Abstract
Core binding factor (CBF) is a heterodimeric protein complex involved in the transcriptional regulation of normal hematopoiesis. Mutations in CBF-encoding genes result in leukemogenic proliferative advantages and impaired differentiation of the hematopoietic progenitors. CBF molecular aberrations are responsible for approximately 20% of all adult acute myeloid leukemia (AML). Although CBF-AMLs are considered to have relatively good prognosis compared to other leukemia subtypes, they are a heterogeneous group of disorders and modern therapy frequently leads to relapse and the associated morbidity and mortality. Improvements in risk stratification and development of targeted therapies are needed for better outcomes. In this review we provide a brief overview of the molecular basis, prognostic categories and the advanced treatment strategies for CBF leukemias.
Molecular basis of CBF leukemia
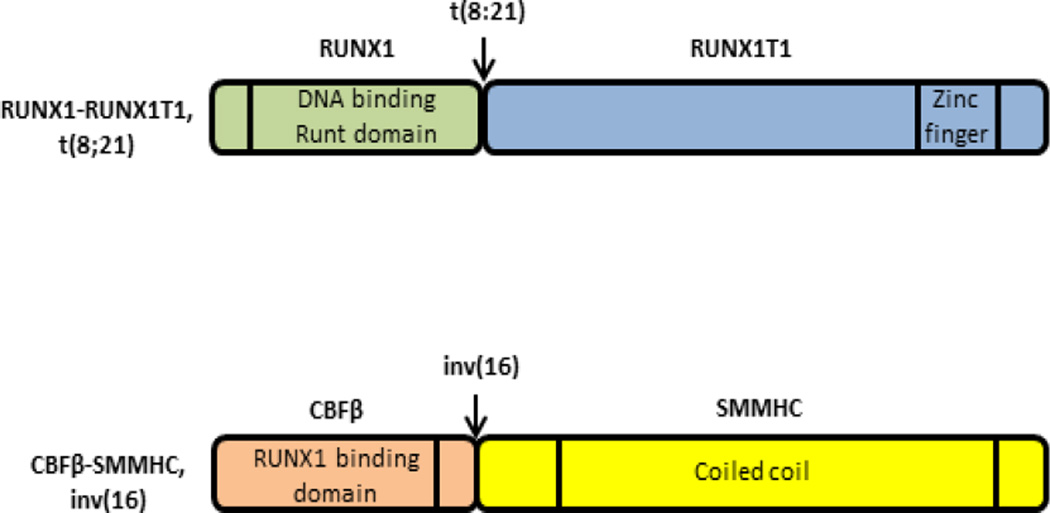
Leukemia is a cancer of the developing blood cells caused by mutations leading to either uncontrolled proliferation (class I) or lack of differentiation (class II) or both. The World Health Organization (WHO) classifies AML into several categories based on underlying genetic alterations to facilitate diagnosis and prognosis1. Recurrent genetic alterations are frequently observed in AML patients. Among them t(8;21)(q22;q22) and inv(16)(p13q22)/t(16;16)(p13q22) are the most common and result in generation of corresponding abnormal fusion genes RUNX1-RUNX1T1 and CBFB-MYH112, respectively (Figure 1).
Figure 1. Illustration of fusion protein products from CBF-AML associated mutations and the domains relevant for leukemogenesis.
Chromosomal aberrations t(8;21) and inv(16) in CBF-AML produce fusion proteins RUNX1-RUNX1T1 and CBFβ-SMMHC. RUNX1-RUNX1T1 lacks the transactivation domain but retains the DNA binding Runt domain of RUNX1, which is fused to the repressor domain of the RUNX1T1 protein. CBFβ-SMMHC retains the RUNX1 binding domain of CBFβ, which is fused with the coiled coil dimerizing domain of SMMHC.
Native RUNX1 and CBFβ form a heterodimeric transcription factor complex CBF that regulates normal hematopoietic ontogeny. Core binding factor is comprised of an alpha subunit and a beta subunit. There are 3 alpha subunits (RUNX1-3) and one beta subunit (CBFβ) identified to date. The alpha subunit binds to a consensus DNA sequence TGT/cGGT and the beta subunit stabilizes the interaction between the alpha subunit and DNA but does not interact with DNA independently3. Association of CBFβ induces a 40-fold increase in the DNA binding affinity of RUNX14. Therefore both subunits are required for maximum transcriptional efficiency of target genes downstream such as lymphocyte-specific protein tyrosine kinase, granulocyte-macrophage colony-stimulating factor-1 receptor, interlukin-3 and myeloperoxidase5. It has been found that RUNX1 also interacts with co-activators p300 and CREB binding protein to mediate transactivation6. Fetal mice null for Runx1 or Cbfb die of CNS hemorrhage and lack of fetal liver hematopoiesis on embryonic day 11.5–12.5, demonstrating that CBF is required for definitive hematopoiesis7–10.
