It is traditionally believed that all somatic cells in the human body contain the same amount of DNA. However, substantial differences in nuclear DNA content have been observed across cell types, and this variation is positively correlated with cell size.
Abstract
Variation in the size of cells, and the DNA they contain, is a basic feature of multicellular organisms that affects countless aspects of their structure and function. Within humans, cell size is known to vary by several orders of magnitude, and differences in nuclear DNA content among cells have been frequently observed. Using published data, here we describe how the quantity of nuclear DNA across 19 different human cell types increases with cell volume. This observed increase is similar to intraspecific relationships between DNA content and cell volume in other species, and interspecific relationships between diploid genome size and cell volume. Thus, we speculate that the quantity of nuclear DNA content in somatic cells of humans is perhaps best viewed as a distribution of values that reflects cell size distributions, rather than as a single, immutable quantity.
Our understanding of the complexity of multicellular organisms, and the diverse cells of which they are comprised, has dramatically increased over the past several decades. Yet, we still lack an understanding of some of the most basic features of the cells that constitute multicellular organisms. For example, the number of different cell types in an organism, or the rate at which different cells grow, divide, and die, remain poorly understood (see Niklas 2015). But perhaps most important, we lack an understanding of the size and abundance of cells that constitute an organism (see Amodeo and Skotheim 2015). Cell size, in particular, affects virtually all structural and functional attributes of multicellular organisms, from the molecular level to the whole organism level.
One key feature of organisms that may vary with cell size is the amount of nuclear DNA. Across species, genome size has long been known to correlate positively with cell and nuclear volume (Price et al. 1973; Szarski 1976; Olmo 1983). But within species, too, the nuclear DNA content of somatic cells has been shown in a few instances to increase with cell size in species such as Daphnia (Beaton and Hebert 1989) and Arabidopsis (Jovtchev et al. 2006). Such increases in nuclear DNA content can have important consequences for cell function, in general, and gene expression, in particular (Hancock et al. 2008; Lee et al. 2009; De Veylder et al. 2011; Marguerat and Bähler 2012).
In the case of humans, substantial differences in DNA content have been observed in many human cell types. Indeed, since Watson and Crick described the structure of DNA, studies of healthy human tissues have reported the presence of polyploid cells (Winkelmann et al. 1987; Biesterfeld et al. 1994). The cell types in which this has been observed appear to have little in common, except that they are generally stable, fully differentiated cells (Winkelmann et al. 1987). Still, these observations have done little to change the traditional view that all healthy somatic cells in the human body hold the same characteristic quantity of DNA (∼7 billion base pairs) based on the long-standing principle of DNA constancy (Mirsky and Ris 1949). Deviations from the diploid quantity of DNA in humans, like other animals, are still often viewed as exceptional, tissue-specific, or indicative of pathology. A more synthetic view of differences in nuclear DNA content across human cell types may provide some clarity on these and other issues.
In this review, we compile and analyze published data to examine the extent to which nuclear DNA content varies across diverse human cell types, and whether such variation is correlated with cell size. We then compare these results with previously reported relationships between nuclear DNA content and cell size within four other species. Finally, we compare these results with the relationships between diploid genome size and cell size observed across species in several broad taxonomic groups. These analyses suggest that systematic variation in nuclear DNA content is a more ubiquitous phenomenon in human cells than was previously appreciated. However, as we later discuss, the mechanisms underlying these patterns remain in question.
THE RELATIONSHIP OF NUCLEAR DNA CONTENT TO CELL SIZE IN HUMANS
Methodology
Our analysis for this work used published data from healthy human cell populations representing 19 different cell types, as designated in the original studies (data provided in Table 1). In the original studies, DNA content was estimated using the Feulgen staining method, and the size of cells or cell nuclei were directly measured. Feulgen staining (Feulgen and Rosenbeck 1942) has been the most widely used method for estimating DNA content for several decades, and is still generally considered a reliable method for making quantitative measurements of DNA content (Chieco and Derenzini 1999; Biesterfeld et al. 2011). Measurement errors are typically <5% using this method (Gregory 2005), which for our analyses is negligible given the orders of magnitude variation in DNA content among the cells we consider. The method works by staining DNA owing to the reaction of Schiff or pseudo-Schiff reagents with aldehydes, which are converted from deoxyribose in DNA after HCl hydrolysis (Chieco and Derenzini 1999). The light absorbance of the stained genetic material is then measured to quantify the relative DNA content of cells.
