Abstract
Anionic lipids act as signals for the recruitment of proteins containing cationic clusters to biological membranes. A family of anionic lipids known as the phosphoinositides (PIPs) are low in abundance, yet play a critical role in recruitment of peripheral proteins to the membrane interface. PIPs are mono-, bis-, or trisphosphorylated derivatives of phosphatidylinositol (PI) yielding seven species with different structure and anionic charge. The differential spatial distribution and temporal appearance of PIPs is key to their role in communicating information to target proteins. Selective recognition of PIPs came into play with the discovery that the substrate of protein kinase C termed pleckstrin possessed the first PIP binding region termed the pleckstrin homology (PH) domain. Since the discovery of the PH domain, more than ten PIP binding domains have been identified including PH, ENTH, FYVE, PX, and C2 domains. Representative examples of each of these domains have been thoroughly characterized to understand how they coordinate PIP headgroups in membranes, translocate to specific membrane docking sites in the cell, and function to regulate the activity of their full-length proteins. In addition, a number of novel mechanisms of PIP-mediated membrane association have emerged, such as coincidence detection – specificity for two distinct lipid headgroups. Other PIP-binding domains may also harbor selectivity for a membrane physical property such as charge or membrane curvature. This review summarizes the current understanding of the cellular distribution of PIPs and their molecular interaction with peripheral proteins.
Keywords: C2 domain; FYVE domain; lipid binding; membrane binding; phosphoinsoitide; PI(3)P; PI(4,5)P2; PI(3,4,5)P3; PH domain; peripheral protein
1. Phosphoinositides
Cellular lipid membranes are dynamic structures that contain >1000 different lipid species (van Meer 2005). This dynamic variety of lipids, which includes the phosphoinositides (PIPs)1, provides spatial and temporal signals to mediate and direct interactions with target proteins. PIPs are derived from phosphatidylinositol (PI), synthesized in the ER by a PI synthase enzyme, which utilizes CDP-diacylglycerol (DAG) and myo-inositol (Agranoff et al. 1958). PI is then transported from the ER by PI transfer proteins (Cockcroft et al. 2007; Ile et al. 2010) and possibly vesicular trafficking to different cellular membranes. Recently, the PI synthase enzyme was found localized in a highly mobile organelle originating from the ER (Kim et al. 2011). The discovery of this organelle harboring PI synthase suggests PI may be dynamically disseminated throughout the cell via this machinery.
Once PI is distributed it can be reversibly phosphorylated on the 3, 4, and/or 5 hydroxyl group yielding seven different PIP species (Figures 1 and 2). This includes the monophosphorylated (PI(3)P, PI(4)P, and PI(5)P) as well as the bis (PI(3,4)P2, PI(3,5)P2, and PI(4,5)P2) and triphosphorylated (PI(3,4,5)P3) forms (Figure 2). PI can vary from ~ 10-20 mol% of total lipid in different cells and tissues (Nasuhoglu et al. 2002; Wenk et al. 2003) while phosphorylated PIPs make up less than 1% of the total cellular lipid pool. Despite such low cellular concentrations of PIPs they regulate a large number of cellular processes including membrane trafficking, cell growth and survival, cytoskeletal dynamics, and chemotaxis (Di Paolo and De Camilli 2006; Falke and Ziemba 2014; Roth 2004; Balla 2013). To date the PIP kinases and PIP phosphatases responsible for maintaining the cellular pools of PIPs have been shown to be expressed in the cytosol or at cellular sites of mammalian cells in a manner that directs the production of distinct pools of PIPs in the cytosolic leaflet of cellular organelles (Balla 2013) as well as in the nucleus (Balla 2013; Shisheva 2013). This distribution enables, at the molecular level, PIPs to selectively regulate membrane trafficking machinery, lipid transfer proteins, enzymes, ion channels, and endocytic and exocytic machinery (Balla 2013; Paolo and De Camilli 2006).
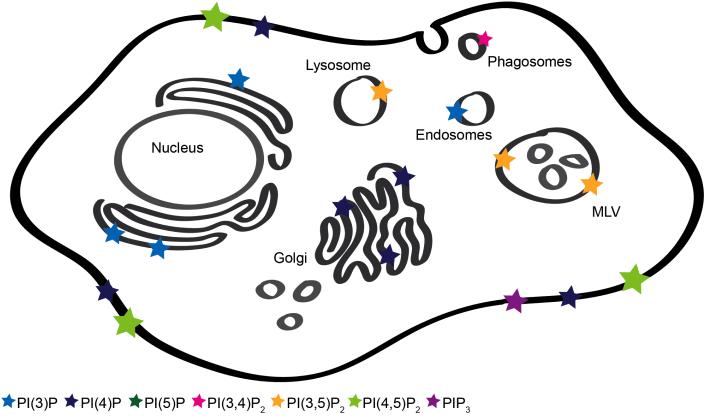
Figure 1. Cellular Distribution of PIPs.
A cartoon of a cell is shown to illustrate the known location of PIPs in mammalian cells. The relative abundance of PIPs is depicted with the size of the star shown.
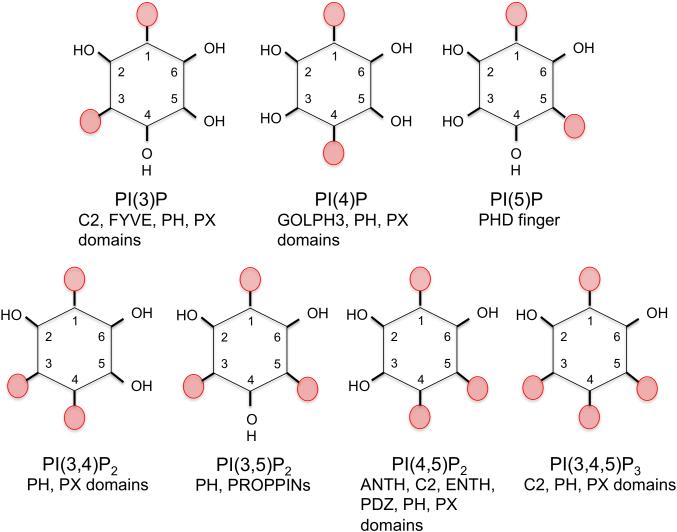
Figure 2. There are seven different PIPs in mammalian cells that mediate recruitment of peripheral proteins to cellular membranes.
The headgroup of each PI is shown with known effector domains listed below each.
1.1 Cellular Distribution of Phosphoinositides
Phosphoinositides are enriched with polyunsaturated fatty acids at the sn-2 position and mainly contain 1-stearoyl and 2-arachidonoyl chains in the cellular setting. The arachidonoyl chain, which has four double bonds, can cause disruption of dense membrane packing as the acyl chain is kinked. The effects of lipid packing density by PIPs may influence interactions at the membrane interface as electrostatic interactions between peripheral proteins and loosely packed lipids have been shown to be stronger than that with more densely packed lipids (Bigay et al. 2012; Mesmin et al. 2007). When PIPs are mixed with other lipids in cellular membranes they can form PIP-enriched regions (Levental et al. 2008; Redfern and Gericke 2004; Wang et al. 2014) that can be further stabilized by peripheral proteins that cluster PIPs (McLaughlin and Murray 2005; Saarikangas et al. 2009). Clustering of PIPs may provide areas of the membrane interface to attract polycationic proteins and proteins with PI-binding domains. Overall, there is a lack of quantitative data in vitro and in cells on the clustering and mixing of lipids on PIP-binding proteins. We feel this is an emerging area of interest to pursue with future biophysical experiments and work by Kooijman and colleagues strongly suggest that lipid-lipid interactions (Graber et al. 2012; Graber et al. 2013) could play a profound role in the ways peripheral proteins associate with membranes. For instance, PI and PI(4,5)P2 cooperatively form mixed domains that may regulate the spatial organization of cell signaling events. Taking into account cellular distribution of PI and its rapid phosphorylation to PIPs, which can then be dephosphorylated back to PI these lipid mixtures may play an important role in regulating lipid kinases and phosphatases as well as peripheral proteins with anionic lipid selectivity of PIP specificity.
PI(4,5)P2 and PI(4)P are the most abundant of the PIPs, enriched in the PM and Golgi, respectively, and make up ~0.2-1% of the cellular lipid pool (Balla et al. 1988; Di Paolo and De Camilli 2006; van Meer 2008; Hammond et al. 2009; Kutateladze 2010) with PI(4,5)P2 estimated to be in the range of 5,000-20,000 molecules/μm2 of the PM cytosolic leaflet (Falkenburger et al. 2010). The other phosphoinositides such as PI(3)P and PI(3,4,5)P3 are found in even smaller quantities at ~20-30% and 2-5% the level of PI(4,5)P2, respectively (See Figure 1). The PM harbors ~20-30 mol% anionic lipids on its inner leaflet (McLaughlin et al. 2002; McLaughlin and Murray 2005; Vance and Steenbergen 2005) that creates an overall negative electric field that attracts peripheral membrane proteins with cationic patches (Olivotto 1996). The electronegativity of the PM is attributed to the enrichment of polyvalent phosphoinositides including PI(4,5)P2 (McLaughlin et al. 2002; McLaughlin and Murray 2005) and PI(3,4,5)P3 (Heo 2006), which make up a small fraction of phospholipids on the inner leaflet of the PM (See Figure 1 for a depiction of the cellular distribution of PIPs) as well as other anionic species. The most abundant anionic lipid in the cytosolic leaflet of the PM is phosphatidylserine (PS), which comprises nearly 15-20% of the inner leaflet (Vance and Steenbergen 2005; Leventis and Grinstein 2010). PS significantly contributes to the recruitment of polycationic proteins (McLaughlin and Murray 2005) as well as proteins containing a specific PS binding motif such as some C2 domains (Cho and Stahelin 2006) and viral proteins (Adu-Gyamfi et al. 2013). The presence of PS in the PM and more recently on the cytosolic face of endosomes and budding vesicles from the Golgi has been detected using the PS specific C2 domain from lactadherin (Yeung et al. 2008, Fairn et al. 2011). The presence of anionic lipids such as PS in combination with other lipid headgroups such as PIPs provides a mode of regulation for receptor proteins that bind weakly to just one anionic lipid. Additive and synergistic effects of either the anionic charge at the PM or the presence of two distinct binding sites on a receptor protein (e.g., PI(4,5)P2 and PS) mediate the selective PM localization of these proteins.
Other examples of selective PI localization include PI(3)P, which is detected in early endosomes and PI(3,5)P2, a PI low in abundance found in early endosomes, late endosomes, and lysosomes (Li et al. 2013; McCartney et al. 2014). Recently, however, novel studies and methods have provided new insights into PIP localization. For instance, PI(4)P was found to be an important component of the PM cytosolic leaflet (Hammond et al. 2012; Nakatsu et al. 2012) while PI(3)P was found in the lumen of the ER in the malaria parasite (Bhattacharjee 2012) and in the smooth ER and Golgi of mammalian cells (Sarkes and Rameh 2010). The more enigmatic PI(5)P, which has emerged as a cytosolic and nuclear signal (Shisheva 2013), has been detected in the plasma membrane and enriched in the smooth ER and/or Golgi (Sarkes and Rameh 2010). Because most PIPs are found both at basal levels in cellular membranes and can be modulated by various cellular stimuli these studies underscore the importance of novel tools and methods to dissect the real-time distribution of PIPs in live cells.
One new quantitative method for cellular PI(4,5)P2 analysis was recently developed by Cho and colleagues (Yoon et al. 2011). The common approach to map cellular localization of anionic lipids such as PS or PIPs has been to fuse a fluorescent protein (e.g., GFP) with a protein module that has high affinity and selectivity for the anionic lipid of interest (Varnai and Balla 2006; Yeung et al. 2006; Yeung et al. 2008; Fairn et al. 2011). The PI(4,5)P2-binding ENTH domain was first engineered for better membrane binding affinity and minimal affinity for cellular proteins. Through labeling with an environmentally sensitive chemical probe (2-dimethylamino-6-acyl-napthalene group) on a free cysteine, the engineered ENTH domain serves as a ‘turn-on’ sensor that undergoes a large increase in fluorescence upon lipid-binding. In addition, the probe undergoes a blueshift upon PI(4,5)P2-dependent membrane binding, which allows for ratiometric detection of PI(4,5)P2 in vitro and in cells. The ratiometric approach allows researchers to overcome problems associated with fluorescently-tagged domains such as photobleaching or varying expression levels. The ENTH probe was able to be delivered to live cells by microinjection or liposome-mediated delivery and could robustly respond to PI(4,5)P2 fluctuations and detect the threshold level of PI(4,5)P2 required to trigger phagocytosis in immune cells. The approach designed by Cho and colleagues could be useful for other PIP signal detection as structural and functional knowledge of other PIP-binding domains are available and should allow engineering of lipid probes for other PIPs. While it may currently be difficult to sense both sides of membrane organelles using this chemical approach, it remains a considerable step forward in studying real-time PI signaling.
1.2 Cellular Manipulation of Phosphoinositides
Manipulation of cellular levels of phosphoinositides has been performed in various ways. These methods include overexpression of PIP phosphatases or kinases, PIP kinase selective inhibitors, monitoring translocation of fluorescently tagged PIP-binding domains, as well as direct chemical analysis of PIP levels (Suh and Hille 2007). Probably the most recent strategy of PIP manipulation has been the development of heterodimerization systems to selectively target cytosolic phosphatase domains to the cytoplasmic interface of cellular membranes to reduce PIPs and study the respective effects on peripheral proteins (Chang-Ileto et al. 2012; Varnai et al. 2006). Below we will highlight the studies that have selectively modulated PIPs at the cytosolic interface of cellular membranes and organelles.
1.2.1 Plasma Membrane
Heo and colleagues demonstrated the effect of modulating PI(4,5)P2 levels in real time on the PM recruitment of certain GTPase proteins with polybasic clusters (Heo et al. 2006). One of the first targetable phosphatase depletion systems consisted of the PM-localizing FRB protein and the 5’-specific Inp54p phosphatase linked to the FKBP12 protein. Inp54p metabolizes PI(4,5)P2 into PI(4)P. Heterodimerization of FRB and FKBP12 was induced by the rapamycin analog, iRap. The authors determined that recruitment of Inp54p by indole rapamycin resulted in the dissociation of the PI(4,5)P2-binding pleckstrin homology (PH) domain from the plasma membrane. However, PI(4,5)P2 depletion did not result in the dissociation of proteins with polybasic motifs, suggesting another PIP plays a role in the docking of these proteins. Using a PI3K inhibitor (LY294002) that prevents PI(3,4,5,)P3 synthesis in concert with the PI(4,5)P2 depletion system, the authors concluded that both PI(4,5)P2 and PI(3,4,5)P3 serve as PM anchors for proteins with polybasic clusters. This strategy has also been used to study the selective gating of KCNQ ion channels by PI(4,5)P2 in the PM (Suh et al. 2006).