The fusion gene CBFB-MYH11 was initially identified in 199311 and the corresponding fusion protein CBFβ-SMMHC (smooth muscle myosin heavy chain) was identified in inv(16) patient samples in 199612. CBFβ-SMMHC forms large nuclear aggegregates13, sequesters the alpha subunit RUNX1 in the cytoplasm14 and arrests differentiation of the inv(16) containing human cell line ME-115. The RUNX1 interacting N-terminal region of CBFβ and the myosin multimerizing C-terminal coiled coil domains of SMMHC direct this sequestration process16. CBFβ-SMMHC also prevents the ubiquitin-mediated proteosomal degradation of RUNX1 and generates a stable complex that dominantly inhibits normal CBF function17. The t(8;21) was first described in 197318 and the RUNX1-RUNX1T1 fusion gene was identified in 199219. The fusion protein product of t(8;21) is comprised of the DNA binding RUNT homology domain of RUNX1 and most of the RUNX1T1 (ETO) except the first 30 amino acids at the N-terminus20. The absence of the C-terminal transactivation domain in the fusion protein RUNX1-RUNX1T1 disrupts normal hematopoiesis in a dominant–negative fashion and therefore specific inactivation of this fusion induces differentiation of the t(8;21) positive Kasumi-1 cell line21. RUNX1-RUNX1T1 has also been shown to silence microRNA-193 resulting in increased leukemogenesis by increasing expression of histone deacetylases (HDAC), DNA-methytransferase1 (DNMT) and ultimately decreasing PTEN expression22. A common potential mechanism of both of these genetic fusion products is the dominant inhibitory effect on native RUNX1 and finally repression of target genes transcription, as mouse embryos heterozygous for RUNX1-RUNX1T1 or CBFB-MYH11 have almost identical phenotypes as the Runx1−/− or Cbfb−/− embryos regarding CNS hemorrhage and hematopoietic defects7,8,23,24.
Cooperating mutations in CBF leukemia
Murine knock-in models have demonstrated that both CBF fusion genes are necessary but not sufficient to cause leukemia and additional mutations are required for the pathogenesis of CBF leukemias2,25. Therefore in preclinical mouse models, mutagenic induction of second mutations are needed for development of AML2. In CBF leukemia patients, frequently detected second mutations are NPM1, c-KIT and FLT3. A study with 300 AML patients (16 to 60 years) showed that 48% of the patients have NPM1 mutations26. Another study with 481 AML patients indicated that 20% of the CBF-AML cytogenetic group had FLT3 mutations27. On the other hand, KIT mutations have been observed for 6.6–46.1% of CBF-AML patients28. NPM1 plays an important role in ribosomal protein assembly, transport, prevents aggregation of nuclear proteins and regulates transcriptional activity of p5326. Leukemogenesis occurs when cytoplasmic mutant NPM1 inactivates the tumor suppressor p19Arf in a p53 dependent or independent manner29. Inactivation of NF-kappaB renders CBF-AML with NPM1 mutation more sensitive to chemotherapy29. Genetic rearrangements that lead to constitutively active hematopoietic receptor tyrosine kinases (RTK) such as FLT3, c-KIT, JAK2 and RAS family members have been identified in CBF-AML patients2. These mutations may be particularly amenable for treatment with specific RTK inhibitors2. Haploinsufficieny of the tumor suppressors TLE1/4 in t(8;21) and overexpression of MN1 in inv(16) have been observed in addition to the epigenetic and posttranslational silencing of differentiation-inducing transcription factor CEBPA in CBF-AML30. There are case reports of rare cooperating mutations such as BCR-ABL14 and TEL-PDGFRβ15 fusion proteins in t(8;21) AML31. Both are examples of constitutively active tyrosine kinases that provide survival and proliferation advantages to progenitor cells without affecting their differentiation. The synergistic effects of these hyperproliferative phenotypes together with the CBF mutation-associated impaired differentiation lead to the multistep pathogenesis of AML (both class I and class II phenotypes).
Prognosis
Although the CBF genetic rearrangements in AML patients are reported to be associated with relatively favorable prognosis32,33, only 40–60% of adult CBF-AML patients exhibit long-term survival28. Additionally all treatment regimens are associated with significant relapse related morbidity and mortality34,35.
Molecularly defined genetic abnormalities are important prognostic factors in AML and important for patient management36. A study with 201 adults with de novo AML indicated the prognostic significance of karyotype on drug resistance, complete remission (CR) and overall survival (OS) at 5 years37. RUNX1-RUNX1T1 had the best 5 year OS of 50% and for CBFB-MYH11 the OS was 43%. Normal karyotype was associated with better prognosis in patients older than 55 years. Another study in the Medical Research Council (MRC) with 1612 patients including children and adults up to 55 years of age investigated the effect of pretreatment karyotype on prognosis and subsequent hematopoietic stem cell transplantation (HSCT) in first CR38. CBF mutations were found to have favorable outcomes without any differences between de novo and secondary AML in the pediatric group and the prognoses were maintained after HSCT in first CR. An additional MRC study with 1065 older patients (median age 66) indicated that inv(16) and t(8;21) are associated with superior CR, OS and lower drug resistance39. Together the findings from these studies suggest that cytogenetically distinct AML subsets are important for risk stratification and prognosis.
Presence of co-operative NPM1 mutation provides favorable overall survival (OS) after intensive double-induction and consolidation therapy only in the absence of FLT326 whereas c-KIT mutations have the worst outcome in CBF-AML patients with 56% relapse rate28,40. Though cooperating KIT mutation have not shown any significant effect on OS in inv(16), poorer OS has been observed for patients with t(8;21)41,42. Additional FLT3 mutations did not have any effect on the prognosis in CBF-AML27. A study by a Japanese group showed an adverse effect of CEBPA mutation on the OS of the patients with CBF-AML43. The prognostic impact of cooperating mutations on inv(16) and t(8;21) is listed in Table 1.
