Drug-resistant forms of Mycobacterium tuberculosis (Mtb) are a major public health threat. Genetic approaches (e.g., RNA profiling) can facilitate the development of new drugs to combat drug-resistant Mtb.
Abstract
Very few chemically novel agents have been approved for antibacterial chemotherapies during the last 50 yr. Yet new antibacterial drugs are needed to reduce the impact on global health of an increasing number of drug-resistant infections, including highly drug-resistant forms of tuberculosis. This review discusses how genetic approaches can be used to study the mechanism of action of whole-cell screening hits and facilitate target-driven strategies for antimicrobial drug development.
Many classes of antibiotics in clinical use today stem from the golden age of antibiotic discovery, which had its most productive period from 1940 to 1960 (Davies 2006; Silver 2011). These drugs are one of the reasons why most bacterial infections can be cured with a few pills, generally without side effects, within a couple of weeks. It is thus easy to forget that bacterial infections have remained a major cause of preventable deaths in developing countries and continue to exert a profound impact on human health. The relative paucity of novel antibacterials discovered after 1960 coincided with the appearance and contributed to the spread of drug-resistant bacterial infections. When resistant to not only one, but multiple drugs, such infections threaten to erode a cornerstone of modern health care.
Tuberculosis (TB) is a prime example of how treatment has succeeded in developed countries while meeting frequent failure in the developing nations. Treatment failure contributed to the reemergence of a thought-to-be-conquered disease that now constitutes as a major public health threat. First-line treatment of TB consists of drugs originally identified during the golden age of antibiotic discovery. Together with social changes, these drugs contributed to a drastic reduction of TB cases in Europe and North America while continuing to kill millions in Africa and Asia. Drug-resistant Mycobacterium tuberculosis (Mtb) appeared shortly after the introduction of TB chemotherapy and evolved into extremely drug-resistant Mtb, which threatens the TB control programs worldwide. Therefore, the need for new therapies for treatment of this deadly disease remains unchanged since the pre-antibiotic era. This review discusses how genetic approaches can facilitate the development of drugs for such new therapies.
GENETICS AND CLASSICAL WHOLE-CELL SCREENING
Whole-cell screening (WCS)-based antibacterial drug discovery begins with identifying small molecules (hits) that inhibit bacterial growth. This inhibitor-first approach provided the foundation of the golden age of antibacterial drug discovery and remains the only development strategy that has delivered antibacterial drugs to the clinic (Brotz-Oesterhelt and Sass 2010). This includes all first-in-class drug candidates currently under clinical development for the treatment of TB (Lechartier et al. 2014). The superior success rate of inhibitor-first strategies is not unique to antibacterial drug development as phenotypic screening has been more productive than target-driven approaches across a spectrum of diseases (Swinney and Anthony 2011).
Several features contribute to the success of classical WCS. These screens allow for selective investment into the chemical structures that can overcome the major challenge of antibacterial drug development: ability to penetrate the bacterial cell envelope. Furthermore, WCS provides an opportunity to discover compounds that inhibit growth by diverse mechanisms, including those that engage multiple targets. The drawback is that the mechanism of all hits remains to be determined.
Whereas it is not required to understand a drug candidate’s mechanism of action (MOA) to obtain regulatory approval for use in humans, knowing how a compound exerts its biological function during development is desirable for several reasons. First, many WCS hits act in a nonspecific manner (e.g., as alkylating agents or detergents) and therefore cannot be developed into drugs (Payne et al. 2007; Roemer and Boone 2013). Second, not all hits that are toxic to the host have to be discarded as toxicity need not necessarily result from a compound’s antibacterial activity. Antibacterial activity and toxicity are linked if they both stem from inhibition of the same target, for example, an enzyme that is conserved in both the pathogen and the host. Such compounds are not suited for further development. However, in cases in which the mechanism of toxicity differs from that of bacterial growth inhibition, chemical modifications can sometimes eliminate a compound’s toxicity without compromising its antibacterial activity. Third, after identifying the target, the structure activity relationship (SAR) of the compound can be explored in more detail. Fourth, and perhaps most importantly, the antibacterial MOA of a compound is a determinant of its in vivo potency. This is exemplified by recently identified pyrimidine-imidazoles, which were active against Mtb in vitro, had appropriate pharmacokinetic properties, but showed no potency in Mtb-infected mice (Pethe et al. 2013). The activity of these molecules relied on the presence of glycerol, which is used in many standard liquid media, but it is not a physiological carbon source. Therefore, lack of in vivo potency in these pyrimidine-imidazoles was mechanism-based, leaving no path to develop these compounds further.
The potential for discovering other compounds with MOAs that intrinsically have no in vivo potency (here referred to as novip compounds) is significant because screening media can, at best, only mimic some of the environments encountered during an infection. More often, they create a physiological context that is quite different from that within the host as many standard media contain nutrients Mtb cannot obtain from the host in the provided form (e.g., iron) (Rodriguez and Smith 2006), cannot obtain from the host at all (e.g., biotin) (Sassetti and Rubin 2003; Woong Park et al. 2011), and lack nutrients that Mtb is apparently able to scavenge from the host (e.g., NAD precursors) (Boshoff et al. 2008). These differences can cause some mutations to either result in death of Mtb in vitro but not during infections (e.g., mutations in nadA [Boshoff et al. 2008]) or prevent growth during infections but not in standard media (e.g., mutations required to scavenge iron from the host [Rodriguez and Smith 2006]). One conceptually straightforward strategy to reduce the risk of identifying novip compounds is to screen against intracellular Mtb (Christophe et al. 2009). However, such screening procedures can also reveal genes that are required ex vivo but not during an infection (Munoz-Elias et al. 2006). Furthermore, Mtb induces a complex and dynamic pathology and can be found in multiple different intracellular and extracellular locations (Barry et al. 2009; Dartois and Barry 2013). Some of these locations permit the pathogen to replicate, whereas others may restrict its growth and yet allow its persistence. Therefore, it seems impossible to design screening conditions that mimic the pathophysiological context of an Mtb infection in humans well enough to completely bias against novips. The best insurance against MOA-based attrition is therefore a solid understanding of a compound’s MOA.
