Summary
Determination of a protein’s N-terminal sequence can be important for the characterization of protein processing. To increase the confidence of protein N-terminal identification, chemical derivatization of the N-terminal amine group by (N-Succinimidyloxycarbonylmethyl)tris(2,4,6-trimethoxyphenyl)phosphonium bromide (TMPP) or dimethyl labeling followed by mass spectrometric analysis is commonly performed. Using this approach, proteins can be separated by SDS-PAGE, and the protein N-terminus of interest is labeled by TMPP or dimethyl in-gel before tryptic digestion and LC-MS analysis. The N-terminus of the protein can thus be easily identified because only the N-terminal tryptic peptide contains the labeling. Peptides with N-terminal derivatization produce a better fragmentation pattern during tandem mass spectrometric analysis, which significantly facilitates sequencing of these peptides.
Keywords: TMPP labeling, dimethyl labeling, N-terminal sequence analysis, mass spectrometry
1. Introduction
Identification of the N terminal sequence of an intact or cleaved protein is crucial for its biochemical and structural characterization [1]. In conventional shotgun proteomic analysis, it is difficult to identify protein N-termini due to infrequent detection of protein N-terminal peptides. Several methods have been developed based on the N-terminal labeling to overcome this limitation, including TMPP and dimethyl labeling [2, 3].
The TMPP labeling approach is straightforward and has been successfully applied to different proteins [4]. Two characteristics of this labeling reagent promote the sensitivity of the method: (i) TMPP labeling introduces a permanent positive charge resulting in an enhanced ionization efficiency and thus a better detection of low-abundance peptides; (ii) the hydrophobic TMPP group shifts the retention time of TMPP derivatized peptides in reversed phase chromatography toward a less complex part of the chromatogram, increasing the sensitivity of detection, especially for short N-terminal peptides that otherwise would not be retained on the column [3, 5]. In addition, TMPP is fully compatible with all standard detergents, chaotropic agents, and reduction conditions used for protein extraction in proteomics [5], which makes TMPP labeling a commonly used method for protein N-terminal sequencing.
An alternative method based on chemical labeling is dimethylation, which labels peptide N-termini and ε-amino groups of lysine with water-soluble formaldehyde via reductive methylation [6-8]. In MS/MS analysis, this labeling strategy provides a signal enhancement for the a1 and yn-1 ions, which are not detectable from most of the nonderivatized fragments [8]. Because of its simplicity, as well as low cost, dimethyl labeling is another promising strategy for protein N-termini identification.
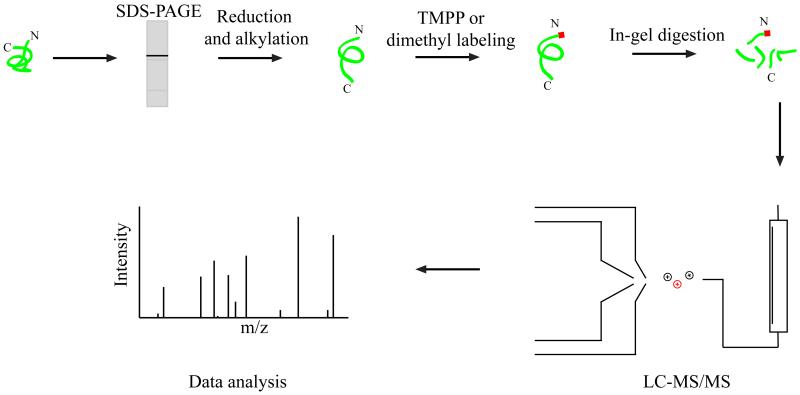
Due to the relatively simple experimental design (classical 1D SDS-PAGE, a single chemical derivatization step performed at the protein level in-gel, followed by protein digestion and LC-MS/MS, as illustrated in Figure 1), the two chemical derivation approaches can be readily used to analyze both purified proteins and highly complex protein mixtures [5]. Here, N-terminal identification using TMPP and dimethyl labeling of purified protein hemoglobin and BSA is described.
Figure 1. Flow chart showing the process of protein separation by SDS-PAGE, N-terminal labeling, in-gel digestion and LC-MS/MS.