Table 1.
Cooperating mutations and prognosis in CBF AML
| Primary mutation | Cooperating mutation | OS | Reference |
|---|---|---|---|
| CBFB-MYH11 | |||
| No cooperating mutation | 43% | 37 | |
| c-KIT+ CBFB-MYH11 | 40% | 54 | |
| NPM1+ CBFB-MYH11 | Better prognosis | 26 | |
| FLT3+ CBFB-MYH11 | No additional effect | 25,27 | |
| N-RAS/K-RAS+ CBFB-MYH11 | No additional effect | 25 | |
| CEBPA+ CBFB-MYH11 | Poorer prognosis | 43 | |
| RUNX1-RUNX1T1 | |||
| No cooperating mutation | 50–60% | 37 | |
| c-KIT+ RUNX1-RUNX1T1 | 14–26% | 41,42 | |
| FLT3+ RUNX1-RUNX1T1 | No additional effect | 25,27 | |
| JAK2+ RUNX1-RUNX1T1 | No additional effect | 25 | |
| CEBPA+ RUNX1-RUNX1T1 | Poorer prognosis | 43 | |
The impact of cytogenetics was also studied in 848 AML patients between 15–83 years of age where patients less than 60 years of age in CR received allo- or auto-SCT44. Data indicated that inv(16) and t(8;21) were associated with favorable outcome and should be treated with an intensive regimen (idarubicin, cytosine arabinoside (Ara-C), etoposide (ICE) plus mitoxantrone and intermediate dose Ara-C) for longer disease free survival and allo-SCT should only be considered as salvage treatment for relapsed or refractory patients28,44.
However, a study with 144 adults with t(8;21) and 168 adults with inv(16) showed better OS and survival after first relapse for the inv(16) group when associated with trisomy 22 and male gender45. This study emphasized that specific features of inv(16) and t(8;21) should be consider separately.
According to French-American-British (FAB) classification t(8;21) is usually M2 (80–90%) but sometimes M1 (10%) whereas inv (16) is usually M4. The t(8;21) are more frequently found in younger and non-white patients. On the other hand inv(16) is often associated with secondary cytogenetic abnormalities, higher WBC and blast percentages and mostly found in patients with median age of 41 years28. The heterogeneity in clinical manifestation and response to treatments demands their consideration as separate entities.
Diagnosis
Several studies noted that the presence of CBF-AML fusion genes are independent indicators for achievement and duration of complete remission (CR) as well as overall survival rate46. Standard cytogenetic analysis can diagnose inv(16) and t(8;21) mutations in metaphase cells for CBF-AML patients. By this method t(8;21) can be easily detected with even suboptimal chromosome preparation whereas inv(16) is hard to detect and frequently misinterpreted as del(16)47.
To identify subtle rearrangement such as inv(16), an alternative reverse transcriptase-polymerase chain reaction (RT-PCR) based analysis is required. Unlike t(8;21) that produces a single transcript easily detectable by both processes, inv(16) can result in multiple variants of the CBFB-MYH11 fusion due to the presence of variable breakpoints in both CBFB and MYH1148. In CBF-AML patient samples these fusion genes have been detected without the presence of visible inv(16) and t(8;21). Therefore RT-PCR can detect most of these CBFB-MYH11 variants and is more sensitive in detecting CBF-AML than cytogenetic analysis48. Sometimes even RT-PCR cannot detect the inv(16) as confirmed by the classical southern blot techniques47. However false positives have not been documented during cytogenetic analysis in a study with 248 newly diagnosed adult primary AML patients and all but one patient was correctly identified47. On the other hand RT-PCR was associated with both false-negative and false-positive results and therefore should not replace cytogenetic analysis for CBF-AML diagnosis.
Cytogenetic analysis can be performed by conventional fluorescent in-situ hybridization (FISH) or by spectral karyotyping (SKY) for multicolor display of different chromosomes36. For these cytogenetic analyses, high quality preparation of chromosomes from the patient’s bone-marrow is desirable. Flurodeoxyuridine or methotrexate synchronization of bone marrow cells provide optimal chromosome length and increased yield of mitosis after culturing for 6–8 hours49. Subsequent cytogenetic analysis is usually performed in at least 20 metaphases according to the International System of Human Cytogenetic Nomenclature50. For accurate evaluation of CBF-AML, RT-PCR and FISH should be performed in conjunction with classical banding techniques regardless of phenotype. Recently developed microarray gene expression profiling (GEP) can also separate patients with inv(16) from patients with t(8;21).
A substantial number of AML patients die because of relapse and therefore evaluation of minimal residual disease (MRD) by RT-PCR is beneficial for proving complete eradication of leukemic blasts. Quantitative RT- PCR assays can efficiently identify the fusion transcripts for patients in early or long–term remission after conventional chemotherapy or HSCT and subsequently predict relapse risk based on critical MRD levels, especially for patients with t(8;21)30. Several recent publications report the importance of MRD monitoring during and after induction and consolidation therapy, through quantitative RT- PCR to detect the residual fusion transcripts in the bone marrow and peripheral blood, which are useful to predict relapse and OS51. Among them a study with 198 CBF-AML patients (age 18–60) indicated that prospective evaluation of MRD is more useful than identification of co-operative mutations for prognosis and treatment stratification to combat relapse52.