Chang-lleto et al. developed a system in COS-7 cells to hydrolyze PI(4,5)P2 at sites of plasma membrane tubule formation induced by the endophilin N-BAR domain (Chang-Ileto et al. 2011). The N-BAR domain was fused to FRB and tagged with EGFP. The 5-phosphatase domain of synaptojanin 1, important for clathrin-mediated endocytosis in the brain, was fused to the FKBP domain with an mRFP tag. This phosphatase converts PI(4,5)P2 into PI(4)P. This approach enabled the demonstration that acute conversion of PI(4,5)P2 to PI(4)P resulted in the fragmentation and condensation of the tubules formed at the plasma membrane by the N-BAR domain. They also determined that dynamin is required for this fission process. These experiments indicate that conversion of PI(4,5)P2 to PI(4)P at these sites of membrane curvature induces membrane fission in a dynamin-dependent manner.
Use of the rapamcyin inducible system has shown that PI(4)P and PI(4,5)P2 are independent determinants of plasma membrane identity with both serving an important signaling role (Hammond et al. 2012). In this study a chimera was generated in which two enzymes were fused to the recruitable FKBP fragment: inositol polyphosphate-5-phosphatase E (INPP5E), which converts PI(4,5)P2 to PI(4)P, and Sac1 phosphatase, which converts PI(4)P to PI. This allowed the authors to metabolize both PIPs at the PM simultaneously upon rapamycin addition. Three additional chimera were made harboring mutations to inactivate either INPP5E or Sac1 or both enzymes so that PI(4,5)P2 and PI(4)P levels in the PM could be modulated independently by these constructs in which only one or neither of the enzymes is active. The authors were able to employ this system to demonstrate that PI4P is not required to maintain the steady state PI(4,5)P2 pool at the PM. Additionally, they determined that PI(4)P and PI(4,5)P2 serve some common roles at the PM, namely, the control of non-specific binding to the PM of polybasic peptides as well as the regulation of certain ion channels (TRPV1).
1.2.2. Endosomal and Lysosomal Pathway
Fili et al. utilized the inducible-dimerization system described above to locally deplete PI(3)P at early endosomes in HeLa cells (Fili et al. 2006). Early endosomes were targeted with Rab5a fused to FKBP and EGFP. Addition of the rapamycin analogue (AP21967) induced the recruitment of the FRB construct containing a myotubularin phosphatase (MTM1) to the Rab5a targeting construct, where PI(3)P was converted to PI. Local depletion of PI(3)P at Rab5a-positive endosomal compartments resulted in microtubule-dependent tubularization of the endosomes and affected the endocytic sorting of certain receptors, providing insight into the specific functions of this lipid in endosome morphology and trafficking (Fili et al. 2006).
Dong et al. developed a rapamycin-induced heterodimerization system for the late endosome/lysosome to study the effects of rapid PI(3,5)P2 depletion on the release of calcium from the mucolipin transient receptor potential (TRPML) channel (Dong et al. 2010). Calcium release is important for proper late endocytic trafficking. Their previous experiments showed that applying exogenous PI(3,5)P2 to isolated endolysosomes increased TRPML activity. However, in order to confirm that endogenous PI(3,5)P2 regulates this channel, they induced the acute depletion of PI(3,5)P2 in Cos-1 cells with an EGFP-FKBP-Rab7 targeting construct and an RFP-FRB-MTM1 (myotubularin) recruitable cytosolic construct. MTM1 converts PI(3,5)P2 into PI(5)P. Upon rapamycin addition, the authors found that TRPML activity was significantly reduced, suggesting PI(3,5)P2 plays a prominent regulatory role in the release of calcium from endolysosomes.
1.2.3 Golgi
A rapamycin-induced system that targets the Golgi has been successfully developed (Szentpeter et al. 2010). To target the Golgi, the C-terminus of the human type I Golgi protein, TGN38, was fused to the N-terminus of a FRB-CFP construct. The human Sac1 phosphatase enzyme that dephosphorylates PI(4)P was fused to an FKBP12-mRFP construct. In confocal microscopy assays, the authors demonstrated that upon addition of rapamycin to cells transfected with these constructs, the Sac1 phosphatase enzyme was recruited to the Golgi where it dephosphorylated PI(4)P within 10 minutes, as evidenced by the concomitant dissociation of a PH domain probe that binds PI(4)P. With this system, the authors were able to confirm that PI(4)P is responsible for the Golgi localization of several clathrin adaptors and that depletion of PI(4)P prevents vesicular transport to both the plasma membrane and to the late endosomes. Additionally, they were able to determine that PI4P contributes to the recovery of PI(4,5)P2 stores at the PM, a hitherto unrecognized function. The authors noted that high expression of the TGN38-FRB-CFP construct affected Golgi structure and integrity. The overexpression of PH domains (OSBP, FAPP1, OSH1, or CERT) also may affect Golgi morphology and required fine-tuned expression with a thymidine kinase promoter. Also of note, the activity of the enzyme prior to recruitment needs to be monitored to account for unintended activity elsewhere. Additionally, a dead kinase should be used to rule out nonspecific effects of recruiting high concentrations of overexpressed protein to the membrane interface. These factors need to be considered when developing a recruitable enzyme system.
1.2.4 Intracellular Organelle Targeting
Komatsu et al. have developed a rapamycin-induced targeting systems to the PM, ER, Golgi, lysosome, and mitochondria (Komatsu et al. 2010). They have tested for targeting motifs that do not distort the targeted organelle of interest upon over-expression and have developed the following constructs: FRB-YFP-Giantin and CFP-FKBP for the Golgi; FRB-MoA (Monoamine Oxidase A) and CFP-FKBP for mitochondria; CFP-FKBP-Cb5 (cytochrome b5) and YFP-FRB for the ER; LAMP-CFP-FRB (lysosomal-associated membrane protein) and YFP-FKBP for lysosome; and Lyn11-FRB and CFP-FKBP for the PM. Experiments were mainly carried out in HeLa cells. The authors changed the constituents of the constructs (e.g., either FKBP or FRB for the targeting construct) or the order of the constituents (e.g., attaching FKBP or FRB to either the N- or C-terminus of the fluorescent or targeting motifs) to optimize translocation efficiency. For application, the authors used targeting systems for the plasma membrane (FKBP-RasGEF and Lyn11-FRB) and the Golgi (FKBP-RasGEF and FRB-Giantin) to study the site-specific activation of the Ras signaling pathway (Komatsu et al. 2010). They showed that Ras activation at the PM, but not at the Golgi, results in actin remodeling. Additionally, they showed that Ras activation at the plasma membrane results in ERK phosphorylation in the nucleus while Ras activation at the Golgi results in ERK phosphorylation primarily at the Golgi, demonstrating a spatial compartmentalization of Ras activity.
1.3 Phosphoinositides in Health and Disease
Since many proteins that interact with PIPs are enzymes, the stereospecific recognition of PI headgroups has important functional consequences with regards to biological activity. Indeed, there are a number of cases where a disease is attributable to the abrogation of these properties (McCrea and DeCamilli 2009). For instance, SHIP2 is a 5-phosphatase that acts on PI(3,4,5)P3 and has been shown to act as a negative regulator of insulin signaling (Wada et al. 2001). Overexpression of SHIP2 has been shown to promote insulin resistance (Kagawa et al. 2008; Wada et al. 2001). Some type 2 diabetic patients have been shown to harbor a 16 base pair deletion of SHIP2 (Marion et al. 2002) that may act to regulate messenger RNA as mechanistic studies have demonstrated that this 16 base pair deletion causes increased expression of SHIP2 (Marion et al. 2002). Additional polymorphisms of SHIP2 have been found correlated with hypertension, obesity, and hyperlipidemia, which are common characteristics of metabolic syndromes (Kaisaki et al. 2008).
Myotubularins are 3-phosphatase enzymes that dephosphorylate PI(3)P and PI(3,5)P2. Mutations in myotubularin 1 have been shown to induce myotubular myopathy, which is characterized by muscle weakness and can often be lethal in infancy (Herman et al. 1999). Charcot-Marie-Tooth Disease has been linked to mutations in two different myotubularin family members, MTMR2 and MTMR13 (Azedine et al. 2003; Bolino et al. 1996; Senderek et al. 2003). Patients with this disease have peripheral neuropathy and leg weakness due to misfolding of myelin sheaths, which presents in childhood and can often lead to the inability to walk by early adulthood.
Perhaps the most well studied cases of alterations in PI metabolism are linked to cancer and mutation of the 3-phosphatase that acts on PI(3,4,5)P3, protein phosphatase and tensin homolog deleted on chromosome ten (PTEN). PTEN is a tumor suppressor known to be mutated in many cancers including melanoma, lung cancer, colon cancer, thyroid cancer, breast cancer, and thyroid cancer (Carracedo and Pandolfi 2008) where its action as a tumor suppressor is diminished. As PTEN acts on PI(3,4,5)P3 to make PI(4,5)P2, the opposite effect of growth activating mutations in the PI3-kinases that convert PI(4,5)P2 to PI(3,4,5)P3 has also been shown in lung, breast, ovarian, colon, and gastric cancer (Bader et al. 2005; Engelmann, J.A. et al. 2006). Accordingly, PI3-kinase inhibitors have shown promise as cancer therapeutics (Osaki et al. 2004).
PTEN is an interesting peripheral protein regulated by unique biophysical properties. PTEN has been shown to harbor a N-terminal binding site for PI(4,5)P2, binding to which induces increased α-helicity of PTEN (Redfern et al. 2008). Moreover, PTEN is selective for PI(4,5)P2 when compared to other PIPs, which helps explain PTEN localization to the inner leaflet of the PM where it dephosphorylates PI(3,4,5)P3. PTEN can also bind PS through its C2 domain (Shenoy et al. 2012; Gericke et al. 2013). Strikingly, the presence of PS and PI(4,5)P2 in the same membrane increases PTEN membrane affinity where they synergize to recruit PTEN to the membrane interface (Shenoy et al. 2012; Gericke et al. 2013). As noted earlier, PS and PI(4,5)P2 are the most abundant PM anionic lipids and their PM spatial organization is expected to regulate PTEN localization. Biophysical studies further suggest PTEN likely has multiple PI(4,5)P2 binding sites (Shenoy et al. 2012) although the distinct role of each site and how they each selective recognition of the PI(4,5)P2 headgroup is still a burning question. Continuations of these biophysical studies will likely yield very useful insight into the mechanisms regulating PTEN catalysis and how individual mutations in cancer patients may dysregulate membrane binding or membrane induced activation of PTEN.
Bipolar disorder and schizophrenia have also been linked to regions of chromosomes that encode lipid kinases and phosphatases including a PI3-kinase (PIK3C2), PI4-kinase type III α (PIK4CA), a PI(4)P 5-kinase (PIP5K2A), and the phosphoinositide phosphatase synaptojanin 1 (Saito et al. 2001; Saito et al. 2003; Stopkova et al. 2003; Stopkova et al. 2004). Alterations of Synaptojanin 1, a PI(4,5)P2 phosphatase, have also been linked to Alzheimer’s disease and Down Syndrome (Berman et al. 2005; Arai et al. 2002). Synaptojanin 1 levels in Down Syndrome patient’s have been shown to be increased while PI(4,5)P2 levels are decreased (Voronov et al. 2008).
Therapeutic intervention at the level of protein-PIP interactions may be useful in improving metabolic disease outcomes. In this vein, computational structural analysis of several PIP-binding domains including C2 and PH domains has suggested that drug-like compounds could be developed to inhibit lipid binding (Jo et al. 2011; Segers et al. 2007). Bioinformatics and computational biology have also begun to play an important role in lipid-binding domain studies yielding predictions of potential lipid-binding proteins (Bhardwaj et al. 2006; Gallego et al. 2010), elucidating the role of electrostatics in lipid binding (Murray et al. 2005; Mulgrew-Nesbitt et al. 2006; Dalton et al. 2007), and enabling the assembly of databases that compile lipid-binding domain data and predictions of membrane binding properties (Bhardwaj et al. 2007; Silkov et al. 2011; Chen et al. 2012).
2. Phosphoinositide-Binding
The specific localization of PIPs provides cellular spatial organization for PIP-binding proteins to recognize specific subcellular locations. PIP-binding proteins are most often peripheral proteins, a collective term for proteins that reversibly translocate to cellular membranes. Peripheral proteins employ different strategies for reversible membrane interactions and many contain one or more modular domains with selective PIP-binding properties. These PIP-binding structural modules, also known as membrane-targeting domains, include protein kinase C (PKC) conserved 2 (C2) (Cho and Stahelin 2006; Corbalan-Garcia and Gomez-Fernandez 2014); pleckstrin homology (PH) (Lemmon 2007; Kutateladze 2010); Fab1, YOTB, Vac1, and EEA1 (FYVE) (Kutateladze 2010); Phox (PX) (Kutateladze 2010); epsin amino-terminal homology (ENTH) (Horvath et al. 2007); AP180 amino-terminal homology (ANTH) (Legendre-Guillemin et al. 2004); Bin amphiphysin Rvs (BAR) (Mim and Unger 2012); band 4.1, ezrin, radixin, moesin (FERM) (Mani et al. 2011); phosphotyrosine binding (PTB) (Alajlouni et al. 2011); postsynaptic density 95, disk large, zonula occludens (PDZ) (Chen et al. 2012; Wawrzyniak et al. 2013); GOLPH3 (Golgi phosphoprotein 3) (Dippold et al. 2009; Wood et al. 2009), PROPPINs (Baskaran et al. 2012); tubby (Santagata et al. 2001), and kinase-associated 1 (KA1) domains (Moravcevic et al. 2012). The PH domain was the first of these shown to bind PIPs (Rebecchi et al. 1992) and the subsequent modules identified above have been shown to have a range of selectivity for the seven PIP species (Cho and Stahelin 2005; Hurley 2006; Kutateladze 2010; Lemmon 2008; Sudhahar et al. 2008; Stahelin 2009; Moravcevic et al. 2012). In addition to PIP-binding regulating several key cellular processes, abnormalities in PIP metabolism (McCrea and De Camilli 2009) or mutation of PIP-binding proteins can contribute to the etiology of disease (Carpten et al. 2007; Duning et al. 2013).
