Abstract
The serine/threonine kinase mammalian target of rapamycin (mTOR) is a key regulator of protein synthesis, cell proliferation and energy metabolism. As constitutive deletion of Mtor gene results in embryonic lethality, the function of mTOR in muscle stem cells (satellite cells) and skeletal muscle regeneration remains to be determined. In this study, we established a satellite cell specific Mtor conditional knockout (cKO) mouse model by crossing Pax7CreER and Mtorflox/flox mice. Skeletal muscle regeneration after injury was severely compromised in the absence of Mtor, indicated by increased number of necrotic myofibers infiltrated by Evans blue dye, and reduced number and size of regenerated myofibers in the Mtor cKO mice compared to wild type (WT) littermates. To dissect the cellular mechanism, we analyzed satellite cell-derived primary myoblasts grown on single myofibers or adhered to culture plates. The Mtor cKO myoblasts exhibited defective proliferation and differentiation kinetics when compared to myoblasts derived from WT littermates. At the mRNA and protein levels, the Mtor cKO myoblasts expressed lower levels of key myogenic determinant genes Pax7, Myf5, Myod, Myog than did the WT myoblasts. These results suggest that mTOR is essential for satellite cell function and skeletal muscle regeneration through controlling the expression of myogenic genes.
Keywords: skeletal muscle, satellite cells, muscle regeneration, mTOR
1. Introduction
The skeletal muscle constitutes ~40% of the animal body mass and plays an important role in locomotion and metabolism. As such, proper muscle growth and homeostasis is a critical determinant of farm animal meat production and human motor performance. Conversely, muscle wasting due to genetic factors (muscular dystrophy), aging (sarcopenia), disuse (atrophy) or cancer (cachexia) severely compromises the life quality of humans. The growth of skeletal muscle mainly depends on the increase of myofiber number (hyperplasia) and myofiber size (hypotrophy)[1]. Generally, the number myofibers in each muscle is fixed at the prenatal stage. At the postnatal stage, muscle growth is accompanied by fusion of myogenic precursor cells to the existing myofibers. In the absence of injury, the rate of myonucleus turnover is at most 1% to 2% per week in adult skeletal muscle[2]. In response to injury, however, an imminent supply of myonuclei is required for the regeneration of the damaged muscle.
Satellite cells located beneath the lamina of myofibers are muscle resident stem cells necessary for postnatal muscle growth and regeneration[3]. Satellite cells are normally quiescent in non-injured mature muscles, and can be marked by the expression of Pax7. In response to cues such as injury and exercise, satellite cells are activated and enter the cell cycle. The proliferating cells (myoblasts) are marked by the coexpression of Pax7 and MyoD. After several rounds of division, myoblasts exit the cell cycle and either return to quiescence or undergo differentiation to fuse with exiting fibers[4]. The differentiating myoblasts rapidly downregulate the expression of Pax7 and upregulate the expression of MyoG, and are thus marked by coexpression of MyoD and MyoG[5,6]. The self-renewal, proliferation and differentiation of satellite cells ensure efficient repair of damaged muscles while maintaining a viable pool of stem cells for future regeneration.
Mammalian (or mechanistic) target of rapamycin (mTOR) is a serine/threonine kinase that serves as the catalytic subunit of two protein complexes: the mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). The mTOR-mediated signaling pathway has been shown to regulate a wide range of cellular processes including protein translation, cell proliferation, apoptosis and autophagy[7]. Recent studies further demonstrate that the mTOR pathway is implicated in maintaining the self-renewal and differentiation of various tissue stem cells, such as epidermal, hematopoietic, germinal and pancreatic cancer stem cells[8,9,10,11].
In the skeletal muscle, mTOR mediates Wnt7a’s pro-proliferation effect on satellite cells and growth promoting effect on myofibers[12,13]. Moreover, a recent study indicates that mTORC1 is necessary and sufficient for the transition of quiescent satellite cells from a dormant G0 to an alerted G0 state[14]. However, whether mTOR is necessary for satellite cell activation and proliferation has yet to be determined through knockout approaches. As Mtor constitutive KO mice die prematurely at the embryonic stage, tissue specific conditional knockout (cKO) mice that bypass embryonic lethality is necessary for dissecting the function of mTOR in myogenesis. Myofiber-specific deletion of Mtor driven by human α-actin-Cre (HSA-Cre) led to severe myopathy, including impaired oxidative metabolism, altered mitochondrial regulation and glycogen accumulation[15]. Whereas conditional deletion of Raptor and Rictor has shed light on the relative role of mTORC1 and mTORC2 in satellite cells[16,17], the role of mTOR (essential for both mTORC1 and mTORC2) in satellite cells remains unknown. Here we used the tamoxifen (TMX) inducible CreER system to address this outstanding question.
2. Materials and methods
2.1 Animals
All procedures involving mice were guided by Purdue University Animal Care and Use Committee. The mouse strains were derived from Jackson Laboratory (Bar Harbor, ME) under these stock numbers: Pax7CreER (#012476) and Mtorflox/flox (#011009). In the Pax7-CreER/Mtorflox/flox (referred to as Mtor cKO) mice, CreER-mediated recombination occurs in Pax7 expressing satellite cells after daily injection of 300 µl of Tamoxifen(10 mg/ml) corn oil solution for 5 days. Mice were housed in the animal facility with free access to standard rodent chow and water. PCR genotyping was done using protocols described by the supplier.
