Abstract
Many, if not most, embryos begin development with extremely short cell cycles that exhibit unusually rapid DNA replication and no gap phases. The commitment to the cell cycle in the early embryo appears to preclude many other cellular processes which only emerge as the cell cycle slows, at a major embryonic transition known as the mid-blastula transition (MBT) just prior to gastrulation. As reviewed here, genetic and molecular studies in Drosophila have identified changes that extend S phase and introduce a post-replicative gap phase, G2, to slow the cell cycle. While many mysteries remain about the upstream regulators of these changes, we review the core mechanisms of the change in cell cycle regulation and discuss advances in our understanding of how these might be timed and triggered. Finally, we consider how the elements of this program may be conserved or changed in other organisms.
Keywords: Drosophila, MBT, maternal zygotic transition, replication, G2, cell cycle
Introduction
The early embryonic cell cycles of most organisms are extensively modified so that they are unusually fast. These unusual cycles slow just as morphogenesis begins at gastrulation. While there are exceptions and differences in the details for each organism (82, 117), this pattern of development is widespread in organisms that lay eggs that develop externally, with many prominent examples, including insects (35, 72), amphibians (77, 78), and fish (54). In studying this process in Xenopus laevis, John Gerhart dubbed the dramatic slowing of in the cell cycle and associated onset of various cellular activities the mid-blastula transition (MBT), after the developmental stage at which the transition occurs in Xenopus laevis embryos (41).
Long before the transition had been named, Boveri, in 1892, recognized a distinctive feature of these early events. He crossed different species of sea urchin, examined the early divisions of resulting embryos, and concluded that the form and rate of cleavage is determined wholly by the mother, while later aspects of development show influences of both parents (115). This early insight exemplifies a widespread feature of early embryogenesis: the mother pre-loads the egg with material that directs the early rapid cell cycles that subdivide the large eggs of nearly all species. Somehow, the preloaded package of gene products executes a precisely timed dynamic program in which the egg goes through a species-specific number of rapid divisions before remodeling the cell cycle.
In this review, we set out to first describe the phenomenon of the mid-blastula transition as it occurs in Drosophila melanogaster, an organism where it has been studied extensively with the aid of excellent genetic and cell biological tools. After describing the cell cycle and development of the early Drosophila embryo, we describe changes in the cell cycle machinery that occur during early development and present a mechanistic model for the slowing of the cell cycle. We then discuss how this transition could potentially be timed in the Drosophila embryo, and then finally, we compare the proposed mechanisms to what is known in other organisms.
Early development in Drosophila
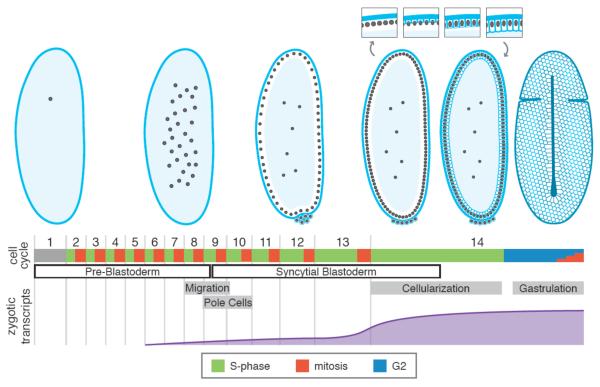
Like other externally developing eggs, the Drosophila egg is large—about 100,000 times the size of an average somatic cell in the adult fly. After fertilization, its nuclei multiply and divide in synchrony in a massive cytoplasm, called a syncytium (86). The nuclei share the cytoplasm and are not separated by plasma membranes; consequently, mitosis occurs without cytokinesis. While this mode of development may seem unfamiliar, it is a paradigm commonly used in insects. The early morphology and cell cycle of the embryo are diagrammed in Figure 1.
Figure 1. Early Development in Drosophila.
A diagram of the first 14 cycles of Drosophila development with notable morphological stages illustrated at the top. Note that while most embryos are displayed as sections through the middle of the embryo with the ventral side to the right, the final illustration is a surface view, with the ventral side up. The process of cellularization is diagrammed in more detail in the insets. The duration of each phase of the cell cycle is below: S phase (green), mitosis (red), and G2 (blue). Mitosis 14 is represented as a series of small bars because the embryo is no longer synchronous at this time and individual groups of cells enter mitosis at different times according to a developmentally programmed schedule. The timing of notable morphological events are demarcated in grey boxes: the migration of the nuclei to the blastoderm, the insulation of the germline by cellularization of the pole cells, the cellularization of the blastoderm nuclei, and the onset of the first gastrulation movement—ventral furrow formation. Below this is diagrammed the approximate number of genes for which zygotic transcripts have been detected over time.
The earliest nuclear cycles are exceptionally rapid—the nuclei divide every 8.6 minutes (35, 86). Thus, while an average tissue culture or somatic cell takes 8–24 hours to go through the cell cycle, a Drosophila embryo manages 14 cell cycles in only 1.5 hours, producing an embryo with thousands of cells. How does the early embryo achieve this notable speed? In short, by modifying the cell cycle, such that the process of DNA replication occurs much more quickly than in most cell cycles (6, 71, 92) and by omitting the gap phases between replication and mitosis (27, 71). So, instead, the nuclei alternate between mitosis and extraordinarily short S phases, during which the entire genome replicates simultaneously in as little as 3.4 minutes (Figure 1) (6). These cell cycles divide the large pre-existing cytoplasm without growth (81). Moreover, the cell cycle is more-or-less synchronous throughout the entire embryo during these cycles.
During interphases 8 and 9, most of the nuclei migrate outwards, forming a shell of nuclei near the plasma membrane by interphase 10, known as the blastoderm (Figure 1) (35, 86). The blastoderm cycles resemble the pre-blastoderm cycles in that they are quick and synchronous, but once the nuclei reach the surface and form the blastoderm, the cell cycle begins to slow gradually, with interphase progressively lengthening slightly each cycle, from about 9 minutes in cycle 10 to 14 minutes in cycle 13 (35, 86). These cycles still lack gap phases.
However, between cycles 13 and 14, a much more dramatic change in the regulation of the cell cycle slows it considerably and desynchronizes it—while interphase 13 was only 20 minutes long and the same length for every nucleus (35), interphase 14 is a minimum of 70 minutes, and its length differs for different groups of cells in a spatially stereotyped pattern (34). This slowing occurs via two mechanisms: first, S phase becomes much longer, but remains synchronous, increasing from 14 minutes in cycle 13 to 50 minutes in cycle 14 (92), and second, interphase 14 exhibits the first G2 gap phase (26, 27) where cells pause for a self-determined, developmentally programmed length of time between the completion of replication and entry into mitosis (Figure1) (34) (27). There is still no G1 gap phase between mitosis and replication (27)—it will not be introduced for several more cell cycles in a separate regulatory change (15, 57, 58).
During interphase 14, more than the cell cycle changes—morphogenesis begins. During the long S phase 14, the process of cellularization pulls membrane down between the nuclei and deposits new membrane to envelop each nucleus in its own plasma membrane, dividing the syncytial blastoderm into over 6,000 cells (Figure 1). Then, following cellularization, the first gastrulation movements begin during G2 of interphase 14. These include formation of a ventral furrow, where the mesodermal precursors internalize, and a cephalic furrow that transiently demarcates the cells that will comprise the cephalon (or head region) of the fly (Figure 1). Throughout these gastrulation movements, stereotyped groups of cells known as “mitotic domains” exit their G2 phases and enter mitosis 14 in a developmentally programmed order, in their first departure from synchronous mitotic cycles (27, 34).
Transitions in embryonic regulation
Early development is extremely fast, and consequently, many events often occur nearly simultaneously and processes with progressive steps can appear abrupt. Often, groups of changes in early embryonic development have been lumped together as abrupt “transitions” that, after more detailed analysis, can be seen to result from several distinct regulatory processes. Here, we try and put the major “transitions” of the early embryo into this context.
At fertilization, the embryonic genome is nearly quiescent, and maternally loaded gene products direct development. As the embryo progresses, however, zygotic gene products become required for developmental events and the progress of the cell cycle; thus, control of development and the cell cycle is handed off from the maternal genome to the zygotic genome, often called the maternal to zygotic transition, MZT. But is it really a single transition? Early definitions of the MZT were made based on the first obvious derangement of an embryo (usually a failure to continue cell division) when the input of the zygotic genome was blocked by inhibiting transcription (44, 66). By focusing on the first obvious deviation from normal, this approach defines a single transition point, but could erroneously collapse a progressive sequence of events into a single moment. While neither the loss of any single zygotic gene nor any large region of the genome perturbs the earliest steps of development, an ever increasing number of mutants display phenotypes as development progresses. This shows that dependency on zygotic gene activity first becomes obvious in cycle 14, but that there is a continuous and progressive emergence of zygotic control of more and more processes (38, 108, 113).
Since transcription must underlie the function of the zygotic genome, molecular assays for the first zygotic gene expression, often called zygotic genome activation, have also been used as a measure of MZT. A major increase in transcription accompanies the progression of early embryonic divisions and experimental measures in numerous systems have defined a point of first activation. Since the first zygotic phenotype does not necessarily become apparent at the time of the changes in zygotic expression that caused it, this has resulted in multiple conflicting times defined as the MZT in Drosophila. For example, even though zygotic phenotypes are first described in cycle 14, cycle 10 has been designated as the first point of zygotic transcription in Drosophila based on the incorporation of radioactive precursors into mature transcripts (28, 120). Additionally, an assay’s ability to define a time of zygotic genome activation is limited by its sensitivity, and several considerations suggest that cycle 10 is simply the point in a progressive process when an increase in transcriptional activity first makes it detectable by this methodology. This is probably further compounded by the increase in nuclear content over the early cycles—when looking at transcriptional output, a similar activation of the genome will be twice as difficult to detect each cycle prior, since there would be half as much template and consequently half as many transcripts produced.
Several findings suggest that transcription can and does occur before cycle 10 in Drosophila. For instance, the regulatory cascade leading to sex determination begins with genes such as sc, which is zygotically expressed by cycle 6 (17) and sisA in cycle 8 (30). The promoters of a number of patterning genes, including, ftz, are also active before cycle 10 (10, 84). Additionally, the transcription of other genes has been detected before cycle 10 (8, 84), and as more sensitive assays have been applied, the list continues to grow (1, 16, 63). It seems as if there is a progressive increase in transcription over the development of the embryo, where the transcription of new genes can be detected in each cycle. Such early transcriptional activity is not unique to Drosophila: when biochemical and autoradiographic methods were pushed to high sensitivity, transcription was detected in Xenopus eggs before the MBT (55), which was previously considered the time of zygotic genome activation (77, 78).