2.1 PIP Binding Assays
Many assays have been developed to qualitatively and quantitatively assess PIP-binding (Cho et al. 2001; Narayan and Lemmon 2006). These techniques have their individual caveats but for the most part can be used to accurately parse out PIP selectivity and affinity. Qualitative assays include lipid blots, which harbor the different PIPs spotted on nitrocellulose and can be used to get a preliminary assessment of PIP selectivity. To more carefully investigate PIP selectivity, a number of centrifugation assays have been used where vesicles or beads containing different PIPs are sedimented to separate free and bound protein. Additionally, membrane flotation assays are available that can similarly isolate free and bound protein using centrifugation and a sucrose gradient. Isothermal titration calorimetry (Bravo et al. 2001), FRET (Nalefski et al. 2001; Landgraf et al. 2008; Lyakhova and Knight 2013) and light scattering (Connell et al. 2008) have also been useful in measuring lipid affinity and selectivity. Kinetic and equilibrium binding analysis has been performed using surface plasmon resonance to elucidate PIP selectivity and affinity (Stahelin et al. 2002; Stahelin et al. 2003a; Stahelin et al. 2003b; Narayan and Lemmon 2006; Silkov et al. 2011; Yoon et al. 2012; Stahelin 2013). Although the above assays have provided robust analysis of PIP binding for peripheral proteins the field is still in need of rapid and stout global approaches for assessing PIP selectivity. This is especially true for newly or previously unidentified PIP-binding proteins. One such approach was recently developed for proteins expressed with a GFP tag (Kim et al. 2013). Synthetic PIPs have also been useful for rapid screening of PIP-binding (Rowland et al. 2012) and cellular identification of PIP-binding proteins using PIP-activity probes and proteomics (Rowland et al. 2011). In fact, this is an emerging area of research and new assays to globally investigate PIP-binding proteins are nicely reviewed by Michael Best in this special issue (Best 2013).
2.2 PIP-Mediated Membrane Association
Membrane-protein recognition is governed by various types of biochemical interactions that are dependent upon the physicochemical properties of both protein and membrane. Subcellular membranes have different compositions of bulk lipids and both constitutive and transient levels of PIPs, which can facilitate the membrane targeting of peripheral proteins. Association of peripheral proteins to membranes containing PIPs can be driven by diffusion and electrostatic forces. Once the protein associates, which is typically through electrostatic interactions with the membrane interface, formation of more tightly bound complexes ensues stabilized by H-bonding, electrostatic bonds, and membrane penetration (Stahelin and Cho 2001; Cho and Stahelin 2005; Lemmon 2008; Moravcevic et al. 2012).
For the majority of PIP-binding proteins, both specific and nonspecific electrostatic interactions with PIPs enhance the ka of lipid-binding (Stahelin and Cho 2001) as observed for the promotion of the protein-protein complexes (Schreiber 2002). Selective PIP binding and membrane penetration stabilize the complex by reducing the kd (Stahelin et al. 2002; Stahelin et al. 2003a; Stahelin et al. 2003b; Hom et al. 2007; He et al. 2009). The initial membrane docking of peripheral proteins facilitates specific interactions with PIPs by reducing the dimensionality of the space through which the protein interacts with its lipid ligand. When a peripheral protein is in the cytoplasm it is in three dimensions whereas when the peripheral protein is bound to the cytosolic face of a membrane bilayer its dimensionality is reduced to two dimensions as it can only effectively diffuse or scoot in the x- and y-plane. PIP-binding domains generally have low affinity for membranes containing anionic lipids devoid of their cognate PIP. Without PIP bound these domains are unable to sufficiently penetrate the membrane due to a high penalty of desolvation (Diraviyam et al. 2003; Stahelin et al. 2002; Stahelin et al. 2003a; Stahelin et al. 2003b) and the PIP acts as an electrostatic switch by reducing the desolvation penalty and inducing the membrane penetration of hydrophobic and aromatic residues that surround the PIP binding pocket. For instance, PI(4,5)P2 binding has been shown to play an important role in inducing the penetration of BAR domains (Yoon et al. 2012). The unique composition of each peripheral protein and/or PIP-binding domain as well as the target membrane provide diverse abilities for protein’s to bind membranes with different specificities, affinities, and orientations.
2.3 Coincidence Detection
Integration of dual lipid signals can provide a mechanism of more fine tuned regulation of peripheral proteins. For instance, the requirement of two distinct lipid-binding sites within the C2, PH, and PX domains has been demonstrated for enhanced localization to the plasma membrane (Gallego et al. 2010; Karathanassis et al. 2002; Lemmon 2008; Lucas and Cho 2011; Moravcevic et al. 2012; Scott et al. 2012). This mode of dual lipid recognition termed “coincidence detection” (Balla 2005; Lemmon 2008; Moravcevic et al. 2012) is seen for plasma membrane PI(4,5)P2 and PS, which coordinate different sites in the C2 domain of PKCα (Guerrero-Valero et al. 2009; Corbalan-Garcia and Gomez-Fernandez 2014) and full-length PTEN (Shenoy et al. 2012; Gericke et al. 2013). PI(4,5)P2 as well as other PIPs (Manna et al. 2008) can associate with a cationic patch that is exposed on the β-groove of some C2 domains (Cho and Stahelin 2006; Guillen et al. 2013; Corbalan-Garcia and Gomez-Fernandez 2014) while PS is engaged in three points of contact with the C2 domain as well as bridging with one of the bound Ca2+ ions (Verdaguer et al. 1999; Corbalan-Garcia and Gomez-Fernandez 2014). The presence of a PS and a PI-binding site in this C2 domain provides higher affinity and selectivity for the PM to which PKCα can rapidly bind when cytosolic Ca2+ levels increase. In addition, the PH domains of Akt1 and PDK-1 bind to both PI(3,4,5)P3 and PS (Huang et al. 2011; Lucas and Cho 2011). Cellular studies have demonstrated that both of these lipid-binding sites are important for their PM recruitment and furthermore, that the PS binding of the PDK1 PH domain is important in enhancing phosphorylation of Akt1 to its active form (Lucas and Cho 2011). Finally, PX domains also have been shown to bind both PIPs and other glycerophospholipids. The p47phox PX domain binds PI(3,4)P2 and uses a shallow pocket lined with basic residues to bind phosphatidic acid (PA) and to a lesser extent PS (Karathanassis et al. 2002).
Dual lipid recognition can enhance specific membrane targeting and may help to explain why a large number of lipid binding domains and peripheral proteins do not have high selectivity for one anionic membrane lipid such as a PIP (Moravcevic et al. 2012). A decade ago, comprehensive binding analysis of PH domains established that ~20% of PH domains from S. cerevisiae had remarkable affinity for PIPs (Yu et al. 2004). However, a recent large-scale analysis of lipid-binding domains in yeast has demonstrated that a number of these modules bind sphingolipids (Gallego et al. 2010). Sphingolipids have often been overlooked and not included in lipid-binding experiments, especially in combination with PIPs. One recent study has demonstrated that the yeast PH domains from Slm1p or Yil105cp bound both PI(4,5)P2 and dihydrosphingosine-1-phosphate where both lipids were required for effective membrane localization (Gallego et al. 2010). For the most part, assays and methodologies have not been employed or developed to discover proteins that may bind PIPs and sphingolipids or those that may bind sphingolipids alone. This suggests that many lipid-binding domains should be carefully rescreened for sphingolipid binding to uncover new modes of membrane recognition that may have been overlooked. Indeed, coincidence detection by the C2 domain of cPLA2α for zwitteroinic lipids and the sphingolipid ceramide-1-phosphate (C1P) has recently been reported (Subramanian et al. 2005; Stahelin et al. 2007) where selective binding to C1P over PA is observed due to H-bonding with the C1P hydroxyl group (Ward et al. 2013). For cellular transport of C1P the recently identified C1P transport protein was shown to recognize the acyl amide moiety of C1P (Simanshu et al. 2013), an interaction that may also provide selectivity for some sphingolipids when compared to their glycerophospholipid counterparts. Additionally, annexin A2 which has long been known to bind PS and PI(4,5)P2 was recently shown to bind C1P with even higher affinity than that of PS or PA (Hankins et al. 2012). Other proteins have also recently been identified that bind ceramide (Saddoughi et al. 2013) as well as phosphorylated sphingolipids (Lamour et al. 2011).
In addition to dual lipid recognition, coincidence detection through a protein-protein interaction site in addition to a protein-lipid interaction site can also occur. Four-phosphate adaptor protein 1 (FAPP1) and OSBP bind PI(4)P and small GTPases at the Golgi membrane. The FAPP1 PH domain binds PIPs, with selectivity for PI(4)P (Lenoir et al. 2010; He et al. 2011; Stahelin et al. 2007) and can also associate with Arf1 through a non-overlapping binding site adjacent to the PIP binding pocket (He et al. 2011). It is thought that engagement of both PI(4)P and Arf1 is necessary for FAPP1 Golgi targeting as the PH domain has moderate affinity for PI(4)P-containing membranes and Arf1 can provide the second recruitment signal. Two or more binding sites provide either a platform to recruit peripheral proteins to specific cellular locations such as the PM where PS and PI(4,5)P2 reside or a point of temporal control such as in the case of Akt1. In most cell types PI(3,4,5)P3 levels increase in the PS rich inner leaflet of the PM in response to various growth inducing stimuli, and thus Akt1 is recruited under specific temporal conditions (Huang et al. 2011). Coincidence detection of lipid and proteins also seems to be an important regulatory mechanism of PDZ (Chen et al. 2012; Wawrzyniak et al. 2013) and some C2 domains (Huang and Szebenyi 2010; Tian et al. 2011).
2.4 Other Considerations Regarding PI Binding
In addition to docking specific lipids, spatial recognition and regulation of membranes can be mediated by the degree of membrane curvature and by inherent differences in bulk lipid compositions among membrane organelles. Temporal regulation can also occur through metabolism of PIPs and DAG, influx of Ca2+ into the cytoplasm, and local membrane curvature changes induced by cell signaling processes. Moreover, a number of peripheral proteins are regulated in both a spatial and temporal fashion. For instance, proper targeting may require a certain degree of membrane curvature in addition to the presence of a temporally regulated lipid or two different lipids with docking sites on a single protein. Recently, it was also demonstrated that some PI-binding domains could be regulated by pH. FYVE domains were the first module shown to target endosomes with higher affinity in an acidic environment due to protonation of histidine residues in the membrane-binding region (Lee et al. 2005; He et al. 2009). Protonation of His residues in acidic pH has been shown to promote higher affinity for lipid vesicles containing PIPs as well as cellular membranes for the PH domain of GRP1 (He et al. 2008) and the Epsin ENTH domain (Hom et al. 2007). Still unclear in pH-dependent PIP-binding, however, is the effect of protonation on PIPs. For instance, Kooijman, Gericke and colleagues have demonstrated that in acidic pH PIPs such as PI(4,5)P2 will undergo protonation (Kooijman et al. 2009). This effectively reduces PIP anionic charge and how His protonation on the peripheral protein may increase affinity for PIPs under these conditions is still not well understood.
3. Phosphoinositide binding modules
To date at least 14 PI-binding modules have been identified (See Table 1). These include: C2, PH, FYVE, PX, ENTH, ANTH, BAR, FERM, PTB, PDZ, GOLPH3, PROPPINs, tubby, and KA1 domains. These different families have varying affinity and selectivity for the seven PIPs. Below we discuss the FYVE, C2, and PH domains and the mechanisms that regulate their PI binding in biological membranes. Readers are also referred to other excellent reviews on PIP binding proteins (Lemmon 2008, Kutateladze 2010; Moravcevic et al. 2012) as well as more in depth reviews on specific PIP-binding domains such as BAR (Qualmann et al. 2011; Rao and Haucke 2011; Mim and Unger 2012), PDZ (Wawrzyniak et al. 2013), and PX (Kutateladze 2010; Teasdale and Collins 2012) domains. Additionally, table 1 has more relevant citations on various PIP binding domains that may be useful to the reader.
Table 1.
PIP-binding domain selectivity, structure, and references.
| Domain | PIP Selectivity | Structure | References |
|---|---|---|---|
| ANTH | PI(4,5)P2 as well as PI(3,4,5)P3, PI(3,4)P2 for some. |
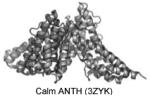
|
Stahelin et al. 2003b; Silkov et al. 2011; Miller et al. 2011 |
| BAR | PI(4,5)P2, PI(4)P |
|
Peter et al. 2004; Mim and Unger 2012; Yoon et al. 2012 |
|
C2 (Conserved
Kinase 2) |
PI(4,5)P2 as well as other PIPs |
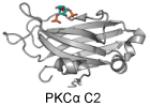
|
Cho and Stahelin 2006; Guerrero-Valero et al. 2009; Ankem et al. 2011; Corbalan-Garcia and Gomez-Fernandez 2014 |
| ENTH | PI(4,5)P2 |
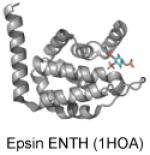
|
Ford et al. 2002; Boucrot et al. 2012 |
| FERM | PI(4,5)P2 |
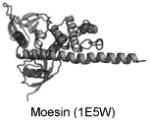
|
Edwards and Keep 2001; Carvalho et al. 2010 |
| FYVE | Majority PI(3)P Protrudin FYVE: PI(4,5)P2, PI(3,4)P2, and PI(3,4,5,)P3 |
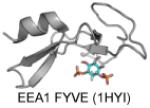
|
Dumas et al. 2001; Kutateladze 2010; Gil et al. 2012 |
| GOLPH | PI(4)P |

|
Wood et al. 2009; Lenoir and Overduin 2013 |
|
KA1 (kinase
associated-1 domain) |
Anionic lipids |
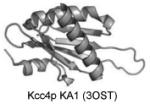
|
Moravcevic et al. 2010 |
| PDZ | PI(4,5)P2 for some but many are nonselective |
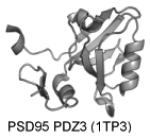
|
Chen et al. 2012; Wawrzyniak et al. 2013 |
|
PH (Pleckstrin
Homology) |
Some selective for one PIP while others are nonselective. PI(3,4,5)P3, PI(4,5)P2, PI(3,4)P2, PI(3,5)P2, PI(3)P, PI(4)P |
|
Lemmon 2007; Lemmon 2008; Kutateladze 2010; Moravcevic et al. 2012 |
| PROPPINS | PI(3,5)P2 and PI(3)P |
|
Baskaran et al. 2012 |
| PTB | PI(4,5)P2 |
|
Stolt et al. 2003 |
| PX | Mostly PI(3)P Some: PI(4)P, PI(4,5)P2, PI(3,4,5)P3, or PI(3,4)P2 |
|
Bravo et al. 2001; Karathanassis et al. 2002; Lemmon 2008; Kutateladze 2010; Moravcevic et al. 2012 |
| TUBBY | PI(4,5)P2 |
|
Santagata et al. 2001 |
3.1 FYVE domains
FYVE domains are small (70 to 80 amino acid) cysteine-rich domains that bind two zinc ions and are found in at least 29 human proteins (Lemmon 2008; Letunic et al. 2009; Kutateladze 2010). These domains are highly homologous, and most FYVE domains specifically bind PI(3)P (Lemmon 2008; Kutateladze 2007; Kutateladze 2010)), although recently the FYVE domain of protrudin was shown to bind PI(4,5)P2, PI(3,4)P2, and PI(3,4,5,)P3 (Gil et al. 2012). Three motifs found in FYVE domains (WxxD, RR/KHHCR, and RVC) shape a cationic patch that interacts with PI(3)P. PI(3)P plays a key role in membrane trafficking and is found in specific subcellular locales, including the cytoplasmic face of early endosomes, multivesicular bodies, and phagosomes (Burd and Emr 1998; Gaullier et al. 1998; Patki V et al. 1998). In addition, PI(3)P has been shown to reside in the ER and Golgi apparatus (Sarkes and Rameh 2010), which may play a role in recruiting FYVE domain containing proteins DFCP1 and Alfy (Ridley et al. 2001; Simonsen et al. 2004). In addition to their role in membrane trafficking, FYVE domain-containing proteins have been implicated in receptor signaling, actin cytoskeleton regulation, and neurite outgrowth. FYVE domains are also found in proteins that are involved in adipocyte differentiation, lipid kinases and phosphatases, ubiquitin ligases, and proteins involved in apoptosis and leukocyte signaling.