Genetic Strategies for Studying the MOA of Bioactive Small Molecules' RNA Profiling
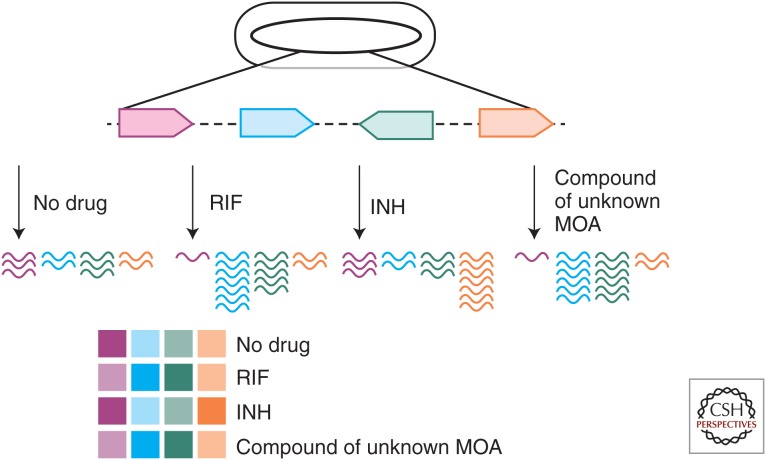
A cell’s genome-wide RNA profile is defined by the abundance of the total RNA produced at a given time. Taking snapshots of these profiles became straightforward after DNA microarrays were invented (Schena et al. 1995; Lashkari et al. 1997), and they can now be reported with even higher resolution by sequencing total RNA extracts (Nagalakshmi et al. 2008; Wilhelm et al. 2008; Arnvig and Young 2012). The foremost study to show the use of genome-wide RNA profiling for characterizing the MOA of bioactive molecules was performed with the anesthetic dyclonine (Hughes et al. 2000). Hughes et al. collected Saccharomyces cerevisiae RNA profiles induced by hundreds of different genetic or chemical treatments. This generated a compendium of reference profiles to which the RNA profile induced by dyclonine could be compared. Pattern matching algorithms revealed that dyclonine caused changes similar to those induced by mutations in the ergosterol pathway. This suggested that dyclonine inhibited sterol biosynthesis and follow-up experiments confirmed the sterol isomerase Erg2p as a target of dyclonine. This pattern matching approach, as illustrated in Figure 1, has since been used to uncover targets and the MOA of many other antimicrobial compounds (Brazas and Hancock 2005; Smith et al. 2010; Wecke and Mascher 2011). Indeed, Mtb microarrays were first used to study the effect of isoniazid on the RNA profile of Mtb (Wilson et al. 1999). Hundreds of compound-induced RNA profiles were reported 5 yr later in a landmark study by Boshoff et al. (2004). These profiles helped to investigate a variety of antimycobacterial small molecules, including capreomycin (Fu and Shinnick 2007), isoniazid (Karakousis et al. 2008), PA-824 (Manjunatha et al. 2009), an inhibitor of menaquinone production (Dhiman et al. 2009), β-lactams (Slayden and Belisle 2009), vancomycin (Provvedi et al. 2009), the benzothioazinone, BTZ043 (Makarov et al. 2009), the natural product chelerythrine (Liang et al. 2011), thioridazine (Thorsing et al. 2013), and several other WCS hits (Stanley et al. 2013; Wang et al. 2013).
Figure 1.
RNA profiling. RNA molecules produced by four genes of the Mtb genome with and without drug treatment are represented as colored lines. The color of the squares depicted below the RNA molecules reflects their abundance, with darker colors identifying more abundant RNAs. The fictional data here would suggest that the compound of unknown MOA functions in a manner similar to RIF.
The principal limitations of RNA profiling are twofold. First, these profiles fail to identify the direct target of a compound as they only reveal the biological pathway(s) that respond to the drug. In part, this is because inhibition of different proteins can produce similar RNA profiles, such as in situations in which the proteins form a complex or are required for the same metabolic pathway. Second, the profiles (and other pattern matching approaches) are most informative for compounds, which induce a signature similar to the compounds with a known MOA. Specific predictions are difficult to make for compounds with an entirely novel MOA as appropriate reference profiles are lacking in such cases. This drawback can be overcome by enriching the reference compendium with RNA profiles from genetically defined mutants (Hughes et al. 2000; Mnaimneh et al. 2004; Freiberg et al. 2005). Unfortunately, too few such profiles are available for Mtb to apply this principle to facilitate the analysis of antimycobacterial compounds.
Even for relatively small bacterial genomes, genome-wide RNA profiles consist of thousands of individual measurements. This complexity offers the potential to generate unique signatures for many bioactive small molecules while making it difficult to identify the most informative RNAs. The amount of data generated by RNA profiling is not only large, but more importantly, the biological responses to growth perturbation are often complex. This foremost applies to RNA profiles collected at time points or at drug concentrations that impact the abundance of hundreds of RNAs. The nonspecific stress responses contained in such profiles can mask a compound-specific response. Reducing the treatment time and/or drug concentration often simplifies the analysis of bioactive molecules by RNA profiling. Furthermore, the ability of genome-wide RNA analyses to detect minute changes in any single gene coupled with a highly dynamic transcriptional response prevalent in bacteria increases the existing complexity by severalfold. Therefore, obtaining very similar RNA profiles in two apparently identical experiments can be a challenge. Description of all experimental details, strict standardization, and avoidance of procedural changes of gene expression (as can occur, for example, by centrifugation of live Mtb) are therefore crucial for producing informative RNA profiles.
Selection and Identification of Resistance Mutations
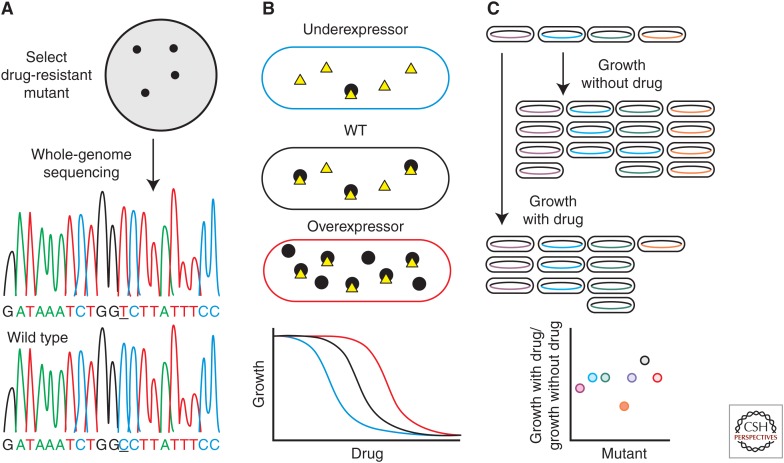
The first indication that bedaquiline (BDQ, formerly known as TMC207 and originally published as R207910) prevents the growth of Mtb by inhibiting the adenosine triphosphate (ATP) synthase was provided by the mutants selected for their ability to grow in the presence of BDQ (Andries et al. 2005). Whole-genome sequencing of these BDQ-resistant strains revealed point mutations in atpE, which encodes subunit c of ATP synthase. Genetic and biochemical studies further established that BDQ inhibits ATP synthesis by binding to AtpE, thus confirming ATP synthase as a direct target (Koul et al. 2007). Selecting for resistance followed by whole-genome sequencing (Fig. 2A) has identified mutations that permit growth in the presence of several other small molecules with antimycobacterial activities. Examples include mutations in genes involved in respiration (qcrB) (Abrahams et al. 2012b; Pethe et al. 2013), fatty acid synthesis (inhA) (Hartkoorn et al. 2012), protein synthesis (aspS) (Ioerger et al. 2013), protein secretion (eccB3) (Ioerger et al. 2013), polyketide biosynthesis (pks13) (Ioerger et al. 2013; Wilson et al. 2013), mycolic acid transport (mmpL3) (Grzegorzewicz et al. 2012; La Rosa et al. 2012; Stanley et al. 2012; Tahlan et al. 2012; Ioerger et al. 2013; Remuinan et al. 2013), and arabinogalactan synthesis (dprE1) (Christophe et al. 2009; Makarov et al. 2009; Magnet et al. 2010; Stanley et al. 2012; Wang et al. 2013).
Figure 2.
Forward and reverse genetic approaches to investigate small molecule MOAs. (A) Selection of resistant mutants. Mutants that are resistant to the compound are selected on agar plates supplemented with the compound of interest. The mutations present in these resistant isolates are identified by whole-genome sequencing. (B) Under/over-expression. Reducing or increasing the expression of a candidate target (black circles) can either decrease or increase the MIC of a compound (yellow triangle) inhibiting the target. (C) Phenotyping pooled mutants. Mutants are labeled with a genetic barcode and their fitness is analyzed in a competitive growth experiment. This principle can be applied to deletion mutants, underexpressors, overexpressors, or a mix of these strains. The orange-label mutant shows increased sensitivity to the tested drug; the black-labeled mutant (which is only shown in the graph) shows decreased sensitivity.