Table 1.
Human nuclear DNA content and cell volume data
| Cell type | Cell/nuclear size (original values) | DNA content (original values) | Size, content (original units) | Cell volume (μ3) | DNA content (pg) | Sources |
|---|---|---|---|---|---|---|
| Amnion epithelial cell (s) | 3.0 | 40.7 | c, a | 1072.1 | 5.9 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.6 | 41.6 | c, a | 958.0 | 6.0 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.9 | 41.7 | c, a | 1048.5 | 6.0 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.0 | 42.4 | c, a | 1437.0 | 6.1 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.4 | 42.7 | c, a | 1227.5 | 6.2 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.5 | 43.0 | c, a | 895.3 | 6.2 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.5 | 43.0 | c, a | 915.8 | 6.2 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.7 | 44.0 | c, a | 968.9 | 6.4 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.8 | 44.6 | c, a | 1002.2 | 6.5 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.9 | 44.6 | c, a | 1036.5 | 6.5 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.2 | 44.7 | c, a | 1147.2 | 6.5 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.0 | 45.4 | c, a | 1437.0 | 6.6 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.6 | 45.5 | c, a | 936.7 | 6.6 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.9 | 45.6 | c, a | 1036.5 | 6.6 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.6 | 46.8 | c, a | 1298.5 | 6.8 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.0 | 47.2 | c, a | 1072.1 | 6.8 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.4 | 47.6 | c, a | 865.5 | 6.9 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.8 | 47.7 | c, a | 1002.2 | 6.9 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.8 | 47.7 | c, a | 1013.5 | 6.9 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.9 | 48.0 | c, a | 1405.1 | 7.0 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.7 | 48.7 | c, a | 1682.4 | 7.1 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.7 | 48.8 | c, a | 968.9 | 7.1 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.9 | 48.8 | c, a | 1036.5 | 7.1 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.1 | 48.9 | c, a | 1109.2 | 7.1 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.8 | 49.1 | c, a | 1373.6 | 7.1 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.6 | 49.3 | c, a | 936.7 | 7.1 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.8 | 49.4 | c, a | 1013.5 | 7.2 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.7 | 49.9 | c, a | 968.9 | 7.2 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.2 | 50.3 | c, a | 1503.2 | 7.3 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.9 | 50.5 | c, a | 1036.5 | 7.3 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.1 | 50.6 | c, a | 1134.2 | 7.3 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.4 | 50.7 | c, a | 1213.8 | 7.3 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.5 | 51.0 | c, a | 905.6 | 7.4 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.0 | 52.0 | c, a | 1437.0 | 7.5 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 2.7 | 52.2 | c, a | 968.9 | 7.6 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.2 | 52.4 | c, a | 1173.3 | 7.6 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.5 | 53.0 | c, a | 1255.3 | 7.7 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 1.8 | 53.0 | c, a | 660.8 | 7.7 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.4 | 53.2 | c, a | 1572.4 | 7.7 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.0 | 53.5 | c, a | 1072.1 | 7.8 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.9 | 53.8 | c, a | 1405.1 | 7.8 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.9 | 54.4 | c, a | 1389.4 | 7.9 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.5 | 54.9 | c, a | 1255.3 | 8.0 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.2 | 57.4 | c, a | 1173.3 | 8.3 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.9 | 58.2 | c, a | 1405.1 | 8.4 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.6 | 62.2 | c, a | 1313.2 | 9.0 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.8 | 64.4 | c, a | 1373.6 | 9.3 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.9 | 71.4 | c, a | 1389.4 | 10.3 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.8 | 80.8 | c, a | 1358.1 | 11.7 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.6 | 82.6 | c, a | 1313.2 | 12.0 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.3 | 85.7 | c, a | 1537.5 | 12.4 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 3.8 | 87.5 | c, a | 1358.1 | 12.7 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.6 | 89.1 | c, a | 1992.2 | 12.9 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.0 | 89.6 | c, a | 1420.9 | 13.0 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.3 | 90.1 | c, a | 1904.3 | 13.1 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.5 | 91.9 | c, a | 1608.4 | 13.3 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.0 | 92.7 | c, a | 1420.9 | 13.4 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.3 | 93.9 | c, a | 1537.5 | 13.6 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.6 | 94.0 | c, a | 1663.6 | 13.6 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.2 | 97.1 | c, a | 1520.3 | 14.1 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.8 | 97.7 | c, a | 2060.6 | 14.2 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.9 | 98.5 | c, a | 1760.2 | 14.3 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.5 | 98.7 | c, a | 1970.0 | 14.3 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.3 | 100.5 | c, a | 1537.5 | 14.6 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.8 | 101.0 | c, a | 2060.6 | 14.6 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.1 | 102.0 | c, a | 1820.4 | 14.8 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 6.0 | 102.2 | c, a | 2131.4 | 14.8 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.6 | 102.9 | c, a | 1645.1 | 14.9 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.7 | 104.5 | c, a | 2014.8 | 15.1 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.1 | 106.7 | c, a | 1820.4 | 15.5 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.3 | 106.8 | c, a | 1883.0 | 15.5 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.0 | 107.9 | c, a | 1780.0 | 15.6 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 4.9 | 109.1 | c, a | 1740.3 | 15.8 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 6.9 | 114.8 | c, a | 2439.8 | 16.6 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.5 | 115.7 | c, a | 1970.0 | 16.8 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 6.7 | 118.7 | c, a | 2358.9 | 17.2 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 7.2 | 120.2 | c, a | 2552.4 | 17.4 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.9 | 124.0 | c, a | 2107.4 | 18.0 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.7 | 126.7 | c, a | 2014.8 | 18.4 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 5.9 | 132.7 | c, a | 2107.4 | 19.2 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 7.0 | 142.5 | c, a | 2467.4 | 20.7 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 7.8 | 149.4 | c, a | 2761.6 | 21.7 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 7.6 | 156.3 | c, a | 2700.3 | 22.7 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 8.7 | 171.5 | c, a | 3056.5 | 24.9 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 7.8 | 185.4 | c, a | 2761.6 | 26.9 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 8.3 | 185.5 | c, a | 2921.7 | 26.9 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 8.9 | 187.9 | c, a | 3126.2 | 27.2 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 8.3 | 192.0 | c, a | 2921.7 | 27.8 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 7.3 | 209.8 | c, a | 2581.4 | 30.4 | Klinger and Schwarzacher 1960 |