2.2 Primary myoblast isolation and culture
Primary myoblasts were isolated as previously described[18]. Briefly, hind limb muscle from 6 weeks old mice were collected, minced and digested with a mixture of collagenase and dispase (Roche). The digestions were stopped with growth medium containing F-10 Ham’s medium, 20% FBS(HyClone, Logan, UT), 4ng/mL basic fibroblast growth factor, and 1% penicillin–streptomycin, filtered and centrifuged at 450g for 5 minutes. Then the cell pellet was suspended and plated on collagen-coated dishes with growth medium. Then the cells were maintained at an incubator at 37 °C with 5% CO2 and the medium was refreshed every 2 days.
2.3 Single myofiber isolation and culture
The entire extensor digitorum longus (EDL) muscles were collected and digested with collagenase A (Sigma) for 1h and triturated. Separated single myofibers were fixed immediately for staining or transferred to a new dish coated with horse serum for culture. The growth medium was Dulbecco’s modified Eagle’s medium supplemented with 20% FBS, 2% chicken embryo extract (Accurate Chemical, Westbury, NY), and 1% penicillin-streptomycin. The myofibers were cultured for 3 days and then used for staining.
2.4 Muscle injury and regeneration
The injury of muscle was performed by intramuscular injection of cardiotoxin (CTX, Sigma) into TA muscle. Briefly, mice were anesthetized using a ketamine-xylazine cocktail, and then shaved to expose the TA belly. Then 50µl of 10µM CTX were injected into TA muscle. Muscles were then harvested at 8 days post injection to assess the completion of regeneration and repair.
2.5 Evans blue (EB) uptake analysis
EB uptake analysis as previous described[19]. EB (20 mg/ml in PBS) was administered to mice intraperitoneally at 24 h prior to collection of muscle samples.
2.6 Hematoxylin-eosin (H&E) and immunostaining
The whole TA muscle were dissected and cut at 10µm thickness. For H&E staining, the sections were rinsed with PBS and stained in haematoxylin for 40 minutes. Then the sections were rinsed with running tap water and stained in eosin for 10 minute. For immunofluorescence staining, sections were fixed with 4% paraformaldehyde and blocked for 1h. Sections were then incubated with indicated primary antibodies diluted in blocking buffer at 4 °C overnight. After that, sections were incubated with secondary antibodies and DAPI for 50 min at room temperature. Fluorescent images were taken using a Leica DM 6000B fluorescent microscope.
2.7 Total RNA extraction, cDNA synthesis and real-time PCR
Total RNA from cells or muscle tissue was extracted using Trizol Reagent according to the manufacturer’s instructions. The quality and concentration of total RNA were measured by a spectrophotometer (Nanodrop 3000, Thermo Fisher). Then total RNA was converted to cDNA using random primers and MMLV reverse transcriptase. Real-time PCR was carried out with a Roche Lightcycler 480 PCR System using SYBR Green Master Mix and gene-specific primers. 18S rRNA was used as internal control. The fold changes of indicated genes were analyzed using 2−ΔΔCT methods.
2.8 Protein Extraction and Western Blot Analysis
Protein extraction and western blot were conducted as previously described[18]. Briefly, total proteins from cells or tissues were homogenized using RIPA buffer. Protein concentrations were determined using Pierce BCA Protein Assay Reagent (Pierce Biotechnology). Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore Corporation), blocked for 1 hour at RT, and then incubated with primary antibodies overnight at 4°C. The phosphorylated S6 (pS6) and S6 antibodies were from Cell signaling, and all the other primary antibodies were from Santa Cruz Biotechnology. Secondary antibodies (anti-rabbit IgG or anti-mouse IgG, Jackson ImmunoResearch) were diluted 8,000-fold. Immunodetection was performed using enhanced chemiluminescence(ECL) substrate (Pierce Biotechnology) and detected with a Gel Logic 2200 imaging system (Carestream).
2.9 Data Analysis
All experimental data are presented as means ± SEM. Comparisons were made by unpaired two-tailed Student’s t-test. A value of p < 0.05 was considered statistically significant.
3 Results
3.1 Establishment of a mouse model for satellite cell-specific deletion of Mtor
To investigate the role of mTOR in satellite cells, we generated the Pax7CreER/Mtorflox/flox mice in which Mtor is specifically deleted in Pax7-expressing satellite cells upon induction with TMX. To verify the deletion efficiency, we first isolated the myoblast from WT and Mtor cKO mice. Myoblasts were cultured for 72h in growth medium supplemented with 4-hydroxy-Tamoxifen (4-OH-TMX, 0.4 µM) or vehicle control. The mRNA level of Mtor in 4-OH-TMX treated myoblasts was about 50% of that in vehicle treated cells, and mTOR protein was almost abolished by 4-OH-TMX(Supplementary Fig. S1A). We further confirmed the deletion of Mtor in vivo. After the Mtor cKO and WT littermate mice were injected with TMX for 5 days and chased for another 5 days, the TA muscles were injected with CTX and samples were collected 5 days later. We detected efficient depletion of mTOR at both protein and mRNA levels in the Mtor cKO compared to WT TA muscles(Supplementary Fig. S1B). These results indicated that the Mtor cKO mouse model results in satellite cell specific deletion of Mtor.