The summarized analyses of specific genes, as well as genome-wide studies of gene expression (63) suggest that different genes initiate expression at different times throughout much of early development. The complexity of transcriptional activation argues for multiple events rather than a discrete genome-wide transition in transcription (84, 116, 117). Furthermore, genome-wide analyses of mutants that affect transcriptional onset have found that some, but not all genes are affected in each mutant, and the affected genes are in partially overlapping groups (3, 61, 64, 79), supporting the idea that each individual gene’s transcriptional onset could be determined by a promoter-specific combination of multiple inputs, leading to numerous programs of transcriptional onset.
Thus, at the genome-wide level, the switch from maternal to zygotic control seems to be a progressive process, rather than a sharp transition, regardless of whether one considers the requirement for zygotic gene function or the onset of zygotic expression. While there is a general and impressive increase in embryonic transcription that culminates in high transcriptional activity in cycle 14, this increase is achieved in multiple increments distributed over early development, combined with an exponential expansion of coding capacity and increased opportunity for transcription as the cell cycle slows. Not surprisingly, use of the term MZT has varied depending on the emphasis of the investigators and the experimental criteria that they use to assess it. However, the concept of the MZT remains useful, so long as one focuses on a very specific process, such as the transition of the control of entry into mitosis, where one can define a definitive time and an associated mechanism for the transition from maternal to zygotic regulation (38).
Like the MZT, the MBT itself was once defined as a transition where seemingly everything that was important in early development changed in a single concerted event. The changes used to define the MBT in the early Xenopus work were the onset of transcription, the initial slowing of the cell cycle, and the onset of cellular movements (77, 78). However, subsequent observations showed that these events could be uncoupled in Xenopus (55), and in Drosophila as well (24). Moreover, the initial departure from “precisely” repeating cell cycles to gradually slowing cycles, which was used to mark the MBT in the original Xenopus literature, was followed by another dramatic change in the cell cycle just prior gastrulation called the Early Gastrula Transition (EGT) (49). In Drosophila, the most dramatic and abrupt changes occur at cycle 14 and this has been conventionally called the MBT in this organism, a convention that we adhere to. However, it is important to note that the changes at cycle 10 in Drosophila embryos, which we call pre-MBT slowing, might be more analogous to the MBT changes of Xenopus, and the changes in Drosophila cycle 14 embryos might be more properly compared to the EGT changes in Xenopus.
The concept of the MBT as a single, concerted transition in the embryo has been eroded by the recognition of numerous steps in early development. However, it remains true that the fourteenth cell cycle of Drosophila marks an important transition in embryogenesis, accompanied by a major change in cell cycle regulation and consequent slowing of the cell cycle, a dramatic up-regulation of zygotic transcription, increased turnover of maternal messages, and the onset of gastrulation movements. Since these concurrent events are not necessarily co-regulated, we suggest that it is important to study the mechanistic underpinnings of each event; here, we focus on the mechanistic basis of the slowing of the cell cycle.
Why have a specialized, exceptionally rapid early cell cycle program?
It’s impossible to be sure of the evolutionary pressures that led to modern biological phenomena. However, a major problem encountered by eggs laid in the external environment could potentially explain the need for rapid cell cycles: eggs cannot eat. Instead, they must subsist entirely on the limited supply of maternal nutrients that are contained in the egg. Thus, in order to support the developmental program until it generates an organism that can feed itself, eggs have evolved to be very large and nutrient-rich. But this creates a new problem—before hatching, the egg has no means of escape or active defense, and it presents a highly nutritious target for predators. The rapid cell cycles of early development are possibly a response to this, as they serve to minimize the pre-hatching vulnerable stage and proceed to hatching as quickly as possible.
Achieving speed
As mentioned briefly in the introduction, the rapidity of the pre-MBT cell cycles is achieved through a potent combination of specializations of the cell cycle: (1) their dependence entirely on maternal contributions, (2) their lack of gap phases, and (3) their unusually speedy replication of the genome.
Dependence only on maternal contributions
During the more leisurely paced cell cycles of later development, stage-specific transcription of genes advances the cycle to the next phase. For example, transcription of S phase genes, promoted by E2F, contributes strongly to the G1 to S transition (21, 22). However, since the rate of production of completed transcripts is limited by the elongation rate and maximal packing of RNA polymerase onto the gene, in an egg with much more cytoplasm per copy of the genome and a very short interphase, it is considerably more difficult to make sufficient numbers of transcripts to control the cytoplasm each cell cycle. Large somatic cells often circumvent this problem often by amplifying their entire genome (37, 48, 99), amplifying selected genes in the genome (11, 101), or carrying repeated arrays of genes that are particularly in demand (9, 52), but these strategies have not been widely adopted in eggs. Instead, the early embryo takes a shortcut and relies primarily on translation of maternally provided transcripts and post-translational modification to direct the early cycles.
Lack of gap phases
In the canonical cell cycle, two gap phases—G1 and G2—serve as pauses before entry into DNA replication and mitosis, respectively. Cells often use them as a time to grow, or to exit the cell cycle if they do not need to undergo further divisions (75). However, the cells of the early embryo, with no external source of nutrition for growth and a clear imperative to proliferate, neither grow nor pause as they subdivide their generous cytoplasms into smaller cells. Thus, the gap phases are dispensed with, thereby hastening the cell cycle, until cycle 14 when a G2 phase—a pause between completion of replication and entry into mitosis—is introduced (27, 71). Pausing in G2 requires that Cdk1 be inactive after the completion of DNA replication—otherwise, it performs its characteristic role and triggers entry into mitosis (26, 27, 74, 75). The appearance of G2 in cell cycle 14 coincides with inhibitory phosphorylation of essentially all the cyclin-dependent kinase 1 (Cdk1) and inactivation of this mitotic activator (Figure 2) (29). Such widespread inhibitory phosphorylation does not occur in the early cycles, but Wee kinase still contributes perhaps by locally constraining Cdk1 activity (104, 105).
Figure 2. Regulation of Cdk1.
Cyclin-dependent kinase 1 has diverse inputs that can regulate its function, which are diagrammed here. First, it requires a cyclin partner. Second, it requires activating phosphorylation in order to be functional. Third, it must be free of inhibitory phosphorylation, which blocks the ATP-binding pocket. This inhibitory phosphorylation is added by Wee and Myt1 kinases (which are thus inhibitors of Cdk1) and removed by String and Twine (Cdc25) phosphatases (which are thus activators of Cdk1)
Rapid S phase
The speed of DNA replication in early Drosophila embryos is truly astounding—in 3.4 minutes, the embryo replicates its entire genome, a process that will take 50 minutes immediately following the MBT, and in differentiated somatic cells can take 8 hours or more (6, 92). Two differences contribute to the extension of the embryonic S phase from 3.4 to 50 minutes. First, replication origins are slightly more tightly packed—on average, every 7.9 kb in the pre-blastoderm embryo (6), compared to 10.6 kb in a cycle 14 embryo (71). Thus, with a slightly longer distance for each replication fork to travel, it would take slightly longer to replicate the DNA between origins in cycle14. However, this ~30% increase in origin spacing does little to explain the ~1500% change in S phase length between the initial rapid cycles and cell cycle 14.
A larger contribution seems to come from a change in replication timing of different sequences. In most S phases, such as those after the MBT, not all sequences in the DNA replicate at the same time. While the euchromatin begins to replicate immediately upon entering S phase, many late replicating sequences, which are generally heterochromatic, wait until later in S phase to replicate (Figure 3a) (92). The prime example of late replicating sequences in Drosophila are the satellite sequences—stretches of megabases of highly repetitive DNA that account for nearly 30% of the genome (62, 92). In the pre-blastoderm cycles, all sequences—both the euchromatin and the satellite sequences—replicate at essentially the same time; since all regions of the DNA are copied simultaneously, the job is completed quite quickly. As development progresses, the satellite sequences shift from being early replicating (and replicating simultaneously with the euchromatin) to being late replicating (Figure 3a) (69, 92, 119). The shift is subtle and gradual in the pre-MBT cycles, where the replication of different sequences still largely overlap, but with slight offsets, and then dramatic after the MBT, when different clusters of satellite sequence replicate in a prolonged sequence (69, 92, 119). In cycle 14, some of the satellite sequences do not begin to replicate until 15 or 30 minutes after S phase begins—a delay that is longer than the entirety of S phase 13 (92). These delays by the late replicating sequences account for most of the lengthening of S phase.
Figure 3. Model for MBT cell cycle slowing.
(a) Cartoon approximations across developmental time (marked at the top), showing replication timing of early and late replicating sequences (92), protein levels of Cyclin (29) and Cdc25 (31), and Cdk1 activity (29). Also listed are the approximate number of genomes and the transcriptional onset of genes discussed extensively in the text. (b) A cartoon of the model espoused in this review for how the cell cycle changes mechanistically during Drosophila early development. Note that cycles 10–12 are omitted and cycle 14 is separated into 3 sections.
Surprisingly, the shift in the satellite sequences from early replicating to late replicating has also been tied to the activity of Cdk1 (32). This was initially unexpected because it is generally thought that Cdk2 is the regulator of S phase, whereas Cdk1 regulates mitosis (74, 75). However, manipulations of Cdk1 levels in early embryos changed the timing of the replication of the satellite sequences (32). In cycle 14, the satellite sequences usually replicate late and Cdk1 activity is usually low (29, 92); increased Cdk1, however, was able to accelerate S phase and promote earlier replication of the satellite sequences (32). Conversely, before the MBT, the satellite sequences usually replicate early, and Cdk1 activity is present throughout the cycle (29, 92); decreasing Cdk1 activity during the early cycles lengthened S phase significantly, it seemed through promoting late replication (32). Despite the thought that Cdk2 usually regulates S phase, injection of activated Cdk2 did not affect the timing of DNA replication (32). This has led to the idea that the satellite sequences are intrinsically late replicating, but that modest Cdk1 activity during S phase can override that preference and cause them to replicate early.