Treatment
Core binding factor leukemias are among the most frequent cytogenetic subtypes and comprise approximately 15% of all adult acute myeloid leukemias28. Although CBF-AML patients have better prognosis, only approximately 40–60% are cured by standard therapy using a backbone of high dose cytarabine treatment in combination with an anthracycline53,54 which is essentially unchanged for the past 40 years55. A study of 285 newly diagnosed patients with AML showed that high dose cytarabine treatment provides the best outcome for CBF-AML patients with 50% demonstrating CR after 5 years56. Another study reported a better outcome after intensive cytarabine therapy in t(8;21) but not in inv(16) positive patients57,58. A study at MD Anderson reported the potential of fludarabine and granulocyte colony-stimulating factor in augmenting the effectiveness of cytarabine against CBF-AML59.
Hematopoietic stem cell transplants (HSCT) and intensive chemotherapy are two well-practiced strategies to prevent relapse for AML patients in first remission. A study at MRC with 1063 AML patients (age under 55 years) showed that allogeneic transplantation after intensive chemotherapy reduced the relapse rate in CBF-AML patients60. Another study at MRC with 381 patients indicated that addition of auto-HSCT with four courses of intensive chemotherapy reduced the relapse rate and improved the OS for patients with inv(16) and t(8;21)61. A study with patients (up to 45 years) in complete remission (CR) showed that though there is no difference in disease free survival rate for patients with inv(16) and t(8;21), when allo-or auto-SCT were performed after intensive consolidation therapy, the OS was better for younger patients receiving allo-SCT62. HSCT is not necessary in first CR for patients with CBF leukemias unless they have relapsed, refractory or otherwise high risk disease54,63. However, patients older than 75 years have very poor prognosis and patients over 60 years may be considered for allo-HSCT28. The poor-prognostic KIT mutation positive CBF-AML patients are still treated with high dose cytarabine and should also be considered for allo-HCT28. For patients who go on to transplant and subsequently present with reduced donor chimerism, reduction of immunosuppression and/or donor lymphocyte infusion (DLI) can sometimes reinduce remission, although there may be risk of graft versus host disease (GVHD) with use of DLI64.
Susceptibility of leukemic cells to T-cell and natural killer cell-mediated immunosurveillance justifies the use of immunotherapy for preventing relapse. In addition to adoptive transfer of native and genetically modified T cells and NK cells, attempts have been made to sensitize AML cells to cytotoxic immune cells and to upregulate T-cell immunity by vaccination or cytokine treatment65,66. However cytokine storm is associated with some types of immunotherapy which may be mitigated by use of cytokine inhibitors or chimeric antigen receptors (CARs) containing engineered NK cells instead66. Immune escape is a potential mechanism by which cancer cells can evade immune surveillance after HSCT or other immunotherapies and ways to combat relapse through this mechanism are urgently needed67.
Approximately 40% of inv(16) and 70% of t(8;21) patients are diagnosed with secondary mutations and therefore treatment specific for those mutations are of particular interest. For example, as constitutively active RTKs are frequently found in patients with RUNX1-RUNX1T1 and CBFB-MYH11fusion genes2, RTK inhibitors such as dasatinib, imatinib and midostaurin can repress the hyperproliferation of leukemic blasts and warrant investigation as potential therapy for CBF-AML30,68. Epigenetic alterations for silencing gene function are often found in CBF-AML and therefore combination therapy with DNMT and HDAC inhibitors are now being used clinically to induce the expression of RUNX1 target genes69–71. A list of currently used drugs for CBF-AML treatment has been provided in Table 2.
Table 2.
Currently available treatment for CBF-AML
| Classification | Drugs | Mechanism of Action | References |
|---|---|---|---|
| Anti-metabolites | Cytarabine | Inhibit DNA and RNA polymerases | 44,53,56 |
| Anthracycline antibiotics | Doxorubicin, Daunorubicin | DNA intercalation | 53 |
| Podophyllotoxin | Etoposide | Inhibit topoisomerase II | 44 |
| DNA -methyltransferase inhibitor | Azacitidine, Decitabine | Inhibit DNA methylation | 71 |
| Histone deacetylase inhibitors | Vorinostat, Valproic acid | Inhibit Histone deacetylation | 30,69,70 |
| Kinase inhibitor | Midostaurin, Dasatinib | Inhibit protein kinases nonspecifically | 30,68 |
| Antibody-drug conjugate | Gemtuzumab ozogamicin (GO) | CD33 targeted DNA damage | 28 |
Mechanistic studies in mice as well as in vitro models have demonstrated that the CBF fusion proteins contribute to leukemogenesis through their interactions with their corresponding normal CBF binding partners (CBFβ for RUNX1-RUNX1T1, and RUNX1 for CBFβ-SMMHC)72,73. We have conducted a small chemical library screen for inhibitors of these interactions74. We identified a benzodiazepine compound, Ro5-3335, which was shown to be effective in suppressing CBF leukemia in animal models74. However further modifications to improve pharmacokinetics are necessary before successful clinical implementation.
Targeted therapy with gemtuzumab ozogamicin (GO), an anti-CD33 antibody conjugated to a calicheamicin derivative, has shown increased OS and reduced relapse rate and can be considered for good risk CBF-AML75. In addition, various tumor suppressors (let-7b/7c and microRNA-127) and myelopoietic microRNAs (microRNA-223) are found to be down regulated in RUNX1-RUNX1T1 and CBFB-MYH11 positive leukemic cells and those microRNAs may become potential therapeutic targets for CBF-AML21,30.