| Amnion epithelial cell (s) | 6.8 | 224.3 | c, a | 2412.5 | 32.5 | Klinger and Schwarzacher 1960 |
| Cardiomyocyte (m) | 17401.0 | 20.0 | v, pg | 5646.7 | 20.0 | Vliegen et al. 1990 |
| Cardiomyocyte (m) | 20794.0 | 21.2 | v, pg | 6339.9 | 21.2 | Vliegen et al. 1990 |
| Cardiomyocyte (m) | 43.8 | 3.6 | cc, p | 1463.2 | 12.6 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 60.7 | 4.1 | cc, p | 2013.5 | 14.4 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 55.5 | 4.5 | cc, p | 1844.6 | 15.8 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 135.5 | 4.6 | cc, p | 4403.8 | 16.1 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 156.4 | 5.1 | cc, p | 5064.6 | 17.9 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 127.0 | 5.8 | cc, p | 4132.1 | 20.2 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 65.1 | 5.9 | cc, p | 2154.9 | 20.7 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 68.1 | 5.9 | cc, p | 2252.3 | 20.7 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 77.3 | 5.9 | cc, p | 2546.1 | 20.7 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 71.1 | 6.0 | cc, p | 2346.7 | 21.0 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 119.2 | 6.6 | cc, p | 3885.0 | 23.1 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 60.3 | 6.8 | cc, p | 2000.2 | 23.8 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 84.1 | 7.2 | cc, p | 2765.2 | 25.2 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 103.7 | 7.9 | cc, p | 3392.5 | 27.6 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 104.0 | 7.9 | cc, p | 3402.4 | 27.7 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 112.1 | 8.4 | cc, p | 3658.9 | 29.4 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 99.8 | 8.5 | cc, p | 3266.2 | 29.8 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 161.5 | 8.6 | cc, p | 5224.0 | 30.1 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 152.5 | 8.8 | cc, p | 4939.6 | 30.8 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 105.1 | 9.0 | cc, p | 3436.8 | 31.5 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 175.4 | 9.5 | cc, p | 5660.7 | 33.3 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 97.6 | 11.9 | cc, p | 3198.5 | 41.7 | Wohlschlaeger et al. 2010 |
| Cardiomyocyte (m) | 129.9 | 13.6 | cc, p | 4225.6 | 47.6 | Wohlschlaeger et al. 2010 |
| Cytotrophoblast (m) | 133.0 | 1.9 | cc, p | 1061.5 | 6.7 | Galton 1962 |
| Cytotrophoblast (m) | 109.0 | 2.1 | cc, p | 932.7 | 7.4 | Galton 1962 |
| Cytotrophoblast (m) | 125.0 | 2.4 | cc, p | 1019.6 | 8.3 | Galton 1962 |
| Cytotrophoblast (m) | 128.0 | 2.4 | cc, p | 1035.4 | 8.5 | Galton 1962 |
| Endocrine pancreatic cell (m) | 98.4 | 2.0 | cc, p | 872.7 | 7.0 | Ehrie and Swartz 1974 |
| Endocrine pancreatic cell (m) | 116.5 | 2.0 | cc, p | 973.9 | 7.0 | Ehrie and Swartz 1974 |
| Endocrine pancreatic cell (m) | 119.0 | 2.0 | cc, p | 987.5 | 7.0 | Ehrie and Swartz 1974 |
| Endocrine pancreatic cell (m) | 205.4 | 4.0 | cc, p | 1408.0 | 14.0 | Ehrie and Swartz 1974 |
| Endocrine pancreatic cell (m) | 217.8 | 4.0 | cc, p | 1462.7 | 14.0 | Ehrie and Swartz 1974 |
| Endocrine pancreatic cell (m) | 221.8 | 4.0 | cc, p | 1480.1 | 14.0 | Ehrie and Swartz 1974 |
| Endocrine pancreatic cell (m) | 435.5 | 8.0 | cc, p | 2294.9 | 28.0 | Ehrie and Swartz 1974 |
| Endocrine pancreatic cell (m) | 447.3 | 8.0 | cc, p | 2335.1 | 28.0 | Ehrie and Swartz 1974 |
| Endocrine pancreatic cell (m) | 453.1 | 8.0 | cc, p | 2354.8 | 28.0 | Ehrie and Swartz 1974 |
| Endothelial cell (m) | 36.5 | 4.2 | cc, pg | 1225.4 | 4.2 | Dorman et al. 1990 |