3.1.1 Membrane-binding mechanism
FYVE domains use multiple mechanisms to achieve nanomolar affinity for PI(3)P-containing membranes. In addition to a cationic pocket that can selectively coordinate the PI(3)P headgroup, FYVE domains also engage in nonspecific electrostatic interactions that enhance their membrane association (Brunecky et al. 2005; Kutateladze et al. 2004; Stahelin et al. 2002). Specifically, the anionic lipid phosphatidylserine, which is found in appreciable concentrations on the cytoplasmic face of endosomes and multivesicular bodies can also contribute to FYVE domain membrane association and the orientation of the domain at the membrane interface (Brunecky et al. 2005). Insertion of hydrophobic residues seems to be a general mechanism of membrane anchoring of the FYVE domain (Brunecky et al. 2005; Diraviyam et al. 2003; Stahelin et al. 2002; Kutateladze et al. 2004; Kutateladze and Overduin 2001; Misra and Hurley 1999). The hydrophobic residues are found in a loop region that can vary in length and amino acid composition adjacent to the PI(3)P binding site. This loop region has also been dubbed the membrane insertion loop (MIL) (Kutateladze 2006; Kutateladze 2010). The MIL penetrates into the membrane after binding the PI(3)P headgroup, the effect of which is to elongate the membrane residence time of the protein (Stahelin et al. 2002). The FYVE domain has been shown to have a high desolvation penalty associated with loss of water molecules surrounding the MIL and in essence the PI(3)P binding to the FYVE domain acts as an electrostatic switch reducing the desolvation penalty associated with membrane insertion of the hydrophobic residues (Stahelin et al. 2002; Diraviyam et al. 2003). The MIL also harbors cationic residues that play a role in electrostatic interactions with acidic lipids at the membrane interface.
Perhaps the most interesting mode of FYVE domain association has to do with the local environmental pH as FYVE domains have two conserved His residues that interact with the inositol 3’-phosphate group. Acidic pH promotes the protonation of these His residues prompting PI(3)P binding in vitro and in cells (Lee et al. 2005; He et al. 2009) through enhanced interctions between protonated His and the PI(3)P headgroup. Additionally, deprotonation of these His residues has been shown to regulate release of the PI(3)P-dependent membrane association (Lee et al. 2005; He et al. 2009). Dimerization of FYVE domain-containing proteins can also play an important role in enhancing the membrane affinity through concurrent interaction with two PI(3)P molecules (Dumas et al. 2001).
The first X-ray structure of a FYVE domain from yeast protein Vps27p (Protein Data Bank (PDB): 1VFY) revealed that it has a shallow pocket that putatively binds PI(3)P (Misra and Hurley 1999). The authors proposed that the FYVE domain binds to the membrane in a perpendicular “side-on” orientation, which would enable the binding of PI(3)P to the pocket and simultaneous interfacial penetration of two Leu residues (Leu185 and Leu186) in the MIL, a mechanism that was later supported by numerous biophysical studies (Brunecky et al. 2005; Diraviyam et al. 2003; Stahelin et al. 2002; Kutateladze et al. 2004; Kutateladze and Overduin 2001). Subsequent NMR studies of the FYVE domain of EEA1 in the presence (PDB: 1HYI) and absence (PDB: 1HYJ) of a soluble PI(3)P and/or lipid micelles suggest a membrane-binding mechanism in which nonspecific interfacial penetration precedes the binding of PI(3)P to its pocket (Kutateladze and Overduin 2001). The X-ray structure of EEA1-FYVE-inositol 1,3-bisphosphate complex demonstrates how the FYVE domain stereospecifically recognizes the PI(3)P headgroup (Dumas et al. 2001). In addition to the aforementioned structures of FYVE domains additional ligand-free structures have been solved for Hrs (PDB: 1DVP), RUFY (PDB: 2YW8 and 2YQM), and Leishmania major Lm5-1 (PDB: 1Z2Q).
All FYVE domain structures that have been solved exhibit a similar overall fold consisting of two double-stranded antiparallel β-sheets and a C-terminal α-helix. The β1 strand harbors the RR/KHHCR motif, which is central to PI(3)P binding. The ligand bound structure of EEA1 (PDB: 1JOC) revealed that the last arginine in the motif (Arg1375) makes hydrogen bonds with the 3-phosphate along with the histidine residues, which used the imidazole ring (His1372) and backbone amide (His1373) to hydrogen bond with the 3-phosphate. The first two arginines of the motif (Arg1370 and Arg1371) interact with the 1-phosphate and important interactions are also observed between the 4-, 5-, and 6-hydroxyl groups of PI(3)P to promote specificity through exclusion of binding other PIPs.
Biophysical and computational studies of the FYVE domains have been used to elucidate the FYVE domain membrane-binding mechanism. Electrostatic potential calculations of Vps27p and Hrs FYVE domains showed that the MIL is surrounded by a positive electrostatic potential because of the presence of cationic residues in the PI(3)P binding site. This positive charge drives the initial membrane association (increasing the rate of association) (Stahelin et al. 2002; Cho and Stahelin 2005), which is followed by coordination of PI(3)P via electrostatic and H-bonds. PI(3)P binding then induces the penetration of the MIL as the desolvation penalty surrounding the MIL is reduced (Kutateladze 2010; Diraviyam et al. 2003; Stahelin et al. 2002; Cho and Stahelin 2005). Biophysical assays using SPR and monolayer penetration indicated that PI(3)P binding is required for the penetration of the MIL into the membrane. Thus, PI(3)P binding serves as an electrostatic switch, reducing the positive potential surrounding the MIL and promoting the penetration of hydrophobic residues (Diraviyam et al. 2003; Stahelin et al. 2002). Coordination of PI(3)P and penetration of hydrophobic residues elongate the membrane residence time (slower dissociation rate) and are thought to stabilize the membrane-bound complex. Computational studies of mulitple FYVE domains demonstrated they have different electrostatic potentials that can contribute to their membrane affinity and orientation at the membrane interface (Diraviyam et al. 2003). The membrane affinity of these different FYVE domains has also been shown to be an important factor in their subcellular localization (Blatner et al. 2004).
3.2 C2 DOMAINS
The C2 domain (~130 residues) (See Figure 3) like the C1 domain was first identified as one of two conserved regulatory domains in PKC (Ono et al. 1989; Osada et al. 1990) Identification of the C2 domain in other proteins, such as synaptotagmins and group IVA cytosolic phospholipase A2 (cPLA2α), which also bind membranes in a Ca2+-dependent manner, led to the postulation that the C2 domain is involved in Ca2+-dependent membrane binding. A large number of proteins containing the C2 domain have been identified since, and most of them are involved in signal transduction or membrane trafficking. The C2 domain represents the second most abundant lipid-binding domain (following the PH domain) with at least 200 examples identified. While most C2 domain proteins are peripheral and bind reversibly to membranes some C2 parent proteins are actually transmembrane proteins involved in membrane trafficking. However, these C2 domains can also bind reversibly in a Ca2+-dependent fashion (Davis et al. 2002).
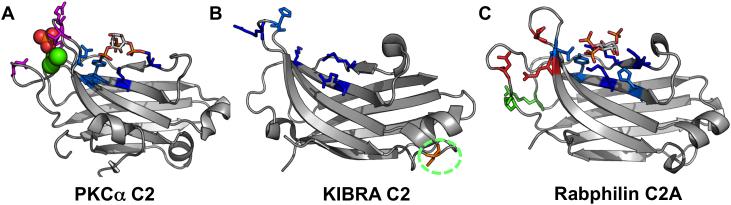
Figure 3. C2 domains have a β-sandwich structure with loop regions adjacent to the “cationic patch”.
These structures have been crystallized with or without inositol headgroups and demonstrate PI-binding through interaction with positively-charged residues in the cationic patch region. PI-binding residues are depicted in blue. Dark blue residues represent positively charged lysines or argininges and light blue are other inositol-headgroup coordinating residues. Residues highlighted in magenta are PS-binding, residues highlighted in green are membrane-inserting, and residues highlighted in red are negative and calcium-coordinating. A. PKCα C2 (3GPE) coordinates PS with Asn189, Arg216, Arg249, and Thr251 where the sulfate is coordinated in the structure (orange and red spheres). In this loop region, negatively charged residues coordinate 3 calcium inos (green spheres) which are crucial to its membrane localization. The PIP2 headgroup is coordinated by Tyr195, Lys197, Lys209, Lys221, Trp245, and Asn253 in the cationic patch region. B. KIBRA C2 (2ZOU) harbors a cationic patch region with Arg753, His755, Arg714, Arg716, Lys692, and Arg696. In addition, Cys771 participates in a disulfide bond between two monomers in the structure. While the molecular basis of WT KIBRA binding to monophosphorylated PIPs such as PI(3)P is unknown, mutation of Met734 or Ser735 alters the C2 domain affinity in such a way it increases the affinity of the C2 domains for PI(4)P, PI(5)P, and PI(4,5)P2 (Duning et al. 2013). C. Rabphilin C2A bound to IP3 (4NP9) coordinates the inositol headgroup in the cationic patch region using Tyr421, Lys423, His425, Lys435, Arg437 and Asn481.
3.2.1 C2 Domain Membrane Binding
The lipid affinity as well as the Ca2+ affinity of the Ca2+-binding C2 domains can vary greatly and some C2 domains are even involved in protein-protein interactions or may play a structural role. Ca2+ can play three different roles in membrane binding of the C2 domain: an electrostatic switch, a Ca2+-bridge, or inducing of intra- or interdomain conformational changes, which in turn trigger lipid binding. On the basis of the diverse Ca2+ affinity of C2 domains, peripheral proteins may be activated in a temporal manner depending upon the extent of the Ca2+ oscillations and their intensities. In other words, does their membrane association and dissociation distinctly follow the cellular Ca2+ oscillations under physiological conditions? While its been shown that enzymes such as PKC follow the Ca2+ spikes (Oancea and Meyer 1998), the disparate targeting of C2 domains under the same cellular conditions are yet to be rigorously measured. A significant number of C2 domains with little to no Ca2+ affinity have also been identified. Some of these Ca2+-independent C2 domains have been reported to bind the membrane or other proteins (Cho and Stahelin 2006; Gericke et al. 2013). The general mechanism of how a number of C2 domains associate with membranes in a Ca2+-dependent fashion is well established (Nalefski et al. 2001; Cho and Stahelin 2006). However, physiological functions and Ca2+- and membrane-binding properties of a large portion of C2 domains still remain unknown. Structural studies have shown that C2 domains have a common fold of conserved eight-stranded antiparallel β-sandwich connected by surface loops (Shao et. al. 1996; Shao et al. 1998). The disparity in C2 domains targeting arises in the surface loops, which are variable in amino acid sequence and conformation and most often involved in lipid binding. Also of functional consequences is a cationic patch in the concave face of the β-sandwich termed the cationic β-groove, which varies in size and electrostatics among C2 domain (Cho and Stahelin 2006). In support of this observation, cationic β-grooves have been shown to bind PIPs including PI(4,5)P2 as well as C1P (Guerrero-Valero et al. 2009; Stahelin et al. 2007; Ward et al. 2013). Thus, many C2 domains are able to coordinate multiple lipids in both a Ca2+-dependent or independent fashion. The preliminary data available on the multiple lipid recognition modes of C2 domains suggests C2 domains may be multiply regulated by different lipids and Ca2+ signals and in some cases may require coincidence detection (e.g., interaction with multiple lipid targets) to achieve high affinity and cellular localization (Corbalan-Garcia and Gomez-Fernandez 2014).
Most lipid binding domains achieve lipid specificity through a specific lipid-binding site formed within a pocket (e.g., C1, PH, PX, and FYVE domains) or with juxtaposed surface cationic residues (e.g., ANTH domain). C2 domains may be unique among lipid-binding domains in that they have neither a well-defined lipid-binding pocket nor a conserved cationic patch. The aforementioned sequence variation in the surface loops as well as the cationic β-groove leads to highly variable and relatively low lipid selectivity for some C2 domains and high affinity and specificity for others (Cho and Stahelin 2006; Corbalan-Garcia and Gomez-Fernandez 2014). Moreover, C2 domains can show different lipid selectivity as a function of Ca2+ concentration because the relative contribution of two lipid-binding sites can vary as PI-binding by the β-groove ligand may lower the Ca2+ requirement for lipid docking acting as an electrostatic switch in a similar mode to Ca2. A majority of Ca2+-dependent C2 domains harbor cationic residues in the Ca2+-binding loops and bind anionic lipids significantly better than zwitterionic ones. Some of these C2 domains bind anionic phospholipids through non-specific electrostatic interactions while others can stereospecifically recognize a lipid headgroup such as PS (Verdaguer et al. 1999; Stahelin et al. 2003c) or PI(3,4,5)P3 (Premkumar et al. 2010).
The cationic β-groove was first shown to interact with inositolpolyphosphates for the C2B domains of synaptotagmin II and IV (Fukuda et al. 1994; Fukuda et al. 1995). Thereafter, the C2B domains of synaptotagmin I (Schiavo et al. 1995) and II (Mehrotra et al. 2000) were shown to bind bis- and tris-phosphoinositides in a Ca2+-independent fashion. These first examples of cationic β-grooves do not have high lipid specificity but play a key role in the vesicle fusion activity of host proteins. The role of the cationic β-groove in PI-binding has been best characterized for PKCα (Guerrero-Valero et al. 2009; Manna et al. 2008). PKCα has been reported to bind PIPs including PI(4,5)P2 with nanomolar affinity (Manna et al. 2008). This binding is attributed to four lysine residues in the β-groove, which at first glance don’t seem to be enough to select PI(4,5)P2 over PI(3,4)P2, PI(3,4,5)P3 or other PIPs. In fact, experimental and computational investigation of the PKCα-C2 under various conditions found that PS and Ca2+ are a prerequisite for the PIP binding, which augments the PS binding by increasing the membrane residence time (Manna et al. 2008). PIP-binding for some C2 domains such as that of Tollip can exhibit broad specificity for PIPs (Ankem et al. 2011) and is regulated by protein-protein interactions with ubiquitin (Mitra et al. 2013).