2. Materials
Prepare all solutions using HPLC grade solvents unless indicated otherwise. Prepare and store all reagents at room temperature unless indicated otherwise. Diligently follow all waste disposal regulations when disposing waste materials.
2.1. SDS-PAGE Components
4-15% precast polyacrylamide gel (Bio-Rad, Hercules, CA, USA). Store at 4°C.
10× SDS-PAGE running buffer: Dissolve 30.0 g of Tris base, 144.0 g of glycine, and 10.0 g of SDS in 1000 ml of H2O (see Note 1). Store the running buffer at room temperature and dilute to 1× with ultrapure water before use.
Laemmli sample buffer (Bio-Rad, Hercules, CA, USA) and ≥98% pure 2-mercaptoethanol (Bio-Rad, Hercules, CA, USA) (see Note 2).
2.2. Labeling Components
Succinimidyloxycarbonylmethyl tris(2,4,6-trimethoxyphenyl)phosphonium bromide (TMPP-Ac-OSu) (Sigma-Aldrich, St. Louis, MO, USA). Dissolve 100 mg TMPP in 1.3 mL of acetonitrile (ACN) to make a 100 mM TMPP solution (see Note 3).
Sodium cyanoborohydride solution (260 mM): Dissolve 16.3mg sodium cyanoborohydride (Sigma-Aldrich, St. Louis, MO, USA) in 1ml water (see Note 4).
Sodium acetate solution (100 mM, pH 5): Mix 14.8 mL 0.2 M acetic acid solution (11.5 mL acetic acid in 1 L H2O) and 35.2 mL 0.2 M sodium acetate solution (16.4 g sodium acetate in 1 L H2O), and fill up to 100 ml with water.
4% formaldehyde: Dilute the 30% formaldehyde (Sigma-Aldrich, St. Louis, MO, USA) with water.
HEPES buffer (0.1M, PH 8.2): Dissolve 23.8mg HEPES in 1ml water. Adjust pH value to 8.2 with sodium hydroxide.
TEAB buffer (0.1M, PH 8.0): Dilute 1 M triethyl ammonium bicarbonate (Sigma-Aldrich, St. Louis, MO, USA) with water.
2.3 Components for sample preparation and mass spectrometric analysis
DTT solution (10 mM): Dilute 1 M DTT with TEAB buffer (see Note 5).
Iodoacetamide solution (55 mM): Dissolve 10.172 mg iodoacetamide (Sigma-Aldrich, St. Louis, MO, USA) in 1 ml TEAB buffer (see Note 6).
C18 Pipette Tips (EMD Millipore Corporation, Billerica, MA, USA)
Self-packed columns (20 cm in length) use Reprosil-Pur C18-AQ beads (3 μm, Dr. Maisch HPLC GmbH, Germany) and 360/75 PicoFrit column (360 μm OD × 75 μm ID, 10 μm Tip ID) (New Objective, USA)
Easy-nLC 1000 Nano HPLC and Q Exactive Mass Spectrometer (Thermo Fisher Scientific)
3. Methods
Carry out all procedures at room temperature unless otherwise specified.
3.1 Sample preparation, reduction and alkylation
Dilute the protein sample with water. Mix the diluted samples with the 2× Laemmli sample buffer (1:1). Heat protein samples at 95°C for 5 minutes. Spin the tubes for a few seconds and load protein samples to the 4–15% gel along with protein standard (2ug/marker/lane). Electrophorese at 15 mA until the sample has entered the gel and then continue at 25 mA till the dye front has reached the bottom of the gel (see Note 7).
Following electrophoresis, open the gel cassette with the lever and gently remove the gel from the cassette to a clean container. Rinse the gel with water for 10 min to remove any particulate matter, and repeat twice (see Note 8).
Coomassie Blue Staining. Stain the gel with approximately 15 - 20 ml EZ-Run™ Protein Gel Staining Solution for 30 min (Bio-Rad, Hercules, CA, USA) (see Note 9).
Replace the staining buffer with water and wash the gel on an orbital shaker for 30-60 min until the background color is weak.
Use a clean scalpel to excise the gel band of interest, cut the gel band into small pieces and transfer the gel band into a clean 0.5 ml Eppendorf tube (see Note 10) (see Note 11).