3.2 Impaired muscle regeneration in Mtor cKO mice
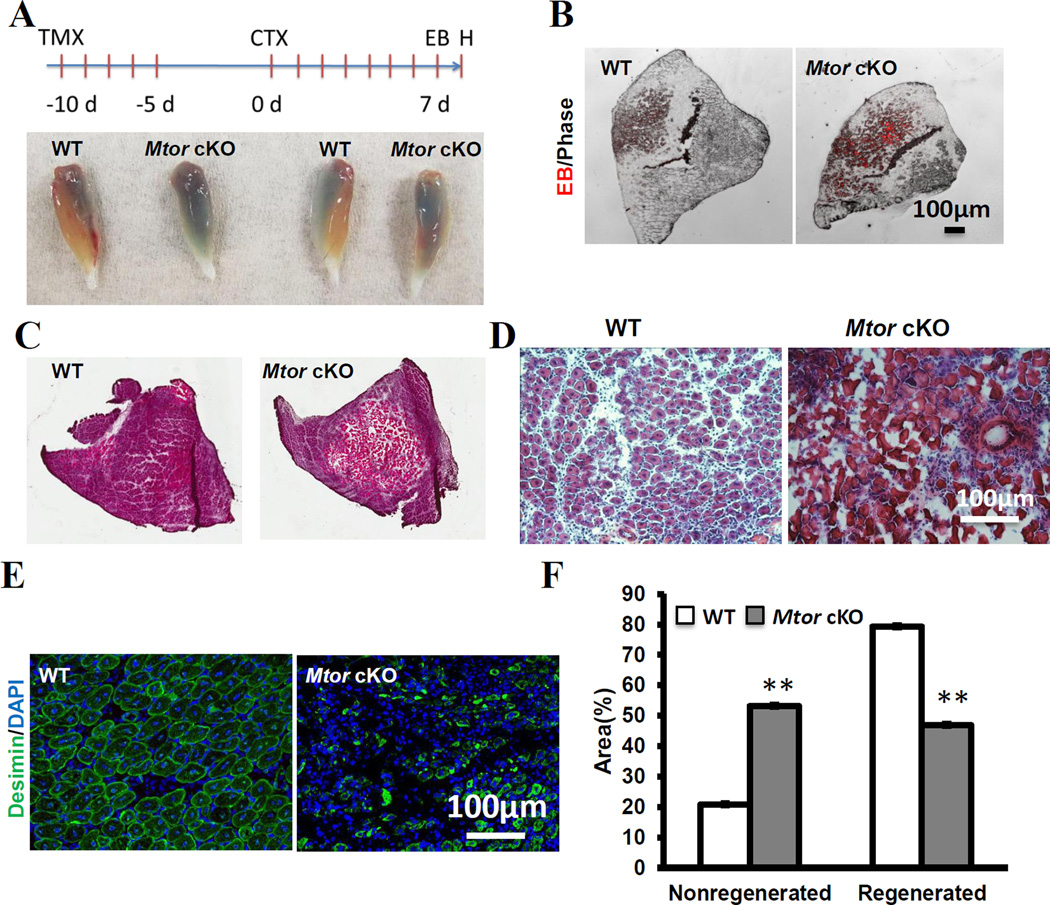
To investigate how satellite specific deletion of Mtor affects muscle regeneration, TA muscles of Mtor cKO and WT mice were injured by intramuscular injection of CTX after TMX induction(Fig. 1A). On Day 7 post injection of CTX (7 dpi), EB was administered intraperitoneally into mice to label injured myofibers, and the TA muscles were collected 24 h after EB injection for histological analysis(Fig. 1A). Notably, injured TA muscles from Mtor cKO mice appeared much bluer than those from WT mice(Fig. 1A), indicating stronger EB dye accumulation. Higher magnification images revealed that there were indeed more EB+ myofibers in the Mtor deleted TA muscles than in the WT TA muscles(Fig. 1B). Next, we performed H&E staining and examined the regenerating regions. Whereas the WT muscles regenerated uniformly with little fibrosis and formed new myofibers with central nuclei, the Mtor cKO muscles were poorly regenerated with extensive scarification and few myofibers with central nuclei(Fig. 1C–D). To calculate the ratio of regenerated to non-regenerated areas, we used desmin to label the newly regenerated myofibers(Fig. 1E). Quantification results showed that the unregenerated areas of Mtor cKO muscles were almost 3 times of that of the WT muscles(Fig. 1F). Taken together these results indicate that Pax7-driven deletion of mTOR lead to severe deficiency in satellite cell-mediated muscle regeneration.
Figure 1. Impaired skeletal muscle regeneration of Mtor cKO mice.
