (Gong et al., 2002), because the siRNAs generated from the transgenic RD29A promoter act in trans to guide de novo methylation of the endoRD29A promoter (Kapoor et al., 2005). The same effect of silenced pRD29A-LUC on endoRD29A was also observed in the Col background ros1 mutant plants. The transcript levels of endoRD29A remain unchanged in dml2+LUC, dml3+LUC, and ibm1+LUC plants but are dramatically reduced in the three ros1 mutant alleles tested (Figure 1B).
The Silenced pRD29A-LUC Allele Behaves like a Paramutagenic Allele
When genetic crosses were made between ros1–4 harboring the pRD29A-LUC transgene (referred to as ros1+LUC) and Col-0 plants, the F1 plants exhibit no LUC signals (Figure 2A). The F2 progeny generated from self crossing of the above F1 plants also show a dark luminescence phenotype, even though 3/4 of the plants that have the LUC transgene should contain functional copies of ROS1 (Figure 2A). These results indicate that maintaining the silenced state of the pRD29A-LUC transgene is independent of the ROS1 gene.
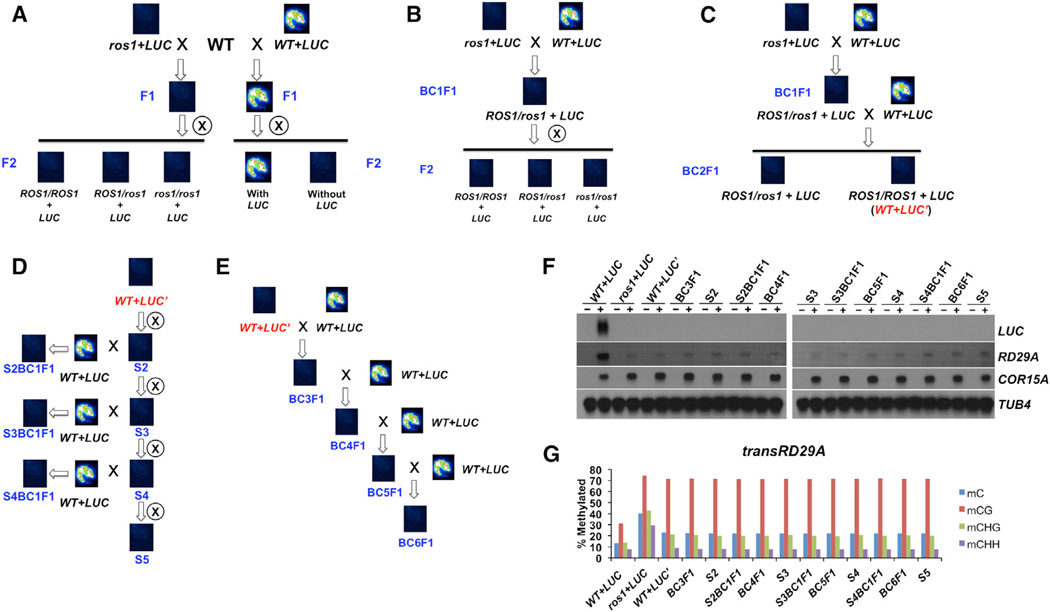
Figure 2. Characterization of the Paramutation-like Phenotype and Molecular Features of the pRD29A-LUC Transgene.
(A–E) Schematic diagrams showing the genetic crosses performed on plants with indicated genotype (black or red italic letter). The luminescence image above each genotype represents the overall LUC phenotype of 45–50 seedlings. Blue letters indicate the generation number of plants used for the analyses: “F” denotes filial generation of crosses; “BC” denotes crosses made with WT+LUC plants; “S” denotes self-crosses.
(F) Northern blotting analyses for pRD29A-LUC and endoRD29A in plants listed in (A)–(E). Two-week-old seedlings before (indicated by −) and after (indicated by +) stress treatment were used for the analyses. TUB8 and COR15A serve as the loading control and the stress-response control, respectively.
(G) DNA methylation levels of the transgenic RD29A promoter region as examined by bisulfite sequencing.
We also found that the silenced pRD29A-LUC transgene is able to transform active pRD29A-LUC alleles into a silenced state. We performed genetic crosses between ros1+LUC and WT plants harboring the active pRD29A-LUC transgene (referred to as WT+LUC). Because this cross resembles the backcross we normally do after genetic screens, the resulting F1 progeny were referred to as BC1F1 plants. No luminescence signals were observed in the BC1F1 plants (Figures 2B and S2A), indicating that either the ros1–4 mutation or the silenced pRD29A-LUC allele behaves dominantly. Interestingly, no luciferase signals were detected in any of the F2 plants either (Figures 2B and S2A). Analyses of transcript levels by qRT-PCR demonstrate cold activation of the transgene was observed only in the WT+LUC plants, but not in ros1+LUC or the F1 and F2 plants (Figure S2B). This non-Mendelian behavior was not observed at other loci, for example, the Pm36 locus. The DNA methylation level at Pm36 in the heterozygous ros1 plants (F2) is indistinguishable from WT plants (Figure S2C). This property of the pRD29A-LUC transgene resembles that of paramutation genes in maize. Thus, we followed the nomenclature of paramutation and referred to the active and silenced alleles as LUC and LUC’, respectively.