Destaining. Add 200 μl 50% ACN in TEAB buffer and agitate (600-1000 rpm) for 20 min. Discard the liquid. Add 200 μl TEAB and agitate (600-1000 rpm) for 20 min. Discard the liquid (see Note 12). Repeat until the gel band is completely destained.
Add 50 μl 100% ACN and agitate (600-1000 rpm) for 10 min. Discard the liquid and dry the gel in a speed vac for 5 min.
Reduction. Add 50 μl freshly prepared 10 mM DTT solution to cover the gel pieces and agitate (600-1000 rpm) for 45 min at 56°C.
Alkylation. Add 100 μl freshly prepared 55 mM iodoacetamide to the tube. React for 30 min in the dark (see Note 13).
Spin briefly and discard the liquid.
Add 100 μl 50% ACN in TEAB buffer and agitate (600-1000 rpm) for 10 min to wash away the reagents. Discard the liquid. Add 100 μl 100% ACN and agitate for 10 min. Spin briefly and discard the liquid. Repeat. Dry the gel piece in a speed vac for 5 min.
3.2 N-terminal labeling and protein digestion
-
For TMPP labeling: add 10 μl TMPP solution in ACN to dry gel piece. Then add 40 μl 100 mM HEPES buffer and incubate overnight (see Note 14).
For dimethyl labeling: add 100 μl 100 mM sodium acetate, 1 μL 4% formaldehye and 1 μL 260 mM sodium cyanoborohydride to the dehydrated gel pieces. Incubate the gel pieces for 20 min while mixing using a bench top test tube mixer (see Note 15).
Discard the liquid. Add 100 μl water and agitate for 10 min. Spin briefly and discard the liquid. Add 100 μl ACN and agitate for 10 min. Spin briefly and discard the liquid. Repeat washing alternately with water and ACN for 3-4 times until excess labeling reagent is clear.
Digestion. Add freshly prepared working solution of Promega trypsin (0.2-0.3 μg Trypisn in 20 μl TEAB buffer), incubate on ice for 30 mins. Add 40 μl TEAB buffer and leave on agitator (300 rpm) overnight at 37°C.
Peptide extraction. Add 50 μl 5% formic acid (FA), agitate for 15 min, collect the liquid; then add 50 μl actonitrile, agitate for 15 min, collect the liquid to the same tube.
Repeat step 4. Concentrate collected sample to dryness in a speed vac. Dissolve peptides with 10 μl 0.1% FA.
Desalt peptides with a C18 ziptip (following the protocol from Millipore), and concentrate sample to dryness in a speed vac. Dissolve peptides in 10 μl 0.1% FA.
Transfer solution to an injection vial for LC-MS/MS (see Note 16).
3.3 Mass spectrometric analysis
A nanoHPLC system was used for separation of the protein digests. Buffer A (0.1% FA in water) and buffer B (0.1% FA in ACN) were used as mobile phases for gradient separation. Protein digests were automatically loaded onto a C18 reversed phase column (column temperature 50°C). Due to the increased retention time of TMPP labeled N-terminal peptides [5], elution was achieved with a gradient of 0-30% B over 40 min, 30%-90% B over 40 min, 90% B for 10 min. For dimethyl labeling (no significant retention time is changed to the dimethyl labeled peptide), elution was achieved with a gradient of 0-30% B over 80 min, 30%-90% B over 1 min, 90% B for 9 min.
The Q-Exactive mass spectrometer was operated in data dependent mode using a top 10 method. Full MS scans were acquired in the Orbitrap mass analyzer over a range of 300-1650 m/z with resolution 70,000 (m/z 200). The target value was 3.00E+06. The ten most intense peaks with charge state ≥ 2 were fragmented in the HCD collision cell with normalized collision energy of 27%, and tandem mass spectra were acquired in the Orbitrap mass analyzer with resolution 17,500 (m/z 200). The target value was 1.00E+06. The ion selection threshold was 1.70E+04 counts, and the maximum allowed ion accumulation times were 20 ms for full MS scans and 60 ms for MS/MS [9].