The consequences of speed
The devotion of the early embryo to cell proliferation quickly generates the cells needed to begin specifying and building the tissues of the embryo, but it also seems to preclude and defer other critical activities. Mitosis is extremely disruptive: it appropriates the cytoskeleton to build a mitotic spindle and cleavage furrow, and during the event, the Golgi is disassembled and translation is suppressed (81). Given that many morphogenetic events require specialized cytoskeletal rearrangements, they must wait for the cell cycle to slow. For example, the movement of plasma membrane during cellularization is directed by massive cytoskeletal structures (35, 87, 97), and it is disrupted and forced to restart if mitosis is induced during cellularization (24). Additionally, the shape changes that drive internalization of cells into the ventral furrow are directed by changes in the cytoskeleton as well (108), and mitosis during the process of formation of the furrow often forces cells back out, preventing their proper internalization (46, 67, 90). Thus, the cell cycle must be slowed to create conditions conducive to morphogenesis.
Furthermore, many morphogenetic events, including cellularization and gastrulation require localized zygotic transcription (60). But, transcription is suppressed during mitosis, and transcripts in the process of being extended—nascent transcripts—are aborted in mitosis and must be restarted in the following interphase. As a result, only short transcripts can be completed within the brief interphases of the early cycles (93). Additionally, intense replication activity may interfere with transcription in interphase—it is notable that in both Xenopus and Drosophila, the earliest observed transcription was seen only in nuclei with early prophase characteristics, suggesting that in these early cycles transcription may be confined to the very end of interphase, as replication is completed and nuclei are preparing for mitosis (55, 84). Thus, when the cell cycle slows at the MBT, it provides an increased opportunity for transcription (24, 93).
A mechanistic view of pre-MBT cell cycle slowing
So, given that it is developmentally critical to end this program of rapid divisions, how does the embryo slow them? There are two phases of cell cycle slowing—first, the gradual slowing of the cycles before the MBT, and then the abrupt slowing at the MBT. We will discuss each of these in turn.
DNA replication and the replication checkpoint time the pre-MBT cycles
Each cell cycle prior to the MBT slows by a slightly bigger factor than the one before, which gives the impression of a progressive process that leads up to the dramatic, switch-like change in cycle 14. This progressive slowing appears to be part of the transition in a number of species, suggesting that it may be an integral component of the early cycles.
Excellent insight into how this slowing occurs comes from studies of mutations in grapes, which encodes the Drosophila homolog of Chk1—a checkpoint kinase that is activated in response to incomplete DNA replication (36, 96). Once activated, Grapes suppresses cyclin:Cdk1 activity by promoting its inhibitory phosphorylation (36, 96). It does this by enhancing the activity of Wee kinase, thereby promoting the addition of the phosphates, and suppressing the activity of Cdc25 phosphatase, which would normally remove them (Figure 2). In most situations, Chk1 kinases are dispensable and perform essential roles only during DNA damage or replication stress; in accord with this, grapes mutants are viable unless so stressed (36, 95, 96).
However, female grapes mutant flies lay eggs that never hatch. Without their maternal supply of Grapes, these embryos (hereafter called grapes embryos) suffer a mitotic catastrophe in mitosis 13 when they attempt to divide prior to completing the replication of their DNA (36, 96, 106, 118). This phenotype seems to result from an inability of grapes embryos to induce inhibitory phosphorylation of Cdk1 (36, 96) and thereby delay mitosis until completion of replication, as it is similar to the phenotype observed in embryos from wee mutant mothers (83, 103). More importantly for the timing of the pre-MBT cycles, grapes embryos fail to extend cycles 11–13 to the degree seen in normal embryos (96). By cycle 13, the mismatch between the increasing duration of S phase and interphase length leads to entry into mitosis with incompletely replicated chromosomes, an event that is visualized as extensive DNA bridging between chromosomes trying to separate in anaphase. This suggests that inhibitory phosphorylation driven by Grapes mediates pre-MBT interphase lengthening. Moreover, when S phase is artificially lengthened by injection of aphidicolin, which inhibits progress of the replication forks, the early cycles are extended in a grapes-dependent manner (96). This suggests that an extended S phase can lengthen interphase by activating Grapes. Indeed, mutations in other genes in the Grapes dependent checkpoint pathway give the same early defects in cell cycle lengthening arguing for a model in which the checkpoint pathway extends interphase (83, 95, 103). Since DNA replication becomes gradually slower during these cycles (92), the model suggests that continuing S phase prevents mitotic entry by activating the checkpoint pathway that inhibits Cdk1 thereby coupling interphase length to S phase length. Accordingly, it is S phase length that sets the pace of these early cycles.
Further confirmation that DNA replication determines the length of the early cycles comes from experiments where S phase was deleted by injection of the inhibitor Geminin (69). Geminin prevents the assembly of pre-replication complexes in the following cycle (70, 85). Thus, upon entry into the next S phase, there are no licensed origins, so no replication begins whatsoever (69). Note that deletion of S phase is different from inhibition of replication (such as with aphidicolin); when replication is inhibited, it begins, but progress of DNA polymerase is slowed, which induces replication stress, activates the checkpoint, and prevents cell cycle progress. In contrast, when S phase is deleted, even though no replication forks are built and no replication occurs, the cell appears to lack signals that replication failed and the cell cycle goes on, though of course abnormally because mitosis occurs with unreplicated chromosomes (69). Deleting S phase during the early cycles shortens the early interphase to the same degree as mutation of grapes, but has no consequence on the duration of interphase in embryos lacking grapes function (69). This further supports the idea that continued DNA replication causes the signal that activates the DNA replication checkpoint in each cycle, and thereby lengthens the pre-MBT cycles slowly.
Why does DNA replication slow during the early cycles?
Initial mechanistic proposals for the gradual slowing of the cell cycle involved titration of a maternal factor; the thought was that as the DNA doubled each cycle, the amount of a maternally loaded component would eventually become insufficient to support continued rapid cell cycles (41). Early models suggested that titration of replication factors could slow S phase by decreasing origin utilization and increasing the distance traversed by each replication fork (78). However, as discussed above, the change in origin frequency is not substantial (6, 71), and it is now apparent that S phase lengthens due to an increasing delay in the initiation of replication of satellite sequences (92, 119). So then, why is there a progressive delay in the timing of initiation of satellite DNA replication in cycles 10–13? The finding that cyclin:Cdk1 drives early replication of satellite sequences prior to the MBT and that down regulation of cyclin:Cdk1 triggers S phase extension at the MBT has led us to view some old findings in a new light (32).
The levels of cyclins and the activation of Cdk1 in the early embryo were examined in detail some years ago (29). A role for cyclin in timing the blastoderm cycles was revealed by genetic reductions in cyclin dose, which modestly increased the lengths of these cycles (29). At the time, the data were interpreted in light of cyclin’s requirement to enter mitosis; it was thought that in these cycles, entry into mitosis required the accumulation of a particular amount of cyclin, and reduction in the amount of cyclin mRNA loaded by the mother would increase the time required to translate that amount. However, further study has revealed that cyclin is not limiting for the entry into mitosis; when RNAi knockdown is used to reduce the level of cyclin mRNA by about two-thirds in grapes mutant embryos, the timing of the pre-blastoderm cycles is not changed (118). Thus, cyclins are in excess, and even with a significant reduction in the level of their message, they accumulate rapidly enough to trigger mitosis in grapes embryos, which occurs even earlier than in wild-type embryos. The finding that cyclin levels do not directly govern mitotic entry is in concert with the more recent findings indicating that S phase duration is the key determinant of interphase length. Thus, we revisit the early studies to consider how cyclin levels and Cdk1 activity might influence S phase duration.
Early observations showed that the cyclins are very abundant in the early embryo, and neither their bulk levels nor the bulk activity of Cdk1 kinase oscillate in the pre-blastoderm cycles (Figure 3a) (29). However, cyclin destruction is required to exit mitosis even in these early cycles without obvious oscillations in cyclin level, which has led to the unconfirmed, but widely accepted assumption that localized cyclin destruction centered around the mitotic spindle allows mitotic exit (105). During the progressively slowing blastoderm cycles, the increased destruction of cyclins becomes apparent and progressively more intense each cycle (29). Thus, if cyclin destruction is coupled to the mitotic apparatus, perhaps the increasing DNA leads to greater destruction of the cyclins each mitosis, and this results in the lower cyclin levels observed at the beginning of each blastoderm interphase. Even though mitotic cyclin destruction is far from complete, once mitotic decline in cyclins becomes apparent, all Cdk1 in the embryo loses its activating phosphorylation and loses activity for a brief period at the beginning of each blastoderm cycle (Figure 3a) (29). The early replication of the satellite sequences depends on Cdk1 activity (32), and in the blastoderm cycles, brief delays in the onset of satellite sequence replication become apparent (92, 119). Thus, we suggest that the increasingly long period of Cdk1 inactivity at the beginning of each blastoderm cycle—seemingly caused by the progressively more thorough cyclin destruction at each mitosis and more prolonged loss of the activating phosphorylation—results in an increasing offset in the time of initiation of satellite replication in each successive cycle (Figure 3b). This gradually makes S phase longer, resulting in more prolonged activation of the replication checkpoint in successive cycles, thereby further delaying the entry into mitosis in each cycle and lengthening it.
The dramatic lengthening of the cell cycle at the MBT
While the gradual lengthening of the pre-MBT cycles seems to be a part of a progressive program, where each successive cycle lengthens slightly, the transition in cycle 14 is much more abrupt. The duration of S phase lengthens from 15 minutes to 50 minutes—a much starker increase than in the preceding cycles—and a G2 phase is introduced (27, 92) (Figure 1). Additionally, for the first time, rather than entering mitosis synchronously, different domains of cells exhibit different schedules of mitosis (34). Both of these changes—the longer S phase and G2—result from the downregulation of Cdk1 (27, 29, 32). But in cycle 14, Cdk1 activity is regulated differently than in the pre-MBT cycles, where progressively increasing cyclin destruction and slight and progressive delays in satellite replication appear to be responsible for the pre-MBT slowing.
Cdc25 phosphatase activity determines the length of interphase 14
Given the apparent ubiquity of the inhibitory kinases, Wee and Myt1, Cdc25 phosphatase, which removes inhibitory phosphates from Cdk1, is necessary for cyclin:Cdk1 activity (Figure 2) (20, 26, 27, 29, 42, 88). Gel shift assays first detect inhibited phospho-isoforms of Cdk1 in cycle 14 and show full conversion of Cdk1 as S phase of cycle 14 progresses (29). In contrast, these inhibited isoforms are rare (though present) in the preceding cycles (29, 103). This suggests a change in the regulation of Cdk1. Indeed, isolation of mutants in string, which encodes one of the two Drosophila homologs of Cdc25 phosphatase (Figure 2), revealed that entry into mitosis 14 required zygotic transcription of cdc25/string (26, 27), and premature string expression from an inducible transgene was sufficient to shorten G2 and trigger early entry into mitosis (27). This showed that progression to mitosis 14 is limited by a requirement for Cdc25 activity. Furthermore, in situs and immunohistochemistry showed that string transcription and subsequent accumulation of String protein foreshadowed the exit from the G2 of cycle 14 and the entry into mitosis for each cell (23, 25, 26). This further supports the idea that developmentally regulated expression of cdc25/string determines the length of G2 for each cell and triggers its entry into mitosis.