We next determined whether the “conversion” of pRD29A-LUC from the active state to the silenced one requires the ros1 mutation. The BC1F1 plants (ROS1 +/−) described earlier were “backcrossed” to WT+LUC (ROS1 +/+) again to generate BC2F1, all of which exhibit no luminescence signals (Figure 2C). We genotyped the BC2F1 population, and ROS1 (+/+) plants (WT+LUC’) were selected for further analyses (Figure 2C). First, WT+LUC’ plants were self-propagated for up to five generations (noted as “S2” to “S5”), and all of them lacked the luminescence phenotype (Figure 2D), indicating that silencing of the transgene is stable and meiotically heritable. When the filial generations from BC2F1 ROS1 (+/+; e.g., S2BC1F1) were also backcrossed to WT+LUC plants, we once again could not detect luciferase signals from the F1 progeny (Figure 2D). Next, we crossed the WT+LUC’ plants (Figure 2C) to WT+LUC to generate BC3F1 (Figure 2E). BC3–6F1 plants all behave like LUC’ and lack luciferase signals (Figure 2E). To rule out the possibility that plants grown at different times may exhibit different phenotype, we examined LUC signals of rosette leaves from plants that were grown in the same batch (Figure S2D). Only leaves from WT+LUC plants exhibit bright luminescence signals upon stress treatment whereas leaves fromall other plants (presumably LUC’) remained dark. These results indicate that LUC’ (the silent allele) is able to convert LUC (the active allele) to LUC’; the new LUC’ is indistinguishable from the original LUC’ in that it is meiotically stable and has the ability to transform active LUC. Consistently, upon stress treatment, significant amount of LUC transcript was detected in WT+LUC plants, but not in any other plants that exhibit the LUC’ phenotype (Figure 2F).
Previous examples of paramutation found that DNA methylation is tightly linked to paramutated loci. Thus, we examined DNA methylation levels at the transgenic RD29A promoter (Figure 2G). WT+LUC plant contains less than 15% total DNA methylation levels at the transgenic RD29A promoter whereas the same sequence was methylated to ~40% in ros1+LUC plants. In the absence of the ros1 mutation, all the other LUC’ plants have similar medium levels of DNA methylation: ~25% (Figure 2G), suggesting their epigenetic states are rather similar and stable.
Paramutation not only occurs between two allelic genes but also occurs between transgenes and homologous endogenous genes at non-allelic positions (Chandler and Stam, 2004). Thus, we determined whether the endogenous RD29A gene also has the paramutation-like phenomenon. Similar to the transgene, stress-induced expression of endoRD29A is repressed in ros1+LUC and WT+LUC’ plants but unaffected in WT+LUC plants (Figure S2E). After crossing WT+LUC’ to Col-0 (WT) plants, however, plants without the LUC transgene in the F2 generation still showed WT levels of endoRD29A expression after stress treatment (Figure S2E), indicating that the active endoRD29A allele is not affected by the silenced endoRD29A or pRD29A-LUC. The LUC phenotype is correlated with the DNA methylation levels at the endoRD29A promoter: WT or ros1–4 plants were not methylated, whereas WT+LUC’ and ros1+LUC plants were heavily methylated (Figure S2F). These results demonstrate the paramutation-like phenomenon only occurs at the transgene.
In summary, establishing the silenced state of the pRD29A-LUC transgene, or LUC’, can be achieved by introducing ros1 mutations. Once established, LUC’ is meiotically stable in the absence of ros1 and is able to transform LUC into LUC’. The interaction between LUC and LUC’ fits the description of paramutation.