These examples of PIP-binding in the β-groove of C2 domains should not only serve to understand the role of all β-grooves in C2 domains but may also be as a site of therapeutic intervention where the β-groove is required for coincidence detection or cellular activity. The disparate roles of this site in C2 domains may also serve as a better target than the more conserved Ca2+-binding sites and surface loop regions. Alternative splicing of some C2 domains may also regulate their lipid and/or Ca2+-binding properties. The C2A domain of Dysferlin has been shown to have a canonical C2A and a variant (C2Av1) these two molecules have different Ca2+-binding properties and different ligand preferences (Fuson et al. 2013). In addition to diverse roles in Ca2+ selectivity and lipid-binding some C2 domains have been shown to induce membrane curvature changes upon membrane association (Martens et al. 2007; Hui et al. 2009; Ward et al. 2012; Yu et al. 2013; Sot et al. 2013). This biophysical property of some C2 domains may be important to vesicle trafficking and exocytosis as well as cellular localization and activity. More rigorous biophysical investigation of how C2 domains mediate this process is warranted to determine how they may be structurally akin to BAR and ENTH domains.
3.2.2. Rasal C2 Domains Generate and Sense Membrane Curvature
Rasal belongs to the Ras GTPase-activating protein (RasGAPs) GAP1 subfamily and has been shown to be epigenetically silenced in some cancers (Sot et al. 2013). Rasal contains tandem C2 domains (C2A and C2B) and its membrane recruitment, which is calcium-dependent has been shown to induce its RasGAP activity (Sot et al. 2013). The C2A domain of Rasal specifically binds phosphatidylserine while the C2B domains interacts with several phosphoinositides including PI(3)P, PI(3,4)P2, PI(4,5)P2, and PI(3,4,5)P3 (Sot et al. 2013). These C2 domains are able to insert hydrophobic residues into the membrane and both generate and sense membrane curvature in a similar fashion to that of synaptotagmin (Hui et al. 2009; Martens et al. 2007) and Doc2b C2 domains (Yu et al. 2013). Lipid binding selectivity is also similar to the tandem C2 domain containing synaptotagmins and Rabphilin 3A where PI binding by the Rasal C2B is most likely provided by cationic residues in the CBL2 in a similar fashion to the C2A domain of Rabphilin 3A C2A domain (Figure 3C). The Rasal C2 domains therefore provide a mode of temporal (Ca2+), spatial (phosphoinositides and the protein target Ras), and conformational (C2A and C2B interaction with membrane lipids) regulation. While Rasal C2 domain binding has been shown to be curvature dependent, its cellular role is not yet well understood. The authors of this study propose that Rasal C2 domain curvature-dependent binding may drive Rasal to cellular regions of high curvature to turn off Ras-GTP although their studies also demonstrated that Rasal activity is not increased in the presence of highly curved membranes (Sot et al. 2013).
3.2.3 KIBRA C2 Domain Phosphoinositide Binding
The human KIBRA gene has been linked to cognition and has been shown to harbor an intronic single-nucleotide polymorphism (SNP) associated with memory performance and the risk of development of Alzheimer’s disease. Strikingly, two common missense SNPs subsequently identified in KIBRA affect cognitive performance and encode variants of the KIBRA C2 domain. KIBRA is a cytoplasmic protein and member of the WWC1 (WW and C2-domain containing protein 1) family that has been shown to interact with dendrin and atypical PKC. The roles of the C2 domain of KIBRA are not well understood to date but several studies have implicated KIBRA in pathways such as vesicle based turnover, endosome vesicle sorting, directing cell migration, and establishment of cell polarity, all of which may require the KIBRA C2 domain to bind membranes. KIBRA C2 binds monophophosphorylated PIPs (PI(3)P) in a Ca2+-dependent fashion (Duning et al. 2013). These newly identified SNPs result in structural changes of the C2 domain (M734I and S735A) (Figure 3B) as well as a link to cognitive performance (Duning et al. 2013). In addition to binding PI(3)P like the WT allele, KIBRA mutants demonstrated increased affinity for PI(4)P and PI(5)P, which was comparable to that of PI(3)P binding. Additionally, KIBRA mutants bound to PI(4,5)P2 in a similar fashion to that of monophosphorylated PIPs. While the cellular effects of these mutations on PIP-binding isn’t known, comparable affinity of KIBRA for PI(4)P, PI(5)P, or PI(4,5)P2 could lead to aberrant localization and temporal signaling.
3.2.4 Toxic Metals and C2 Domain Phosphoinositide Binding
Recently, the C2 domain of PKCα was shown to bind Pb2+ with high affinity, which can compete with Ca2+ for an interaction with the C2 domain (Morales et al. 2011). Further structural studies using NMR revealed that Pb2+ and PI(4,5)P2 synergistically enhanced each others affinity for the C2 domain (Morales et al. 2012). PI(4,5)P2 binding to the PKCα C2 domain was able to enhance the C2 domain’s affinity for Pb2+ and the association of Pb2+ with membrane anionic sites (Morales et al. 2012). These results may have important implications to lead poisoning where excess lead in cells could lead to enhanced interactions with some C2 domains and biological membranes at low or resting concentrations of cytosolic Ca2+. On the other hand, Cd2+, which is also a toxic metal can form a strong complex with the membrane binding loops of the PKCα C2 domain but it is not sufficient for promoting membrane binding (Morales et al. 2013). In this case, toxic Cd2+ would inhibit proper regulation of PKCα by Ca2+ and membrane lipids.
3.3. PH Domains
The PH domain is composed of ~100 amino acids and is the most abundant lipid-binding domain with >250 examples identified and we refer you to several more detailed reviews on PH domains (Lemmon 2007; Lemmon 2008; Kutateladze 2010) (Figure 4). Of the seven PIs in the mammalian cell, the PH domain binds specifically to PI(3,4,5)P3, PI(4,5)P2, PI(3,4)P2 (Lemmon 2007; Lemmon 2008; Kutateladze 2010; Moravcevic et al. 2012). Binding of PH domains to monophosphorylated PIPs seems to be less selective and have lower affinity than to that of bisphosphorylated PIPs (Yu et al. 2004; Lemmon et al. 2007; Stahelin et al. 2007; Prashek et al. 2013) and others may be coincident detectors for PIPs and sphingolipids or bind sphingolipids alone (Gallego et al. 2010). The membrane binding of PH domains is initially driven by non-specific electrostatic interactions, which is followed by specific PIP binding to achieve selective targeting and increase the membrane residence time. Like FYVE and some C2 domains, several PH domains anchor to the membrane through aliphatic residues adjacent to the PI-binding site (Kutateladze 2010; Lenoir et al. 2010; Moravcevic et al. 2012).
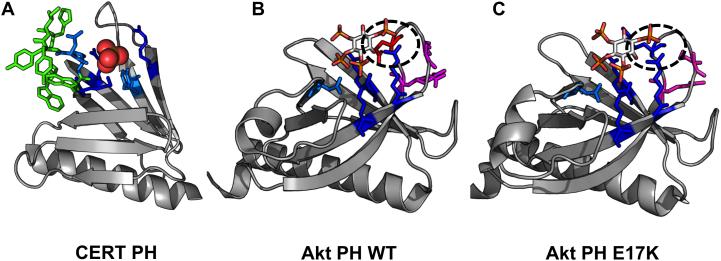
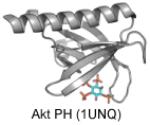
Figure 4. Pleckstrin Homology (PH) domains have a β-barrel structure with an alpha helical cap.
These structures have been crystallized with either sulfate of inositol headgroups and demonstrate PI-binding at one end of the barrel with positively-charged residues and hydrophobic residues which insert into the membrane from the loop regions of the protein. PI-binding residues are depicted in blue. Dark blue residues are positively charged lysines or argininges and light blue are other inositol-headgroup coordinating residues. Residues highlighted in green insert themselves into the membrane from the loop regions, and residues highlighted in magenta are PS-binding. A. CERT PH (4HHV) coordinates a sulfate ion (orange and yellow spheres) in the inositol-binding region with Lys32, Thr34, Asn35, Arg43, Tyr54, Arg66 and inserts into the membrane with Trp33, Tyr36, Ile37, His38, Trp40. B. Akt PH (1UNQ) coordinates IP4 with Glu17, Arg23, Arg25, Asn52, and Arg86. It coordinates PS with Arg15 and Lys20. C. Akt PH (2UZS) is known to be converted to a constitutively active form in some cancers via mutation of Glu17 to Lys17 (highlighted by the black ovals in B and C).
The majority of PH domains have a conserved basic motif (K-Xn-(K/R)-X-R) in which the basic lysines and arginines play an important role in forming H-bonds with the head group of the PI. Other basic residues located within the domain vary from domain to domain and can provide a stronger binding affinity and create a unique binding pocket. Two distinct members of the PH domain family (TIAM1 and ARHGAP9) (Ceccarelli et al. 2007) bind membranes through a site on the opposite side of the α1-αd2 loop suggesting that there are still novel PH domains to be discovered within the genome.
The importance of PH domains and disease was recently highlighted in an elegant study demonstrating an Glu to Lys (position 17) mutation in the AKT1 PH domain causes cancer (Figure 4). The E17K mutant was constitutively active due to pathological localization of E17K to the PM (Carpten et al. 2007). Falke and colleagues have shed some light on this pathological mechanism demonstrating that the PIP specificity of the AKT1 PH domain is drastically altered by the E17K mutation (Landgraf et al. 2008). Their biophysical analysis demonstrated E17K binds PI(4,5)P2 with even greater affinity than PIP3 and the constitutive PM localization of E17K may be due to binding to pools of PI(4,5)P2 often found in high concentration on the inner leaflet of the PM.
4. CONCLUSIONS AND FUTURE PERSPECTIVES
PIPs play important roles in the regulation of many cellular processes, including cell signaling and membrane trafficking (McCrea and De Camilli 2009). When these interactions are abolished or over-stimulated a number of life-threatening diseases can occur. Therapeutics aimed at the protein-lipid interface may serve as a viable means of achieving high selectivity among targets. Achieving efficacy in abolishing the PI-dependent activation of the drug target or acting allosterically to promote protein ubiquitylation (Jo et al. 2011) are two avenues that have been explored. However, designing small molecule inhibitors of PIP-binding proteins is a challenging task, in part because the driving forces of protein-PIP association are a composite of many chemical interactions. Electrostatic forces, cation-π interactions, van der Waals forces, and hydrogen bonding all play a role in maintaining proper membrane function and in the association of proteins with biological membranes. We still have much to learn about the infusion of these forces into the processes of molecular design that will ultimately produce the compounds necessary to control biological events at the membrane interface. Nonetheless, recent studies demonstrate lipid-binding inhibitors can be designed that inhibit lipid-binding of C2 and PH domains (Jo et al. 2011; Segers et al. 2007). In the case of the Akt1 PH domain inhibitor the effects are very unique (Jo et al. 2011). This drug, SC66, is able to block PI(3,4,5)P3 binding by the PH domain but also facilitates an allosteric effect on Akt1 that promotes its ubiquitylation and destruction (Jo et al. 2011). The advent of better high-throughput screening, computational design, as well as enzyme and cell assays to assess drug efficacy will likely be necessary to expedite the discovery of small molecules in this field.
Progress in our understanding of the membrane binding mechanisms of PIP-binding proteins has been considerable over the past decade thanks to rapid progress in bioinformatics, computational biology, in vitro biophysical studies, structural biology, cellular imaging, and single molecule studies. Furthermore, proteomics and lipidomics has increased our awareness of the large number of PIP-binding proteins in nature (van Meer 2005; Clark et al. 2011; Rowland et al. 2011; Best 2013; Catimel et al. 2013; Oxley et al. 2013) some of which have potential therapeutic value or unique PIP-binding properties. The future research on PIP-binding interactions will encompass more interdisciplinary studies of both computational predictions and validations of these conjectures. Because many PIP-binding proteins have been discovered to act as coincident detectors, which often have low affinity for one lipid ligand and target cellular membranes through multivalent mechanisms it can be difficult to parse out the true lipids that mediate cellular recruitment and activation of PIP-binding proteins. Some of the challenges that remain are the development of methods to resolve mechanisms of coincidence detection for peripheral proteins including those proteins that may bind sphingolipids in addition to PIPs.
ACKNOWLEDGEMENTS
PI-binding domain related research in the author’s lab is funded by the NSF (7112361) and American Heart Association (12GRNT12080254).