Resistance mutations can directly identify high-value drug targets. Selection and identification of such mutations requires no target-specific assay development, can be fast, and is comprehensive as it surveys the entire genome. However, this approach is not without limitations and fails when a resistant strain cannot be obtained. Furthermore, alteration of the target responsible for the growth inhibitory effect of a small molecule constitutes only one of the several mechanisms by which mutations can impart resistance. Alternatively, methods such as chemical inactivation, efflux, or failure to transform an inactive prodrug into its active derivative can form the basis of resistance. The knowledge of mutation frequencies that cause resistance by these mechanisms proves highly useful to determine the value of a compound. A high frequency of resistance owing to such mechanisms significantly devalues the compound. However, this knowledge rarely plays a role in defining a compound’s MOA. Nevertheless, in scenarios in which a mutation can be mapped to a true target, resistance mutation provides one of the simplest ways to define a compound’s MOA.
Linking Small Molecules to Candidate Targets by Increasing or Reducing Gene Activity
To be directly involved in the MOA of a bioactive small molecule, a protein has to physically interact with that small molecule. That a candidate protein has the potential for such an interaction within live bacteria can be confirmed indirectly by showing an immediate correlation between the protein’s abundance and the minimal inhibitor concentration required to inhibit growth (MIC) (Fig. 2B). This type of experiment has helped to clarify the MOA of INH (isoniazid), which was debated to impair one of two enzymes involved in fatty acid biosynthesis, InhA or KasA. Strains that overexpressed these proteins were analyzed for their susceptibilities to INH and the KasA inhibitor thiolactomycin (TLM). Overexpression of InhA increased the MIC of INH but did not change the MIC of TLM; overexpression of KasA had the opposite effect (Larsen et al. 2002). These results supported the view that InhA and KasA are primary targets of INH and TLM, respectively. Similar overexpression studies have often been performed with Mycobacterium smegmatis in which the MOA of different inhibitors was linked to one or more targets, including d-cycloserine to alanine racemase (Alr) and d-ala-d-ala ligase (Ddl) (Feng and Barletta 2003), platensimycin to KasA and KasB (Brown et al. 2009), ethambutol to the arabinosyltransferases EmbB and EmbC (Goude et al. 2009), and certain diphenylurea compounds to inosine monophosphate dehydrogenase (Olaleye et al. 2010; Usha et al. 2011).
In mycobacteria, overexpression has primarily been used to evaluate the role of specific candidate target proteins for compound activity, but it can also provide a means to find candidate targets for a compound with an entirely unknown MOA. To achieve this, libraries of overexpression clones are systematically screened for those that improve growth in the presence of the inhibitor. Such a screen, which used an ordered library of overexpression clones for all essential Escherichia coli genes, identified the lipoprotein chaperon, LolA, as a candidate target of the novel antibacterial compound, MAC12343. Biochemical studies confirmed that MAC12343 binds LolA and perturbs lipoprotein trafficking (Pathania et al. 2009). Similar overexpression libraries are under construction for Mtb and will hopefully be available soon.
A complication of overexpression screens, especially when using random libraries, is the frequent isolation of efflux pumps. These pumps increase resistance by lowering the intracellular concentration of the compound under investigation, and their isolation does not reveal a compound’s MOA. Difficult to interpret MIC shifts can also occur if the candidate target belongs to a protein family of which several members are encoded in the same genome. If the inhibitor binding site is conserved, overexpression of any member of such a protein family will increase the MIC even when only one family member is the physiological target. This has been discussed for inosine-5′-monophosphate (IMP) dehydrogenases (GuaB1/2/3) (Usha et al. 2011), ketoacyl-ACP synthases (KasA/B) (Brown et al. 2009), and methionine aminopeptidases (MapA/B) (Olaleye et al. 2010). In some of these cases, the MIC may have increased because the overproduced protein sequestered the inhibitor from its physiological target.
As target overexpression should increase a compound’s MIC, so should target underexpression decrease the MIC (Fig. 2B). This was indeed observed for several conditional M. smegmatis knockdown mutants described recently. Underexpression of gyrase subunit A (GyrA, target of quinolones [Sugino et al. 1977], Alr [Caceres et al. 1997], dihydrofolate reductase [DHFR, target of trimethoprim] [Schweitzer et al. 1990], RNA polymerase subunit B [RpoB, target of RIF] [Lancini et al. 1969]), and protein biotin ligase (BirA, target of BioAMS [Duckworth et al. 2011]) sensitized M. smegmatis to ciprofloxacin, d-cycloserine, trimethoprim, RIF, and BioAMS, respectively (Duckworth et al. 2011; Kim et al. 2010; Wei et al. 2011). Similar observations were made with Mtb, in which partial genetic inactivation of the biotin synthesis enzyme BioA or DHFR increased its susceptibility toward the novel inhibitors of these enzymes (Shi et al. 2011; Kumar et al. 2012). For many of these mutants, it was also shown that they were most strongly sensitized to inhibitors of the partially depleted protein. These and other data (Giaever et al. 1999; DeVito et al. 2002; Lum et al. 2004; Yin et al. 2004; Donald et al. 2009; Abrahams et al. 2012a; Ollinger et al. 2012) confirmed that partial depletion of an in vitro essential protein often causes compound-specific MIC shifts and can thus provide information on the MOA of a bioactive small molecule. However, as is the case for overexpression, underexpression studies by themselves are not sufficient to claim the underproduced protein as the MIC determining target of an inhibitor, as underexpression could also decrease the MIC by sensitizing other enzymes in the same pathway to chemical inhibition.
That underexpression nevertheless provides a powerful tool for identifying candidate targets is apparent from work with S. cerevisiae. Construction of underexpressors is relatively straightforward in S. cerevisiae and is often achieved with deletion of one gene copy. Such heterozygous deletion strains are available for almost all S. cerevisiae genes including those that are essential for normal growth. This mutant collection has been used extensively to study the MOA of bioactive small molecules by drug-induced haploinsufficiency profiling (HIP). HIP measures the effect of sublethal drug concentrations on the relative growth rates of individual mutants (Fig. 2C). Primarily, these studies confirmed that the mutants of known targets of antimicrobial agents are among those most susceptible to growth inhibition by the respective drug (Giaever et al. 1999, 2004; Lum et al. 2004) and were later implemented in identification of novel candidate targets for established drugs. This included the rRNA processing exosome, which was predicted by HIP (Giaever et al. 2004; Lum et al. 2004) and confirmed by various experiments (Fang et al. 2004; Hoskins and Scott Butler 2007; Hoskins and Butler 2008; Kammler et al. 2008) to be involved in the MOA of the anticancer drug 5-fluorouracil. In the opportunistic pathogen Candida albicans, HIP helped to identify the guanine monophosphate synthase (Rodriguez-Suarez et al. 2007), a fatty acid desaturase (Xu et al. 2009), and poly(A) polymerase (Jiang et al. 2008) as targets of novel inhibitors. Experiments conceptually similar to HIP have been performed in bacteria where gene expression can be efficiently reduced by antisense RNAs (Donald et al. 2009; Huber et al. 2009; Xu et al. 2010). Unfortunately, antisense RNAs have only rarely been efficient in Mtb. Currently, the generation of a large collection of Mtb underexpressors depends on approaches that require homologous recombination during mutant construction, such as transcriptional or proteolytic silencing (Ehrt et al. 2005; Wei et al. 2011).