Multiple Gene-Silencing Pathways Are Required to Maintain the Silenced State of LUC’
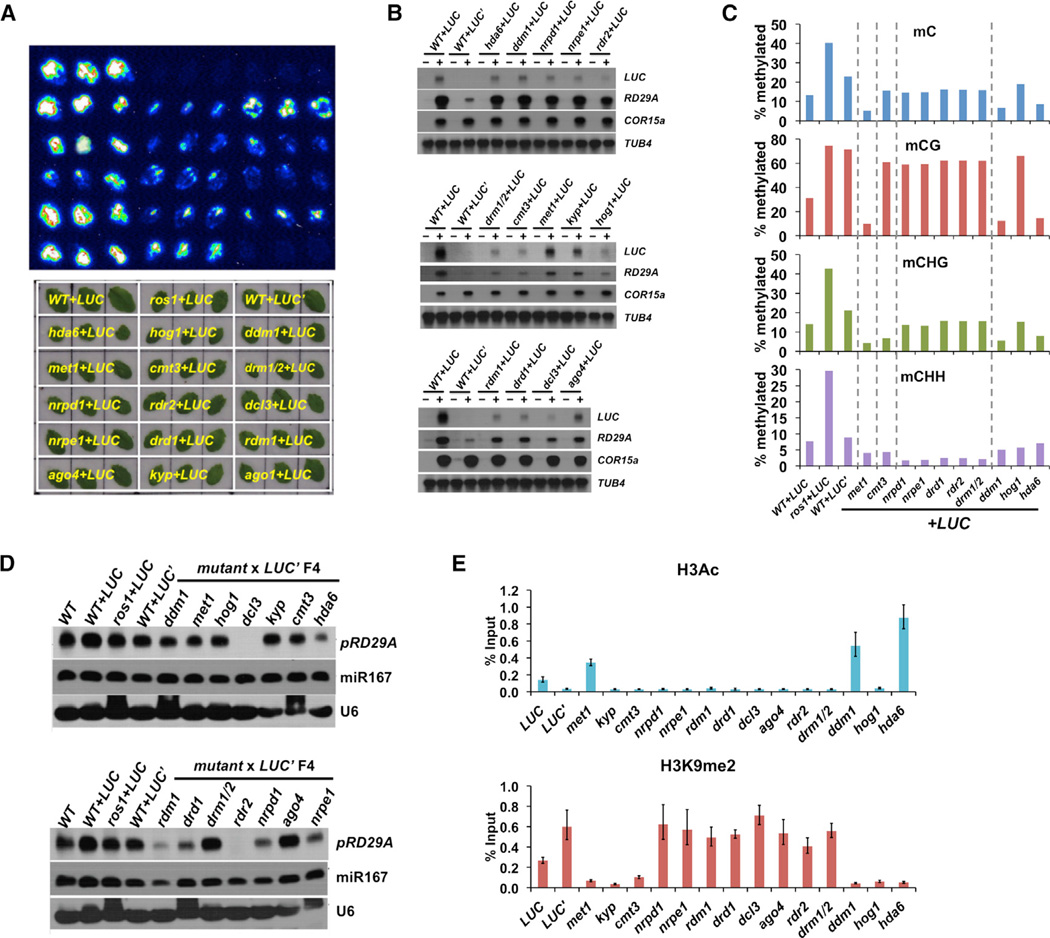
By utilizing the available mutants in the Col ecotype that affect DNA methylation and/or histone modifications, we next determined which epigenetic marks are required to maintain the silenced state of LUC’. We crossed WT+LUC’ plants (Figure 2C) to mutants involved in gene silencing, and luminescence signals were examined in filial generations. None of the F1 or F2 plants showed any LUC signals (Figure S3A). In the F3 plants, however, transgene silencing was released to different degrees in those homozygous mutants except for ago1 (Figure 3A). Whereas all the other genes are known to affect DNA methylation or histone modifications, AGO1 is required for microRNA production and post-transcriptional gene silencing (PTGS) (Vaucheret et al., 2004). Thus, the results suggest PTGS is not involved in LUC’ silencing.
Figure 3. Multiple Epigenetic Pathways Are Required to Maintain LUC’ Silencing.
(A) Bioluminescence and bright field imaging results using rosette leaves from mutant+LUC plants. Genotypes of the plants are marked in yellow on the bright field image. The F3 plants used for the analyses were pre-screened for the presence of pRD29A-LUC transgene.
(B) Transcript levels of pRD29A-LUC and endoRD29A genes in the F3 mutant plants are examined by northern blotting. Please note that the signals from WT+LUC plants differ on different blots due to different exposure time, which serve as a positive control. Stress-treated and control plants are indicated by − and +, respectively. TUB4 and COR15A each serve as the loading control and the control for normal cold response.
(C) DNA methylation levels at the transgenic RD29A promoter in the mutant+LUC’ crosses F4 plants as measured by bisulfite sequencing.
(D) Northern blotting analyses of 24-nt siRNAs generated from the RD29A promoter (endogenous + transgenic). U6 snoRNA and miR167 each serves as the loading control and microRNA pathway control.
(E) Chromatin immunoprecipitation followed by quantitative PCR was used to examine histone H3 acetylation and H3K9me2 levels at the transgenic RD29A promoter. Error bars indicate SD calculated from qPCR reactions of three technical replicates.