Peak lists were created and searched by Mascot Distiller (Matrix Science, London, UK). Software setting: semiTrypsin with 1 missed cleavage; carbamidomethyl (C) as fixed modification; oxidation (M) as variable modification. For TMPP labeling, a mass addition of 572.1811(C29H33O10P) (N-Terminus, K and Y) must be included as a variable moditication. For dimethyl labeling, a mass addition of 28.0313 (C2H4) (N-terminus, K, P and R) as a variable modification must be included. Peptide tolerance was set to 10 ppm for peptides and MS/MS tolerance was 0.03 Da (see Note 17).
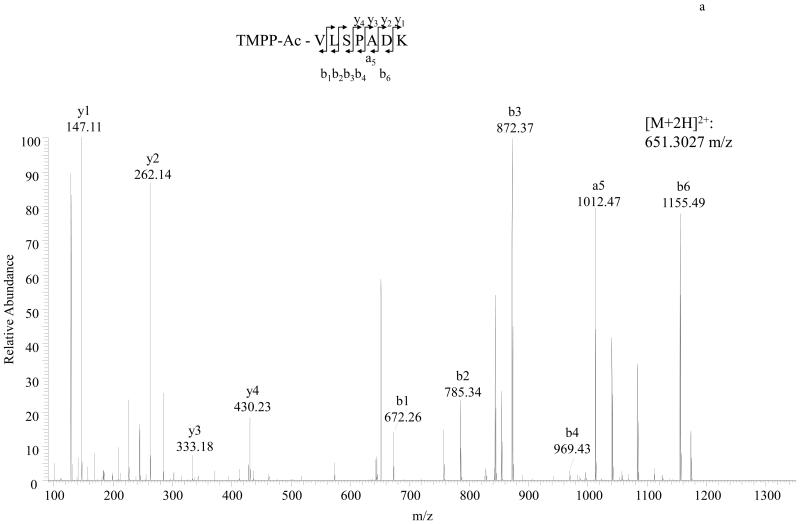
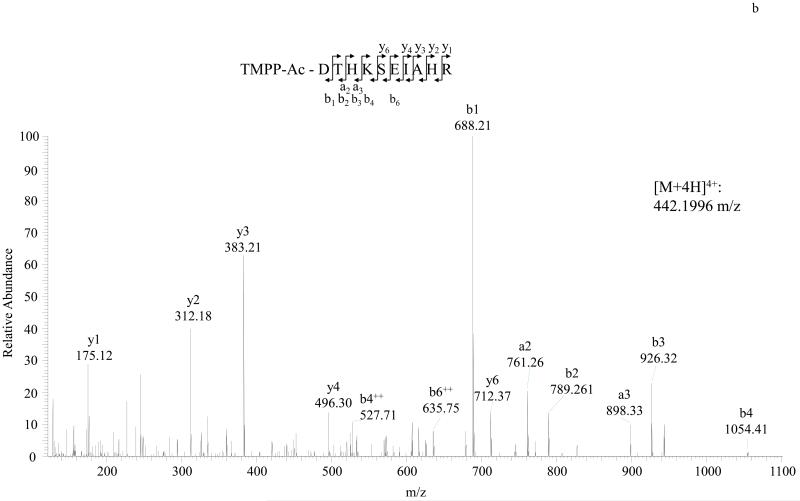
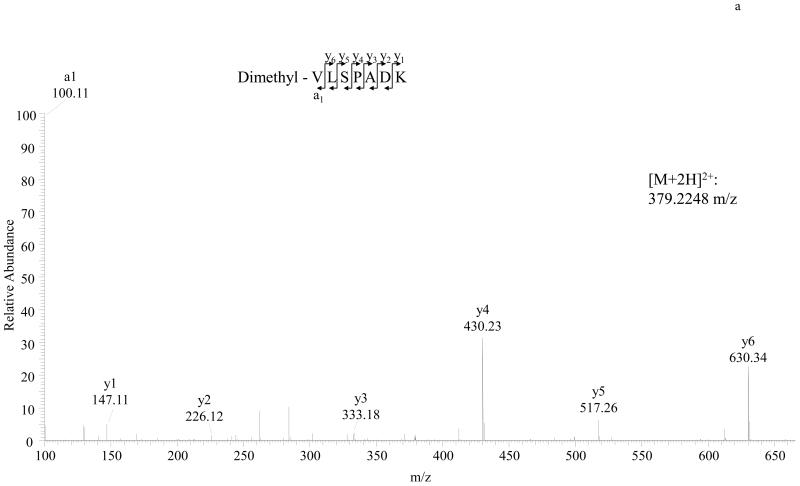
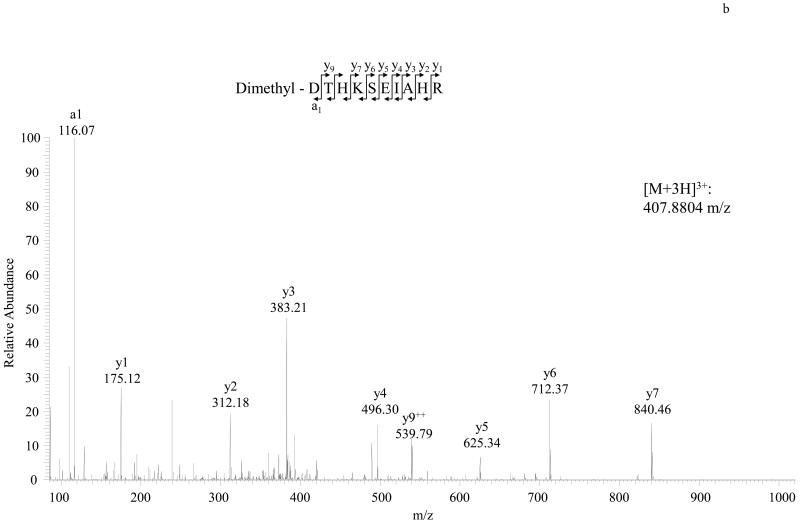
MS/MS spectra of TMPP labeled peptide VLSPADK (Hemoglobin A) and TMPP labeled peptide DTHKSEIAHR (BSA) are shown in Figure 2. As illustrated in Figure 3, dimethyl labeled peptide VLSPADK or DTHKSEIAHR identified the same N-terminal sequences of Hemoglobin A and BSA respectively. We also found dimethyl labeling to be more sensitive than TMPP labeling (Detection limit: 0.1 pmol BSA for dimethyl; 3 pmol BSA for TMPP).
Figure 2.
Detailed characterization of TMPP labeled N-terminal peptides of Hemoglobin A and BSA. A, MS/MS spectrum of the doubly charged N-terminal peptide VLSPADK of Hemoglobin A. Peptide sequence is shown at the top of the spectrum, with annotation of the matched ions. B, MS/MS spectrum of the triply charged N-terminal peptide DTHKSEIAHR of BSA. The fragmentation of the derivatized peptide produces a- and b-type ion series in addition to y-type ions.