Injection of cdc25 mRNA in early cycle 14 showed that Cdc25 activity could have a second effect on shortening the cell cycle, aside from triggering exit from G2. Cdc25 activity in cycle 14 triggered early replication of the satellite sequences and thereby vastly shortened S phase (32). This effect was phenocopied when mRNA encoding a variant of Cdk1 that is resistant to inhibitory phosphorylation was injected (32). Conversely, lowering Cdk1 activity in cycle 13 lengthened the normally short pre-MBT S phase and seemed to trigger sequences to become prematurely late replication (32). This suggested that a reduction in Cdc25 and the consequent reduction in Cdk1 activity between interphase 13 and 14 lengthened cycle 14 both through slowing DNA replication and promoting G2.
Maternal Cdc25 is depleted via destruction of Cdc25/Twine protein
But, why does Cdc25 suddenly become limiting in cycle 14, so as to permit inhibitory phosphorylation of Cdk1? Early work had shown that the egg has significant maternal RNA stores (23) encoding two different Cdc25 proteins, String (26) and Twine (2, 13). Additionally, immunoblots showed protein levels of String (29) and Twine (18, 31) were also high during the pre-MBT cycles. While dramatic destruction of both maternal transcripts occurs in cycle 14 (23), RNAi knockdown of string and twine mRNA in the early embryo did not cause cell cycle arrest (31), suggesting that downregulation of these messages is not a sufficient explanation for the cell cycle change, though it likely acts to reinforce the change (23, 76). String protein declines slowly during the blastoderm cycles and becomes essentially undetectable in cycle 13 (29), but Twine protein is relatively stable before the MBT, and its level remains high until early in cycle 14, when it is abruptly destabilized and destroyed (18, 31). Thus, it seems that Twine protein destruction likely allows accumulation of the inhibitory phosphorylation on Cdk1—catalyzed by inhibitory kinases, Wee and Myt1 (Figure 2) (51, 83, 103)—to inhibit its activity, lengthen S phase and add G2 (Figure 3b).
Inhibitory phosphorylation collaborates with an inhibitor of Cdk1
Genetic studies further support involvement of Twine and other regulators of Cdk1 in the trigger that prolongs the cell cycle at the MBT. Embryos laid by mothers that were germline deficient for string and heterozygous for twine (and so had about 0.25x the normal cdc25 gene dose) often cellularized one cycle early (23). Reciprocally, embryos from mothers with two additional copies of twine (and so about 1.5x the normal cdc25 gene dose) occasionally executed the MBT one cycle late (23). The penetrance of this extra pre-MBT division increased dramatically when embryos were mutant for frühstart, a Cdk kinase inhibitor (CKI) that is transcribed in early cycle 14 (40, 45, 46). These results further confirm that the amount of maternal Cdc25 and embryonic Cdk1 activity influence the timing of the MBT and that Cdc25 and Cdk1 downregulation are most likely the key timer of cell cycle slowing (Figure 3b).
The transition is active and dependent on transcription
While the changes to cell cycle regulators that result in the extension of interphase at the MBT are described above, it does not explain why the transition happens in cycle 14. A major historical interest has been the dependence of the cell cycle on transcription, which also seems to be a major component involved in triggering the MBT. Requirement for transcription has been probed in many systems by testing the consequence of its inhibition by the alpha-amanitin, an inhibitor of RNA polymerase II (14, 23, 77, 92). In Drosophila, injection of alpha-amanitin shortly before the MBT prevents the prolongation of S phase and the MBT slowing of the cell cycle (23, 92). As described above, Cdc25 elimination is critical for these processes, and alpha-amanitin also prevented the programmed destruction of Twine protein in cycle 14 (31). The exact transcriptional cascade that triggers destruction of Twine has not been fully elucidated, though candidate factors have been identified, including tribbles, which acts through an unknown mechanism (31, 107). Regardless, these results argue that Twine destruction and the MBT-associated prolongation of the cell cycle are active processes that require transcription in Drosophila.
Timing the mid-blastula transition
The N/C ratio
Early and influential experiments that physically manipulated the number of nuclei in frog egg fragments provided evidence that changing the ratio of nuclei to cytoplasm (hereafter, “the N/C ratio”) alters the time of MBT events (77, 78), suggesting that some events require accumulation of particular amounts of DNA to occur. Studies of Drosophila strains that produce haploid embryos support the idea that the N/C ratio is important for determining the time of slowing the cell cycle in Drosophila as well (24). These embryos, which have half as much DNA, require one more cell cycle (presumably for one more round of replication) to trigger slowing of the cell cycle (24). An even more detailed analysis that used aneuploidy to generate embryos with varying amounts of DNA suggests that there is a threshold around 70% of the DNA that is normally present in a cycle 14 embryo—embryos with less than 75% the normal amount of DNA regularly went through an extra cycle before the MBT, and embryos with more than 134% often went through one fewer cycle before the MBT (64). However, it does not seem likely that the N/C is read out at a single time—the N/C ratio influences the activation of transcription of the patterning gene ftz in cycle 9 (84), but also detailed analyses of aneuploid embryos indicate that it also is monitored in cycle 14 (64).
Multiple timers for multiple events
Summaries of the early work exploring the influence of N/C on the MBT tend to ignore an important aspect of the studies of haploid Drosophila embryos—not all MBT associated events were delayed a cycle. Cellularization actually began in cycle 14 as normal, but the abnormally early mitosis aborted it and forced it to restart and complete in the now long cycle 15 (24). Similarly, an analysis of transcriptional activation suggested that a dramatic upregulation of zygotic transcription still occurred in cycle 14 (24). Thus, some MBT-associated events do not depend on reaching a threshold N/C ratio. Instead, it appeared that the lower N/C ratio, by allowing a continuation of rapid cell cycles, might indirectly interfere with many other processes normally scheduled to begin as the cell cycle slowed.
A recent experiment emphasizes that N/C does not affect all MBT events. Embryos whose cell cycles were prematurely arrested in interphase 12 or 13 using RNAi against the mitotic cyclins initiated cellularization, an MBT event, at the normal time—neither advanced nor delayed (68). This bypass of the normal N/C threshold suggests that the N/C threshold is required only to downregulate Cdk1 activity, which was done here experimentally, or for some consequence of this downregulation, such as the arrest of the cell cycle. Furthermore, the normal timing of MBT events that occurred in this bypass argues that there are other timers that are independent of N/C and cell cycle progress. Recognition that there is such a timer is somewhat disquieting because we have no idea how it works. However, numerous events occur with amazing temporal precision in the early embryo—even before the approach of the MBT—that are not significantly affected by inhibition of transcription or deletion of large segments of the genome (23, 73, 114). These events, such as the migration of the nuclei to the syncytial blastoderm (35, 102), give an independent and compelling argument for timers that are distinct from the MBT and use maternally provided products to both time and execute developmental events.
Could N/C-dependent transcription time the MBT cell cycle transition?
As argued above, the onset of zygotic transcription seems to be a progressive process, where different genes exhibit different requirements for their onset. A study that used high-throughput methods to compare expression of several hundred genes in wild-type embryos and haploid mutants has shown that the expression of some genes is deferred for one cycle in haploid embryos, while others initiate transcription in cycle 14 regardless of the accumulated DNA (64). These were classified as N/C-dependent and time-dependent genes, respectively. Thus, at least some transcripts expressed at cycle 14 are regulated by the N/C ratio. Given that transcription is required for cell cycle slowing, one attractive model is that N/C-dependent transcription governs onset of Twine destruction, and delayed Twine destruction in haploid embryos supports this idea (31). Furthermore, the tribbles transcript, though provided maternally, is upregulated by N/C-dependent zygotic expression in cycle 13 (64), and it functions by unknown mechanisms to promote destruction of String and Twine (46, 67, 90). RNAi experiments suggest that it contributes to, but is not essential, for the onset of destruction of Twine at the MBT (31). Thus, N/C-dependent tribbles transcription could contribute to timing Twine destruction, though there appear to be other transcriptional inputs into its destruction, since alpha-amanitin is more effective than tribbles RNAi at suppressing Twine destruction (31). Additionally, transcription of the cyclin:Cdk inhibitor, frühstart, begins in cycle 14 (40, 45, 46) and seems to be dependent on the N/C ratio (64). Its action as an inhibitor of Cdk1 seems to further contribute to and reinforce slowing of the cell cycle. Thus, the onset of a small collection of N/C-dependent transcripts could be the trigger for the slowing of the cell cycle (Figure 3b).
While N/C-dependent genes that are expressed before the MBT are excellent candidates for the regulation of the cell cycle, many N/C dependent genes are expressed after the MBT, and their dependence on N/C might be indirect if their expression depends on an MBT event such as cell cycle slowing to enable sufficient time for elongation of their transcripts. Thus, a few directly N/C-dependent genes could create a positive feedback loop by slowing the cell cycle and enabling the transcription of other indirectly N/C-dependent genes.
While a model based on N/C-dependent transcription as a timer of cell cycle slowing is attractive, several cautions are in order. First, a demonstration that transcription is required for cell cycle slowing does not require that transcription be the the timing signal. Second, thorough experiments that created aneuploid embryos missing different parts of the genome have failed to identify any single locus that is required for cell cycle slowing at the MBT (73, 114). Third, at present it has not been determined whether N/C-dependent transcripts respond directly or indirectly to the N/C ratio. Thus, it remains formally possible that the cell cycle fails to slow in cycle 14 in haploids because of the failure to express N/C-dependent transcripts, or that the cell cycle might fail to slow in cycle 14 in haploids for an independent reason and the appearance of the transcripts is then indirectly delayed due to the altered developmental program. Nonetheless, the most attractive model at present is that increasing transcription of several different loci that are dependent on the N/C ratio, including tribbles and frühstart, are involved in the change in regulation of cell cycle and its slowing in cycle 14 (Figure 3b).
The mysterious identities of the timers
The question of how the N/C is detected and could result in this transcription that affects or triggers the MBT remodeling of the cell cycle is still a major question in the field. Several prominent proposals have been made, but they are all unsatisfying in some way. We present them below, as well as their caveats. However, we consider this question to be one of the foremost remaining questions in understanding the timing of the Drosophila mid-blastula transition.