All of the RdDM mutants tested showed increased luminescence signals to a certain extent in the F3 plants, including nrpd1, rdr2, dcl3, rdm1, drd1, nrpe1, ago4, and drm1/2 (Figure 3A). Increased LUC transcript levels in those mutants are correlated with elevated luciferase signals (Figure 3B). As expected, all of the RdDM mutants showed a substantial decrease in the CHH methylation level at the transgene promoter and to a less extent in CG or CHG methylation levels (Figure 3C). siRNAs generated from the RD29A promoter were also examined using a pRD29A-specific probe (Figure 3D). RD29A-specific 24-nt siRNAs can be detected in Col-0 plants and are elevated in WT+LUC, ros1+LUC, and WT+LUC’ plants. Consistent with the current RdDM model (Matzke and Mosher, 2014), mutations in genes involved in siRNA production, including NRPD1, RDR2, and DCL3, lead to strong decreases in 24-nt siRNA levels at the RD29A promoter, whereas ago4 or drm1/2 had no effect on siRNA levels (Figure 3D). Interestingly, mutations in genes involved in generating scaffold RNAs, such as RDM1, DRD1, and NRPE1, also result in strong decreases of 24-nt siRNAs, suggesting that Pol V function contributes to 24-nt siRNA accumulation at the RD29A promoter (Figure 3D).
We also tested two genes, KYP and CMT3, involved in the regulation of CHG methylation. KYP is a histone methyltransferase that binds to methylated CHG and specifically methylates H3K9 (Johnson et al., 2007). CMT3 is a CHG-specific DNA methyltransferase that binds to H3K9me1/2 (Du et al., 2012). The two enzymes form a positive feedback loop and maintain CHG methylation levels. Thus, mutation in either gene usually leads to decreases in CHG methylation and H3K9 methylation simultaneously. Surprisingly, they had different effects on the silencing of the LUC’ allele: cmt3+LUC’ exhibited relatively weaker luminescence compared to kyp+LUC’ (Figures 3A and 3B). Both plants exhibited slightly reduced siRNA levels compared to WT+LUC’, which was correlated with their slightly reduced CHH methylation (Figure 3D). The difference in LUC signals is correlated with their effects on H3K9me2: the decrease in H3K9me2 was greater in kyp+LUC’ plants than in cmt3+LUC’ plants (Figure 3E).
CG methylation is also required for LUC’ silencing. MET1 is the major DNA methyltransferase responsible for CG methylation maintenance in Arabidopsis. Mutations in MET1 lead to significantly decreased DNA methylation levels in all three contexts and strong derepression of the LUC’ transgene (Figure 3).
We also tested HDA6, DDM1, and HOG1. HDA6 is a broad-specificity histone deacetylase that is required for the silencing of many RdDM target loci as well as rDNA repeats (To et al., 2011). DDM1 is an ATP-dependent chromatin remodeling factor that acts mainly on histone H1-containing transposons and repetitive sequences (Jeddeloh et al., 1998; Zemach et al., 2013). HOG1 encodes an S-adenosyl-L-homocysteine (SAH) hydrolase and is required for the generation of S-adenosyl-methionine (SAM), the methyl-group donor for both DNA and histone methyltransferases (Rocha et al., 2005). The common characteristic of the three genes is that a loss-of-function mutation changes the levels of DNA methylation as well as histone modifications. We found that hda6 and ddm1 lead to strong derepression of the LUC’ gene whereas hog1 has only weak effects (Figures 3A and 3B). This is correlated with their effects on DNA methylation levels at the transgenic promoter: hda6 and ddm1 reduced DNA methylation to the same levels as met1, whereas hog1 had only a small effect (Figure 3C). Among all the mutants tested, hda6, ddm1, and met1 are the only ones that show an increase in histone acetylation at the pRD29A-LUC promoter (Figure 3E). Thus, decreases in DNA methylation levels, but not changes in H3K9me2 levels or in histone H3 acetylation levels, are correlated with derepression of LUC’.