Footnotes
- ANTH
- AP180 amino-terminal homology
- BAR
- Bin amphiphysin Rvs (BAR)
- C2
- protein kinase C conserved 2
- ENTH
- epsin amino-terminal homology
- FAPP1
- Four-phosphate adaptor protein 1
- FERM
- band 4.1, ezrin, radixin, moesin
- FYVE
- Fab1, YOTB, Vac1, and EEA1
- GOLPH3
- Golgi phosphoprotein 3
- KA1
- kinase associated-1 domains
- MIL
- membrane insertion loop
- PA
- phosphatidic acid
- PDZ
- postsynaptic density 95, disk large, zonula occludens
- PH
- pleckstrin homology domain
- PI
- phosphatidylinositol
- PIP
- phosphoinositide
- PKC
- protein kinase C
- PDB
- Protein Data Bank
- PM
- plasma membrane
- PS
- phosphatidylserine
- PTB
- phosphotyrosine binding (PTB)
- PX
- Phox domain
- TGN
- trans-Golgi network
LITERATURE CITED
- Adu-Gyamfi E, Soni SP, Xue Y, Digman MA, Gratton E, Stahelin RV. The Ebola virus matrix protein penetrates into the plasma membrane: a key step in viral protein 40 (VP40) oligomerization and viral egress. J. Biol. Chem. 2013;288:5779–5789. doi: 10.1074/jbc.M112.443960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agranoff BW, Bradley RM, Brady RO. The enzymatic synthesis of inositol phosphatide. J. Biol. Chem. 1958;233:1077–1083. [PubMed] [Google Scholar]
- Alajlouni R, Drahos KE, Finkielstein CV, Capelluto DG. Lipid-mediated membrane binding properties of Disabled-2. Biochim. Biophys. Acta. 2011;1808:2734–2744. doi: 10.1016/j.bbamem.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Ankem G, Mitra S, Sun F, Moreno AC, Chutvirasakul B, Azurmend HF, Li L, Capelluto DGS. The C2 domain of Tollip, a toll-like receptor signaling regulator, exhibits broad preference for phosphoinositides. Biochem. J. 2011;435:597–608. doi: 10.1042/BJ20102160. [DOI] [PubMed] [Google Scholar]
- Arai Y, Ijuin T, Takenawa T, Becker LE, Takashima S. Excessive expression of synaptojanin in brains with Down syndrome. Brain Development. 2002;24:67–72. doi: 10.1016/s0387-7604(01)00405-3. [DOI] [PubMed] [Google Scholar]
- Azzedine H, Bolino A, Taieb T, Birouk N, Di Duca M, Bouhouche A, Benamou S, Mrabet A, Hammadouche T, Chkili T, Goudier R, Ravazzolo R, Brice A, Laporte J, LeGuern E. Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early onset glaucoma. Am. J. Hum. Genet. 2003;72:1141–1153. doi: 10.1086/375034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat. Rev. Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- Balla T. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J. Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- Balla T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T, Baukal AJ, Guillemette G, Catt KJ. Multiple pathways of inositol polyphosphate metabolism in angiotensin-stimulated adrenal glomerulosa cells. J. Biol. Chem. 1988;263:4083–4091. [PubMed] [Google Scholar]
- Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol. Cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best MD. Global approaches for the elucidation of phosphoinositide-binding proteins. Chem. Phys. Lipids. 2013 doi: 10.1016/j.chemphyslip.2013.10.014. in press. [DOI] [PubMed] [Google Scholar]
- Bhardwaj N, Stahelin RV, Langois RE, Cho W, Lui H. Structural bioinformatics prediction of membrane-binding proteins. J. Mol. Biol. 2006;359:486–495. doi: 10.1016/j.jmb.2006.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N, Stahelin RV, Cho W, Lu H. MeTaDoR: a comprehensive resource for membrane targeting domains and their host proteins. Bioinformatics. 2007;23:3110–3112. doi: 10.1093/bioinformatics/btm395. [DOI] [PubMed] [Google Scholar]
- Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatner N, Stahelin RV, Diraviyam K, Hawkins PT, Hong W, Murray D, Cho W. The molecular basis of the differential subcellular localization of FYVE domains. J. Biol. Chem. 2004;279:53818–53827. doi: 10.1074/jbc.M408408200. [DOI] [PubMed] [Google Scholar]
- Bolino A, Brancolini V, Bono F, Bruni A, Gambardella A, Romeo G, Quattrone A, Devoto M. Localization of a gene responsible for autosomal recessive demyelinating neuropathy with focally folded myelin sheaths to chromosome 11q23 by homozygosity mapping and haplotype sharing. Hum. Mol. Genet. 1996;5:1051–1054. doi: 10.1093/hmg/5.7.1051. [DOI] [PubMed] [Google Scholar]
- Borman DE, Dall’Armi C, Voronov SV, McIntire LB, Zhang H, Moore AZ, Staniszewski A, Arancio O, Kim TW, Di Paolo G. Oligomeric amyloid-beta peptide disrupts phosphatidylinositol-4,5-bisphosphate metabolism. Nat. Neurosci. 2008;11:547–554. doi: 10.1038/nn.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E, Pick A, Camdere G, Liska N, Evergren E, McMahon HT, Kozlov MM. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149:124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJ, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol. Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- Brunecky R, Lee S, Rzepecki PW, Overduin M, Prestwich GD, Kutateladze AG, Kutateladze TG. Investigation of the binding geometry of a peripheral membrane protein. Biochemistry. 2005;44:16064–16071. doi: 10.1021/bi051127+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- Buser CA, Sigal CT, Resh MD, McLaughlin S. Membrane binding of myristoylated peptides corresponding to the NH2 terminus of Src. Biochemistry. 1994;33:13093–13101. doi: 10.1021/bi00248a019. [DOI] [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27:5527–5541. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- Carvalho K, Khalifat N, Maniti O, Nicolas C, Arold S, Picart C, Ramos L. Phosphatidylinositol 4,5-bisphosphate-induced conformational change of ezrin and formation of ezrin oligomers. Biochemistry. 2010;49:9318–9327. doi: 10.1021/bi101141d. [DOI] [PubMed] [Google Scholar]
- Catimel B, Kapp E, Yin MX, Gregory M, Wong LS, Condron M, Church N, Kershaw N, Holmes AB, Burgess AW. The PI(3)P interactome from a colon cancer cell. J Proteomics. 2013;82:35–51. doi: 10.1016/j.jprot.2013.01.031. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Blasutig IM, Goudreault M, Li Z, Ruston J, Pawson T, Sicheri F. Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J. Biol. Chem. 2007;282:13864–13874. doi: 10.1074/jbc.M700505200. [DOI] [PubMed] [Google Scholar]
- Chang-Ileto B, Frere SG, Chan RB, Voronov SV, Roux A, Di Paolo G. Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission. Dev. Cell. 2011;20:206–218. doi: 10.1016/j.devcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang-Ileto B, Frere SG, Di Paolo G. Acute manipulation of phosphoinositide levels in cells. Methods Cell Biol. 2012;108:187–207. doi: 10.1016/B978-0-12-386487-1.00010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sheng R, Kallberg M, Silkov A, Tun MP, Bhardwaj N, Kurilova S, Hall RA, Honig B, Lu H, Cho W. Genome-wide functional annotation of dual-specificity protein- and lipid-binding modules that regulate protein interactions. Mol. Cell. 2012;46:226–237. doi: 10.1016/j.molcel.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W, Bittova L, Stahelin RV. Membrane binding assays for peripheral proteins. Anal. Biochem. 2001;296:153–161. doi: 10.1006/abio.2001.5225. [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Clark J, Anderson KE, Juvin V, Smith TS, Karpe F, Wakelam MJO, Stephens LR, Hawkins PT. Quantification of PtdInsP3 molecular speciesin cells and tissues by mass spectrometry. Nature Methods. 2011;8:267–272. doi: 10.1038/nmeth.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell E, Scott P, Davletov B. Real-time assay for monitoring membrane association for lipid-binding domains. Anal. Biochem. 2008;377:83–88. doi: 10.1016/j.ab.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbalan-Garcia S, Gomez-Fernandez JC. Signaling through C2 domains: More than one lipid target. Biochim. Biophys. Acta. 2014 doi: 10.1016/j.bbamem.2014.01.008. http://dx.doi.org/10.1016/j.bbamem.2014.01.008 [DOI] [PubMed]
- Dalton AK, Ako-Adjei D, Murray PS, Murray D, Vogt VM. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J. Virol. 2007;81:6434–6445. doi: 10.1128/JVI.02757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DB, Doherty KR, Delmonte AJ, McNally EM. J. Biol. Chem. 2002;277:22883–22888. doi: 10.1074/jbc.M201858200. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Dippold HC, et al. GOLPH3 bridges phosphatidylinositol-4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diraviyam K, Stahelin RV, Cho W, Murray D. Computer modeling of the membrane interaction of FYVE domains. J Mol. Biol. 2003;328:721–736. doi: 10.1016/s0022-2836(03)00325-5. [DOI] [PubMed] [Google Scholar]
- Dong XP, Shen D, Wang X, Dawson T, Li X, Zhang Q, Cheng X, Zhang Y, Weisman LS, Delling M, Xu H. PI(3,5)P2 controls membrane trafficking by direct activation of mucolipion Ca2+ release channels in the endolysosome. Nat. Commun. 2010;1:38. doi: 10.1038/ncomms1037. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright DG. Multivalent endosome targeting by homodimeric EEA1. Mol. Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- Duning K, et al. Common exonic missense variants in the C2 domain of the human KIBRA protein modify lipid binding and cognitive performance. Transl. Psychiatry. 2013;3:e272. doi: 10.1038/tp.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SD, Keep NH. The 2.7 A crystal structure of the activated FERM domain of moesin: an analysis of structural changes on activation. Biochemistry. 2001;40:7061–7068. doi: 10.1021/bi010419h. [DOI] [PubMed] [Google Scholar]
- Ellson CD, et al. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox) Nat. Cell Biol. 2001;3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- Endo M, Shirouzu M, Yokoyama S. The Cdc42 binding and scaffolding activities of the fission yeast adaptor protein Scd2. J. Biol. Chem. 2003;278:843–852. doi: 10.1074/jbc.M209714200. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. High resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J. Cell Biol. 2011;194:257–275. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Ziemba B. Interplay between phosphatidylinositol lipid and calcium signaling at the leading edge of chemotaxing ameboid cells. Chem. Phys. Lipids. 2014 doi: 10.1016/j.chemphyslip.2014.01.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburger BH, Jensen JB, Hille B. Kinetics of M1 muscarinic receptor and G protein signaling to phospholipase C in living cells. J. Gen. Physiol. 2010;135:81–97. doi: 10.1085/jgp.200910344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fili N, Calleja V, Woscholski R, Parker PJ, Larijani B. Compartmental signal modulation: Endosomal phosphatidylinositol 3-phosphate controls endosome morphology and selective cargo sorting. Proc. Natl. Acad. Sci. U S A. 2006;103:15473–15478. doi: 10.1073/pnas.0607040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Aruga J, Niinobe M, Aimoto S, Mikoshiba K. J Biol. Chem. 1994;269:29206–29211. [PubMed] [Google Scholar]
- Fukuda M, Kojima T, Aruga J, Niinobe M, Mikoshiba K. J. Biol. Chem. 1995;270:26523–26527. doi: 10.1074/jbc.270.44.26523. [DOI] [PubMed] [Google Scholar]
- Fuson K, Rice A, Mahling R, Snow A, Nayak K, Shanbhogue P, Meyer AG, Redpath GM, Hinderliter A, Cooper ST, Sutton RB. Alternate splicing of dysferlin C2A confers Ca2+-dependent and Ca2+-independent binding for membrane repair. Structure. 2013 doi: 10.1016/j.str.2013.10.001. doi: 10.1016/j.str.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego O, Betts MJ, Gvozdenovic-Jeremic J, Maeda K, Matezki C, Aguilar-Gurrieri C, Beltran-Alvarez P, Bonn S, Fernandez-Tornero C, Jensen LJ, Kuhn M, Trott J, Rybin V, Muller CW, Bork P, Kaksonen M, Russell RB, Gavin AC. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol. Syst. Biol. 2010;6:430. doi: 10.1038/msb.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier JM, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- Gericke A, Leslie NR, Losche M, Ross AH. PtdIns(4,5)P2-mediated cell signaling: emerging principles and PTEN as a paradigm for regulatory mechanism. Adv. Exp. Med. Biol. 2013;991:85–104. doi: 10.1007/978-94-007-6331-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil JE, Kim E, Kim IS, Ku B, Park WS, Oh BH, Ryu SH, Cho W, Heo WD. Phosphoinositides differentially regulate protruding localization through the FYVE domain. J. Biol. Chem. 2012;287:41268–41276. doi: 10.1074/jbc.M112.419127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber ZT, Gericke A, Kooijman EE. Phosphatidylinositol-4,5-bisphosphate ionization in the presence of cholesterol, calcium or magnesium ions. Chem. Phys. Lipids. 2013 doi: 10.1016/j.chemphyslip.2013.11.004. in press. [DOI] [PubMed] [Google Scholar]
- Graber ZT, Jiang Z, Gericke A, Kooijman EE. Phosphatidylinositol-4,5-bisphosphate ionization and domain formation in the presence of lipids with hydrogen bond donor capabilities. Chem. Phys. Lipids. 2012;165:696–704. doi: 10.1016/j.chemphyslip.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Guillen J, Ferrer-Orta C, Buxaderas M, Perez-Sanchez D, Guerrero-Valero M, Luengo-Gil G, Pous J, Guerra P, Gomez-Fernandez JC, Verdaguer N, Corbalan-Garcia S. Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1. Proc. Natl. Acad. Sci. USA. 2013;110:20503–20508. doi: 10.1073/pnas.1316179110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Valero M, Ferrer-Orta C, Querol-Audi J, Marin-Vicente C, Fita I, Gomez-Fernandez JC, Verdaguer N, Corbalan-Garcia S. Proc. Natl. Acad. Sci. 2009;106:6603–6607. doi: 10.1073/pnas.0813099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Schiavo G, Irvine RF. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P2. Biochem. J. 2009;422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- Hankins JL, Ward KE, Linton SS, Barth BM, Stahelin RV, Fox TE, Kester M. Ceramide 1-phosphate mediates endothelial cell invasion via the annexin a2-p11 heterotetrameric protein complex. J. Biol. Chem. 2013;288:19726–19738. doi: 10.1074/jbc.M113.481622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Haney RM, Vora M, Verkhusha VV, Stahelin RV, Kutateladze TG. Molecular mechanism of membrane targeting by the GRP1 PH domain. J. Lipid Res. 2008;49:1807–1815. doi: 10.1194/jlr.M800150-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Scott JL, Heroux A, Roy S, Lenoir M, Overduin M, Stahelin RV, Kutateladze TG. Molecular basis of phosphatidylinositol 4-phosphate and ARF1 GTPase recognition by the FAPP1 pleckstrin homology (PH) domain. J. Biol. Chem. 2011;286:18650–18657. doi: 10.1074/jbc.M111.233015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Vora M, Haney RM, Filonov GS, Musselman CA, Burd CG, Kutateladze AG, Verkhusha VV, Stahelin RV, Kutateladze TG. Membrane insertion of the FYVE domain is modulated by pH. Proteins. 2009;76:852–860. doi: 10.1002/prot.22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman GE, Finegold M, Zhao W, de Guoyon B, Metzenberg A. Medical complications in long-term survivors with X-linked myotubular myopathy. J Pediatr. 1999;134:206–216. doi: 10.1016/s0022-3476(99)70417-8. [DOI] [PubMed] [Google Scholar]