(A) Schematics of experimental design and representative images of whole TA muscles 8 days after CTX injection. Blue color indicates Evans Blue (EB) uptake. (B) Representative image of damaged TA muscle sections showing EB fluorescence (Red color). (C, D) Representative H&E staining image of TA muscle sections. (E) Representative image of regenerated TA muscle labeled with desmin. (F) Quantification the ratio of regenerated and undegenerated areas in WT and Mtor cKO TA muscles. n=4 for each group, ** p < 0.01.
3.3 Deletion of Mtor inhibited satellite cell proliferation and differentiation
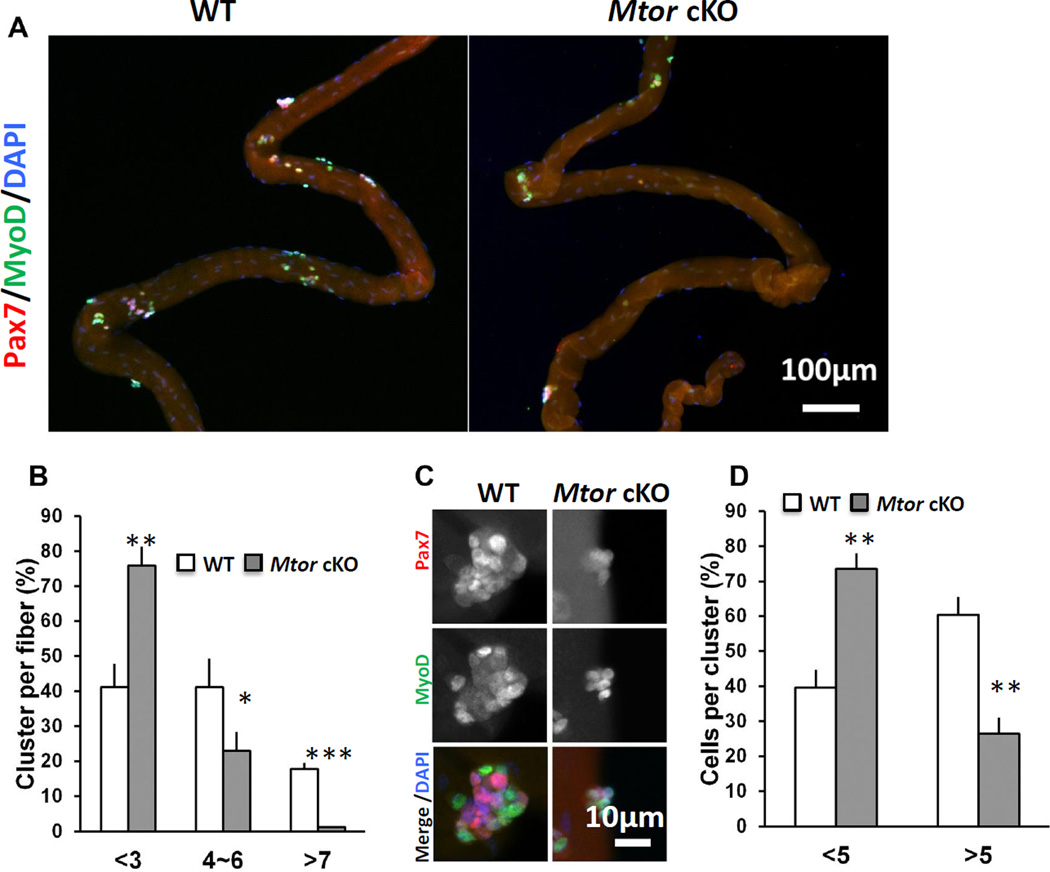
To understand how mTOR regulates satellite cells, we treated adult Mtor cKO and WT mice with TMX for five days, chased for another 5 days and then collected single EDL myofibers for analysis. The number of satellite cells per EDL myofiber was identical between WT and Mtor cKO mice (data not shown), suggesting that mTOR is not necessary for short-term maintenance of satellite cells. EDL myofibers were subsequently cultured for 72 h to allow satellite cell activation and proliferation. Notably, the number of clusters per EDL myofiber was decreased in the Mtor cKO myofibers compared to WT myofibers(Fig. 2A). A majority of EDL myofibers (75%) from the Mtor cKO mice had less than 3 clusters of myoblasts per myofiber, whereas 60% WT myofiber had more than 3 clusters per myofiber(Fig. 2B). The average number of myoblasts/cluster was also lower in the Mtor knockout group compared to WT(Fig. 2C). Over 60% of WT clusters had more than 5 myoblasts, whereas only 25% of clusters in the Mtor knockout fibers had more than 5 myoblasts(Fig 2D). These results suggested deletion of Mtor inhibited the activation and proliferation of satellite cells.
Figure 2. Single myofiber analysis of WT and Mtor cKO EDL muscles.