The pRD29A-LUC Transgene Is Likely Composed of Multi-copy Repeats
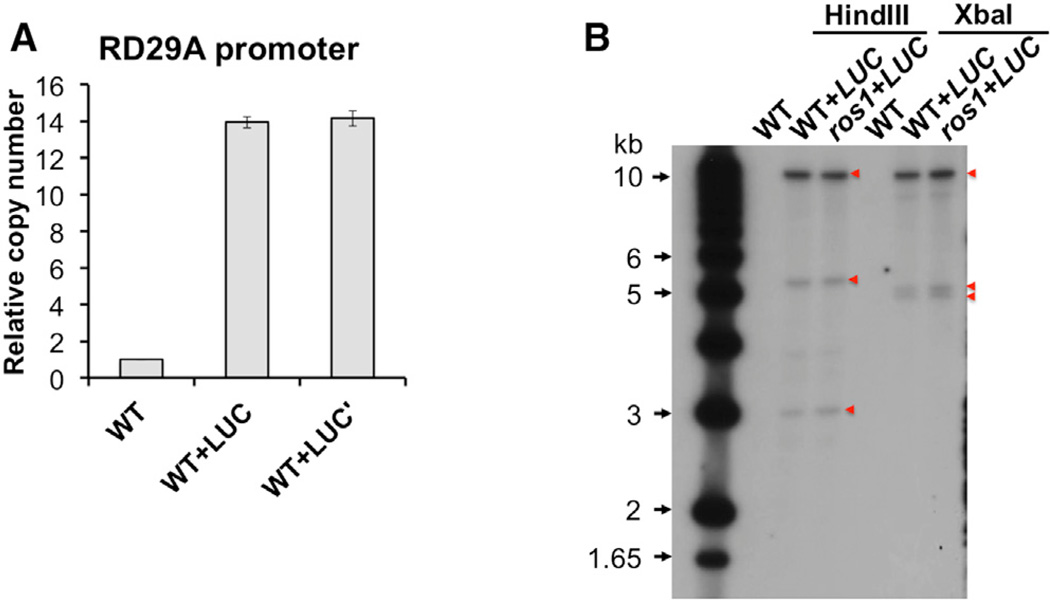
Next, we tested the possibility that the pRD29A-LUC transgene exists in the genome as DNA repeats. First, we used real-time PCR to quantify the number of RD29A promoter sequences in the genome. By using the TUB8 gene as the internal control, the numbers of RD29A promoter sequences in WT+LUC and ros1+LUC plants were quantified and normalized to Col-0 plants without the transgene. We found that both WT+LUC and ros1+LUC plants contain 14 copies of RD29A promoter sequences (Figure 4A). Because the genome contains only one copy of the endogenous RD29A gene, data suggest that the pRD29A-LUC transgene is a 13-copy repeat (Figure 4A).
Figure 4. The pRD29A-LUC Transgene Is Likely a 13-Copy Repeat.
(A) Number of RD29A promoter sequences in WT+LUC measured by qPCR. Non-transgenic WT plants were used as a reference of one. Error bars indicate SD calculated from qPCR reactions of three technical replicates.
(B) Southern blotting of HindIII- and XbaI-digested genomic DNA using a LUC-specific probe (Figure S1A). DNA size markers were indicated on the left side of the membrane. LUC-specific bands were indicated by red triangles.
We next carried out Southern blotting using a probe that targets the 30 portion of the LUC coding sequence (Figure S1A). For a single-repeat insertion, a single band larger than 2.6 kb would be expected, assuming the probe was specific. The size of the band should equal to 2.6 kb plus the distance between the closest restriction endonuclease site in the genome and the right border of the T-DNA insertion (Figure S1A). Because the 9.8-kb T-DNA is the unit that is inserted into the genomic DNA, if T-DNA is repeated multiple times in a head-to-tail manner, then a strong band of 9.8 kb plus another band representing the right border fragment would be expected. What we observed are three bands with one strong band runs around 10 kb (corresponds to the 9.8-kb T-DNA) and two weaker bands that are >3 kb (correspond to two right border fragments) in either HindIII- or XbaI-digested genomic DNA samples (Figure 4B); these are consistent with the hypothesis that the T-DNA were inserted into two sites of the genome. Alternatively, because the structure of repeats is unknown, one of the two shorter bands could be due to complex structures such as inverted repeats or a truncated repetitive unit.
To find out whether the T-DNA repeats were inserted into the genome in two separate locations, we mapped the pRD29A-LUC transgene using F2 population generated from C24+LUC and Col+LUC’ crosses. Because the lengths of LUC (C24) and LUC (Col) coding sequences are slightly different, the two transgenes can be distinguished using PCR. We found that the pRD29A-LUC transgene was mapped to two locations in the genome: one in a 253-kp region on chromosome 1 and the other in a 280-kb region on chromosome 2 (Figure S4A).
We next asked whether each single T-DNA locus exhibited the paramutation phenomenon or whether the interaction between the two T-DNA loci was required for paramutation to occur. To address this question, the T-DNA repeats on two different chromosomes were isolated in the F2 plants generated from WT+LUC and WT (Col-0) crosses. The two T-DNA loci were arbitrarily named LUC1 and LUC2, respectively. We found that LUC1/2 plants emit bright luminescence upon stress treatment (Figure S4B). We again used quantitative PCR to measure the copy number of RD29A promoter sequence in LUC1/2 plants and found that the LUC1 and LUC2 loci contain six and seven copies of RD29A promoter sequences, respectively (Figure S4C).