WCS hits are growth inhibitory by definition and thus should primarily target the essential gene products. Nevertheless, mutations in nonessential genes often modulate a compound’s activity. Evidence for this came from several studies including those that used the E. coli Keio collection. This library contains almost 4000 mutants each of which has a single gene deleted. Of these 4000 mutants, 283 showed greater sensitivity than wild type to at least one of the 14 different antibiotics tested (Tamae et al. 2008; Liu et al. 2010). Whereas some strains were more susceptible to several classes of antibiotics, others were only sensitized to compounds with the same MOA (Girgis et al. 2009; Liu et al. 2010). The drug sensitivity profiles of the latter mutants can be used to classify a compound’s MOA with strategies similar to those implemented in RNA profiling. The discriminative power of these profiles can be enriched with mutations that decrease antibiotic susceptibility, which in E. coli are as frequent as those that increase susceptibility (Girgis et al. 2009).
The homozygous deletion mutants in S. cerevisiae are equivalent to the E. coli Keio mutants. Chemical profiling of such mutants (homozygous deletion profiling, HOP) has been particularly useful for compounds whose primary target is not a protein. For example, HOP of ∼4700 S. cerevisiae mutants revealed that fitness defects caused by deletion of genes involved in DNA repair or in survival during DNA damage can be diagnostic for DNA-damaging agents. In fact, the profiles of these mutants were distinct for chemicals that damage DNA by different mechanisms as they could distinguish compounds that generate interstrand cross-links from those that do not (Lee et al. 2005). The interpretation of HOP experiments can be facilitated by data from genetic interaction studies. Two genes are defined as interacting if the phenotype of their double mutant deviates from the expected phenotypes of the two single gene mutants (Dixon et al. 2009). In S. cerevisiae, many such digenic interactions have been defined. These interactions are valuable for analyses of bioactive small molecules as the genetic interaction profile of a gene can be similar to the HOP profile of a small molecule and thus link the small molecule to the respective gene (Parsons et al. 2004, 2006; Costanzo et al. 2010).
FACILITATING TARGET-BASED DRUG DISCOVERY
Target-based approaches typically begin with a screen for inhibitors of a purified enzyme. Unfortunately, most inhibitors identified in several high-throughput screenings (HTSs) against bacterial enzymes had no activity against live bacteria (Payne et al. 2007). Nevertheless, target-based approaches have their own advantages. Primarily, MOA studies are more straightforward for a target-based approach than for WCS hits. For example, a bisubstrate inhibitor (BioAMS) designed to inactivate the protein biotin ligase (BirA) was recently found to inhibit the growth of Mtb. The study conducted by Duckworth et al. (2011) examined the impact of BioAMS on growth and biotinylation of proteins in M. smegmatis and Mtb. As expected, the activity of BioAMS (but not of INH or ethambutol) against Mtb and M. smegmatis directly correlated with expression of BirA, and growth inhibition coincided with the depletion of biotinylated proteins. Together, these straightforward observations confirmed BirA as the primary target of BioAMS (Duckworth et al. 2011). The second main advantage of target-based approaches is that they can survey a much larger chemical space than what can be covered in phenotypic screens, especially if fragment-based approaches are included (Scott et al. 2012). This is significant because the physicochemical properties of most molecules in a typical WCS library are different from those of known antibacterials (Brotz-Oesterhelt and Sass 2010).
The Power and Limits of Genetic Target Validation
Choosing a poor target will doom a target-based drug discovery program from the start, yet this might become obvious only after years of work and millions of dollars have been invested. Identifying appropriate targets is thus crucial. Ideally, the selected protein should be (1) conserved among all isolates of the targeted pathogen(s), (2) susceptible to inhibition by a small molecule, (3) different enough from any human homolog to allow development of low toxicity inhibitors, (4) amenable to biochemical HTS, and (5) required for bacterial growth (or survival) in a manner that its partial inactivation suffices to improve a patient’s health. Showing that a target fulfills all these criteria requires a small-molecule inhibitor that is safe for use in humans (i.e., a drug). Trying to develop drugs against new targets, therefore, always bears the additional risk that comes from working against a target that is yet to be fully validated. This additional risk applies to inhibitors identified from either WCS or biochemical screens and is one of the factors causing the high rate of attrition of compounds with entirely novel MOAs (Kola and Landis 2004).
Druggability assessments evaluate potential binding pockets by computational or chemical methods (Perot et al. 2010; Barelier and Krimm 2011; Fauman et al. 2011); sequence conservation—within bacteria and between bacterial and human homologs—can be assessed with genome sequence data. An important contribution of genetics to the evaluation of potential targets is its ability to determine the importance of a potential target for growth and survival. The most complete source for this information stems from transposon mutagenesis. Many of the genes that Mtb requires to grow normally in vitro (Sassetti et al. 2001, 2003), in macrophages (Rengarajan et al. 2005), or in mouse spleens (Sassetti and Rubin, 2003) were defined by transposon site hybridization (TraSH). TraSH uses DNA microarrays to simultaneously map and analyze the phenotypic consequences of thousands of transposon insertions. Recently, Tn-seq, which uses next-generation DNA sequencing instead of microarrays, has improved these analyses by providing single-nucleotide resolution and a larger dynamic range (Griffin et al. 2012; Zhang et al. 2012, 2013). TraSH and Tn-seq have contributed more to the phenotypic characterization of gene functions in Mtb than any other genetic approach. However, they are not ideal to validate gene products as targets for drug development for two reasons. First, TraSH and Tn-seq can identify in vitro essential genes, but neither approach generates mutants for such genes that could be analyzed under a variety of conditions. In the same manner as WCS can identify novip compounds, genetic screens can identify novip targets (i.e., targets whose inactivation kills Mtb under a specific in vitro condition but does not prevent persistence or even growth in animals or humans). It is therefore important to evaluate the role of a gene product for survival under a variety of conditions including those that induce nonreplicating persistence. For genes that Mtb requires to grow under standard laboratory conditions, which are among the most attractive targets for drug development, these analyses require conditional knockdown mutants.
The construction of conditional knockdown mutants in Mtb became possible with the development of chemically controlled genetic switches for mycobacteria (Schnappinger and Ehrt 2014). Many of the switches developed for mycobacteria depend on placing the gene of interest downstream from a modified promoter so that its transcription can be regulated by a tetracycline repressor and a tetracycline derivative (Blokpoel et al. 2005; Carroll et al. 2005; Ehrt et al. 2005; Klotzsche et al. 2009; Boldrin et al. 2010). These regulatory systems facilitated the analyses of several genes in Mtb (Schnappinger and Ehrt 2014), but they depend on secondary events, most notably cell division, to deplete the protein of interest. They are therefore less suited to study the importance of gene functions for survival of nonreplicating Mtb. Analyses of nonreplicating bacteria can be performed more efficiently with regulatory systems that directly increase protein turnover. In Mtb, this can be achieved by modifying a gene’s 3′ end so that it encodes the sequence of the DAS+4 degradation tag (Kim et al. 2010). Conditionality is achieved by controlling the expression of a protein (SspB) that activates the degradation tag. Recently, transcriptional and proteolytic modes of regulation have been combined in a dual-control (DUC) switch, which can achieve efficient and robust gene inactivation in growing and nonreplicating Mtb (Kim et al. 2013). Application of the DUC switch to analyses of the NAD synthetase (NadE) identified this enzyme to be essential for survival of both replicating and nonreplicating bacteria. Its inactivation was further sufficient to rapidly eliminate culturable bacteria from mouse lungs and spleens during acute and chronic infections (Kim et al. 2013). Essentiality of NadE for viability of Mtb under all conditions tested so far strongly suggests that this enzyme is also required for growth and survival under most (if not all) conditions that Mtb encounters in humans. Whether it allows the development of small-molecule inhibitors that would be safe for use in humans remains to be shown.