Titration models
The most influential models for how the N/C ratio could be detected and trigger a transition have been titration models, where some maternally loaded component in the embryo is limiting (41). In their initial forms, they revolved around limiting cell cycle components and the idea that the increasing DNA in the embryo would eventually become insufficient for the maternal cell cycle component to continue to support rapid cell cycles, thus slowing the cell cycle (78). Recent results have shown that overexpression of some DNA replication components can delay the onset of the MBT in Xenopus, which could potentially be consistent with such a mechanism (12). However, results in Drosophila argue that the titration of a cell cycle component is not directly responsible for lengthening cycle 14. Reintroduction of Cdc25 at the MBT is sufficient to drive a short cycle, which shows that it is the only component limiting for a short cell cycle (32). It, however, does not seem to be passively titrated by increased DNA, but rather actively destroyed in cycle 14 (31). Thus, if a component is titrated in the Drosophila embryo, it is probably not a cell cycle component that mechanistically slows the cell cycle. However, titration of a maternal component could still act as a sensor for N/C, only now the new findings argue that the signal generated by this sensor must trigger an active process to bring on the destruction of Twine and the MBT. One potential candidate could be an inhibitor of transcription of some kind whose titration triggers transcription of N/C-dependent transcripts. The inhibitor tramtrack is thought to act in this fashion to allow transcription of a small collection of genes in cycle 11 (84); perhaps different inhibitors act in cycles 13 and 14 that remain to be identified.
Inhibition by the short length of the rapid cycles
Another popular model has been that the sheer shortness of the early cycles made transcription impossible, since entry into mitosis aborts nascent transcription (93). Accordingly, N/C might activate transcription indirectly by slowing the cell cycle and making interphase long enough that there is sufficient time to complete the transcription of the genes important for triggering the MBT. This certainly contributes to the the reinforcement of the onset of transcription after the cell cycle is actively slowed; some genes transcribed in cycle 14 are too long to be completed during a short cell cycle, and so require the slowing of the cell cycle as a permissive condition to allow their transcription (93). However, experiments using RNAi against cyclins to prematurely lengthen the cell cycle suggest that an inability to complete elongation does not limit the onset of MBT events—it did not advance the time of Twine destruction, which suggests that the transcripts involved in Twine destruction were not advanced (31). Moreover, it did not advance the timing of cellularization or gastrulation, indicating that perhaps no MBT events are timed by such a model (68).
Zelda or Vielfaltig, a DNA-binding protein
A mutant of great interest for the timing of the MBT is the zinc-finger DNA-binding protein Zelda (also known as Vielfaltig), which is found highly enriched at genes that are expressed in cycle 14 and also those genes expressed during the pre-MBT cycles (which typically have more Zelda binding sites) (47, 79). Indeed, increasing or decreasing the number of Zelda binding sites near a gene can advance or delay (respectively) the time of onset of transcription of that gene (8), and adding Zelda binding motifs to a GFP reporter can confer pre-MBT expression on the reporter (16). A percentage of zelda embryos go through an extra rapid division, suggesting that Zelda regulates the transcription of one or more genes involved in timing the MBT (61, 107). However, Zelda binds to most of its target genes in cycle 8, long before they will become activated (47), and the current model for its function is that it assists the binding of other transcription factors (89). Thus, it seems less likely that Zelda is the switch that changes at the time of transcription to turn on expression of particular genes, but more likely it instead creates a permissive condition that allows those genes to be activated by another trigger—perhaps the binding of other transcription factors.
Smaug, an RNA-binding protein
One mutant whose role at the MBT is rather confusing is the smaug mutant. Embryos from mothers mutant for smaug (hereafter “smaug embryos”) do not slow their cell cycle or execute the MBT. Additionally, expression of smaug in a gradient results in a gradient of cellularization, so Smaug has been proposed as a timer of the MBT (3). smaug embryos exhibit defects in activation of the DNA replication checkpoint—they do not exhibit pre-MBT slowing or respond to aphidicolin, like grapes embryos (3). As discussed previously, the DNA replication checkpoint has an important role in regulating the cell cycle of the early embryo, so it is possible that Smaug’s role in regulating the cell cycle acts through Chk1. However, smaug embryos also have penetrant defects in the onset of zygotic transcription of a number of genes (3). Indeed, tribbles and frühstart expression are thought to be decreased in smaug embryos, as well some regulators of cellularization (3). The basis of Smaug’s connection to the DNA replication checkpoint these phenotypes are not understood, however. Smaug is an RNA-binding protein, thought to repress translation of its targets through its binding and promote their destruction by recruiting the CCR4/POP2/NOT deadenylase complex and thereby shortening their poly(A) tail (91, 98, 109). There is no understood connection, however, between Smaug’s known biochemical function in repressing translation of or destabilizing transcripts and the onset of zygotic transcription or the activity of the DNA replication checkpoint. Moreover, there is no explanation for how Smaug would detect or be regulated by the N/C ratio. Clearly, more investigation into Smaug’s potential role in the transition is needed.
An evolutionary perspective on the MBT
The genetics and detailed developmental studies of Drosophila have offered us a fairly detailed view of the MBT, but how does it relate to events in other organisms? As presented at the outset of this article, rapid cleavage divisions are a widespread feature of early embryos (82), as if there has been a consistent benefit to the basic strategy of rapid early cell proliferation throughout the evolution of animals with externally deposited eggs. Indeed, while Drosophila goes through 13 rapid cell cycles of lengths ranging from 8–20 minutes prior to dramatic slowing and desynchronization, Xenopus laevis goes through 12 rapid cell cycles of approximately 30 minutes each, and zebrafish undergo 10 rapid cell cycles that are about 15 minutes each (54, 56). Thus, for all of these organisms, within three hours of fertilization, they have subdivided the massive maternally provided cytoplasm into thousands of cells.
However, convergent evolution might have created this behavior independently in different lineages, and even if the programs are related by descent, it is unlikely that the regulatory program would be conserved unchanged. Organisms have dramatically different eggs with associated distinctions in early embryogenesis. Thus, we might expect the control of embryonic cell cycle slowing to have undergone considerable modification in parallel with the changes in organization of early embryogenesis (59, 122). In fact, sequence homology comparison suggests that Twine and String diverged quite recently, and Twine may be unique to Drosophila, as the only other Drosophila species have Cdc25 homologs that are more similar to Twine than String (authors’ personal observation). This suggests that at least some details of control of the MBT must be different in other other Diptera, and one might expect more dramatic differences over larger evolutionary time spans.
Xenopus
The Xenopus early cycles show superficial similarity to those in Drosophila. The Xenopus embryo undergoes 11 rapid cycles each approximately 30 minutes long, and a twelfth cycle that is slightly longer and is defined as the MBT. Cycles 13–15 get progressively longer (50, 99 and 253 min, respectively) (49). The early gastrula transition (EGT) occurs early in the greatly extended cycle 15 and is associated with onset of gastrulation. The Xenopus MBT is dependent on the N/C ratio, as determined by ligation of eggs, and is not dependent on zygotic transcription, based on injection of alpha-amanitin (77). The detailed cell cycle changes associated with cycle prolongation immediately after the Xenopus MBT are somewhat unclear. An early report that both G1 and G2 were introduced at the MBT (43), was contradicted by two later reports (39, 50), however, it is not clear that any of these studies have the temporal resolution and sensitivity to define the specific cycle at which either a G1 or G2 is introduced. Nonetheless, all of the studies agree that extension of S phase is an early change, and that by the time of gastrulation, embryos acquire both a G1 and G2. At the time of the Xenopus MBT, Chk1 becomes activated and is required for cell cycle lengthening (94), similar to Drosophila. Moreover, Cdk1 shows dramatic inhibitory phosphorylation at the MBT (33), which is dependent on Chk1 and accumulates between the time of the MBT and EGT (94). This involvement of the checkpoint pathway in the initial slowing of the cell cycle is similar to Grapes/Chk1 dependent “pre-MBT” slowing of the cell cycle in Drosophila. Perhaps, as in Drosophila, Chk1 activation in Xenopus acts to defer mitosis as S phase prolongs. However, in Xenopus, Chk1 phosphorylation both inhibits Cdc25 activity and promotes its destruction (94, 112), while in Drosophila the abrupt destruction of Twine/Cdc25 does not require Chk1 activity (31).
Recent work in Xenopus has found that overexpression of four DNA replication factors—Dbp11/Cut5, Treslin/SLD3, Drf1, and RecQ4—can trigger additional short, pre-MBT like cycles in Xenopus, but does not accelerate the cycle before the MBT (12). These factors contribute to the maturation of the pre-replication complex necessary to build functional replication forks. They promote a key transition that appears to be the pivotal step at which cyclin:Cdk is required for activation of replication (7). One or a combination of these proteins might be titrated by increasing DNA to trigger the MBT transition to longer S phase (12). Even if none of these proteins is actually titrated by additional DNA at the MBT, acceleration of S phase by their joint overproduction identifies a step in the maturation of the pre-replication complex that limits post-MBT replication. Presumably a regulatory change influencing this step creates the sensitivity to overproduction of these proteins at the MBT, and it is notable that the downregulation of cyclin:Cdk1, which is the event triggering cycle prolongation in Drosophila, would affect this same step, albeit by a different input.
Zebrafish
The zebrafish embryo goes through 9 rapid cycles, slows very slightly in interphases 10–11, and exhibits an extreme slowing in interphase 12. It first begins to exhibit cell cycle asynchrony in cycle 11 (54). Partial enucleation experiments suggest that the MBT slowing of the cell cycle is also timed by the N/C ratio in zebrafish, as in Drosophila and Xenopus (54). The dramatic slowing of the cell cycle does not seem to depend on transcription, as embryos injected with alpha-amanitin did not exhibit signs of continued cell division (14, 53). The embryos acquire a G1 phase at the MBT, but in a transcription-dependent fashion, suggesting that it is not the entry into G1 that regulates the dramatic slowing of the cell cycle at the MBT in zebrafish (121). Instead, as in Drosophila, slowing of the cell cycle seems to result from a longer S phase at the MBT (121) and acquisition of a G2 phase at the MBT, though this G2 phase does not require zygotic transcription (14). Cdc25 and Cdk1 activity become limiting at the MBT, like in Drosophila, as overexpression of either Cdc25a or inhibitory phosphorylation-resistant Cdk1 cause continued rapid divisions, while phosphorylation-resistant Cdk2 does not (14). However, upregulation of Cdk1 activity is not capable of preventing the lengthening of S phase in zebrafish, and so inhibitory-resistant Cdk1 caused cells to enter mitosis with unreplicated DNA and undergo mitotic catastrophe (14). This suggests that, while downregulation of Cdc25 and Cdk1 may be responsible for adding a G2 phase and for the observed change in cell cycle timing at the MBT, similar to Drosophila, the mechanism that causes S phase to get longer is different and not dependent on Cdk1. Neither the connection between the N/C ratio and the lengthening of S phase, nor the connection between the N/C ratio and the downregulation of Cdc25 and Cdk1 is known in zebrafish.