The LUC1 and LUC2 plants were then crossed to WT+LUC’, and the luminescence phenotype were examined in the F1 and F2 progeny. No LUC signals were detected in the F1 or F2 plants (data not shown; Figure S4B). In the F2 progenies, we isolated homozygous LUC1 or LUC2 plants based on their difference in pRD29A copy number, and they were named LUC1’ and LUC2’. We tested whether LUC1’ or LUC2’ is able to convert LUC1 or LUC2 allele into a silenced state. Indeed, the F1 plants from crosses between LUC1 and LUC1’, or between LUC2 and LUC2’, lack luminescence upon stress treatment (Figure S4B), indicating LUC1’ or LUC2’ individually are sufficient to silence a homologous allele.
The T-DNA insertion contains pRD29A-LUC as well as the kanamycin-resistance gene p35S-NPT II (Figure S1A). We examined whether the NPT II locus also exhibits paramutation-like properties. Whereas ros1+LUC plants are sensitive to kanamycin and show clearly decreased NPT II transcript levels, the F1 plants from crosses between WT+LUC and ros1+LUC, or between WT+LUC and WT+LUC’, are resistant to kanamycin (Figure S4D). NPT II transcript levels in the F2 plants of WT+LUC/ros1+LUC crosses also follow Mendelian genetics (Figure S4E). These results indicate paramutation-like phenomenon is only observed for pRD29A-LUC, but not for the NPT II gene in the vicinity, even though the NPT II gene also exists as 13 copies in the genome (Figure S4F).
DISCUSSION
A Paramutation-like Phenomenon in Arabidopsis
Paramutation is an unusual epigenetic phenomenon that has been observed in plants, fungi, Drosophila, and mammals. Previous studies indicate that siRNAs and DNA methylation likely play important roles in paramutation, but a full explanation is still lacking (Hollick, 2012). The large genome size, limited availability of mutants, and long generation time are hurdles for studying the molecular mechanism of paramutation in maize. In this report, we describe a T-DNA transgene in Arabidopsis that behaves like a classical paramutation gene. Establishment of the silenced state of the pRD29A-LUC transgene is induced by mutations in the DNA glycosylase gene ROS1. Once generated, the silenced pRD29A-LUC (LUC’) allele can be meiotically transmitted in the absence of ros1 and is able to transform an active pRD29A-LUC (LUC) allele into a silenced allele (LUC’). The newly transformed LUC’ is indistinguishable from the original LUC’.
Limited cases of transgene silencing have been reported to exhibit paramutation-like properties in Arabidopsis. One example is the hygromycin phosphotransferase (HPT) transgene that is stably silenced in tetraploid Arabidopsis plants (Mittelsten Scheid et al., 2003). The HPT system, however, differs from the LUC system in several ways. Crossing tetraploids containing the silenced HPT gene to diploid WT plants (without transgene) generates Arabidopsis with two copies of the silenced HPT gene. Although the silencing of the two copies of HPT gene is stable for multiple generations, they are apparently insufficient to silence an active HPT allele (Mittelsten Scheid et al., 2003). If crosses are made between tetraploid Arabidopsis plants that harbor the silenced and active HPT gene, the F1 plants show uniform hygromycin resistance, indicating that the active HPT allele is expressed normally (Mittelsten Scheid et al., 2003). The active HPT alleles lose their transcriptional activity in only some of the F2 progeny of the above crosses.
Another two examples of paramutation-like phenomenon in Arabidopsis involve T-DNA insertions in the intron of an actively transcribed gene. T-DNA insertion into the middle of a gene is a common way to disrupt gene function in Arabidopsis. In the case of cob-6, where a SALK T-DNA is in the first intron of the COBRA gene, its phenotype is suppressed by crossing with another T-DNA mutant srf6-1 or other randomly selected SALK T-DNA lines (Xue et al., 2012). Similarly, ag-TD, which contains a T-DNA in the second intron of the AGAMOUS gene, is suppressed by other mutants that contain the same T-DNA sequence, such as yuc1-1 (Gao and Zhao, 2013). In both cases, suppression of the mutant phenotype is not due to the other loss-of-function mutant but due to the interaction among the same T-DNA sequences. Like paramutation, the suppressed mutant is relatively stable for generations and can convert the original loss-of-function mutant to a suppressed mutant allele (Gao and Zhao, 2013; Xue et al., 2012). Restoration of gene function (or suppression of mutant phenotype) is correlated with silencing of the selection marker gene within the T-DNA, and DNA methylation is likely involved (Xue et al., 2012). However, the nature of epigenetic changes in the T-DNA and how those changes lead to restoration of host gene function remains unclear.