| Endothelial cell (m) | 42.4 | 4.2 | cc, pg | 1418.2 | 4.2 | Dorman et al. 1990 |
| Endothelial cell (m) | 41.1 | 4.6 | cc, pg | 1375.8 | 4.6 | Dorman et al. 1990 |
| Endothelial cell (m) | 63.5 | 5.3 | cc, pg | 2102.6 | 5.3 | Dorman et al. 1990 |
| Exocrine pancreatic cell (m) | 100.1 | 2.0 | cc, p | 882.5 | 7.0 | Ehrie and Swartz 1974 |
| Exocrine pancreatic cell (m) | 107.7 | 2.0 | cc, p | 925.5 | 7.0 | Ehrie and Swartz 1974 |
| Exocrine pancreatic cell (m) | 112.8 | 2.0 | cc, p | 953.7 | 7.0 | Ehrie and Swartz 1974 |
| Hepatic parenchymal cell (m) | 34.7 | 1.8 | cc, a | 1124.6 | 4.8 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 34.4 | 2.0 | cc, a | 1118.2 | 5.4 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 45.1 | 2.1 | cc, a | 1333.5 | 5.6 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 34.8 | 2.3 | cc, a | 1126.7 | 6.1 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 26.1 | 2.3 | cc, a | 934.6 | 6.2 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 38.9 | 2.3 | cc, a | 1211.3 | 6.2 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 37.9 | 2.3 | cc, a | 1191.0 | 6.3 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 36.6 | 2.4 | cc, a | 1164.2 | 6.5 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 33.9 | 2.5 | cc, a | 1107.7 | 6.6 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 33.2 | 2.5 | cc, a | 1092.8 | 6.6 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 32.9 | 2.5 | cc, a | 1086.3 | 6.7 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 41.7 | 2.5 | cc, a | 1267.3 | 6.7 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 27.6 | 2.5 | cc, a | 969.1 | 6.8 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 51.2 | 2.5 | cc, a | 1448.1 | 6.8 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 35.2 | 2.6 | cc, a | 1135.1 | 6.8 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 41.8 | 2.6 | cc, a | 1269.2 | 6.9 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 29.5 | 2.6 | cc, a | 1012.0 | 7.0 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 29.4 | 2.6 | cc, a | 1009.7 | 7.1 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 40.4 | 2.6 | cc, a | 1241.5 | 7.1 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 47.4 | 2.7 | cc, a | 1377.3 | 7.2 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 39.9 | 2.7 | cc, a | 1231.4 | 7.2 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 32.5 | 2.7 | cc, a | 1077.7 | 7.2 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 55.3 | 2.7 | cc, a | 1522.5 | 7.2 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 30.6 | 2.7 | cc, a | 1036.3 | 7.4 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 36.9 | 2.8 | cc, a | 1170.5 | 7.4 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 22.3 | 2.8 | cc, a | 843.7 | 7.5 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 36.4 | 2.8 | cc, a | 1160.1 | 7.5 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 28.1 | 2.8 | cc, a | 980.5 | 7.5 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 33.9 | 2.8 | cc, a | 1107.7 | 7.5 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 54.5 | 2.8 | cc, a | 1508.1 | 7.5 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 38.6 | 2.9 | cc, a | 1205.2 | 7.7 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 49.3 | 2.9 | cc, a | 1413.0 | 7.7 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 36.1 | 2.9 | cc, a | 1153.9 | 7.7 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 44.6 | 2.9 | cc, a | 1323.9 | 7.8 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 33.3 | 2.9 | cc, a | 1094.9 | 7.9 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 31.4 | 3.1 | cc, a | 1053.9 | 8.4 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 36.0 | 3.2 | cc, a | 1151.8 | 8.5 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 71.4 | 3.4 | cc, a | 1797.6 | 9.1 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 98.7 | 3.8 | cc, a | 2218.6 | 10.1 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 78.1 | 4.4 | cc, a | 1905.5 | 11.8 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 44.5 | 4.8 | cc, a | 1321.9 | 12.8 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 65.8 | 4.8 | cc, a | 1704.6 | 12.9 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 71.6 | 4.9 | cc, a | 1800.8 | 13.1 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 65.0 | 5.0 | cc, a | 1691.1 | 13.3 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 69.0 | 5.1 | cc, a | 1758.1 | 13.7 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 64.1 | 5.2 | cc, a | 1675.9 | 13.9 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 72.9 | 5.3 | cc, a | 1822.0 | 14.1 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 70.7 | 5.3 | cc, a | 1786.1 | 14.1 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 91.7 | 5.3 | cc, a | 2115.0 | 14.2 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 87.0 | 5.4 | cc, a | 2043.9 | 14.5 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 78.4 | 5.4 | cc, a | 1910.2 | 14.5 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 147.2 | 5.4 | cc, a | 2876.9 | 14.5 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 70.3 | 5.5 | cc, a | 1779.5 | 14.7 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 81.6 | 5.6 | cc, a | 1960.6 | 15.0 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 72.9 | 5.7 | cc, a | 1822.0 | 15.2 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 73.6 | 6.1 | cc, a | 1833.4 | 16.4 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 65.3 | 6.4 | cc, a | 1696.2 | 17.2 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 177.5 | 7.4 | cc, a | 3249.1 | 19.9 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 106.7 | 9.3 | cc, a | 2333.9 | 24.9 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 129.4 | 9.8 | cc, a | 2645.7 | 26.2 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 141.9 | 9.8 | cc, a | 2809.1 | 26.4 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 136.8 | 9.9 | cc, a | 2743.1 | 26.5 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 142.1 | 11.6 | cc, a | 2811.7 | 31.1 | Swartz 1956 |
| Hepatic parenchymal cell (m) | 144.7 | 13.6 | cc, a | 2845.0 | 36.6 | Swartz 1956 |
| Hepatocyte (s) | 35.7 | 18.3 | cc, a | 1200.9 | 8.7 | Meek and Harbison 1967 |
| Hepatocyte (s) | 46.1 | 20.7 | cc, a | 1537.3 | 9.9 | Meek and Harbison 1967 |
| Hepatocyte (s) | 41.9 | 21.0 | cc, a | 1400.8 | 10.0 | Meek and Harbison 1967 |
| Hepatocyte (s) | 59.3 | 22.4 | cc, a | 1965.6 | 10.7 | Meek and Harbison 1967 |
| Hepatocyte (s) | 39.0 | 22.6 | cc, a | 1306.2 | 10.8 | Meek and Harbison 1967 |
| Hepatocyte (s) | 42.2 | 23.6 | cc, a | 1411.4 | 11.3 | Meek and Harbison 1967 |
| Hepatocyte (s) | 73.4 | 23.8 | cc, a | 2422.6 | 11.4 | Meek and Harbison 1967 |
| Hepatocyte (s) | 64.7 | 24.4 | cc, a | 2142.5 | 11.6 | Meek and Harbison 1967 |
| Hepatocyte (s) | 46.7 | 25.6 | cc, a | 1558.2 | 12.2 | Meek and Harbison 1967 |
| Hepatocyte (s) | 68.0 | 25.8 | cc, a | 2246.3 | 12.3 | Meek and Harbison 1967 |
| Hepatocyte (s) | 64.1 | 26.7 | cc, a | 2121.7 | 12.7 | Meek and Harbison 1967 |
| Hepatocyte (s) | 89.5 | 29.5 | cc, a | 2939.3 | 14.1 | Meek and Harbison 1967 |
| Hepatocyte (s) | 54.1 | 30.6 | cc, a | 1798.8 | 14.6 | Meek and Harbison 1967 |
| Hepatocyte (s) | 73.1 | 32.1 | cc, a | 2412.3 | 15.3 | Meek and Harbison 1967 |
| Hepatocyte (s) | 89.5 | 32.5 | cc, a | 2939.3 | 15.5 | Meek and Harbison 1967 |
| Hepatocyte (s) | 64.1 | 33.3 | cc, a | 2121.7 | 15.9 | Meek and Harbison 1967 |
| Hepatocyte (s) | 73.1 | 34.4 | cc, a | 2412.3 | 16.4 | Meek and Harbison 1967 |
| Hepatocyte (s) | 96.3 | 35.8 | cc, a | 3155.6 | 17.1 | Meek and Harbison 1967 |