Other connections
There are additional hints in the literature that some of the mechanisms uncovered in Drosophila may be important in the early development of other organisms. For instance, there is a suggestion that, in urochordate embryos, transcription may change the length of S phase and therefore introduce the first asynchrony into the cell cycle (19). Furthermore, there are suggestions that Cdk1 modulation of replication timing may occur in mammalian systems as well, as tested in cell culture (111). Additionally, specific requirement for Chk1 kinase in early embryogenesis extends all the way to mammals (110), suggesting that this may be a critically conserved component of slowing embryonic cell cycles.
The MBT might not look the same in every organism
There are numerous organisms that do not seem to exhibit a classic MBT. However, perhaps some of these organisms may retain elements of the program, but in somewhat differing contexts. For instance, in segmented annelids and short germband insects, cells do not all slow rapid cell cycles synchronously. Instead, the posterior cells continue proliferation and begin morphogenesis in a protracted sequence. For instance, studies in the leech, Helobdella triserialis, have detailed cell cycle slowing in a very different context (5). The major slowing of the cell cycle that can be observed in leech occurs during the asymmetric divisions of the teloblast cells, which produce the segmented ectodermal structures. The teloblast cells are large and divide rapidly and asymmetrically about once an hour, with an 11 minute S phase and 21 minute G2. The large daughter of the division, in a stem cell pattern, continues the rapid teloblast cell cycle program. The much smaller daughter, however, is called a primary blast cell and has a very different cell cycle—a 4.7 hour S phase and a 16–28 hour G2 (5). Thus, these smaller cells exhibit a dramatic extension of S phase and acquisition of a long G2, in conjunction with a massive upregulation in zygotic transcription, like the cycle 14 cells of a Drosophila embryo. This cell cycle slowing and transcriptional activation are viewed as representing the parallel with the MBT in leeches (4, 5). Because the change occurs repeatedly in a continuing lineage, the regulation—especially its connection with timing—differs markedly from that in Drosophila. Nonetheless, these changes have a striking association with the abrupt change in N/C that occurs with the birth of the small blast cell. Thus, even if the MBT is not always a synchronous, embryo-wide event, it may nonetheless retain a fundamental input from N/C and exhibit similar changes in cell cycle phases to prolong the cycle.
Mammalian embryos are also often viewed as not having an MBT, due to their initially slow cycles upon fertilization. However, a broader consideration of chordates, suggests that the rapid early cycles and abrupt slowing of the cycle in association with gastrulation is widespread. It is seen in the proto-chordates such as Ciona, as well as in fish, amphibians and birds. Did it suddenly disappear in parallel with the late evolution of eutharian development, or was it changed in ways that make it less recognizable? The early stages of mammalian development cannot be compared to the development of even relatively close non-mammalian vertebrates because new steps were added at the beginning of development to generate the extra-embryonic tissues that are a central feature of mammalian development. This insertion into early development displaced gastrulation and its associated processes to a later time; at gastrulation and afterward, many features of gene expression and mammalian morphogenesis can be compared to other vertebrates (82). Exceedingly rapid cell cycles occur just prior to gastrulation in mammals (65, 100), and several features argue that these rapid cycles represent the evolutionary vestige of the pre-MBT cycles, and that slowing of the cycle at gastrulation may have parallels to the MBT (82). But just as we have seen in other organisms, the regulation of MBT-associated events is likely to have changed in conjunction with the evolution of mammalian program of early development. One type of change stands out. In externally developing eggs, the transition from maternal control to zygotic control is more or less coincident with the slowing of the cell cycle, but the mammalian embryo transitions to dependence on zygotic gene expression during the early cleavage cycles before it begins to generate the extra-embryonic tissues, and long before gastrulation (80). Nonetheless, after completion of the uniquely eutherian early program, onset of expression of particular zygotic genes, independent of zygotic genome activation, could generate conditions that recapitulate the pre-gastrulation programs that are part of the legacy imprinted on metazoan development over hundreds of millions of years of evolution.
Summary Points.
In early development, many embryos exhibit a period of rapid cell division, enabled by a modified cell cycle with an unusually rapid S phase and a lack of gap phases.
In Drosophila, the cell cycle first slows gradually, as late replicating sequences begin to delay replication slightly, lengthening S phase and triggering the DNA replication checkpoint, which delays entry into mitosis.
Upon achieving a particular N/C ratio, in a transcription-dependent process, the Drosophila embryo actively triggers inhibition of Cdk1 via destruction of the Cdc25 isoform Twine, elimination of cdc25 mRNAs, and transcription of a CDK inhibitor, frühstart.
This inhibition of Cdk1 triggers the cell cycle slowing at the MBT—late replicating sequences begin to delay replication significantly and a G2 gap phase is introduced where cells now depend on the transcription of cdc25 in order to enter mitosis.
It is not fully clear how the embryo determines when to initiate this program to inhibit Cdk1, but we propose that the best current model is that transcription of specific zygotic transcripts times it, and thus links the process to the major activation of the Drosophila genome in cycles 13 and 14.
A number of other changes in the embryo, including the onset of morphogenesis are timed by an unknown mechanism other than the N/C ratio.
Future Issues.
How does the Drosophila embryo detect the N/C ratio and transduce it into a change in transcriptional activity?
What are the many mechanisms that trigger the onset of zygotic transcription?
Does transcription truly time the changes to the cell cycle at the MBT, or is it merely permissive?
How does Cdk1 activity affect the timing of late replicating sequences?
Acknowledgments
We thank members of the O’Farrell lab for helpful discussions, and Alexander Schier, Katherine Rogers, and James Gagnon for comments on the manuscript. This research was supported by National Institutes of Health grant GM037193 to P.H.O’F and a National Science Foundation Graduate Research Fellowship and Jane Coffin Childs Memorial Fund Postdoctoral Fellowship to J.A.F.
Definitions
- morphogenesis
biological processes that cause an organism to take its shape, including cell shape changes and cell movements
- mid-blastula transition (MBT)
a change in the cell cycle in early embryos where specialized synchronous rapid cycles slow down and become asynchronous
- syncytium
in embryology, a large cytoplasm shared by many nuclei, which is a common developmental paradigm used in insects
- cytokinesis
division of the cytoplasm by plasma membrane at the end of mitosis, separating the cell into two daughters
- gap phases
phases of the cell cycle where cells wait before beginning replication (G1) or before entering mitosis (G2)
- S phase
the phase of the cell cycle during which the DNA is replicated
- blastoderm
the region of the egg that will give rise to embryonic tissues. In Drosophila, a shell of nuclei surrounding the yolk.
- cellularization
the process of dividing the syncytial cytoplasm into many cells and enveloping each nucleus in its own cell membrane
- gastrulation
the process of cell internalization that converts the single-layered blastula into the gastrula, with three different germ layers
- maternal-zygotic transition (MZT)
the switch of developmental control from maternal gene products to zygotic gene products, defined either by functional requirements or zygotic transcription
- zygotic genome activation (ZGA)
the stage when significant levels of transcription are observed from the initially relatively quiescent zygotic genome
- replication origin
genomic location where DNA replication will begin by building a replication fork; “licensed” by assembling pre-replication complexes on the DNA prior to replication
- euchromatin
DNA (or chromatin) that is generally not compacted, except during mitosis, and which can be transcriptionally active
- heterochromatin
DNA (or chromatin) that is often compacted during interphase, tends to be transcriptionally silent, and is bound by heterochromatin-binding proteins
- aphidicolin
a reversible inhibitor of eukaryotic nuclear DNA replication that slows or stalls the progress of DNA polymerase
- pre-replication complex
a protein complex formed on origins that, after maturation through additional protein recruitment, will subsequently direct assembly of replication forks and initiation of replication
- CKI
a cyclin-dependent kinase inhibitor—a protein that bind to specific Cdks or cyclin:Cdk complexes and inhibits their activity
- alpha-amanitin
a drug used to inhibit RNA polymerase II (which is most sensitive to it) and thus transcription of mRNAs
- Diptera
the order of insects characterized by having two wings that includes flies, mosquitos, gnats, and midges, among others
- Eutharia
the considerably larger clade of mammals that includes the placental mammals (versus the Metatheria which is predominantly marsupials)
Literature Cited
- 1.Ali-Murthy Z, Lott SE, Eisen MB, Kornberg TB. An essential role for zygotic expression in the pre-cellular Drosophila embryo. PLoS Genet. 2013;9(4):e1003428. doi: 10.1371/journal.pgen.1003428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM. Twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell. 1992;69(6):977–88. doi: 10.1016/0092-8674(92)90616-k. [DOI] [PubMed] [Google Scholar]
- 3.Benoit B, He CH, Zhang F, Votruba SM, Tadros W, et al. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development. 2009;136(6):923–32. doi: 10.1242/dev.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bissen ST, Weisblat DA. Early differences between alternate n blast cells in leech embryo. J. Neurobiol. 1987;18(3):251–69. doi: 10.1002/neu.480180302. [DOI] [PubMed] [Google Scholar]
- 5.Bissen ST, Weisblat DA. The durations and compositions of cell cycles in embryos of the leech, Helobdella triserialis. Development. 1989;106(1):105–18. doi: 10.1242/dev.106.1.105. [DOI] [PubMed] [Google Scholar]
- 6.Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb. Symp. Quant. Biol. 1974;38:205–23. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- 7.Boos D, Sanchez-Pulido L, Rappas M, Pearl LH, Oliver AW, et al. Regulation of DNA Replication through Sld3-Dpb11 Interaction Is Conserved from Yeast to Humans. Current Biology. 2011;21(13):1152–57. doi: 10.1016/j.cub.2011.05.057. [DOI] [PubMed] [Google Scholar]