Factors that Contribute to Maintaining the Silenced State of LUC’
We selected four groups of genes to test whether they are required for LUC’ silencing: a gene involved in miRNA function; genes in the RdDM pathway; genes involved in the maintenance of CHG and CG methylation; and genes indirectly involved in DNA methylation regulation, including DDM1, HDA6, and HOG1. Surprisingly, we found all the genes, except for AGO1, are required to maintain the silenced state of LUC’. The observation that H3Ac and H3K9me2 are decreased in only selected mutants suggests the two histone marks are not direct causes of silencing (Figure 3E). Significant decrease in non-CG methylation was observed in all the tested mutants (Figure 3C), but non-CG methylation only contributes to a small fraction of the total DNA methylation levels at the transgenic RD29A promoter. Without knowing how the DNA methylation information is quantitatively read out and translated into changes in the chromatin, it is difficult to understand the result, because significant decrease in non-CG methylation is also observed in WT+LUC’ plants compared to ros1+LUC plants (Figure 3C). It is possible that specific proteins bind to methylated DNA and higher-than-threshold levels of non-CG methylation can trigger changes in the chromatin structure and gene silencing. Alternatively, other factors besides DNA methylation also contribute to silencing of LUC’. For example, long noncoding RNAs have been shown to play important roles in structure maintenance and nuclear organization (Rinn and Guttman, 2014). Disruption of RdDM genes affects noncoding RNA production by RNA polymerase IV or V, which in turn may affect the silenced state of LUC’.
On Establishing Paramutation
In maize, six genes were identified in genetic screen that search for factors necessary to maintain silenced states of paramutation loci, but not all of them are required for the establishment of paramutation (Barbour et al., 2012; Hale et al., 2007). All of the six genes are required for siRNA accumulation, but whether other RdDM components also participate in paramutation remains elusive. We found not only the RdDM pathway but also CG and CHG methylation maintenance are required to maintain silencing of LUC’. The next step is to test whether they are also required for the conversion process, by which LUC becomes LUC’.
DNA repeats is a feature that is closely linked to maize paramutation (for reviews, see Chandler, 2010). It was found that the number of repeats upstream of the B’ allele is positively correlated with the degree of silencing and paramutation (Stam et al., 2002). We also found the pRD29A-LUC transgene exists in multiple copies in the genome. They are likely distributed in two locations with six and seven tandem repeats of T-DNA, respectively (Figure S4). It was proposed that the junction sequences of tandem repeats create features that are distinct from the single repeat unit and may be important for paramutation or small RNA production (Chandler, 2010). However, in our case, pRD29A-LUC exists as dispersed repeats and the nearby p35S-NPT II gene does not show similar paramutation-like phenotype (Figure S4D), suggesting that the repeats of specific sequences contribute to paramutation.
It will be interesting to examine whether homologs of genes identified in our study also promote paramutation in maize. If it is confirmed that the same paramutation factors identified in Arabidopsis also play a role in maize, the LUC/LUC’ system has the advantage of a much-simpler genome and abundant genetic resources. Further studies of the system may help with characterization of the epigenetic nature of paramutation as well as quick identification of the core paramutation factors.
EXPERIMENTAL PROCEDURES
Plant Material and Genetic Analyses
The pRD29A-LUC transgenic line used in this study was obtained by agrobacterium transformation using the floral-dip protocol. For each type of cross indicated in Figure 2, typically two reciprocal crosses were made. Then, the luminescence phenotype of 45–50 seedlings from each cross is assayed on the plate.
To identify genes that are required to maintain silencing of LUC’, the pRD29A-LUC transgene (LUC’) was introduced into all the mutants (Table S2) by genetic crosses. To confirm the function of mutants used in the study, genomic DNA was digested using a DNA methylation sensitive restriction enzyme and amplified using specific sets of primers (chop-PCR) targeting 5S rDNA repeats (Figure S3B) and AtSN1 (Figure S3C) to assay for the DNA methylation status at these loci. Semiquantitative PCR was performed on the F3 plants generated from the crosses, and only seedlings with the highest LUC transgene signals and homozygous mutations were selected. Quantitative real-time PCR was then used to identify F3 plants that contain 14 copies of the pRD29A sequences (Col WT plant was used as control; Figure S3D), and only those plants were used for luminescence analyses (Figure 3A).
RD29A Copy Number Analyses
Genomic DNA was extracted from each plant using the standard CTAB protocol. qPCR was then performed using gene-specific primers (Table S1) and the SYBR Green qPCR kit (New England Biolabs). Relative quantification of RD29A sequence in the transgenic plants was performed using TUB8 as the internal control and the Col-0 plant as a reference of one.
Supplementary Material
Highlights.