Conclusions
Despite favorable prognosis, modern therapy for CBF-AML is still associated with significant morbidity and mortality due to relapse34 and infection during intensive chemotherapy76. Cytogenetic characterization and discovery of oncogenic molecular events not only enhance our understanding of CBF-AML but also improve risk stratification and development of targeted therapies. Secondary cooperating mutations frequently serve as potential therapeutic targets and occasionally as additional prognostic factors for patients with inv(16) and t(8;21). Further studies are required to develop more effective and targeted therapy to achieve a 100% cure rate for CBF-AML patients.
Summary.
-
➢
CBF-AML is associated with t(8:21) or inv(16) which results in the abnormal fusion gene RUNX1-RUNX1T1 or CBFB-MYH11 respectively. These aberrant fusion genes lead to impaired differentiation of hematopoietic progenitors.
-
➢
Advanced FISH based cytogenetic characterization and RT-PCR based detection of CBF leukemia fusion genes are important for correct diagnosis and risk stratification of CBF-AML patients as well as for estimating minimal residual disease (MRD).
-
➢
CBF-AMLs are considered to have favorable prognosis but still only approximately 40–60% of patients are cured by standard therapy and relapse remains a major post-treatment complication.
-
➢
CBF-AML fusion genes are necessary but not sufficient to cause leukemia. Secondary cooperative mutations promote additional pathogenesis such as hyperproliferation and abrogated differentiation.
-
➢
Along with standard cytarabine-doxorubicin therapy, mutation specific targeted therapy, HSCT and immunotherapy hold promise for eradicating relapse and improving cure rates for CBF-AML.
Acknowledgments
Support: Supported by St. Jude Children’s Research Hospital, ALSAC and the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health
Abbreviations
- ABL
abelson murine leukemia viral oncogene homolog
- AML
acute myeloid leukemia
- BCR
breakpoint cluster region protein
- CAR
chimeric antigen receptors
- CBF
core binding factor
- CEBPA
CCAAT/enhancer-binding protein alpha
- CR
complete remission
- CREB
cAMP response element-binding protein
- DLI
donor lymphocyte infusion
- DNMT
DNA-methyltransferase
- FISH
fluorescent in-situ hybridization
- FLT3
Fms-like tyrosine kinase 3
- GO
gemtuzumab ozogamicin
- GVHD
graft versus host disease
- HDAC
histone deacetylases
- HSCT
hematopoietic stem cell transplantation
- JAK2
janus kinase 2
- MN1
meningioma (disrupted in balanced translocation) 1
- MRC
Medical Research Council
- MRD
minimal residual disease
- MYH11
myosin, heavy chain 11
- NF-kappaB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NK
natural killer
- NPM1
nucleophosmin1
- OS
overall survival
- p53
tumor protein p53
- PDGFRβ
beta-type platelet-derived growth factor receptor
- PTEN
phosphatase and tensin homolog
- RTK
receptor tyrosine kinases
- RT-PCR
reverse transcriptase-polymerase chain reaction
- RUNX1
runt-related transcription factor 1
- RUNX1T1
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
- SKY
spectral karyotyping
- SMMHC
smooth muscle myosin heavy chain
- TLE1/4
transducin-like enhancer protein 1/4
- WHO
World Health Organization
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Falini B, Tiacci E, Martelli MP, Ascani S, Pileri SA. New classification of acute myeloid leukemia and precursor-related neoplasms: changes and unsolved issues. Discov Med. 2010;10(53):281–292. [PubMed] [Google Scholar]
- 2.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2(7):502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Wang Q, Crute BE, et al. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13(6):3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu TL, Goetz TL, Graves BJ, Speck NA. Auto-inhibition and partner proteins, core-binding factor beta (CBFbeta) and Ets-1, modulate DNA binding by CBFalpha2 (AML1) Mol Cell Biol. 2000;20(1):91–103. doi: 10.1128/mcb.20.1.91-103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaiman AL, Lewis AF, Crute BE, Speck NA, Lenz J. Transcriptional activity of core binding factor-alpha (AML1) and beta subunits on murine leukemia virus enhancer cores. J Virol. 1995;69(5):2898–2906. doi: 10.1128/jvi.69.5.2898-2906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17(11):2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Stacy T, Binder M, et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93(8):3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, Stacy T, Miller JD, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87(4):697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki K, Yagi H, Bronson RT, et al. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci U S A. 1996;93(22):12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 11.Liu P, Tarle SA, Hajra A, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261(5124):1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 12.Liu PP, Wijmenga C, Hajra A, et al. Identification of the chimeric protein product of the CBFB-MYH11 fusion gene in inv(16) leukemia cells. Genes Chromosomes Cancer. 