The second limitation of TraSH and Tn-seq is that they do not allow measurement of the extent to which a gene product needs to be inactivated to impair growth. Although the impact of a transposon insertion on gene function can vary with its insertion site, the phenotypes reported by TraSH/Tn-seq are often attributable to insertions that create a null allele. Complete inactivation of a protein in live bacteria is rarely, if ever, achieved by small molecules. The phenotypes that are caused by complete deletion are thus less relevant to evaluating a new target for drug development than those that result from its partial depletion. All other factors being equal, a protein that needs to be inactivated by >90% (or less) is a more attractive target than one that needs to be inhibited by 90% before growth is inhibited. That vulnerability of mycobacteria to partial depletion indeed varies among different essential proteins was first shown in M. smegmatis. For example, HIV-protease-induced degradation depleted M. smegmatis of >97% of DHFR and Alr, but this depletion only slowed growth down. In contrast, even modest depletion of RpoB was sufficient to stop growth entirely (Wei et al. 2011). Unfortunately, the extent of functional inactivation that is necessary to impair the growth of Mtb is known only for a few proteins, including BioA and PptT, which are required for the synthesis of biotin and polyketides, respectively. Mtb is not particularly vulnerable to partial inactivation of either enzyme, as both require to be depleted by >90% to affect growth (Woong Park et al. 2011; Leblanc et al. 2012). Applying conditional gene inactivation systems to the characterization of more genes will hopefully identify proteins that need to be depleted >90% to stop Mtb from growing.
Target-Directed WCS
Classical WCS often requires complex studies to define the MOA of hits; biochemical screens rarely identify inhibitors that can penetrate the bacterial cell envelope. Both difficulties can be overcome with target-directed WCS. The principle of this approach was first established in Staphylococcus aureus. Overexpression of a fabF antisense RNA caused the 5′ end of the fabF mRNA to be degraded. This decreased the expression of FabF, an enzyme required for fatty acid biosynthesis, and increased the sensitivity of S. aureus to FabF inhibitors (Young et al. 2006). Agar diffusion assays were then used to screen small-molecule libraries against wild-type S. aureus and the FabF underexpressor. Molecules that were more active against the underexpressor included platencin and platensimycin, two novel inhibitors of fatty acid biosynthesis with broad-spectrum activity against Gram-positive bacteria (Wang et al. 2006, 2007; Jayasuriya et al. 2007; Fischbach and Walsh 2009). In Mtb, transcriptional repression instead of antisense RNA has been used to generate mutants expressing lower than wild-type levels of pantothenate synthase (PanC), diaminopimelate decarboxylase (LysA), isocitrate lyase (Icl1), or the type I signal peptidase (LepB). In addition, these recently constructed mutants show target-specific changes in their susceptibility to different small-molecule inhibitors (Abrahams et al. 2012a; Ollinger et al. 2012). WCS with these strains promises to identify new inhibitors that are able to reach the cytoplasm of Mtb and inhibit a desired target.
CONCLUDING REMARKS
None of the approaches discussed in this review existed during the golden age of antibiotic discovery, and antibacterial drug discovery can be pursued without them. However, modern genetics can help improve a process that has delivered few novel molecular entities during the last 50 yr. One important contribution would be to help reduce late-stage attrition (i.e., failure in clinical trials). The cost of introducing a new drug to the market varies between US$500 and 2000 million (Adams and Brantner 2006; Paul et al. 2010). Late-stage attrition can therefore drain the resources of commercially unattractive areas, like antibacterial drug development, and cause them to fail entirely over a long period of time. The two main causes of late-stage attrition are lack of safety and efficacy in people (Kola and Landis 2004; Bennani 2012). Predicting clinical efficacy will remain imprecise. However, if we understand the MOA of the drug candidates that are being tested in the clinic, it should be possible to avoid trials with chemically different compounds that fail for the same mechanistic reasons. The genetic approaches discussed here have helped to characterize the MOA of several antibacterial compounds. However, the MOA of many antibacterials is complex and new insights are still gained into the MOA of drugs that have been in clinical use for decades (Chakraborty et al. 2013). No single approach will therefore be sufficient to define the MOA of most antibacterials. Studies that combine several approaches—for example, underexpression, overexpression, resistance selection, and biochemical studies (Huber et al. 2009)—promise to be the most successful. Drug candidates with novel MOAs are attractive because they could improve TB chemotherapy fundamentally (e.g., by reducing its duration). But they bear a higher risk of late-stage failure if they inactivate targets that have not yet been clinically validated. Measuring the effects that can be achieved by inactivating such novel targets (e.g., by genetic means) in a variety of assays, including animal models, seems prudent.
Second, genetics can also contribute to drug development by providing single-gene underexpressors that help increase the sensitivity of WCS and bias hits toward a desired target. Underexpressors can furthermore be used in a secondary assay to evaluate biochemical HTS hits. This distinguishes compounds that penetrate the bacterial cell envelope poorly (and are not active against wild type but active against an underexpressor) from those that do not penetrate at all (and are active neither against wild type nor the underexpressor). Such compounds could be starting points for medicinal chemistry efforts to improve compound efficacy against wild-type bacteria, which could expand the chemical space into the area that is effective against bacteria and underrepresented in most screening libraries.
ACKNOWLEDGMENTS
I thank Kathryn O’Brien, Shipra Grover, and Sabine Ehrt for critically reading the manuscript. I am grateful for past and present support from the National Institutes of Health, the Bill & Melinda Gates Foundation, The Wellcome Trust, and the Ellison Medical Foundation.