Figure 3.
Detailed characterization of dimethyl labeled N-terminal peptides of Hemoglobin A and BSA. A, MS/MS spectrum of the doubly charged N-terminal peptide VLSPADK of Hemoglobin A. B, MS/MS spectrum of the triply charged N-terminal peptide DTHKSEIAHR of BSA. Unlike TMPP labeling, the dimethyl labeling strategy enhances a1 and yn-1 ions.
Acknowledgement
This work was supported by NIH Shared Instrumentation Grant S10RR027990 and NINDS grant P30 NS050276 to T.A.N.
Footnotes
SDS is a detergent. Handle it with care and take measures to prevent inhalation of air born SDS when weighing SDS powder- it is best weighed in a fume hood. Check the pH of the 10X solution. The pH of the buffer should be between 8.1-8.5. Do not adjust PH with acid or base.
Sample buffer: Add 50 μl 2-mercaptoethanol to 950 μl Laemmli sample buffer. Protein sample is mixed 1:1 with the sample buffer.
TMPP solution is made freshly, kept at −20°C and not stored longer than 48 h to ensure high labeling efficiency.
Cyanoborohydride solutions should be made freshly, kept at 4°C and not stored longer than 24 h to ensure high labeling efficiency [10].
DTT solution (1 M): dissolve 0.15 g DTT in 1ml water. Store at −20°C.
Iodoacetamide powder is stored at −5 to −30°C and protected from light. Iodoacetamide solution is made freshly before use.
Pull the green tape gently to remove it from the bottom of the cassette and remove the comb by pulling upward in one smooth motion to keep the lane straight.