1996;16(2):77–87. doi: 10.1002/(SICI)1098-2264(199606)16:2<77::AID-GCC1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Wijmenga C, Gregory PE, Hajra A, et al. Core binding factor beta-smooth muscle myosin heavy chain chimeric protein involved in acute myeloid leukemia forms unusual nuclear rod-like structures in transformed NIH 3T3 cells. Proc Natl Acad Sci U S A. 1996;93(4):1630–1635. doi: 10.1073/pnas.93.4.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol. 1998;18(12):7432–7443. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanno Y, Kanno T, Sakakura C, Bae SC, Ito Y. Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor alpha (CBFalpha) subunit by the leukemia-related PEBP2/CBFbeta-SMMHC fusion protein inhibits PEBP2/CBF-mediated transactivation. Mol Cell Biol. 1998;18(7):4252–4261. doi: 10.1128/mcb.18.7.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kundu M, Liu PP. Function of the inv(16) fusion gene CBFB-MYH11. Curr Opin Hematol. 2001;8(4):201–205. doi: 10.1097/00062752-200107000-00004. [DOI] [PubMed] [Google Scholar]
- 17.Huang G, Shigesada K, Ito K, et al. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20(4):723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowley JD. Identificaton of a translocation with quinacrine fluorescence in a patient with acute leukemia. Ann Genet. 1973;16(2):109–112. [PubMed] [Google Scholar]
- 19.Erickson P, Gao J, Chang KS, et al. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80(7):1825–1831. [PubMed] [Google Scholar]
- 20.Miyoshi H, Kozu T, Shimizu K, et al. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12(7):2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcucci G, Caligiuri MA, Bloomfield CD. Molecular and clinical advances in core binding factor primary acute myeloid leukemia: a paradigm for translational research in malignant hematology. Cancer Invest. 2000;18(8):768–780. doi: 10.3109/07357900009012209. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Gao L, Luo X, et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121(3):499–509. doi: 10.1182/blood-2012-07-444729. [DOI] [PubMed] [Google Scholar]
- 23.Castilla LH, Wijmenga C, Wang Q, et al. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87(4):687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 24.Yergeau DA, Hetherington CJ, Wang Q, et al. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15(3):303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 25.Renneville A, Roumier C, Biggio V, et al. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008;22(5):915–931. doi: 10.1038/leu.2008.19. [DOI] [PubMed] [Google Scholar]
- 26.Dohner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106(12):3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 27.Santos FP, Jones D, Qiao W, et al. Prognostic value of FLT3 mutations among different cytogenetic subgroups in acute myeloid leukemia. Cancer. 2011;117(10):2145–2155. doi: 10.1002/cncr.25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solh M, Yohe S, Weisdorf D, Ustun C. Core-binding factor acute myeloid leukemia: Heterogeneity, monitoring, and therapy. Am J Hematol. 2014;89(12):1121–1131. doi: 10.1002/ajh.23821. [DOI] [PubMed] [Google Scholar]
- 29.Hatzimichael E, Georgiou G, Benetatos L, Briasoulis E. Gene mutations and molecularly targeted therapies in acute myeloid leukemia. Am J Blood Res. 2013;3(1):29–51. [PMC free article] [PubMed] [Google Scholar]
- 30.Mrozek K, Marcucci G, Paschka P, Bloomfield CD. Advances in molecular genetics and treatment of core-binding factor acute myeloid leukemia. Curr Opin Oncol. 2008;20(6):711–718. doi: 10.1097/CCO.0b013e32831369df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojima K, Yasukawa M, Ishimaru F, et al. Additional translocation (8;21)(q22;q22) in a patient with Philadelphia-positive chronic myelogenous leukaemia in the blastic phase. Br J Haematol. 1999;106(3):720–722. doi: 10.1046/j.1365-2141.1999.01588.x. [DOI] [PubMed] [Google Scholar]
- 32.von Neuhoff C, Reinhardt D, Sander A, et al. Prognostic impact of specific chromosomal aberrations in a large group of pediatric patients with acute myeloid leukemia treated uniformly according to trial AML-BFM 98. J Clin Oncol. 2010;28(16):2682–2689. doi: 10.1200/JCO.2009.25.6321. [DOI] [PubMed] [Google Scholar]
- 33.Appelbaum FR, Kopecky KJ, Tallman MS, et al. The clinical spectrum of adult acute myeloid leukaemia associated with core binding factor translocations. Br J Haematol. 2006;135(2):165–173. doi: 10.1111/j.1365-2141.2006.06276.x. [DOI] [PubMed] [Google Scholar]
- 34.Hospital MA, Prebet T, Bertoli S, et al. Core-binding factor acute myeloid leukemia in first relapse: a retrospective study from the French AML Intergroup. Blood. 2014;124(8):1312–1319. doi: 10.1182/blood-2014-01-549212. [DOI] [PubMed] [Google Scholar]
- 35.Paschka P, Dohner K. Core-binding factor acute myeloid leukemia: can we improve on HiDAC consolidation? Hematology Am Soc Hematol Educ Program. 2013;2013:209–219. doi: 10.1182/asheducation-2013.1.209. [DOI] [PubMed] [Google Scholar]