| Hepatocyte (s) | 53.8 | 35.9 | cc, a | 1788.3 | 17.1 | Meek and Harbison 1967 |
| Hepatocyte (s) | 68.0 | 38.3 | cc, a | 2246.3 | 18.2 | Meek and Harbison 1967 |
| Hepatocyte (s) | 84.1 | 39.1 | cc, a | 2763.9 | 18.6 | Meek and Harbison 1967 |
| Hepatocyte (s) | 78.9 | 39.7 | cc, a | 2598.6 | 18.9 | Meek and Harbison 1967 |
| Hepatocyte (s) | 63.8 | 39.9 | cc, a | 2111.3 | 19.0 | Meek and Harbison 1967 |
| Hepatocyte (s) | 68.3 | 40.3 | cc, a | 2256.7 | 19.2 | Meek and Harbison 1967 |
| Hepatocyte (s) | 72.8 | 40.9 | cc, a | 2401.9 | 19.5 | Meek and Harbison 1967 |
| Hepatocyte (s) | 113.4 | 41.9 | cc, a | 3699.8 | 19.9 | Meek and Harbison 1967 |
| Hepatocyte (s) | 126.2 | 43.3 | cc, a | 4109.2 | 20.6 | Meek and Harbison 1967 |
| Hepatocyte (s) | 90.2 | 43.7 | cc, a | 2959.9 | 20.8 | Meek and Harbison 1967 |
| Hepatocyte (s) | 106.9 | 44.1 | cc, a | 3494.7 | 21.0 | Meek and Harbison 1967 |
| Hepatocyte (s) | 101.1 | 45.7 | cc, a | 3309.8 | 21.8 | Meek and Harbison 1967 |
| Hepatocyte (s) | 96.0 | 47.0 | cc, a | 3145.3 | 22.4 | Meek and Harbison 1967 |
| Hepatocyte (s) | 90.2 | 47.6 | cc, a | 2959.9 | 22.7 | Meek and Harbison 1967 |
| Hepatocyte (s) | 140.1 | 48.0 | cc, a | 4548.1 | 22.9 | Meek and Harbison 1967 |
| Hepatocyte (s) | 95.3 | 48.7 | cc, a | 3124.7 | 23.2 | Meek and Harbison 1967 |
| Hepatocyte (s) | 107.2 | 49.1 | cc, a | 3504.9 | 23.4 | Meek and Harbison 1967 |
| Hepatocyte (s) | 120.8 | 49.8 | cc, a | 3935.3 | 23.7 | Meek and Harbison 1967 |
| Hepatocyte (s) | 90.2 | 50.3 | cc, a | 2959.9 | 23.9 | Meek and Harbison 1967 |
| Hepatocyte (s) | 140.1 | 50.9 | cc, a | 4548.1 | 24.3 | Meek and Harbison 1967 |
| Hepatocyte (s) | 101.4 | 52.7 | cc, a | 3320.1 | 25.1 | Meek and Harbison 1967 |
| Hepatocyte (s) | 113.7 | 53.4 | cc, a | 3710.1 | 25.4 | Meek and Harbison 1967 |
| Hepatocyte (s) | 156.2 | 56.6 | cc, a | 5057.1 | 27.0 | Meek and Harbison 1967 |
| Hepatocyte (s) | 113.0 | 58.7 | cc, a | 3689.5 | 27.9 | Meek and Harbison 1967 |
| Hepatocyte (s) | 120.8 | 63.0 | cc, a | 3935.3 | 30.0 | Meek and Harbison 1967 |
| Hepatocyte (s) | 140.7 | 79.6 | cc, a | 4568.4 | 37.9 | Meek and Harbison 1967 |
| Hepatocyte (s) | 170.4 | 91.6 | cc, a | 5503.9 | 43.6 | Meek and Harbison 1967 |
| Hepatocyte (s) | 177.5 | 97.9 | cc, a | 5726.9 | 46.6 | Meek and Harbison 1967 |
| Lymphocyte (m) | 17.9 | 4.0 | cc, pg | 611.8 | 4.0 | Dorman et al. 1990 |
| Lymphocyte (m) | 19.8 | 4.0 | cc, pg | 675.0 | 4.0 | Dorman et al. 1990 |
| Lymphocyte (m) | 18.5 | 4.2 | cc, pg | 631.8 | 4.2 | Dorman et al. 1990 |
| Lymphocyte (m) | 37.6 | 5.8 | cc, pg | 1261.4 | 5.8 | Dorman et al. 1990 |
| Lymphocyte (s) | 18.1 | 17.2 | cc, a | 618.7 | 6.3 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 19.9 | 17.4 | cc, a | 679.4 | 6.3 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 18.6 | 17.7 | cc, a | 633.9 | 6.4 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 23.5 | 18.5 | cc, a | 796.5 | 6.7 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 21.2 | 18.6 | cc, a | 721.0 | 6.7 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 22.8 | 18.8 | cc, a | 773.8 | 6.8 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 21.4 | 18.9 | cc, a | 728.5 | 6.9 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 22.9 | 18.9 | cc, a | 777.6 | 6.9 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 24.9 | 19.0 | cc, a | 845.4 | 6.9 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 22.4 | 19.0 | cc, a | 762.5 | 6.9 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 24.3 | 19.0 | cc, a | 822.9 | 6.9 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 23.0 | 19.1 | cc, a | 781.4 | 6.9 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 22.8 | 19.3 | cc, a | 773.8 | 7.0 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 23.8 | 19.7 | cc, a | 807.8 | 7.1 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 28.0 | 19.9 | cc, a | 946.9 | 7.2 | Bedi and Goldstein 1976 |