- Hom RA, Vora M, Regner M, Subach OM, Cho W, Verkhusha VV, Stahelin RV, Kutateladze TG. pH-independent binding of the Epsin ENTH domain and the AP180 ANTH domain to PI(4,5)P2-containing bilayers. J. Mol. Biol. 2007;373:412–423. doi: 10.1016/j.jmb.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath CA, Vanden Broeck D, Boulet GA, Bogers J, De Wolf MJ. Epsin: inducing membrane curvature. Int. J. Biochem. Cell Biol. 2007;39:1765–1770. doi: 10.1016/j.biocel.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Huang BX, Akbar M, Kevala K, Kim HY. Phosphatidylserine is a critical modulator for Akt activation. J. Cell Biol. 2011;192:979–992. doi: 10.1083/jcb.201005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J. Biol. Chem. 285:42130–42139. doi: 10.1074/jbc.M110.143412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca2+-regulated fusion. Cell. 2009;138:709–721. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. Membrane binding domains. Biochim. Biophys. Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Heo WD, Grimely JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo H, Lo PK, Li Y, Loison F, Green S, Wang J, Silbers LE, Ye K, Chen H, Luo HR. Deactivation of Akt by a small molecule inhibitor targeting pleckstrin homology domain and facilitating Akt ubiquitination. Proc. Natl. Acad. Sci. U S A. 2011;108:6486–6491. doi: 10.1073/pnas.1019062108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaisaki PJ, Delphine M, Woon PY, Sebag-Montefiore L, Wilder SP, Menzel S, Vionnet N, Marion E, Riveline JP, Charpentier G, Schurmans S, Levy JC, Lathrop M, Farrall M, Gauguier D. Polymorphisms in type II SH2-domain containing inositol 5-phosphatase (INPPL1, SHIP2) are associated with physiological abnormalities of the metabolic syndrome. Diabetes. 2004;53:1900–1904. doi: 10.2337/diabetes.53.7.1900. [DOI] [PubMed] [Google Scholar]
- Kagawa S, Soeda Y, Ishihara H, Oya T, Sasahara M, Yaguchi S, Oshita R, Wada T, Tsuneki H, Sasaoka T. Impact of transgenic overexpression of SH2-containing inositol 5’-phosphatase 2 on glucose metabolism and insulin signaling mice. Endocrinology. 2008;149:642–650. doi: 10.1210/en.2007-0820. [DOI] [PubMed] [Google Scholar]
- Karathanassis D, Stahelin RV, Bravo J, Perisic O, Pacold CM, Cho W, Williams RL. Binding of the PX domain or p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 2002;21:5057–5068. doi: 10.1093/emboj/cdf519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Afsari HS, Cho W. High-throughput fluorescence assay for membrane-protein interaction. J. Lipid Res. 2013;54:3531–3538. doi: 10.1194/jlr.D041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Guzman-Hernandez ML, Balla T. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev. Cell. 2011;21:813–824. doi: 10.1016/j.devcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T, Kukelyansky I, McCaffery JM, Ueno T, Varela LC, Inoue T. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat. Methods. 2010;7:206–208. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman EE, King KE, Gangoda M, Gericke A. Ionization properties of phosphatidylinositol polyphosphates in mixed model membranes. Biochemistry. 2009;48:9360–9371. doi: 10.1021/bi9008616. [DOI] [PubMed] [Google Scholar]
- Kutateladze TG. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim. Biophys. Acta. 2006;1761:868–877. doi: 10.1016/j.bbalip.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze TG. Mechanistic similarities in docking of the FYVE and PX domains to phosphatidylinositol-3-phosphate containing membranes. Prog. Lipid Res. 2007;46:315–327. doi: 10.1016/j.plipres.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze TG. Translation of the phosphoinositide code by PI effectors. Nat. Chem. Biol. 2010;6:507–513. doi: 10.1038/nchembio.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze TG, Capelluto DG, Ferguson CG, Cheever ML, Kutateladze AG, Prestwich GD, Overduin M. Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 2004;279:3050–3057. doi: 10.1074/jbc.M309007200. [DOI] [PubMed] [Google Scholar]
- Kutateladze TG, Overduin M. Structural mechanism of endosome docking by the FYVE domain. Science. 2001;291:1793–1796. doi: 10.1126/science.291.5509.1793. [DOI] [PubMed] [Google Scholar]
- Lamour NF, Wijesinghe DS, Mietla JA, Ward KE, Stahelin RV, Chalfant CE. Ceramide kinase regulates the production of tumor necrosis factor α (TNFα) via inhibition of TNFα -converting enzyme. J. Biol. Chem. 2011;286:42808–42817. doi: 10.1074/jbc.M111.310169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf KE, Pilling C, Falke JJ. Molecular Mechanism of an Oncogenic Mutation That Alters Membrane Targeting: Glu17Lys Modifies the PIP Lipid Specificity of the AKT1 PH Domain. Biochemistry. 2008;47:12260–12269. doi: 10.1021/bi801683k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SA, Eyeson R, Cheever ML, Geng J, Verkhusha VV, Burd C, Overduin M, Kutateladze TG. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. USA. 2005;102:13052–13057. doi: 10.1073/pnas.0503900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp. 2007:81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lenoir M, Coskun U, Grzybek M, Cao X, Buschhorn SB, James J, Simons K, Overduin M. Structural basis of wedging the Golgi membrane by FAPP pleckstrin homology domains. EMBO Rep. 2010;11:279–284. doi: 10.1038/embor.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir M, Overduin M. PtdIns(4)P signaling and recognition systems. Adv. Exp. Med. Biol. 2013;991:59–83. doi: 10.1007/978-94-007-6331-9_5. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acid Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I, Cebers A, Janmey PA. Combined electrostatics and hydrogen bonding determine intermolecular interactions between polyphosphoinositides. J. Am. Chem. Soc. 2008;130:9025–9030. doi: 10.1021/ja800948c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang X, Zhang X, Zhao M, Tsang WL, Zhang Y, Yau RG, Weisman LS, Xu H. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc. Natl. Acad. Sci. U S A. 2013;110:21165–21170. doi: 10.1073/pnas.1311864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyakhova TA, Knight JD. The C2 domains of granuphilin are high-affinity sensors for plasma membrane lipids. Chem. Phys. Lipids. 2013 doi: 10.1016/j.chemphyslip.2013.10.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani T, Hennigan RF, Foster LA, Conrady DG, Herr AB, Ip W. FERM domain phosphoinositide binding targets merlin to the membrane and is essential for its growth-suppressive function. Mol. Cell Biol. 2011;31:1983–1996. doi: 10.1128/MCB.00609-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna D, Bhardwaj N, Vora MS, Stahelin RV, Lu H, Cho W. Differential roles of phosphatidylserine, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 in plasma membrane targeting of C2 domains. Molecular dynamics simulation, membrane binding, and cell translocation studies of the PKCalpha C2 domain. J. Biol. Chem. 2008;283:26047–26058. doi: 10.1074/jbc.M802617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion E, Kaisaki PJ, Pouillon V, Gueydan C, Levy JC, Bodson A, Krzentowski G, Daubresse JC, Mockel J, Behrends J, Servais G, Szpirer C, Kruys V, Gauguier D, Schurmans S. The gene INPPL1, encoding the lipid phosphatase SHIP2, is a candidate for type 2 diabetes in rat and man. Diabetes. 2002;51:2012–2017. doi: 10.2337/diabetes.51.7.2012. [DOI] [PubMed] [Google Scholar]
- McCrea HJ, De Camilli P. Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology. 2009;24:8–16. doi: 10.1152/physiol.00035.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- McCartney AJ, Zhang Y, Weisman LS. Phosphatidylinositol 3,5-bisphosphate: Low abundance, high significance. Bioessays. 2014;36:52–64. doi: 10.1002/bies.201300012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Wang J, Gambhir A, Murray D. PIP2 and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- Mehrotra B, Myszka DG, Prestwich GD. Binding kinetics and ligand specificity for the interactions of the C2B domain of synaptotagmin II with inositol polyphosphates and phosphoinositides. Biochemistry. 2000;39:9679–9686. doi: 10.1021/bi000487o. [DOI] [PubMed] [Google Scholar]
- Mesmin B, Drin G, Levi S, Rawet M, Cassel D, Bigay J, Antonny B. Two-lipid packing sensor motifs contribute to the sensitivity of ArfGAP1 to membrane curvature. Biochemistry. 2007;46:1779–1790. doi: 10.1021/bi062288w. [DOI] [PubMed] [Google Scholar]
- Mim C, Unger VM. Membrane curvature and its generation by BAR proteins. Trends Biochem. Sci. 2012;37:526–533. doi: 10.1016/j.tibs.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SE, Sahlender DA, Graham SC, Honing S, Robinson MS, Peden AA, Owen DJ. The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell. 2011;147:1118–1131. doi: 10.1016/j.cell.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- Mitra S, Traughber CA, Brannon MK, Gomez S, Capelluto DGS. Ubiquitin interacts with the Tollip C2 and CUE domains and inhibits binding of Tollip to phosphoinositides. J. Biol. Chem. 2013;288:25780–25791. doi: 10.1074/jbc.M113.484170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Mendrola JM, Schmitz KR, Wang YH, Slochower D, Janmey PA, Lemmon MA. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins, facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales KA, Igumenova TI. Synergistic effect of Pb(2+) and phosphatidylinositol 4,5-bisphosphate on C2 domain-membrane interactions. Biochemistry. 2012;51:3349–3360. doi: 10.1021/bi201850h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales KA, Yang Y, Long Z, Li P, Taylor AB, Hart PJ, Igumenova TI. Cd2+ as a Ca2+ surrogate in protein-membrane interactions: isostructural but not isofunctional. J. Am. Chem. Soc. 2013;135:12980–12983. doi: 10.1021/ja406958k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales KA, Lasagna M, Gribenko AV, Yoon Y, Reinhart GD, Lee JC, Cho W, Li P, Igumenova TI. Pb2+ as modulator of protein-membrane interactions. J. Am. Chem. Soc. 2011;133:10599–10611. doi: 10.1021/ja2032772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulgrew-Nesbitt A, Diraviyarm K, Wang J, Singh S, Murray P, Li Z, Rogers L, Mirkovic N, Murray D. The role of electrostatics in protein-membrane interactions. Biochim. Biophys. Acta. 2006;1761:812–826. doi: 10.1016/j.bbalip.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Murray D, Honig B. Electrostatic control of the membrane targeting of C2 domains. Mol. Cell. 2002;9:145–154. doi: 10.1016/s1097-2765(01)00426-9. [DOI] [PubMed] [Google Scholar]
- Murray PS, Li Z, Wang J, Tang CL, Honig B, Murray D. Retroviral matrix domains share electrostatic homology: models for membrane binding function throughout the viral life cycle. Structure. 2005;13:1521–1531. doi: 10.1016/j.str.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Nalefski EA, Wisner MA, Chen JZ, Sprang SR, Fukuda M, Mikoshiba K, Falke JJ. Biochemistry. 2001;40:3089–3100. doi: 10.1021/bi001968a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY, Pirruccello M, Hao M, Ingolia NT, Wenk MR, De Camilli P. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 2012;199:1003–1016. doi: 10.1083/jcb.201206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasuhoglu C, Feng S, Mao J, Yamamoto M, Yin HL, Earnest S, Barylko B, Albanesi JP, Hilgemann DW. Nonradioactive analysis of phosphatidylinositides and other anionic phospholipids by anion-exchange high-performance liquid chromatography with suppressed conductivity detection. Anal. Biochem. 2002;301:243–254. doi: 10.1006/abio.2001.5489. [DOI] [PubMed] [Google Scholar]
- Oancea E, Teruel MN, Quest AF, Meyer T. Green fluorescent protein (GFP)-tagged cysteine rich domains from protein kinase C as fluorescent indicators of diacylglycerol signaling in living cells. J. Cell Biol. 1998;140:485–498. doi: 10.1083/jcb.140.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivotto M, Arcangeli A, Carla M, Wanke E. Electric fields at the plasma membrane level: a neglected element in the mechanisms of cell signaling. Bioessays. 1996;18:495–504. doi: 10.1002/bies.950180612. [DOI] [PubMed] [Google Scholar]
- Ono Y, Fujii T, Igarashi K, Kuno T, Tanaka C, Kikkawa U, Nishizuka Y. Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc. Natl. Acad. Sci. U S A. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada S, Mizuno K, Saido TC, Akita Y, Suzuki K, Kuroki T, Ohno S. A phorbol ester receptor/protein kinase, nPKC eta, a new member of the protein kinase C family predominantly expressed in lung and skin. J. Biol. Chem. 1990;265:22434–22440. [PubMed] [Google Scholar]
- Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- Oxley D, Ktistakis N, Farmaki T. Differential isolation and identification of PI(3)P and PI(3,5)P2 binding proteins from Arabidopsis thaliana using an agarose phoshpatidylinositol-phoshpate affinity chromatogaphy. J. Proteomics. 2013;8:580–594. doi: 10.1016/j.jprot.2013.08.020. [DOI] [PubMed] [Google Scholar]
- Patki V, Lawe DC, Corvera S, Virbasius JV, Chawla A. A functional PtdIns(3)P-binding motif. Nature. 1998;394:433–434. doi: 10.1038/28771. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Ponting CP. Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: binding partners of SH3 domains? Protein Sci. 1996;5:2353–2357. doi: 10.1002/pro.5560051122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashek J, Truong T, Yao X. Crystal structure of the pleckstrin homology domain from ceramide transfer protein: implications for conformational change upog ligand binding. PLoS One. 2013;8:e79590. doi: 10.1371/journal.pone.0079590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L, Bobkov AA, Patel M, Jaroszewski L, Bankston LA, Stec B, Vuori K, Cote JF, Liddington RC. Structural basis of membrane targeting by the DOCK180 family of Rho family guanine exchange factors (Rho-GEFs) J. Biol. Chem. 2010;285:13211–13222. doi: 10.1074/jbc.M110.102517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylypenko O, Lundmark R, Rasmuson E, Carlsson SR, Rak A. The PX-BAR membrane remodeling unit of sorting nexin 9. EMBO J. 2007;26:4788–4800. doi: 10.1038/sj.emboj.7601889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Koch D, Kessels MM. Let’s go bananas: revisiting the endocytic BAR code. EMBO J. 2011;30:3501–3515. doi: 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Haucke V. Membrane shaping by the Bin/amphiphysin/Rvs (BAR) domain protein superfamily. Cell Mol. Life Sci. 2011;68:3983–3993. doi: 10.1007/s00018-011-0768-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecchi M, Peterson A, McLaughlin S. Phosphoinositide-specific phospholipase C-delta 1 binds with high affinity to phospholipid vesicles containing phosphatidylinositol 4,5-bisphosphate. Biochemistry. 1992;31:12742–12747. doi: 10.1021/bi00166a005. [DOI] [PubMed] [Google Scholar]
- Redfern RA, Gericke A. Domain formation in phosphatidylinositol monophosphate/phosphatidylcholine mixed vesicles. Biophys. J. 2004;86:2980–2992. doi: 10.1016/S0006-3495(04)74348-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern RE, Redfern D, Furgason ML, Munson M, Ross AH, Gericke A. PTEN phosphatase selectivity binds phosphoinositides and undergoes structural changes. Biochemistry. 2008;47:2162–2171. doi: 10.1021/bi702114w. [DOI] [PubMed] [Google Scholar]