- 36.Mrozek K, Heinonen K, Bloomfield CD. Prognostic value of cytogenetic findings in adults with acute myeloid leukemia. Int J Hematol. 2000;72(3):261–271. [PubMed] [Google Scholar]
- 37.Dastugue N, Payen C, Lafage-Pochitaloff M, et al. Prognostic significance of karyotype in de novo adult acute myeloid leukemia. The BGMT group. Leukemia. 1995;9(9):1491–1498. [PubMed] [Google Scholar]
- 38.Grimwade D, Walker H, Oliver F, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92(7):2322–2333. [PubMed] [Google Scholar]
- 39.Grimwade D, Walker H, Harrison G, et al. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001;98(5):1312–1320. doi: 10.1182/blood.v98.5.1312. [DOI] [PubMed] [Google Scholar]
- 40.Paschka P, Marcucci G, Ruppert AS, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J Clin Oncol. 2006;24(24):3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 41.Manara E, Bisio V, Masetti R, et al. Core-binding factor acute myeloid leukemia in pediatric patients enrolled in the AIEOP AML 2002/01 trial: screening and prognostic impact of c-KIT mutations. Leukemia. 2014;28(5):1132–1134. doi: 10.1038/leu.2013.339. [DOI] [PubMed] [Google Scholar]
- 42.Park SH, Chi HS, Min SK, et al. Prognostic impact of c-KIT mutations in core binding factor acute myeloid leukemia. Leuk Res. 2011;35(10):1376–1383. doi: 10.1016/j.leukres.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 43.Matsuo H, Kajihara M, Tomizawa D, et al. Prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia: a report from the Japanese Pediatric Leukemia/Lymphoma Study Group. Blood Cancer J. 2014;4:e226. doi: 10.1038/bcj.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visani G, Bernasconi P, Boni M, et al. The prognostic value of cytogenetics is reinforced by the kind of induction/consolidation therapy in influencing the outcome of acute myeloid leukemia--analysis of 848 patients. Leukemia. 2001;15(6):903–909. doi: 10.1038/sj.leu.2402142. [DOI] [PubMed] [Google Scholar]
- 45.Marcucci G, Mrozek K, Ruppert AS, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23(24):5705–5717. doi: 10.1200/JCO.2005.15.610. [DOI] [PubMed] [Google Scholar]
- 46.Strout MP, Marcucci G, Caligiuri MA, Bloomfield CD. Core-binding factor (CBF) and MLL-associated primary acute myeloid leukemia: biology and clinical implications. Ann Hematol. 1999;78(6):251–264. doi: 10.1007/s002770050511. [DOI] [PubMed] [Google Scholar]
- 47.Mrozek K, Prior TW, Edwards C, et al. Comparison of cytogenetic and molecular genetic detection of t(8;21) and inv(16) in a prospective series of adults with de novo acute myeloid leukemia: a Cancer and Leukemia Group B Study. J Clin Oncol. 2001;19(9):2482–2492. doi: 10.1200/JCO.2001.19.9.2482. [DOI] [PubMed] [Google Scholar]
- 48.Mitterbauer M, Laczika K, Novak M, et al. High concordance of karyotype analysis and RT-PCR for CBF beta/MYH11 in unselected patients with acute myeloid leukemia. A single center study. Am J Clin Pathol. 2000;113(3):406–410. doi: 10.1309/D94U-351N-HT3D-F1F3. [DOI] [PubMed] [Google Scholar]
- 49.Webber LM, Garson OM. Fluorodeoxyuridine synchronization of bone marrow cultures. Cancer Genet Cytogenet. 1983;8(2):123–132. doi: 10.1016/0165-4608(83)90044-4. [DOI] [PubMed] [Google Scholar]
- 50.Raimondi SC, Chang MN, Ravindranath Y, et al. Chromosomal abnormalities in 478 children with acute myeloid leukemia: clinical characteristics and treatment outcome in a cooperative pediatric oncology group study-POG 8821. Blood. 1999;94(11):3707–3716. [PubMed] [Google Scholar]
- 51.Hourigan CS, Karp JE. Minimal residual disease in acute myeloid leukaemia. Nat Rev Clin Oncol. 2013;10(8):460–471. doi: 10.1038/nrclinonc.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jourdan E, Boissel N, Chevret S, et al. Prospective evaluation of gene mutations and minimal residual disease in patients with core binding factor acute myeloid leukemia. Blood. 2013;121(12):2213–2223. doi: 10.1182/blood-2012-10-462879. [DOI] [PubMed] [Google Scholar]
- 53.Burnett AK. New induction and postinduction strategies in acute myeloid leukemia. Curr Opin Hematol. 2012;19(2):76–81. doi: 10.1097/MOH.0b013e3283500a92. [DOI] [PubMed] [Google Scholar]
- 54.Wolach O, Stone RM. Is it time to change conventional consolidation chemotherapy for acute myeloid leukemia in CR1? Curr Opin Hematol. 2015;22(2):123–131. doi: 10.1097/MOH.0000000000000119. [DOI] [PubMed] [Google Scholar]
- 55.Yates JW, Wallace HJ, Jr, Ellison RR, Holland JF. Cytosine arabinoside (NSC-63878) and daunorubicin (NSC-83142) therapy in acute nonlymphocytic leukemia. Cancer Chemother Rep. 1973;57(4):485–488. [PubMed] [Google Scholar]
- 56.Bloomfield CD, Lawrence D, Byrd JC, et al. Frequency of prolonged remission duration after high-dose cytarabine intensification in acute myeloid leukemia varies by cytogenetic subtype. Cancer Res. 1998;58(18):4173–4179. [PubMed] [Google Scholar]
- 57.Nguyen S, Leblanc T, Fenaux P, et al. A white blood cell index as the main prognostic factor in t(8;21) acute myeloid leukemia (AML): a survey of 161 cases from the French AML Intergroup. Blood. 2002;99(10):3517–3523. doi: 10.1182/blood.v99.10.3517. [DOI] [PubMed] [Google Scholar]