- 8.Bosch ten JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133(10):1967–77. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- 9.Brown DD, Wensink PC, Jordan E. Purification and some characteristics of 5S DNA from Xenopus laevis. Proc Natl Acad Sci USA. 1971;68(12):3175–79. doi: 10.1073/pnas.68.12.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JL, Sonoda S, Ueda H, Scott MP, Wu C. Repression of the Drosophila fushi tarazu (ftz) segmentation gene. EMBO J. 1991;10(3):665–74. doi: 10.1002/j.1460-2075.1991.tb07995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvi BR, Lilly MA, Spradling AC. Cell cycle control of chorion gene amplification. Genes Dev. 1998;12(5):734–44. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science. 2013;341(6148):893–96. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courtot C, Fankhauser C, Simanis V, Lehner CF. The Drosophila cdc25 homolog twine is required for meiosis. Development. 1992;116(2):405–16. doi: 10.1242/dev.116.2.405. [DOI] [PubMed] [Google Scholar]
- 14.Dalle Nogare DE, Pauerstein PT, Lane ME. G2 acquisition by transcription-independent mechanism at the zebrafish midblastula transition. Dev Biol. 2009;326(1):131–42. doi: 10.1016/j.ydbio.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 15.de Nooij JC, Letendre MA, Hariharan IK. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell. 1996;87(7):1237–47. doi: 10.1016/s0092-8674(00)81819-x. [DOI] [PubMed] [Google Scholar]
- 16.De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5(5):e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshpande G, Stukey J, Schedl P. Scute (sis-b) function in Drosophila sex determination. Mol Cell Biol. 1995;15(8):4430–40. doi: 10.1128/mcb.15.8.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Talia S, She R, Blythe SA, Lu X, Zhang QF, Wieschaus EF. Posttranslational Control of Cdc25 Degradation Terminates Drosophila’s Early Cell-Cycle Program. Curr Biol. 2013;23(2):127–32. doi: 10.1016/j.cub.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dumollard R, Hebras C, Besnardeau L, McDougall A. Beta-catenin patterns the cell cycle during maternal-to-zygotic transition in urochordate embryos. Dev Biol. 2013;384(2):331–42. doi: 10.1016/j.ydbio.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Dunphy WG, Kumagai A. The cdc25 protein contains an intrinsic phosphatase activity. Cell. 1991;67(1):189–96. doi: 10.1016/0092-8674(91)90582-j. [DOI] [PubMed] [Google Scholar]
- 21.Duronio RJ, O’Farrell PH. Developmental control of the G1 to S transition in Drosophila: cyclin E is a limiting downstream target of E2F. Genes Dev. 1995;9(12):1456–68. doi: 10.1101/gad.9.12.1456. [DOI] [PubMed] [Google Scholar]
- 22.Dyson N. PRB, p107 and the regulation of the E2F transcription factor. J Cell Sci Suppl. 1994;18:81–87. doi: 10.1242/jcs.1994.supplement_18.12. [DOI] [PubMed] [Google Scholar]
- 23.Edgar BA, Datar SA. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila’s early cell cycle program. Genes Dev. 1996;10(15):1966–77. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- 24.Edgar BA, Kiehle CP, Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 1986;44(2):365–72. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- 25.Edgar BA, Lehman DA, O’Farrell PH. Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle. Development. 1994;120(11):3131–43. doi: 10.1242/dev.120.11.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar BA, O’Farrell PH. Genetic control of cell division patterns in the Drosophila embryo. Cell. 1989;57(1):177–87. doi: 10.1016/0092-8674(89)90183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar BA, O’Farrell PH. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62(3):469–80. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986;44(6):871–77. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- 29.Edgar BA, Sprenger F, Duronio RJ, Leopold P, O’Farrell PH. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev. 1994;8(4):440–52. doi: 10.1101/gad.8.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erickson JW, Cline TW. Key aspects of the primary sex determination mechanism are conserved across the genus Drosophila. Development. 1998;125(16):3259–68. doi: 10.1242/dev.125.16.3259. [DOI] [PubMed] [Google Scholar]
- 31.Farrell JA, O’Farrell PH. Mechanism and regulation of Cdc25/Twine protein destruction in embryonic cell-cycle remodeling. Curr Biol. 2013;23(2):118–26. doi: 10.1016/j.cub.2012.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farrell JA, Shermoen AW, Yuan K, O’Farrell PH. Embryonic onset of late replication requires Cdc25 down-regulation. Genes Dev. 2012;26(7):714–25. doi: 10.1101/gad.186429.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrell JE, Wu M, Gerhart JC, Martin GS. Cell cycle tyrosine phosphorylation of p34cdc2 and a microtubule-associated protein kinase homolog in Xenopus oocytes and eggs. Mol Cell Biol. 1991;11(4):1965–71. doi: 10.1128/mcb.11.4.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foe VE. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 1989;107(1):1–22. [PubMed] [Google Scholar]
- 35.Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- 36.Fogarty P, Campbell SD, Abu-Shumays R, Phalle BS, Yu KR, et al. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr Biol. 1997;7(6):418–26. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- 37.Follette PJ, Duronio RJ, O’Farrell PH. Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Curr Biol. 1998;8(4):235–38. doi: 10.1016/s0960-9822(98)70089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Follette PJ, O’Farrell PH. Cdks and the Drosophila cell cycle. Curr. Opin. Genet. Dev. 1997;7(1):17–22. doi: 10.1016/s0959-437x(97)80104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frederick DL, Andrews MT. Cell cycle remodeling requires cell-cell interactions in developing Xenopus embryos. J. Exp. Zool. 1994;270(4):410–16. doi: 10.1002/jez.1402700411. [DOI] [PubMed] [Google Scholar]
- 40.Gawliński P, Nikolay R, Goursot C, Lawo S, Chaurasia B, et al. The Drosophila mitotic inhibitor Frühstart specifically binds to the hydrophobic patch of cyclins. EMBO Rep. 2007;8(5):490–96. doi: 10.1038/sj.embor.7400948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerhart JC. Mechanisms Regulating Pattern Formation in the Amphibian Egg and Early Embryo. In: Goldberger RF, editor. Biological Regulation and Development. 2nd ed Vol. 2, Molecular Organization and Cell Function. Plenum Press; New York: 1980. pp. 133–316. [Google Scholar]
- 42.Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342(6245):39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 43.Graham CF, Morgan RW. Changes in the cell cycle during early amphibian development. Dev Biol. 1966;14(3):439–60. [Google Scholar]
- 44.Gross PR, Cousineau GH. Macromolecule synthesis and the influence of actinomycin on early development. Exp. Cell Res. 1964;33:368–95. doi: 10.1016/0014-4827(64)90002-3. [DOI] [PubMed] [Google Scholar]
- 45.Grosshans J, Müller HAJ, Wieschaus E. Control of cleavage cycles in Drosophila embryos by frühstart. Dev Cell. 2003;5(2):285–94. doi: 10.1016/s1534-5807(03)00208-9. [DOI] [PubMed] [Google Scholar]
- 46.Grosshans J, Wieschaus E. A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila. Cell. 2000;101(5):523–31. doi: 10.1016/s0092-8674(00)80862-4. [DOI] [PubMed] [Google Scholar]
- 47.Harrison MM, Li X-Y, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7(10):e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayashi S. A Cdc2 dependent checkpoint maintains diploidy in Drosophila. Development. 1996;122(4):1051–58. doi: 10.1242/dev.122.4.1051. [DOI] [PubMed] [Google Scholar]
- 49.Howe JA, Howell M, Hunt T, Newport JW. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes Dev. 1995;9(10):1164–76. doi: 10.1101/gad.9.10.1164. [DOI] [PubMed] [Google Scholar]
- 50.Iwao Y, Uchida Y, Ueno S, Yoshizaki N, Masui Y. Midblastula transition (MBT) of the cell cycles in the yolk and pigment granule-free translucent blastomeres obtained from centrifuged Xenopus embryos. Dev Growth Differ. 2005;47(5):283–94. doi: 10.1111/j.1440-169X.2005.00802.x. [DOI] [PubMed] [Google Scholar]
- 51.Jin Z, Homola EM, Goldbach P, Choi Y, Brill JA, Campbell SD. Drosophila Myt1 is a Cdk1 inhibitory kinase that regulates multiple aspects of cell cycle behavior during gametogenesis. Development. 2005;132(18):4075–85. doi: 10.1242/dev.01965. [DOI] [PubMed] [Google Scholar]
- 52.Kafatos FC, Mitsialis SA, Spoerel N, Mariani B, Lingappa JR, Delidakis C. Studies on the developmentally regulated expression and amplification of insect chorion genes. Cold Spring Harb. Symp. Quant. Biol. 1985;50:537–47. doi: 10.1101/sqb.1985.050.01.066. [DOI] [PubMed] [Google Scholar]
- 53.Kane DA, Hammerschmidt M, Mullins MC, Maischein HM, Brand M, et al. The zebrafish epiboly mutants. Development. 1996;123:47–55. doi: 10.1242/dev.123.1.47. [DOI] [PubMed] [Google Scholar]
- 54.Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119(2):447–56. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- 55.Kimelman D, Kirschner M, Scherson T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 1987;48(3):399–407. doi: 10.1016/0092-8674(87)90191-7. [DOI] [PubMed] [Google Scholar]
- 56.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 57.Knoblich JA, Sauer K, Jones L, Richardson H, Saint R, Lehner CF. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77(1):107–20. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 58.Lane ME, Sauer K, Wallace K, Jan YN, Lehner CF, Vaessin H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87(7):1225–35. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- 59.Laugsch M, Schierenberg E. Differences in maternal supply and early development of closely related nematode species. Int. J. Dev. Biol. 2004;48(7):655–62. doi: 10.1387/ijdb.031758ml. [DOI] [PubMed] [Google Scholar]
- 60.Leptin M. Gastrulation Movements: the Logic and the Nuts and Bolts. Curr Opin Cell Biol. 2005;8(3):305–20. doi: 10.1016/j.devcel.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Liang H-L, Nien C-Y, Liu H-Y, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456(7220):400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lohe AR, Hilliker AJ, Roberts PA. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134(4):1149–74. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lott SE, Villalta JE, Schroth GP, Luo S, Tonkin LA, Eisen MB. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 2011;9(2):e1000590. doi: 10.1371/journal.pbio.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu X, Li JM, Elemento O, Tavazoie S, Wieschaus EF. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development. 2009;136(12):2101–10. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mac Auley A, Werb Z, Mirkes PE. Characterization of the unusually rapid cell cycles during rat gastrulation. J Embryol Exp Morphol. 1993;117(3):873–83. doi: 10.1242/dev.117.3.873. [DOI] [PubMed] [Google Scholar]