REFERENCES
- Abrahams GL, Kumar A, Savvi S, Hung AW, Wen S, Abell C, Barry CE III, Sherman DR, Boshoff HI, Mizrahi V. 2012a. Pathway-selective sensitization of Mycobacterium tuberculosis for target-based whole-cell screening. Chem Biol 19: 844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams KA, Cox JA, Spivey VL, Loman NJ, Pallen MJ, Constantinidou C, Fernandez R, Alemparte C, Remuinan MJ, Barros D, et al. 2012b. Identification of novel imidazo[1,2-a]pyridine inhibitors targeting M. tuberculosis QcrB. PloS ONE 7: e52951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams CP, Brantner VV. 2006. Estimating the cost of new drug development: Is it really 802 million dollars? Health Aff (Millwood) 25: 420–428. [DOI] [PubMed] [Google Scholar]
- Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, et al. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307: 223–227. [DOI] [PubMed] [Google Scholar]
- Arnvig K, Young D. 2012. Non-coding RNA and its potential role in Mycobacterium tuberculosis pathogenesis. RNA Biol 9: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barelier S, Krimm I. 2011. Ligand specificity, privileged substructures and protein druggability from fragment-based screening. Curr Opin Chem Biol 15: 469–474. [DOI] [PubMed] [Google Scholar]
- Barry CE III, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. 2009. The spectrum of latent tuberculosis: Rethinking the biology and intervention strategies. Nat Rev Microbiol 7: 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennani YL. 2012. Drug discovery in the next decade: Innovation needed ASAP. Drug Discov Today 17: S31–S44. [DOI] [PubMed] [Google Scholar]
- Blokpoel MC, Murphy HN, O’Toole R, Wiles S, Runn ES, Stewart GR, Young DB, Robertson BD. 2005. Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res 33: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrin F, Casonato S, Dainese E, Sala C, Dhar N, Palu G, Riccardi G, Cole ST, Manganelli R. 2010. Development of a repressible mycobacterial promoter system based on two transcriptional repressors. Nucleic Acids Res 38: e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE III. 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: Novel insights into drug mechanisms of action. J Biol Chem 279: 40174–40184. [DOI] [PubMed] [Google Scholar]
- Boshoff HI, Xu X, Tahlan K, Dowd CS, Pethe K, Camacho LR, Park TH, Yun CS, Schnappinger D, Ehrt S, et al. 2008. Biosynthesis and recycling of nicotinamide cofactors in Mycobacterium tuberculosis. An essential role for NAD in nonreplicating bacilli. J Biol Chem 283: 19329–19341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazas MD, Hancock RE. 2005. Using microarray gene signatures to elucidate mechanisms of antibiotic action and resistance. Drug Discov Today 10: 1245–1252. [DOI] [PubMed] [Google Scholar]
- Brotz-Oesterhelt H, Sass P. 2010. Postgenomic strategies in antibacterial drug discovery. Future Microbiol 5: 1553–1579. [DOI] [PubMed] [Google Scholar]
- Brown AK, Taylor RC, Bhatt A, Futterer K, Besra GS. 2009. Platensimycin activity against mycobacterial β-ketoacyl-ACP synthases. PloS ONE 4: e6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres NE, Harris NB, Wellehan JF, Feng Z, Kapur V, Barletta RG. 1997. Overexpression of the d-alanine racemase gene confers resistance to d-cycloserine in Mycobacterium smegmatis. J Bacteriol 179: 5046–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll P, Muttucumaru DG, Parish T. 2005. Use of a tetracycline-inducible system for conditional expression in Mycobacterium tuberculosis and Mycobacterium smegmatis. Appl Environ Microbiol 71: 3077–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Gruber T, Barry CE III, Boshoff HI, Rhee KY. 2013. Para-aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacterium tuberculosis. Science 339: 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophe T, Jackson M, Jeon HK, Fenistein D, Contreras-Dominguez M, Kim J, Genovesio A, Carralot JP, Ewann F, Kim EH, et al. 2009. High content screening identifies decaprenyl-phosphoribose 2′ epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathog 5: e1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. 2010. The genetic landscape of a cell. Science 327: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dartois V, Barry CE III. 2013. A medicinal chemists’ guide to the unique difficulties of lead optimization for tuberculosis. Bioorg Med Chem Lett 23: 4741–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. 2006. Where have all the antibiotics gone? Can J Infect Dis Med Microbiol 17: 287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVito JA, Mills JA, Liu VG, Agarwal A, Sizemore CF, Yao Z, Stoughton DM, Cappiello MG, Barbosa MD, Foster LA, et al. 2002. An array of target-specific screening strains for antibacterial discovery. Nat Biotechnol 20: 478–483. [DOI] [PubMed] [Google Scholar]
- Dhiman RK, Mahapatra S, Slayden RA, Boyne ME, Lenaerts A, Hinshaw JC, Angala SK, Chatterjee D, Biswas K, Narayanasamy P, et al. 2009. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol Microbiol 72: 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. 2009. Systematic mapping of genetic interaction networks. Annu Rev Genet 43: 601–625. [DOI] [PubMed] [Google Scholar]
- Donald RG, Skwish S, Forsyth RA, Anderson JW, Zhong T, Burns C, Lee S, Meng X, LoCastro L, Jarantow LW, et al. 2009. A Staphylococcus aureus fitness test platform for mechanism-based profiling of antibacterial compounds. Chem Biol 16: 826–836. [DOI] [PubMed] [Google Scholar]
- Duckworth BP, Geders TW, Tiwari D, Boshoff HI, Sibbald PA, Barry CE III, Schnappinger D, Finzel BC, Aldrich CC. 2011. Bisubstrate adenylation inhibitors of biotin protein ligase from Mycobacterium tuberculosis. Chem Biol 18: 1432–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res 33: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Hoskins J, Butler JS. 2004. 5-fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol Cell Biol 24: 10766–10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauman EB, Rai BK, Huang ES. 2011. Structure-based druggability assessment—Identifying suitable targets for small molecule therapeutics. Curr Opin Chem Biol 15: 463–468. [DOI] [PubMed] [Google Scholar]
- Feng Z, Barletta RG. 2003. Roles of Mycobacterium smegmatis d-alanine:d-alanine ligase and d-alanine racemase in the mechanisms of action of and resistance to the peptidoglycan inhibitor d-cycloserine. Antimicrob Agents Chemother 47: 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT. 2009. Antibiotics for emerging pathogens. Science 325: 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiberg C, Fischer HP, Brunner NA. 2005. Discovering the mechanism of action of novel antibacterial agents through transcriptional profiling of conditional mutants. Antimicrob Agents Chemother 49: 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu LM, Shinnick TM. 2007. Genome-wide exploration of the drug action of capreomycin on Mycobacterium tuberculosis using Affymetrix oligonucleotide GeneChips. J Infect 54: 277–284. [DOI] [PubMed] [Google Scholar]
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW. 1999. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet 21: 278–283. [DOI] [PubMed] [Google Scholar]
- Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. 2004. Chemogenomic profiling: Identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci 101: 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgis HS, Hottes AK, Tavazoie S. 2009. Genetic architecture of intrinsic antibiotic susceptibility. PloS ONE 4: e5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goude R, Amin AG, Chatterjee D, Parish T. 2009. The arabinosyltransferase EmbC is inhibited by ethambutol in Mycobacterium tuberculosis. Antimicrob Agents Chemother 53: 4138–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. 2012. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol 19: 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzegorzewicz AE, Pham H, Gundi VA, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SE, Kordulakova J, et al. 2012. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol 8: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartkoorn RC, Sala C, Neres J, Pojer F, Magnet S, Mukherjee R, Uplekar S, Boy-Rottger S, Altmann KH, Cole ST. 2012. Towards a new tuberculosis drug: Pyridomycin—Nature’s isoniazid. EMBO Mol Med 4: 1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins J, Butler JS. 2008. RNA-based 5-fluorouracil toxicity requires the pseudouridylation activity of Cbf5p. Genetics 179: 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins J, Scott Butler J. 2007. Evidence for distinct DNA- and RNA-based mechanisms of 5-fluorouracil cytotoxicity in Saccharomyces cerevisiae. Yeast 24: 861–870. [DOI] [PubMed] [Google Scholar]