The ros1 mutation induces transcriptional silencing of a pRD29A-LUC transgene
The transgene exhibits a ros1-independent paramutation-like phenotype
The transgene exists as 13 copies in the genome
Maintaining silencing of the transgene requires multiple epigenetic pathways
ACKNOWLEDGMENTS
We thank Dong Wang, Donglei Yang, Ge Bai, Jay B. Hollick, Damon R. Lisch, and Ortrun Mittelsten Scheid for helpful communications and discussions. We thank Kunwu Li, Xianglin Cao, and Rebecca Stevenson for technical assistance. We also thank the following researchers for gifts of mutant seeds: Robert Fisher (ros1–3, dml2-1, and dml3-1); Tetsuji Kakutani (ibm1-1); Eric Richards (ddm1–10); Hervé Vaucheret (ago1–27); Honggui La (ros1–5); Judith Bender (suvh4/kyp); Ortrun Mittelsten Scheid (hog1); and Steve Jacobsen (ddc). This work was supported by NIGMS grants to J.-K.Z. and by the Chinese Academy of Sciences.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.04.034.
REFERENCES
- Arteaga-Vazquez M, Sidorenko L, Rabanal FA, Shrivistava R, Nobuta K, Green PJ, Meyers BC, Chandler VL. RNA-mediated transcommunication can establish paramutation at the b1 locus in maize. Proc. Natl. Acad. Sci. USA. 2010;107:12986–12991. doi: 10.1073/pnas.1007972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour J-ER, Liao IT, Stonaker JL, Lim JP, Lee CC, Parkinson SE, Kermicle J, Simon SA, Meyers BC, Williams-Carrier R, et al. required to maintain repression2 is a novel protein that facilitates locus-specific paramutation in maize. Plant Cell. 2012;24:1761–1775. doi: 10.1105/tpc.112.097618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink RA. A Genetic Change Associated with the R Locus in Maize Which Is Directed and Potentially Reversible. Genetics. 1956;41:872–889. doi: 10.1093/genetics/41.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska K, Brzeski J, Smith J, Chandler VL. Transgenic expression of CBBP, a CXC domain protein, establishes paramutation in maize. Proc. Natl. Acad. Sci. USA. 2010;107:5516–5521. doi: 10.1073/pnas.1001576107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler VL. Paramutation’s properties and puzzles. Science. 2010;330:628–629. doi: 10.1126/science.1191044. [DOI] [PubMed] [Google Scholar]
- Chandler VL, Stam M. Chromatin conversations: mechanisms and implications of paramutation. Nat. Rev. Genet. 2004;5:532–544. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- de Vanssay A, Bougé AL, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, Ronsseray S. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–115. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard KF, Jr, Stonaker JL, Parkinson SE, Lim JP, Hale CJ, Hollick JB. RNA polymerase IV functions in paramutation in Zea mays. Science. 2009;323:1201–1205. doi: 10.1126/science.1164508. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhao Y. Epigenetic suppression of T-DNA insertion mutants in Arabidopsis. Mol. Plant. 2013;6:539–545. doi: 10.1093/mp/sss093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Hale CJ, Stonaker JL, Gross SM, Hollick JB. A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 2007;5:e275. doi: 10.1371/journal.pbio.0050275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick JB. Paramutation: a trans-homolog interaction affecting heritable gene regulation. Curr. Opin. Plant Biol. 2012;15:536–543. doi: 10.1016/j.pbi.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Bender J, Richards EJ. The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 1998;12:1714–1725. doi: 10.1101/gad.12.11.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Agius F, Zhu JK. Preventing transcriptional gene silencing by active DNA demethylation. FEBS Lett. 2005;579:5889–5898. doi: 10.1016/j.febslet.2005.08.039. [DOI] [PubMed] [Google Scholar]
- Lee TF, Gurazada SG, Zhai J, Li S, Simon SA, Matzke MA, Chen X, Meyers BC. RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics. 2012;7:781–795. doi: 10.4161/epi.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid O, Afsar K, Paszkowski J. Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat. Genet. 2003;34:450–454. doi: 10.1038/ng1210. [DOI] [PubMed] [Google Scholar]
- Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T. An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 2009;28:1078–1086. doi: 10.1038/emboj.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J, Guttman M. RNA Function. RNA and dynamic nuclear organization. Science. 2014;345:1240–1241. doi: 10.1126/science.1252966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha PS, Sheikh M, Melchiorre R, Fagard M, Boutet S, Loach R, Moffatt B, Wagner C, Vaucheret H, Furner I. The Arabidopsis HOMOLOGY-DEPENDENT GENE SILENCING1 gene codes for an S-adenosyl-L-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell. 2005;17:404–417. doi: 10.1105/tpc.104.028332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M, Belele C, Dorweiler JE, Chandler VL. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 2002;16:1906–1918. doi: 10.1101/gad.1006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T, et al. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 2011;7:e1002055. doi: 10.1371/journal.pgen.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Ruprecht C, Street N, Hematy K, Chang C, Frommer WB, Persson S, Niittylä T. Paramutation-like interaction of T-DNA loci in Arabidopsis. PLoS ONE. 2012;7:e51651. doi: 10.1371/journal.pone.0051651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.