(A) Representative images of single EDL myofibers cultured for 72 h. Myoblasts were labeled with Pax7 (red) and MyoD (green). (B) Abundance of satellite cell-derived myoblast clusters grown on EDL myofibers after cultured for 72 h. n = 4 pairs of mice, 20 myofibers from each mouse were analyzed. (C) Representative images of clusters of myoblasts labeled with Pax7 (red) and MyoD (green) after grown for 72 h. (D) Relative abundance of large (containing >5 myoblasts) and small (containing 5 or less myoblasts) clusters grown on cultured EDL myofibers of Mtor cKO and WT mice. n=4 pairs of mice, 50 clusters for each mouse. * p < 0.05, ** p < 0.01, *** p < 0.001.
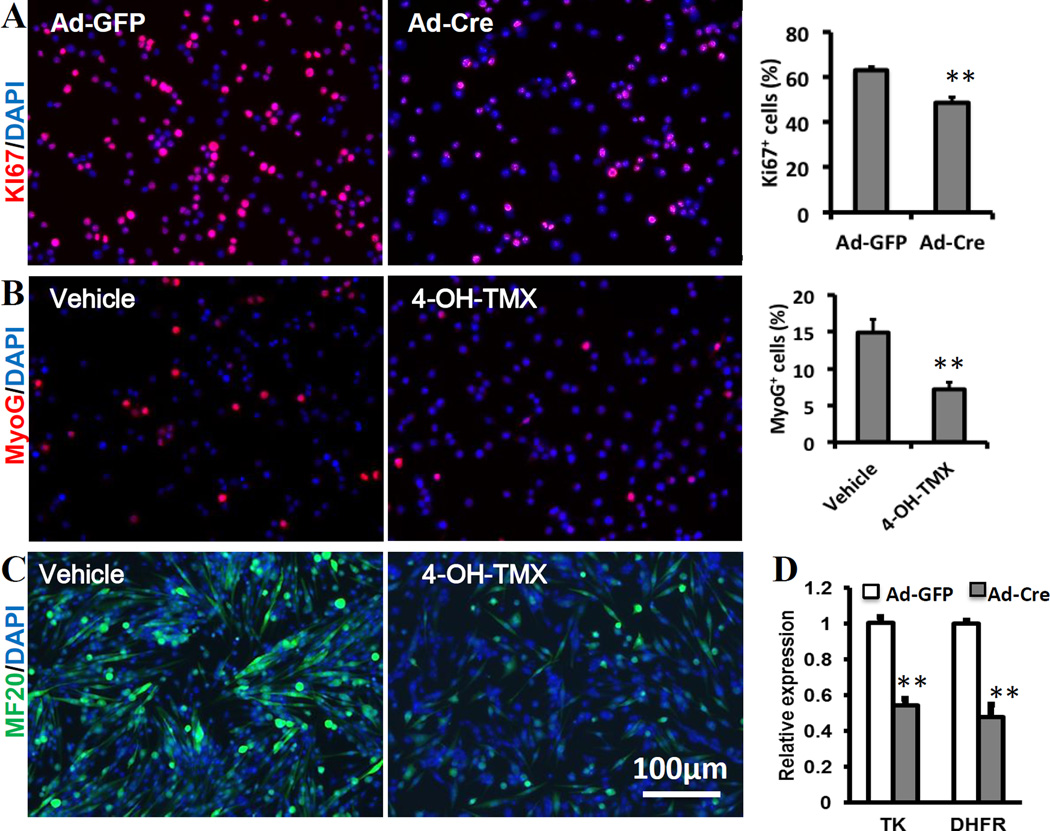
To further characterize the effect of Mtor ablation on satellite cells. We cultured primary myoblasts from Mtor flox/flox mice. The myoblasts were treated with adenovirus-Cre (Ad-Cre) to delete Mtor or treat with Ad-GFP as the control. Myoblasts were then labeled with the proliferation marker Ki67(Fig. 3A). A significant reduction of Ki67+ myoblasts was observed from the Mtor deleted sample(Fig. 3A). The relative mRNA levels of TK and DHFR, two proliferation related genes, in the Mtor deleted myoblasts were about 50% of those in control myoblasts(Fig. 3D). These results confirm that deletion of Mtor inhibits myoblast proliferation.
Fig. 3. Defective satellite cell proliferation and differentiation in Mtor cKO mice.
(A) Representative images of Ki67 staining that specifically labels proliferating myoblasts and the ratio of Ki67+ cells. N=6. (B) Representative images of MyoG staining that specifically labels differentiation myoblasts and the ratio of MyoG+ cells. N=6. (C) Representative images of MF20 staining that specifically labels sarcomeric myosin heavy chain in newly formed myotubes. (D) Relative mRNA expression of TK and DHFR in proliferating myoblasts. ** p < 0.01.
We next determined whether Mtor ablation affects myoblast differentiation. Primary myoblasts isolated from Mtor cKO mice were pretreated with 4-OH-TMX to delete Mtor or vehicle control for 48h. Then the myoblasts were seeded on Matrigel-coated plates at the same density and differentiated for 4d. The differentiated myoblasts were labeled with MyoG. Of note, fewer myoblasts was MyoG positive in the Mtor -deleted myoblasts(Fig. 3B), suggesting impaired myoblasts differentiation after Mtor deletion. In addition, fewer cells were labeled with MF20, which marks sarcomeric myosin heavy chain in newly formed myotubes, in the Mtor deleted sample(Fig. 3C), indicating impaired formation of mature fibers. Together, these data indicate that mTOR pathway is not only necessary for myoblast proliferation, but also necessary for myoblast differentiation.