- Huber J, Donald RG, Lee SH, Jarantow LW, Salvatore MJ, Meng X, Painter R, Onishi RH, Occi J, Dorso K, et al. 2009. Chemical genetic identification of peptidoglycan inhibitors potentiating carbapenem activity against methicillin-resistant Staphylococcus aureus. Chem Biol 16: 837–848. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. 2000. Functional discovery via a compendium of expression profiles. Cell 102: 109–126. [DOI] [PubMed] [Google Scholar]
- Ioerger TR, O’Malley T, Liao R, Guinn KM, Hickey MJ, Mohaideen N, Murphy KC, Boshoff HI, Mizrahi V, Rubin EJ, et al. 2013. Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PloS ONE 8: e75245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasuriya H, Herath KB, Zhang C, Zink DL, Basilio A, Genilloud O, Diez MT, Vicente F, Gonzalez I, Salazar O, et al. 2007. Isolation and structure of platencin: A FabH and FabF dual inhibitor with potent broad-spectrum antibiotic activity. Angew Chem Int Ed Engl 46: 4684–4688. [DOI] [PubMed] [Google Scholar]
- Jiang B, Xu D, Allocco J, Parish C, Davison J, Veillette K, Sillaots S, Hu W, Rodriguez-Suarez R, Trosok S, et al. 2008. PAP inhibitor with in vivo efficacy identified by Candida albicans genetic profiling of natural products. Chem Biol 15: 363–374. [DOI] [PubMed] [Google Scholar]
- Kammler S, Lykke-Andersen S, Jensen TH. 2008. The RNA exosome component hRrp6 is a target for 5-fluorouracil in human cells. Mol Cancer Res 6: 990–995. [DOI] [PubMed] [Google Scholar]
- Karakousis PC, Williams EP, Bishai WR. 2008. Altered expression of isoniazid-regulated genes in drug-treated dormant Mycobacterium tuberculosis. J Antimicrob Chemother 61: 323–331. [DOI] [PubMed] [Google Scholar]
- Kim JH, Wei JR, Wallach JB, Robbins RS, Rubin EJ, Schnappinger D. 2010. Protein inactivation in mycobacteria by controlled proteolysis and its application to deplete the beta subunit of RNA polymerase. Nucleic Acids Res 39: 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, O’Brien KM, Sharma R, Boshoff HI, Rehren G, Chakraborty S, Wallach JB, Monteleone M, Wilson DJ, Aldrich CC, et al. 2013. A genetic strategy to identify targets for the development of drugs that prevent bacterial persistence. Proc Natl Acad Sci 110: 19095–19100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotzsche M, Ehrt S, Schnappinger D. 2009. Improved tetracycline repressors for gene silencing in mycobacteria. Nucleic Acids Res 37: 1778–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola I, Landis J. 2004. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov 3: 711–715. [DOI] [PubMed] [Google Scholar]
- Koul A, Dendouga N, Vergauwen K, Molenberghs B, Vranckx L, Willebrords R, Ristic Z, Lill H, Dorange I, Guillemont J, et al. 2007. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat Chem Biol 3: 323–324. [DOI] [PubMed] [Google Scholar]
- Kumar A, Zhang M, Zhu L, Liao RP, Mutai C, Hafsat S, Sherman DR, Wang MW. 2012. High-throughput screening and sensitized bacteria identify an M. tuberculosis dihydrofolate reductase inhibitor with whole cell activity. PloS ONE 7: e39961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa V, Poce G, Canseco JO, Buroni S, Pasca MR, Biava M, Raju RM, Porretta GC, Alfonso S, Battilocchio C, et al. 2012. MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob Agents Chemother 56: 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancini G, Pallanza R, Silvestri LG. 1969. Relationships between bactericidal effect and inhibition of ribonucleic acid nucleotidyltransferase by rifampicin in Escherichia coli K-12. J Bacteriol 97: 761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MH, Vilcheze C, Kremer L, Besra GS, Parsons L, Salfinger M, Heifets L, Hazbon MH, Alland D, Sacchettini JC, et al. 2002. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol Microbiol 46: 453–466. [DOI] [PubMed] [Google Scholar]
- Lashkari DA, DeRisi JL, McCusker JH, Namath AF, Gentile C, Hwang SY, Brown PO, Davis RW. 1997. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci 94: 13057–13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc C, Prudhomme T, Tabouret G, Ray A, Burbaud S, Cabantous S, Mourey L, Guilhot C, Chalut C. 2012. 4′-Phosphopantetheinyl transferase PptT, a new drug target required for Mycobacterium tuberculosis growth and persistence in vivo. PLoS Pathog 8: e1003097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechartier B, Rybniker J, Zumla A, Cole ST. 2014. Tuberculosis drug discovery in the post-post-genomic era. EMBO Mol Med 6: 158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G. 2005. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet 1: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Zeng F, Guo A, Liu L, Guo N, Li L, Jin J, Wu X, Liu M, Zhao D, et al. 2011. Microarray analysis of the chelerythrine-induced transcriptome of Mycobacterium tuberculosis. Curr Microbiol 62: 1200–1208. [DOI] [PubMed] [Google Scholar]
- Liu A, Tran L, Becket E, Lee K, Chinn L, Park E, Tran K, Miller JH. 2010. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: Generating an antibiotic bar code. Antimicrob Agents Chemother 54: 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A, et al. 2004. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116: 121–137. [DOI] [PubMed] [Google Scholar]
- Magnet S, Hartkoorn RC, Szekely R, Pato J, Triccas JA, Schneider P, Szantai-Kis C, Orfi L, Chambon M, Banfi D, et al. 2010. Leads for antitubercular compounds from kinase inhibitor library screens. Tuberculosis 90: 354–360. [DOI] [PubMed] [Google Scholar]
- Makarov V, Manina G, Mikusova K, Mollmann U, Ryabova O, Saint-Joanis B, Dhar N, Pasca MR, Buroni S, Lucarelli AP, et al. 2009. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324: 801–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunatha U, Boshoff HI, Barry CE. 2009. The mechanism of action of PA-824: Novel insights from transcriptional profiling. Commun Integr Biol 2: 215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, et al. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44. [DOI] [PubMed] [Google Scholar]
- Munoz-Elias EJ, Upton AM, Cherian J, McKinney JD. 2006. Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol Microbiol 60: 1109–1122. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaleye O, Raghunand TR, Bhat S, He J, Tyagi S, Lamichhane G, Gu P, Zhou J, Zhang Y, Grosset J, et al. 2010. Methionine aminopeptidases from Mycobacterium tuberculosis as novel antimycobacterial targets. Chem Biol 17: 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollinger J, O’Malley T, Ahn J, Odingo J, Parish T. 2012. Inhibition of the sole type I signal peptidase of Mycobacterium tuberculosis is bactericidal under replicating and nonreplicating conditions. J Bacteriol 194: 2614–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C. 2004. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol 22: 62–69. [DOI] [PubMed] [Google Scholar]
- Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, et al. 2006. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126: 611–625. [DOI] [PubMed] [Google Scholar]
- Pathania R, Zlitni S, Barker C, Das R, Gerritsma DA, Lebert J, Awuah E, Melacini G, Capretta FA, Brown ED. 2009. Chemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targeting. Nat Chem Biol 5: 849–856. [DOI] [PubMed] [Google Scholar]
- Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. 2010. How to improve R&D productivity: The pharmaceutical industry’s grand challenge. Nat Rev Drug Discov 9: 203–214. [DOI] [PubMed] [Google Scholar]
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6: 29–40. [DOI] [PubMed] [Google Scholar]
- Perot S, Sperandio O, Miteva MA, Camproux AC, Villoutreix BO. 2010. Druggable pockets and binding site centric chemical space: A paradigm shift in drug discovery. Drug Discov Today 15: 656–667. [DOI] [PubMed] [Google Scholar]
- Pethe K, Bifani P, Jang J, Kang S, Park S, Ahn S, Jiricek J, Jung J, Jeon HK, Cechetto J, et al. 2013. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat Med 19: 1157–1160. [DOI] [PubMed] [Google Scholar]