- Ridley SH, et al. FENS-1 and DFCP1 are FYVE domain containing proteins with distinct functions in the endosomal and Golgi compartments. J. Cell Sci. 2001;114:3991–4000. doi: 10.1242/jcs.114.22.3991. [DOI] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC. C2-domains, structure and function of a universal Ca2+binding domain. J. Biol. Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- Roth MG. Phosphoinositides in cell regulation and membrane dynamics. Physiol. Rev. 2004;84:699–730. doi: 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- Rowland MM, Bostic HE, Gong D, Speers AE, Lucas N, Cho W, Cravatt BF, Best MD. Phosphatidylinositol 3,4,5-trisphosphate activity probes for the labeling and proteomic characterization of protein binding partners. Biochemistry. 2011;50:11143–11161. doi: 10.1021/bi201636s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland MM, Gong D, Bostic HE, Lucas N, Cho W, Best MD. Microarray analysis of Akt PH domain binding employing synthetic biotinylated analogues of all seven phosphoinositide headroup isomers. Chem. Phys. Lipids. 2012;165:207–215. doi: 10.1016/j.chemphyslip.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Pykalainen A, Laurinmaki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Saddoughi SA, Gencer S, Peterson YK, Ward KE, Mukhopadhyay A, Oaks J, Bielawski J, Szulc ZM, Thomas RJ, Selvam SP, Senkal CE, Garrett-Mayer E, De Palma RM, Fedarovich D, Liu A, Habib AA, Stahelin RV, Perrotti D, Ogretmen B. Sphingosine analogue drug FTY720 targets I2PP2A/SET and mediates lung tumor suppression via activation of PP2A-RIPK1-dependent necrosis. EMBO Mol. Med. 2013;5:105–121. doi: 10.1002/emmm.201201283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Guan F, Papolos DF, Lau S, Klein M, Fann CS, Lachman HM. Mutation analysis of SYNJ1: a possible candidate gene for chromosome 21q22-linked bipolar disorder. Mol Psychiatry. 2001;6:387–395. doi: 10.1038/sj.mp.4000871. [DOI] [PubMed] [Google Scholar]
- Saito T, Stopkova P, Diaz L, Papolos DF, Boussemart L, Lachman HM. Polymorphism screening of PIK4CA: possible candidate gene for chromosome 22q11-linked psychiatric disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2003;116:77–83. doi: 10.1002/ajmg.b.10042. [DOI] [PubMed] [Google Scholar]
- Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shaprio L. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- Sarkes D, Rameh LE. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphinositides. Biochem. J. 2010;428:375–384. doi: 10.1042/BJ20100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavo G, Gu QM, Prestwich GD, Sollner TH, Rothman JE. Calcium-dependent switching of the specificity of phosphoinositide binding to synaptotagmin. Proc. Natl. Acad. Sci. U S A. 1996;93:13327–32. doi: 10.1073/pnas.93.23.13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G. Kinetic studies of protein-protein interactions. Curr. Opin. Struct. Biol. 2002;12:41–47. doi: 10.1016/s0959-440x(02)00287-7. [DOI] [PubMed] [Google Scholar]
- Segers K, Sperandio O, Sack M, Fischer R, Miteva MA, Rosing J, Nicoleas GA, Villoutreix BO. Design of protein membrane interaction inhibitors by virtual ligand screening , proof of concept with the C2 domain of factor V. Proc. Natl. Acad. Sci. U S A. 2007;104:1269–1702. doi: 10.1073/pnas.0701051104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senderek J, Bergmann C, Weber S, Ketelsen UP, Schorle H, Rudnik-Schoneborn S, Buttner R, Buchheim E, Zerres K. Mutation of the SBF2 gene, encoding a novel member of the myotubularin family, in Charcot-Marie-Tooth neuropathy type 4B2/11p15. Hum. Mol. Genet. 2003;12:349–356. doi: 10.1093/hmg/ddg030. [DOI] [PubMed] [Google Scholar]
- Seet LF, Hong W. The Phox (PX) domain proteins and membrane traffic. Biochim. Biophys. Acta. 2006;1761:878–896. doi: 10.1016/j.bbalip.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Scott JL, Musselman CA, Adu-Gyamfi E, Kutateladze TG, Stahelin RV. Emerging methodologies to investigate lipid-protein interactions. Integr. Biol. 2012;4:247–258. doi: 10.1039/c2ib00143h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seykora JT, Myat MM, Allen LA, Ravetch JV, Aderem A. Molecular determinants of the myristoyl-electrostatic switch of MARCKS. J. Biol. Chem. 1996;271:18797–18802. doi: 10.1074/jbc.271.31.18797. [DOI] [PubMed] [Google Scholar]
- Shao X, Davletov BA, Sutton RB, Sudhof TC, Rizo J. Bipartite Ca2+-binding motif in C2 domains of synaptotagmin and protein kinase c. Science. 1996;273:248–251. doi: 10.1126/science.273.5272.248. [DOI] [PubMed] [Google Scholar]
- Shao X, Fernandez I, Sudhof TC, Rizo J. Solution structure ofs of the Ca2+-free and Ca2+-bound C2A domain of synaptotagmin I: does Ca2+ induce a conformational change? Biochemistry. 1998;37:16106–16115. doi: 10.1021/bi981789h. [DOI] [PubMed] [Google Scholar]
- Shenoy S, Shekar P, Heinrich F, Daou MC, Gericke A, Ross AH, Losche M. Membrane association of the PTEN tumor suppressor: molecular details of the protein-membrane complex from SPR binding studies and neutron reflection. PLoS One. 2012;7:e32591. doi: 10.1371/journal.pone.0032591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisheva A. PtdIns5P: News and views of its appearance, disappearance, and deeds. Arch. Biochem. Biophys. 2013;538:171–180. doi: 10.1016/j.abb.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkov A, Yoon Y, Lee H, Gokhale N, Adu-Gyamfi E, Stahelin RV, Cho W, Murray D. Genome-wide structural analysis reveals novel membrane binding properties of AP180 N-terminal homology (ANTH) domains. J. Biol. Chem. 2011;286:34155–34163. doi: 10.1074/jbc.M111.265611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanshu DK, Kamlekar RK, Wijesinghe DS, Zou X, Zhai X, Mishra SK, Molotkovsky JG, Malinina L, Hinchcliffe EH, Chalfant CE, Brown RE, Patel DJ. Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids. Nature. 2013;500:463–467. doi: 10.1038/nature12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Birkeland HC, Gilooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- Sot B, Behrmann E, Raunser S, Wittinghofer A. Ras GTPase activating (RasGAP) activity of the dual specificity GAP protein Rasal requires colocalization and C2 domain binding to lipid membranes. Proc. Natl. Acad. Sci. U S A. 2013;110:111–116. doi: 10.1073/pnas.1201658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin RV. Lipid binding domains: more than simple lipid effectors. J. Lipid Res. 2009;50:S299–S304. doi: 10.1194/jlr.R800078-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin RV. Surface plasmon resonance: a useful technique for cell biologists to characterize biomolecular interactions. Mol. Biol. Cell. 2013;24:883–886. doi: 10.1091/mbc.E12-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin RV, Burian A, Bruzik KS, Murray D, Cho W. Membrane binding mechanisms of the PX domains of NADPH p40phox and p47phox. J. Biol. Chem. 2003a;278:14469–14479. doi: 10.1074/jbc.M212579200. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Cho W. Differential roles of ionic, aliphatic, and aromatic residues in membrane-protein interactions, a surface plasmon resonance study on phospholipases A2. Biochemistry. 2001;40:4672–4678. doi: 10.1021/bi0020325. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Cho W. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Karathanassis D, Bruzik KS, Waterfield MD, Bravo J, Williams RL, Cho W. Structural and membrane binding analysis of the Phox homology domain of phosphoinositide kinase-C2alpha. J. Biol. Chem. 2006;281:39396–39406. doi: 10.1074/jbc.M607079200. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Karathanassis D, Murray D, Williams RL, Cho W. Structural and membrane binding analysis of the Phox homology domain of Bem1p: basis of phosphatidylinositol 4-phosphate specificity. J. Biol. Chem. 2007;282:25737–25747. doi: 10.1074/jbc.M702861200. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. Phosphatidylinositol 3-phosphate induces the membrane penetration of FYVE domains of Vps27p and Hrs. J. Biol. Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Long F, Peter BJ, Murray D, De Camilli P, McMahon HT, Cho W. Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J. Biol. Chem. 2003b;278:28993–28999. doi: 10.1074/jbc.M302865200. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Rafter JD, Das S, Cho W. The molecular basis of differential subcellular localization of C2 domains of protein kinase C-alpha and group IVa cytosolic phospholipase A2. J. Biol. Chem. 2003c;278:12452–12460. doi: 10.1074/jbc.M212864200. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Subramanian P, Vora M, Cho W, Chalfant CE. Ceramide-1-phosphate binds group IVA cytosolic phospholipase a2 via novel site in the C2 domain. J. Biol. Chem. 2007;282:20467–20474. doi: 10.1074/jbc.M701396200. [DOI] [PubMed] [Google Scholar]
- Stopkova P, Saito T, Fann CS, Papolos DF, Vevera J, Paclt I, Zukov I, Stryjer R, Strous RD, Lachman HM. Polymorphism screening of PIP5K2A: a candidate gene for chromosome 10p-linked psychiatric disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2003;123:50–58. doi: 10.1002/ajmg.b.20012. [DOI] [PubMed] [Google Scholar]
- Stopkova P, Saito T, Papolos DF, Vevera J, Paclt I, Zukov I, Bersson YB, Margolis BA, Strous RD, Lachman HM. Identification of PIK3C3 promoter variant associated with bipolar disorder and schizophrenia. Biol. Psychiatry. 2004;55:981–988. doi: 10.1016/j.biopsych.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Subramanian P, Stahelin RV, Szulc Z, Bielawska A, Cho W, Chalfant CE. Ceramide 1-phosphate acts as a positive allosteric activator of group IVA cytosolic phospholipase A2 alpha and enhances the interaction of the enzyme with phosphatidylcholine. J. Biol. Chem. 2005;280:17601–17607. doi: 10.1074/jbc.M414173200. [DOI] [PubMed] [Google Scholar]
- Sudhahar CG, Haney RM, Xue Y, Stahelin RV. Cellular membranes and lipid-binding domains as attractive targets for drug development. Curr. Drug Targets. 2008;9:603–613. doi: 10.2174/138945008785132420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh BC, Hille B. Regulation of KCNQ channels by manipulation of phosphoinositides. J. Physiol. 2007;582:911–916. doi: 10.1113/jphysiol.2007.132647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh BC, Inoue T, Meyer T, Hille B. Rapid chemically inducible changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentpeter Z, Varnai P, Balla T. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc. Natl. Acad. Sci. U S A. 2010;107:8225–8230. doi: 10.1073/pnas.1000157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale RD, Collins BM. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions, and roles in disease. Biochem. J. 2012;441:39–59. doi: 10.1042/BJ20111226. [DOI] [PubMed] [Google Scholar]
- Terrillon S, Bouvier M. Receptor activity-independent recruitment of βarrestin2 reveals specific signaling modes. EMBO J. 2004;23:3950–3961. doi: 10.1038/sj.emboj.7600387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien C, Di Fulvio S, Pickles S, Sinnreich M. Characterization of lipid binding specificities of dysferlin C2 domains reveals novel interactions with phosphoinositides. Biochemistry. 2009;48:2377–2384. doi: 10.1021/bi802242r. [DOI] [PubMed] [Google Scholar]
- Tian M, Bai C, Lin Q, Lin H, Liu M, Ding F, Wang HR. Binding of RhoA by the C2 domain of E3 ligase Smurf1 is essential for Smurf1-regulated RhoA ubiquitination and cell protrusive activity. FEBS Lett. 2011;585:2199–2204. doi: 10.1016/j.febslet.2011.06.016. [DOI] [PubMed] [Google Scholar]
- Vance JE, Steenbergen R. Metabolism and functions of phosphatidylserine. Prog. Lipid Res. 2005;44:207–234. doi: 10.1016/j.plipres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Balla T. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim. Biophys. Acta. 2006;1761:915–921. doi: 10.1016/j.bbalip.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Varnai P, Thyagarajan B, Rohacs T, Balla T. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J. Cell Biol. 2006;175:377–382. doi: 10.1083/jcb.200607116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC. Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. EMBO J. 1999;18:6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronov SV, Frere SG, Giovedi S, Pollina EA, Borel C, Zhang H, Schmidt C, Akeson EC, Wenk MR, Cimasoni L, Arancio O, Davisson MT, Antonarakis SE, Gardner K, De Camilli P, Di Paolo G. Synaptojanin 1-linked phosphoinositide dyshomeostasis and cognitive defects in mouse models of Down’s syndrome. Proc. Natl. Acad. Sci. U S A. 2008;105:9415–9420. doi: 10.1073/pnas.0803756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Sasaoka T, Funaki M, Hori H, Murakami S, Ishiki M, Haruta T, Asano T, Ogawa W, Ishihara H, Kobayashi M. Overexpression of SH-2 containing inositol phosphatase 2 results in negative regulation of insulin-induced metabolic actions in 3T3: L1 adipocytes via its 5’-phosphatase catalytic activity. Mol. Cell Biol. 2001;21:1633–1646. doi: 10.1128/MCB.21.5.1633-1646.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Slochower DR, Janmey PA. Counterion-mediated cluster formation by polyphosphoinositides. Chem. Phys. Lipids. 2014 doi: 10.1016/j.chemphyslip.2014.01.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward KE, Bhardwaj N, Vora M, Chalfant CE, Lu H, Stahelin RV. The molecular basis of ceramide-1-phosphate recognition by C2 domains. J. Lipid Res. 2013;54:636–648. doi: 10.1194/jlr.M031088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward KE, Ropa JP, Adu-Gyamfi E, Stahelin RV. C2 domain membrane penetration by group IVA cytosolic phospholipase A2 induces membrane curvature changes. J. Lipid Res. 2012;53:2656–2666. doi: 10.1194/jlr.M030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzyniak AM, Kashyap R, Zimmermann P. Phosphoinositides and PDZ domain scaffolds. Adv. Exp. Med. Biol. 2013;991:41–57. doi: 10.1007/978-94-007-6331-9_4. [DOI] [PubMed] [Google Scholar]
- Wenk MR, Lucast L, Di Paolo G, Romanelli AJ, Suchy SF, Nussbaum RL, Cline GW, Shulman GI, De Camilli P. PIP kinase Igamma is the major PI(4,5)P(2) synthesizing enzyme at the synapse. Neuron. 31:79–88. doi: 10.1016/s0896-6273(01)00456-1. [DOI] [PubMed] [Google Scholar]
- Wood CS, Schmitz KR, Bessman NJ, Setty TG, Ferguson KM, Burd CG. PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J. Cell Biol. 2009;187:967–975. doi: 10.1083/jcb.200909063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Gilbert GE, Shi J, Silvus J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S. Receptor activation alters inner surface potential during phagocytosis. Science. 2006;313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Lee PJ, Kurilova S, Cho W. In situ quantitative imaging of cellular lipids using molecular sensors. Nat. Chem. 2011;3:868–874. doi: 10.1038/nchem.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Zhang Z, Cho W. Phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) specifically induces membrane penetration and deformation by Bin/amphiphysin/Rvs (BAR) domains. J. Biol. Chem. 2012;287:34078–34090. doi: 10.1074/jbc.M112.372789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Rathore SS, Davis EM, Ouyang Y, Shen J. Doc2b promotes GLUT4 exocytosis by activating the SNARE-mediated fusion reaction in a calcium and membrane bending-dependent manner. Mol. Biol. Cell. 2013;24:1176–1184. doi: 10.1091/mbc.E12-11-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;81:917–924. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]