3.4 Mtor knockout inhibited myogenic gene expression
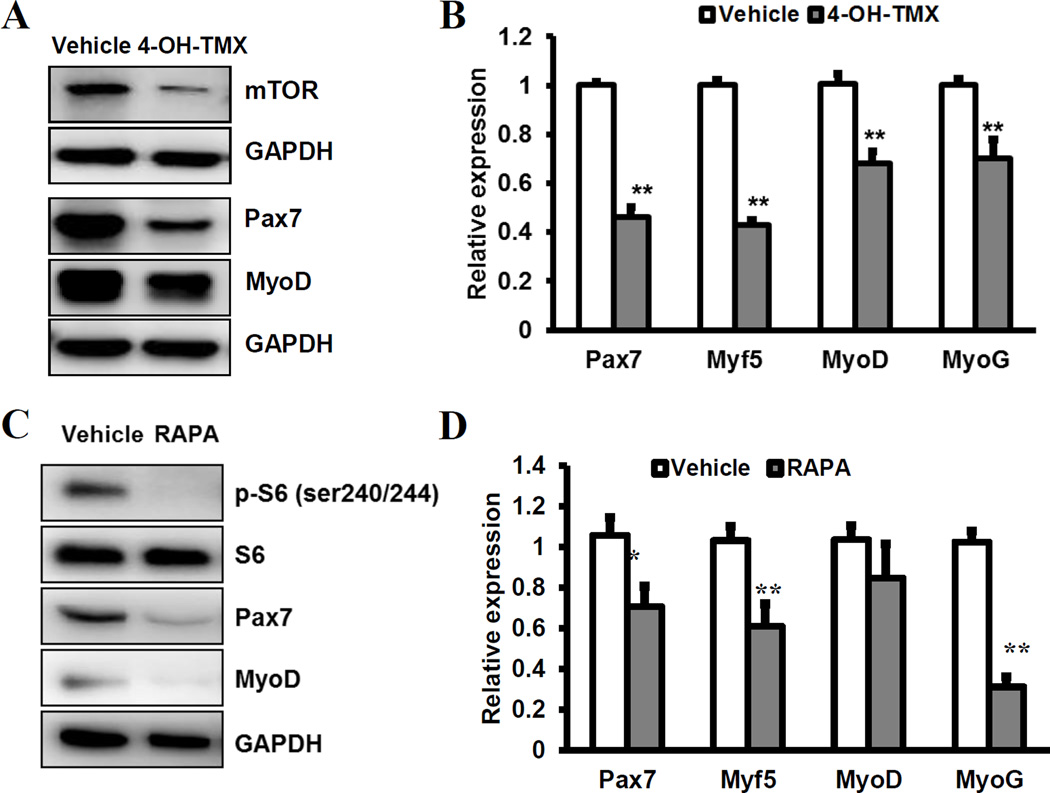
We finally determined how mTOR affect expression of myogenic genes. First, we checked the mRNA and protein levels of various myogenic genes in mTORflox/flox myoblast pretreated with vehicle or 4-OH-TMX. In the 4-OH-TMX treated myoblasts, we observed a robust reduction of Pax7 and a slight reduction of MyoD at the protein level(Fig. 4A). In addition, Pax7, Myf5, MyoD and MyoG mRNA levels were significantly lower in the 4-OH-TMX treated myoblasts compared to control myoblasts(Fig. 4B). Pharmacological inhibition of mTOR in WT myoblasts using rapamycin similarly reduced the protein level of Pax7 and MyoD(Fig. 4C). The concomitant reduction of phosphorylated S6 (pS6) but not total S6(Fig. 4C) demonstrate the effectiveness of rapamycin in blocking mTOR signaling. In addition, rapamycin treatment reduced the mRNA levels of myogenic genes Pax7, Myf5 and Myog, but not Myod(Fig. 4D). Taken together, these results demonstrate that genetic and pharmacological inhibition of mTOR similarly reduce the expression of key myogenic genes at both mRNA and protein levels.
Fig. 4. mTOR signaling regulates the expression of key myogenic genes.
(A, B) Relative levels of mTOR, Pax7, MyoD and MyoG proteins (A) and mRNA (B) in myoblasts depleted of Mtor (4-OH-TMX) and control myoblasts (vehicle treated). N=4. (C, D) Relative levels of phosphorylated S6 (pS6), total S6, Pax7, MyoD and MyoG proteins (C) and mRNA (D) in WT myoblasts treated with rapamycin (RAPA) and vehicle control. N=4. * p < 0.05, ** p < 0.01.