- 58.Delaunay J, Vey N, Leblanc T, et al. Prognosis of inv(16)/t(16;16) acute myeloid leukemia (AML): a survey of 110 cases from the French AML Intergroup. Blood. 2003;102(2):462–469. doi: 10.1182/blood-2002-11-3527. [DOI] [PubMed] [Google Scholar]
- 59.Borthakur G, Kantarjian H, Wang X, et al. Treatment of core-binding-factor in acute myelogenous leukemia with fludarabine, cytarabine, and granulocyte colony-stimulating factor results in improved event-free survival. Cancer. 2008;113(11):3181–3185. doi: 10.1002/cncr.23927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burnett AK, Wheatley K, Goldstone AH, et al. The value of allogeneic bone marrow transplant in patients with acute myeloid leukaemia at differing risk of relapse: results of the UK MRC AML 10 trial. Br J Haematol. 2002;118(2):385–400. doi: 10.1046/j.1365-2141.2002.03724.x. [DOI] [PubMed] [Google Scholar]
- 61.Burnett AK, Goldstone AH, Stevens RM, et al. Randomised comparison of addition of autologous bone-marrow transplantation to intensive chemotherapy for acute myeloid leukaemia in first remission: results of MRC AML 10 trial. UK Medical Research Council Adult and Children's Leukaemia Working Parties. Lancet. 1998;351(9104):700–708. doi: 10.1016/s0140-6736(97)09214-3. [DOI] [PubMed] [Google Scholar]
- 62.Willemze R, Suciu S, Mandelli F, et al. Autologous versus allogeneic stem cell transplantation in acute myeloid leukemia. Ann Hematol. 2004;83(Suppl 1):S134. doi: 10.1007/s00277-004-0850-2. [DOI] [PubMed] [Google Scholar]
- 63.Ossenkoppele G, Lowenberg B. How I treat the older patient with acute myeloid leukemia. Blood. 2015;125(5):767–774. doi: 10.1182/blood-2014-08-551499. [DOI] [PubMed] [Google Scholar]
- 64.Rujkijyanont P, Morris C, Kang G, et al. Risk-adapted donor lymphocyte infusion based on chimerism and donor source in pediatric leukemia. Blood Cancer J. 2013;3:e137. doi: 10.1038/bcj.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barrett AJ, Le Blanc K. Immunotherapy prospects for acute myeloid leukaemia. Clin Exp Immunol. 2010;161(2):223–332. doi: 10.1111/j.1365-2249.2010.04197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tettamanti S, Magnani CF, Biondi A, Biagi E. Acute myeloid leukemia and novel biological treatments: monoclonal antibodies and cell-based gene-modified immune effectors. Immunol Lett. 2013;155(1–2):43–46. doi: 10.1016/j.imlet.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 67.Vago L, Perna SK, Zanussi M, et al. Loss of mismatched HLA in leukemia after stem-cell transplantation. N Engl J Med. 2009;361(5):478–488. doi: 10.1056/NEJMoa0811036. [DOI] [PubMed] [Google Scholar]
- 68.Stone RM, Fischer T, Paquette R, et al. Phase IB study of the FLT3 kinase inhibitor midostaurin with chemotherapy in younger newly diagnosed adult patients with acute myeloid leukemia. Leukemia. 2012;26(9):2061–2068. doi: 10.1038/leu.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fredly H, Gjertsen BT, Bruserud O. Histone deacetylase inhibition in the treatment of acute myeloid leukemia: the effects of valproic acid on leukemic cells, and the clinical and experimental evidence for combining valproic acid with other antileukemic agents. Clin Epigenetics. 2013;5(1):12. doi: 10.1186/1868-7083-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schaefer EW, Loaiza-Bonilla A, Juckett M, et al. A phase 2 study of vorinostat in acute myeloid leukemia. Haematologica. 2009;94(10):1375–1382. doi: 10.3324/haematol.2009.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomas X. DNA methyltransferase inhibitors in acute myeloid leukemia: discovery, design and first therapeutic experiences. Expert Opin Drug Discov. 2012;7(11):1039–1051. doi: 10.1517/17460441.2012.722618. [DOI] [PubMed] [Google Scholar]
- 72.Roudaia L, Cheney MD, Manuylova E, et al. CBFbeta is critical for AML1-ETO and TEL-AML1 activity. Blood. 2009;113(13):3070–3079. doi: 10.1182/blood-2008-03-147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shigesada K, van de Sluis B, Liu PP. Mechanism of leukemogenesis by the inv(16) chimeric gene CBFB/PEBP2B-MHY11. Oncogene. 2004;23(24):4297–4307. doi: 10.1038/sj.onc.1207748. [DOI] [PubMed] [Google Scholar]
- 74.Cunningham L, Finckbeiner S, Hyde RK, et al. Identification of benzodiazepine Ro5-3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFbeta interaction. Proc Natl Acad Sci U S A. 2012;109(36):14592–14597. doi: 10.1073/pnas.1200037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hills RK, Castaigne S, Appelbaum FR, et al. Addition of gemtuzumab ozogamicin to induction chemotherapy in adult patients with acute myeloid leukaemia: a meta-analysis of individual patient data from randomised controlled trials. Lancet Oncol. 2014;15(9):986–996. doi: 10.1016/S1470-2045(14)70281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Creutzig U, Zimmermann M, Bourquin JP, et al. Randomized trial comparing liposomal daunorubicin with idarubicin as induction for pediatric acute myeloid leukemia: results from Study AML-BFM 2004. Blood. 2013;122(1):37–43. doi: 10.1182/blood-2013-02-484097. [DOI] [PubMed] [Google Scholar]