- 66.Manes C. The participation of the embryonic genome during early cleavage in the rabbit. Dev Biol. 1973;32(2):453–59. doi: 10.1016/0012-1606(73)90254-6. [DOI] [PubMed] [Google Scholar]
- 67.Mata J, Curado S, Ephrussi A, Rørth P. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating String/CDC25 proteolysis. Cell. 2000;101(5):511–22. doi: 10.1016/s0092-8674(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 68.McCleland ML, O’Farrell PH. RNAi of Mitotic Cyclins in Drosophila Uncouples the Nuclear and Centrosome Cycle. Current Biology. 2008;18(4):245–54. doi: 10.1016/j.cub.2008.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCleland ML, Shermoen AW, O’Farrell PH. DNA replication times the cell cycle and contributes to the mid-blastula transition in Drosophila embryos. J Cell Biol. 2009;187(1):7–14. doi: 10.1083/jcb.200906191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93(6):1043–53. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 71.McKnight SL, Miller OL. Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell. 1977;12(3):795–804. doi: 10.1016/0092-8674(77)90278-1. [DOI] [PubMed] [Google Scholar]
- 72.McKnight SL, Miller OL. Post-replicative nonribosomal transcription units in D. melanogaster embryos. Cell. 1979;17(3):551–63. doi: 10.1016/0092-8674(79)90263-0. [DOI] [PubMed] [Google Scholar]
- 73.Merrill PT, Sweeton D, Wieschaus E. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. Development. 1988;104(3):495–509. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- 74.Morgan D. Cyclin-Dependent Kinases: Engines, Clocks, and Microprocessors. Annu Rev Cell Dev Biol. 1997;13(1):261–91. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 75.Morgan DO. The Cell Cycle. 1st ed New Science Press Ltd.; London: 2007. [Google Scholar]
- 76.Nabel-Rosen H, Toledano-Katchalski H, Volohonsky G, Volk T. Cell divisions in the Drosophila embryonic mesoderm are repressed via posttranscriptional regulation of string/cdc25 by HOW. Curr Biol. 2005;15(4):295–302. doi: 10.1016/j.cub.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 77.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30(3):675–86. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 78.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30(3):687–96. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 79.Nien C-Y, Liang H-L, Butcher S, Sun Y, Fu S, et al. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7(10):e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nothias JY, Majumder S, Kaneko KJ. Regulation of gene expression at the beginning of mammalian development. J Biol Chem. 1995;270(38):22077–80. doi: 10.1074/jbc.270.38.22077. [DOI] [PubMed] [Google Scholar]
- 81.O’Farrell PH. How metazoans reach their full size: the natural history of bigness. In: Hall MN, Raff M, Thomas G, editors. Cell Growth: Control of Cell Size. Cold Spring Harbor Press; New York: 2004. pp. 1–22. [Google Scholar]
- 82.O’Farrell PH, Stumpff J, Su TT. Embryonic cleavage cycles: how is a mouse like a fly? Curr Biol. 2004;14(1):R35–R45. doi: 10.1016/j.cub.2003.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Price D, Rabinovitch S, O’Farrell PH, Campbell SD. Drosophila wee1 has an essential role in the nuclear divisions of early embryogenesis. Genetics. 2000;155(1):159–66. doi: 10.1093/genetics/155.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pritchard DK, Schubiger G. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 1996;10(9):1131–42. doi: 10.1101/gad.10.9.1131. [DOI] [PubMed] [Google Scholar]
- 85.Quinn LM, Herr A, McGarry TJ, Richardson H. The Drosophila Geminin homolog: roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis. Genes Dev. 2001;15(20):2741–54. doi: 10.1101/gad.916201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rabinowitz M. Studies on the cytology and early embryology of the egg of Drosophila melanogaster. Journal of Morphology. 1941;69(1):1–49. [Google Scholar]
- 87.Rose LS, Wieschaus E. The Drosophila cellularization gene nullo produces a blastoderm-specific transcript whose levels respond to the nucleocytoplasmic ratio. Genes Dev. 1992;6(7):1255–68. doi: 10.1101/gad.6.7.1255. [DOI] [PubMed] [Google Scholar]
- 88.Russell P, Nurse P. Cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45(1):145–53. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- 89.Satija R, Bradley RK. The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res. 2012;22(4):656–65. doi: 10.1101/gr.130682.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Seher TC, Leptin M. Tribbles, a cell-cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation. Curr Biol. 2000;10(11):623–29. doi: 10.1016/s0960-9822(00)00502-9. [DOI] [PubMed] [Google Scholar]
- 91.Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15(4):284–94. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 92.Shermoen AW, McCleland ML, O’Farrell PH. Developmental control of late replication and S phase length. Curr Biol. 2010;20(23):2067–77. doi: 10.1016/j.cub.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shermoen AW, O’Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67(2):303–10. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shimuta K, Nakajo N, Uto K, Hayano Y, Okazaki K, Sagata N. Chk1 is activated transiently and targets Cdc25A for degradation at the Xenopus midblastula transition. EMBO J. 2002;21(14):3694–3703. doi: 10.1093/emboj/cdf357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sibon OC, Laurençon A, Hawley R, Theurkauf WE. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr Biol. 1999;9(6):302–12. doi: 10.1016/s0960-9822(99)80138-9. [DOI] [PubMed] [Google Scholar]
- 96.Sibon OC, Stevenson VA, Theurkauf WE. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388(6637):93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- 97.Simpson L, Wieschaus E. Zygotic activity of the nullo locus is required to stabilize the actin-myosin network during cellularization in Drosophila. Development. 1990;110(3):851–63. doi: 10.1242/dev.110.3.851. [DOI] [PubMed] [Google Scholar]
- 98.Smibert CA, Wilson JE, Kerr K, Macdonald PM. Smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10(20):2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- 99.Smith AV, Orr-Weaver TL. The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development. 1991;112(4):997–1008. doi: 10.1242/dev.112.4.997. [DOI] [PubMed] [Google Scholar]
- 100.Snow MHL. Gastrulation in the mouse: Growth and regionalization of the epiblast. J Embryol Exp Morphol. 1977;42(1):293–303. [Google Scholar]
- 101.Spradling AC. The organization and amplification of two chromosomal domains containing Drosophila chorion genes. Cell. 1981;27(1 Pt 2):193–201. doi: 10.1016/0092-8674(81)90373-1. [DOI] [PubMed] [Google Scholar]
- 102.Stiffler LA, Ji JY, Trautmann S, Trusty C, Schubiger G. Cyclin A and B functions in the early Drosophila embryo. Development. 1999;126(23):5505–13. doi: 10.1242/dev.126.23.5505. [DOI] [PubMed] [Google Scholar]
- 103.Stumpff J, Duncan T, Homola E, Campbell SD, Su TT. Drosophila Wee1 kinase regulates Cdk1 and mitotic entry during embryogenesis. Curr Biol. 2004;14(23):2143–48. doi: 10.1016/j.cub.2004.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Su TT, Campbell SD, O’Farrell PH. Drosophila grapes/CHK1 mutants are defective in cyclin proteolysis and coordination of mitotic events. Curr Biol. 1999;9(16):919–22. doi: 10.1016/s0960-9822(99)80399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Su TT, Sprenger F, DiGregorio PJ, Campbell SD, O’Farrell PH. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes Dev. 1998;12(10):1495–1503. doi: 10.1101/gad.12.10.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sullivan W, Fogarty P, Theurkauf W. Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development. 1993;118(4):1245–54. doi: 10.1242/dev.118.4.1245. [DOI] [PubMed] [Google Scholar]
- 107.Sung H-W, Spangenberg S, Vogt N, Grosshans J. Number of nuclear divisions in the Drosophila blastoderm controlled by onset of zygotic transcription. Curr Biol. 2013;23(2):133–38. doi: 10.1016/j.cub.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 108.Sweeton D, Parks S, Costa M, Wieschaus E. Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development. 1991;112(3):775–89. doi: 10.1242/dev.112.3.775. [DOI] [PubMed] [Google Scholar]
- 109.Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, et al. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12(1):143–55. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 110.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, et al. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev. 2000;14(12):1439–47. [PMC free article] [PubMed] [Google Scholar]
- 111.Thomson AM, Gillespie PJ, Blow JJ. Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels. J Cell Biol. 2010;188(2):209–21. doi: 10.1083/jcb.200911037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Uto K, Inoue D, Shimuta K, Nakajo N, Sagata N. Chk1, but not Chk2, inhibits Cdc25 phosphatases by a novel common mechanism. EMBO J. 2004;23(16):3386–96. doi: 10.1038/sj.emboj.7600328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wieschaus E. Embryonic transcription and the control of developmental pathways. Genetics. 1996;142(1):5–10. doi: 10.1093/genetics/142.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wieschaus E, Sweeton D. Requirements for X-linked zygotic gene activity during cellularization of early Drosophila embryos. Development. 1988;104(3):483–93. doi: 10.1242/dev.104.3.483. [DOI] [PubMed] [Google Scholar]
- 115.Wilson EB. The cell in development and heredity. 3rd ed Macmillan; New York: 1925. p. 1107. [Google Scholar]
- 116.Yasuda GK, Baker J, Schubiger G. Temporal regulation of gene expression in the blastoderm Drosophila embryo. Genes Dev. 1991;5(10):1800–1812. doi: 10.1101/gad.5.10.1800. [DOI] [PubMed] [Google Scholar]
- 117.Yasuda GK, Schubiger G. Temporal regulation in the early embryo: is MBT too good to be true? Trends Genet. 1992;8(4):124–27. doi: 10.1016/0168-9525(92)90369-F. [DOI] [PubMed] [Google Scholar]
- 118.Yuan K, Farrell JA, O’Farrell PH. Different cyclin types collaborate to reverse the S-phase checkpoint and permit prompt mitosis. J Cell Biol. 2012;198(6):973–80. doi: 10.1083/jcb.201205007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yuan K, Shermoen AW, O’Farrell PH. Illuminating DNA replication during Drosophila development using TALE-lights. Current Biology. 2014 doi: 10.1016/j.cub.2014.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zalokar M. Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Dev Biol. 1976;49(2):425–37. doi: 10.1016/0012-1606(76)90185-8. [DOI] [PubMed] [Google Scholar]
- 121.Zamir E, Kam Z, Yarden A. Transcription-dependent induction of G1 phase during the zebrafish midblastula transition. Mol Cell Biol. 1997;17(2):529–36. doi: 10.1128/mcb.17.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhurov V, Terzin T, Grbić M. In)discrete charm of the polyembryony: evolution of embryo cloning. Cell Mol Life Sci. 2007;64(21):2790–98. doi: 10.1007/s00018-007-7047-5. [DOI] [PMC free article] [PubMed] [Google Scholar]