4 Discussion
In this study we illustrated an essential role of mTOR in satellite cells and skeletal muscle regeneration. It has been reported that constitutive deletion of Mtor lead to fatality at the embryonic stage[20], thus precluding the analysis of how mTOR regulates postnatal satellite cells. Since Pax7 is specifically expressed in muscle stem cells[21], we used Pax7CreER knockin allele to specifically delete Mtor in satellite cells. The Mtor cKO mouse model was confirmed by reduced mTOR protein and mRNA expression in satellite cells and newly regenerated skeletal muscles. Previous study used human skeletal muscle α-actin (HSA)-Cre mice to knockout Mtor in post mitotic myofibers but not in satellite cells[15]. Compared to that established muscle specific mouse model, our model not only allows Mtor deletion in satellite cells but also allows stage specific deletion of Mtor depending on the time of TMX administration.
It has been reported that rapamycin impairs muscle regeneration by blocking the mTOR pathway[22]. In contrast, activation of mTOR pathway by leucine supplementation improves skeletal muscle regeneration[23], indicating the mTOR pathway is important in the regeneration process. Due to potential off-target effect of rapamycin and leucine, the specific role of mTOR in satellite cells has yet to be determined by direct knockout approaches. Herein we used a genetic approach to knockout Mtor in satellite cells. No obvious reduction in satellite cells and muscle phenotype were observed in the absence of muscle injury in short-term. This can be explained by a non-essential role of mTOR in the maintenance of quiescent satellite cells in adult skeletal muscles. However, skeletal muscle regeneration was severely affected by Mtor deletion in satellite cells. This observation suggests that mTOR is essential for the regenerative capacity of satellite cells, including their activation, proliferation and differentiation.
Recent studies have reported that mTOR plays a key role in stem cell activity. Suppression of the mTOR pathway led to hematopoietic stem cells depletion in both mice and humans[24]. In contrast, hyperactivity of mTOR induced pre-mature differentiation and subsequent depletion of germline stem cells[10]. Thus the role of mTOR in various stem cells is cell type specific. Our analysis of Mtor deficient satellite cells demonstrates that mTOR is essential for the proliferation and differentiation of satellite cells, manifested by reduced number of myoblasts and fewer Ki67+ and MyoG+ cells.
To determine the pathways through which mTOR regulates satellite cell activity. We first checked the expression of Pax7, a marker for quiescent satellite cells and proliferating myoblasts. Mtor deletion and rapamycin treatment both reduced Pax7 expression, which explains the proliferation defects of Mtor cKO satellite cells. Myogenic regulatory factors Myf5, MyoD and MyoG play distinct and overlapping roles in myogenesis. Myf5 support the transient amplification of myoblasts prior to differentiation[25], while MyoD is involved in satellite cell activation and promotes early differentiation[26]. MyoG is essential for the terminal differentiation of myoblasts[27]. The expression of Myf5, MyoD and MyoG was all reduced in the Mtor knockout myoblasts. These results are consistent with a previous report showing that rapamycin reduces the expression of Myf5, MyoG and MyoD in C2C12 myoblasts[28,29].
In summary, our data provided novel insights into the role of mTOR in postnatal satellite cells and skeletal muscle regeneration. We established a satellite cell specific knockout of Mtor model and provided direct genetic evidence demonstrating that mTOR is essential for proper satellite cell activity and skeletal muscle regeneration. These results may have implications in the treatment of muscular diseases and promoting meat growth in farm animals by targeting the mTOR pathway.
Supplementary Material
Highlights.
Pax7CreER was used to delete Mtor gene in satellite cells
Satellite cell specific deletion of Mtor impairs muscle regeneration
mTOR is necessary for satellite cell proliferation and differentiation
Deletion of Mtor leads to reduced expression of key myogenic genes
Acknowledgements
We thank Jun Wu for lab management and maintaining mouse colonies, and other members of the Kuang lab for technical assistance and discussion. This work was partially sponsored by National Institutes of Health of USA (AR060652 to SK); national key technology support program of China (2014BAD20B01 to RZ); Science and Technology Department of Hubei Province, China (2013BBA057 to CD); and a scholarship from China Scholarship Council (201206760022 to PZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology. 2008;23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- 2.Schmalbruch H, Lewis D. Dynamics of nuclei of muscle fibers and connective tissue cells in normal and denervated rat muscles. Muscle & nerve. 2000;23:617–626. doi: 10.1002/(sici)1097-4598(200004)23:4<617::aid-mus22>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 3.Kuang S, Rudnicki MA. The emerging biology of satellite cells and their therapeutic potential. Trends in molecular medicine. 2008;14:82–91. doi: 10.1016/j.molmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Cornelison D, Wold BJ. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Developmental biology. 1997;191:270–283. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- 5.Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates a mechanism for self-renewal? The Journal of cell biology. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes & Development. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 8.Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell stem cell. 2009;5:279–289. doi: 10.1016/j.stem.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Nguyen-McCarty M, Hexner EO, Danet-Desnoyers G, Klein PS. Maintenance of hematopoietic stem cells through regulation of Wnt and mTOR pathways. Nat Med. 2012;18:1778–1785. doi: 10.1038/nm.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun P, Quan Z, Zhang B, Wu T, Xi R. TSC1/2 tumour suppressor complex maintains Drosophila germline stem cells by preventing differentiation. Development. 2010;137:2461–2469. doi: 10.1242/dev.051466. [DOI] [PubMed] [Google Scholar]