Do not touch the gel with ungloved hands and always use a new or acid-cleaned dish to avoid contamination.
Make sure the staining buffer covers the gel completely.
Be sure to use extremely clean surfaces and new scalpels. This should be done in a flow hood to minimize the possibility of contamination by dust, hair, flakes of skin, or other forms of dirt.
Use polypropylene tubes and low-retention tips to minimize protein loss by adsorption to tube walls.
An increase of temperature promotes the destaining process.
Iodoacetamide is unstable and light-sensitive. Prepare iodoacetamide solutions immediately before use and perform alkylation in the dark. Excess reaction time will cause other functional groups to be labeled.
For TMPP labeling, do not use ammonium bicarbonate or Tris buffer that can decrease the labeling efficiency.
For dimethyl labeling, during sample preparation, no buffers and solutions containing primary amines (such as ammonium bicarbonate and Tris) should be used that can decrease the labeling efficiency, as formaldehyde can react with these reagent [10].
Before loading peptide samples for LC-MS/MS, avoid sample contamination by keratin, polymers and detergents. Try to work as cleanly as possible, because contamination with other proteins can prevent identification of the protein of interest. The most frequent contaminants are BSA and human keratin. The keratin comes from dust, small hairs and fingerprints. Even a small hair contains overwhelming amounts of keratin compared to the amount of your sample.
Selective N-terminal TMPP derivatization can be achieved by keeping the solution at pH 8.2, exploiting the weaker basicity of the N-terminal amine relative to the ε-amino group of the lysine side chain [5]. In order to minimize derivatization of tyrosine residues, 0.1 M hydroxylamine can be added to the solution to the quench derivatizing reagent for 1 h after TMPP labeling reaction [5][11].
References
- 1.Speicher KD, Gorman N, Speicher DW. N-terminal sequence analysis of proteins and peptides. Curr Protoc Protein Sci. 2009 doi: 10.1002/0471140864.ps1110s57. Chapter 11: p. Unit11 10. [DOI] [PubMed] [Google Scholar]
- 2.Gallien S, et al. Ortho-proteogenomics: multiple proteomes investigation through orthology and a new MS-based protocol. Genome Res. 2009;19(1):128–35. doi: 10.1101/gr.081901.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armengaud J. A perfect genome annotation is within reach with the proteomics and genomics alliance. Curr Opin Microbiol. 2009;12(3):292–300. doi: 10.1016/j.mib.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Huang ZH, et al. A picomole-scale method for charge derivatization of peptides for sequence analysis by mass spectrometry. Anal Chem. 1997;69(2):137–44. doi: 10.1021/ac9608578. [DOI] [PubMed] [Google Scholar]
- 5.Bertaccini D, et al. An improved stable isotope N-terminal labeling approach with light/heavy TMPP to automate proteogenomics data validation: dN-TOP. J Proteome Res. 2013;12(6):3063–70. doi: 10.1021/pr4002993. [DOI] [PubMed] [Google Scholar]
- 6.Hsu JL, et al. Beyond quantitative proteomics: signal enhancement of the a1 ion as a mass tag for peptide sequencing using dimethyl labeling. J Proteome Res. 2005;4(1):101–8. doi: 10.1021/pr049837+. [DOI] [PubMed] [Google Scholar]
- 7.Hsu JL, Huang SY, Chen SH. Dimethyl multiplexed labeling combined with microcolumn separation and MS analysis for time course study in proteomics. Electrophoresis. 2006;27(18):3652–60. doi: 10.1002/elps.200600147. [DOI] [PubMed] [Google Scholar]
- 8.Hsu JL, et al. Stable-isotope dimethyl labeling for quantitative proteomics. Anal Chem. 2003;75(24):6843–52. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 9.Sun L, Zhu G, Dovichi NJ. Comparison of the LTQ-Orbitrap Velos and the Q-Exactive for proteomic analysis of 1-1000 ng RAW 264.7 cell lysate digests. Rapid Commun Mass Spectrom. 2013;27(1):157–62. doi: 10.1002/rcm.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boersema PJ, et al. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4(4):484–94. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 11.Boersema PJ, et al. Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates. Proteomics. 2008;8(22):4624–32. doi: 10.1002/pmic.200800297. [DOI] [PubMed] [Google Scholar]