- Provvedi R, Boldrin F, Falciani F, Palu G, Manganelli R. 2009. Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology 155: 1093–1102. [DOI] [PubMed] [Google Scholar]
- Remuinan MJ, Perez-Herran E, Rullas J, Alemparte C, Martinez-Hoyos M, Dow DJ, Afari J, Mehta N, Esquivias J, Jimenez E, et al. 2013. Tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide and N-benzyl-6′,7′-dihydrospiro[piperidine-4,4’-thieno[3,2-c]pyran] analogues with bactericidal efficacy against Mycobacterium tuberculosis targeting MmpL3. PloS One 8: e60933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J, Bloom BR, Rubin EJ. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci 102: 8327–8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez GM, Smith I. 2006. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J Bacteriol 188: 424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Suarez R, Xu D, Veillette K, Davison J, Sillaots S, Kauffman S, Hu W, Bowman J, Martel N, Trosok S, et al. 2007. Mechanism-of-action determination of GMP synthase inhibitors and target validation in Candida albicans and Aspergillus fumigatus. Chem Biol 14: 1163–1175. [DOI] [PubMed] [Google Scholar]
- Roemer T, Boone C. 2013. Systems-level antimicrobial drug and drug synergy discovery. Nat Chem Biol 9: 222–231. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci 100: 12989–12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci 98: 12712–12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48: 77–84. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470. [DOI] [PubMed] [Google Scholar]
- Schnappinger D, Ehrt S. 2014. Regulated expression systems for mycobacteria and their applications. Microbiol Spectr 2: 10.1128/microbiolspec.MGM1122-0018-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer BI, Dicker AP, Bertino JR. 1990. Dihydrofolate reductase as a therapeutic target. FASEB J 4: 2441–2452. [DOI] [PubMed] [Google Scholar]
- Scott DE, Coyne AG, Hudson SA, Abell C. 2012. Fragment-based approaches in drug discovery and chemical biology. Biochemistry 51: 4990–5003. [DOI] [PubMed] [Google Scholar]
- Shi C, Geders TW, Park SW, Wilson DJ, Boshoff HI, Abayomi O, Barry CE III, Schnappinger D, Finzel BC, Aldrich CC. 2011. Mechanism-based inactivation by aromatization of the transaminase BioA involved in biotin biosynthesis in Mycobacterium tuberculosis. J Am Chem Soc 133: 18194–18201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver LL. 2011. Challenges of antibacterial discovery. Clin Microbiol Rev 24: 71–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayden RA, Belisle JT. 2009. Morphological features and signature gene response elicited by inactivation of FtsI in Mycobacterium tuberculosis. J Antimicrob Chemother 63: 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Ammar R, Nislow C, Giaever G. 2010. A survey of yeast genomic assays for drug and target discovery. Pharmacol Ther 127: 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Grant SS, Kawate T, Iwase N, Shimizu M, Wivagg C, Silvis M, Kazyanskaya E, Aquadro J, Golas A, et al. 2012. Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS Chem Biol 7: 1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Kawate T, Iwase N, Shimizu M, Clatworthy AE, Kazyanskaya E, Sacchettini JC, Ioerger TR, Siddiqi NA, Minami S, et al. 2013. Diarylcoumarins inhibit mycolic acid biosynthesis and kill Mycobacterium tuberculosis by targeting FadD32. Proc Natl Acad Sci 110: 11565–11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A, Peebles CL, Kreuzer KN, Cozzarelli NR. 1977. Mechanism of action of nalidixic acid: Purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci 74: 4767–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney DC, Anthony J. 2011. How were new medicines discovered? Nat Rev Drug Discov 10: 507–519. [DOI] [PubMed] [Google Scholar]
- Tahlan K, Wilson R, Kastrinsky DB, Arora K, Nair V, Fischer E, Barnes SW, Walker JR, Alland D, Barry CE III, et al. 2012. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother 56: 1797–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamae C, Liu A, Kim K, Sitz D, Hong J, Becket E, Bui A, Solaimani P, Tran KP, Yang H, et al. 2008. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J Bacteriol 190: 5981–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsing M, Klitgaard JK, Atilano ML, Skov MN, Kolmos HJ, Filipe SR, Kallipolitis BH. 2013. Thioridazine induces major changes in global gene expression and cell wall composition in methicillin-resistant Staphylococcus aureus USA300. PloS ONE 8: e64518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usha V, Gurcha SS, Lovering AL, Lloyd AJ, Papaemmanouil A, Reynolds RC, Besra GS. 2011. Identification of novel diphenyl urea inhibitors of Mt-GuaB2 active against Mycobacterium tuberculosis. Microbiology 157: 290–299. [DOI] [PubMed] [Google Scholar]
- Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, et al. 2006. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441: 358–361. [DOI] [PubMed] [Google Scholar]
- Wang J, Kodali S, Lee SH, Galgoci A, Painter R, Dorso K, Racine F, Motyl M, Hernandez L, Tinney E, et al. 2007. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc Natl Acad Sci 104: 7612–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Sambandan D, Halder R, Wang J, Batt SM, Weinrick B, Ahmad I, Yang P, Zhang Y, Kim J, et al. 2013. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. Proc Natl Acad Sci 110: E2510–E2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wecke T, Mascher T. 2011. Antibiotic research in the age of omics: From expression profiles to interspecies communication. J Antimicrob Chemother 66: 2689–2704. [DOI] [PubMed] [Google Scholar]
- Wei JR, Krishnamoorthy V, Murphy K, Kim JH, Schnappinger D, Alber T, Sassetti CM, Rhee KY, Rubin EJ. 2011. Depletion of antibiotic targets has widely varying effects on growth. Proc Natl Acad Sci 108: 4176–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243. [DOI] [PubMed] [Google Scholar]
- Wilson M, DeRisi J, Kristensen HH, Imboden P, Rane S, Brown PO, Schoolnik GK. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci 96: 12833–12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Kumar P, Parashar V, Vilcheze C, Veyron-Churlet R, Freundlich JS, Barnes SW, Walker JR, Szymonifka MJ, Marchiano E, et al. 2013. Antituberculosis thiophenes define a requirement for Pks13 in mycolic acid biosynthesis. Nat Chem Biol 9: 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woong Park S, Klotzsche M, Wilson DJ, Boshoff HI, Eoh H, Manjunatha U, Blumenthal A, Rhee K, Barry CE III, Aldrich CC, et al. 2011. Evaluating the sensitivity of Mycobacterium tuberculosis to biotin deprivation using regulated gene expression. PLoS Pathog 7: e1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Sillaots S, Davison J, Hu W, Jiang B, Kauffman S, Martel N, Ocampo P, Oh C, Trosok S, et al. 2009. Chemical genetic profiling and characterization of small-molecule compounds that affect the biosynthesis of unsaturated fatty acids in Candida albicans. J Biol Chem 284: 19754–19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HH, Trawick JD, Haselbeck RJ, Forsyth RA, Yamamoto RT, Archer R, Patterson J, Allen M, Froelich JM, Taylor I, et al. 2010. Staphylococcus aureus TargetArray: Comprehensive differential essential gene expression as a mechanistic tool to profile antibacterials. Antimicrob Agents Chemother 54: 3659–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D, Fox B, Lonetto ML, Etherton MR, Payne DJ, Holmes DJ, Rosenberg M, Ji Y. 2004. Identification of antimicrobial targets using a comprehensive genomic approach. Pharmacogenomics 5: 101–113. [DOI] [PubMed] [Google Scholar]
- Young K, Jayasuriya H, Ondeyka JG, Herath K, Zhang CW, Kodali S, Galgoci A, Painter R, Brown-Driver V, Yamamoto R, et al. 2006. Discovery of FabH/FabF inhibitors from natural products. Antimicrob Agents Chemother 50: 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Ioerger TR, Huttenhower C, Long JE, Sassetti CM, Sacchettini JC, Rubin EJ. 2012. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog 8: e1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, et al. 2013. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155: 1296–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]