- 11.Matsubara S, Ding Q, Miyazaki Y, Kuwahata T, Tsukasa K, Takao S. mTOR plays critical roles in pancreatic cancer stem cells through specific and stemness-related functions. Sci. Rep. 2013;3 doi: 10.1038/srep03230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentzinger CF, von Maltzahn J, Dumont NA, Stark DA, Wang YX, Nhan K, Frenette J, Cornelison D, Rudnicki MA. Wnt7a stimulates myogenic stem cell motility and engraftment resulting in improved muscle strength. The Journal of cell biology. 2014;205:97–111. doi: 10.1083/jcb.201310035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Maltzahn J, Bentzinger CF, Rudnicki MA. Wnt7a-Fzd7 signalling directly activates the Akt/mTOR anabolic growth pathway in skeletal muscle. Nature cell biology. 2012;14:186–191. doi: 10.1038/ncb2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodgers JT, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai C-R. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to GAlert. Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Risson V, Mazelin L, Roceri M, Sanchez H, Moncollin V, Corneloup C, Richard-Bulteau H, Vignaud A, Baas D, Defour A, Freyssenet D, Tanti JF, Le-Marchand-Brustel Y, Ferrier B, Conjard-Duplany A, Romanino K, Bauche S, Hantai D, Mueller M, Kozma SC, Thomas G, Ruegg MA, Ferry A, Pende M, Bigard X, Koulmann N, Schaeffer L, Gangloff YG. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. Journal of Cell Biology. 2009;187:859–874. doi: 10.1083/jcb.200903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung C-M, Calejman Camila M., Sanchez-Gurmaches J, Li H, Clish Clary B, Hettmer S, Wagers Amy J, Guertin David A. Rictor/mTORC2 Loss in the Myf5 Lineage Reprograms Brown Fat Metabolism and Protects Mice against Obesity and Metabolic Disease. Cell Reports. 2014;8:256–271. doi: 10.1016/j.celrep.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia JY, Casanova E, Costa CF, Brink M, Zorzato F, Hall MN, Ruegg MA. Skeletal Muscle-Specific Ablation of raptor, but Not of rictor, Causes Metabolic Changes and Results in Muscle Dystrophy. Cell Metabolism. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Shan T, Zhang P, Liang X, Bi P, Yue F, Kuang S. Lkb1 Is Indispensable for Skeletal Muscle Development, Regeneration, and Satellite Cell Homeostasis. STEM CELLS. 2014;32:2893–2907. doi: 10.1002/stem.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P, Shan T, Liang X, Deng C, Kuang S. Mammalian target of rapamycin is essential for cardiomyocyte survival and heart development in mice. Biochemical and biophysical research communications. 2014;452:53–59. doi: 10.1016/j.bbrc.2014.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S. mTOR Is Essential for Growth and Proliferation in Early Mouse Embryos and Embryonic Stem Cells. Molecular and Cellular Biology. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 22.Ge YJ, Wu AL, Warnes C, Liu JM, Zhang CB, Kawasome H, Terada N, Boppart MD, Schoenherr CJ, Chen J. mTOR regulates skeletal muscle regeneration in vivo through kinase-dependent and kinase-independent mechanisms. American Journal of Physiology-Cell Physiology. 2009;297:C1434–C1444. doi: 10.1152/ajpcell.00248.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira MG, Baptista IL, Carlassara EOC, Moriscot AS, Aoki MS, Miyabara EH. Leucine Supplementation Improves Skeletal Muscle Regeneration after Cryolesion in Rats. Plos One. 2014;9 doi: 10.1371/journal.pone.0085283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J, Li D, Wang F. Assessing the function of mTOR in human embryonic stem cells, mTOR. Springer. 2012:361–372. doi: 10.1007/978-1-61779-430-8_23. [DOI] [PubMed] [Google Scholar]
- 25.Ustanina S, Carvajal J, Rigby P, Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25:2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- 26.Montarras D, Lindon C, Pinset C, Domeyne P. Cultured myf5 null and myoD null muscle precursor cells display distinct growth defects. Biology of the Cell. 2000;92:565–572. doi: 10.1016/s0248-4900(00)01110-2. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama S, Ito Y, Ueno-Kudoh H, Shimizu H, Uchibe K, Albini S, Mitsuoka K, Miyaki S, Kiso M, Nagai A, Hikata T, Osada T, Fukuda N, Yamashita S, Harada D, Mezzano V, Kasai M, Puri PL, Hayashizaki Y, Okado H, Hashimoto M, Asahara H. A Systems Approach Reveals that the Myogenesis Genome Network Is Regulated by the Transcriptional Repressor RP58. Developmental Cell. 2009;17:836–848. doi: 10.1016/j.devcel.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun YT, Ge YJ, Dmevich J, Zhao Y, Band M, Chen J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. Journal of Cell Biology. 2010;189:1157–1169. doi: 10.1083/jcb.200912093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatfield I, Harvey I, Yates ER, Redd JR, Reiter LT, Bridges D. The role of TORC1 in muscle development in Drosophila. Sci. Rep. 2015:5. doi: 10.1038/srep09676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.