| Lymphocyte (s) | 7.4 | 2.7 | n, a | 1437.3 | 4.1 | Petrakis 1953 |
| Lymphocyte (s) | 5.5 | 3.0 | n, a | 793.2 | 4.5 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 3.0 | n, a | 1293.6 | 4.6 | Petrakis 1953 |
| Lymphocyte (s) | 5.2 | 3.0 | n, a | 712.0 | 4.6 | Petrakis 1953 |
| Lymphocyte (s) | 7.7 | 3.0 | n, a | 1562.6 | 4.6 | Petrakis 1953 |
| Lymphocyte (s) | 7.5 | 3.1 | n, a | 1462.0 | 4.7 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 3.1 | n, a | 1278.2 | 4.8 | Petrakis 1953 |
| Lymphocyte (s) | 7.1 | 3.1 | n, a | 1340.8 | 4.8 | Petrakis 1953 |
| Lymphocyte (s) | 5.6 | 3.2 | n, a | 824.1 | 4.8 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 3.3 | n, a | 1293.6 | 5.0 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 3.3 | n, a | 1666.4 | 5.0 | Petrakis 1953 |
| Lymphocyte (s) | 7.5 | 3.3 | n, a | 1462.0 | 5.0 | Petrakis 1953 |
| Lymphocyte (s) | 8.2 | 3.3 | n, a | 1746.7 | 5.0 | Petrakis 1953 |
| Lymphocyte (s) | 7.8 | 3.4 | n, a | 1597.1 | 5.2 | Petrakis 1953 |
| Lymphocyte (s) | 6.5 | 3.4 | n, a | 1118.1 | 5.2 | Petrakis 1953 |
| Lymphocyte (s) | 6.9 | 3.5 | n, a | 1239.8 | 5.3 | Petrakis 1953 |
| Lymphocyte (s) | 7.7 | 3.5 | n, a | 1562.6 | 5.3 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 3.5 | n, a | 1666.4 | 5.3 | Petrakis 1953 |
| Lymphocyte (s) | 7.5 | 3.5 | n, a | 1495.0 | 5.3 | Petrakis 1953 |
| Lymphocyte (s) | 5.0 | 3.5 | n, a | 680.8 | 5.4 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 3.5 | n, a | 953.5 | 5.4 | Petrakis 1953 |
| Lymphocyte (s) | 7.4 | 3.5 | n, a | 1453.6 | 5.4 | Petrakis 1953 |
| Lymphocyte (s) | 6.5 | 3.6 | n, a | 1118.1 | 5.4 | Petrakis 1953 |
| Lymphocyte (s) | 5.0 | 3.6 | n, a | 671.6 | 5.5 | Petrakis 1953 |
| Lymphocyte (s) | 8.2 | 3.6 | n, a | 1737.6 | 5.5 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 3.6 | n, a | 950.4 | 5.5 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 3.6 | n, a | 1293.6 | 5.5 | Petrakis 1953 |
| Lymphocyte (s) | 7.5 | 3.7 | n, a | 1462.0 | 5.6 | Petrakis 1953 |
| Lymphocyte (s) | 7.8 | 3.7 | n, a | 1579.6 | 5.6 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 3.7 | n, a | 1286.0 | 5.7 | Petrakis 1953 |
| Lymphocyte (s) | 5.0 | 3.7 | n, a | 671.6 | 5.7 | Petrakis 1953 |
| Lymphocyte (s) | 6.2 | 3.8 | n, a | 1028.4 | 5.8 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 3.8 | n, a | 956.9 | 5.8 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 3.8 | n, a | 953.5 | 5.8 | Petrakis 1953 |
| Lymphocyte (s) | 6.6 | 3.8 | n, a | 1142.7 | 5.8 | Petrakis 1953 |
| Lymphocyte (s) | 7.1 | 3.8 | n, a | 1317.1 | 5.8 | Petrakis 1953 |
| Lymphocyte (s) | 6.5 | 3.8 | n, a | 1113.7 | 5.8 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 3.9 | n, a | 953.5 | 5.9 | Petrakis 1953 |
| Lymphocyte (s) | 10.1 | 3.9 | n, a | 2627.9 | 5.9 | Petrakis 1953 |
| Lymphocyte (s) | 6.8 | 3.9 | n, a | 1217.0 | 5.9 | Petrakis 1953 |
| Lymphocyte (s) | 6.1 | 3.9 | n, a | 980.3 | 5.9 | Petrakis 1953 |
| Lymphocyte (s) | 6.3 | 3.9 | n, a | 1035.5 | 6.0 | Petrakis 1953 |
| Lymphocyte (s) | 7.2 | 3.9 | n, a | 1364.7 | 6.0 | Petrakis 1953 |
| Lymphocyte (s) | 7.5 | 3.9 | n, a | 1495.0 | 6.0 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 3.9 | n, a | 1299.0 | 6.0 | Petrakis 1953 |
| Lymphocyte (s) | 7.2 | 4.0 | n, a | 1370.2 | 6.1 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 4.0 | n, a | 1657.9 | 6.1 | Petrakis 1953 |
| Lymphocyte (s) | 8.1 | 4.0 | n, a | 1711.1 | 6.1 | Petrakis 1953 |
| Lymphocyte (s) | 10.0 | 4.0 | n, a | 2594.9 | 6.1 | Petrakis 1953 |
| Lymphocyte (s) | 6.5 | 4.0 | n, a | 1113.7 | 6.1 | Petrakis 1953 |
| Lymphocyte (s) | 6.8 | 4.0 | n, a | 1209.7 | 6.1 | Petrakis 1953 |
| Lymphocyte (s) | 5.6 | 4.0 | n, a | 836.6 | 6.2 | Petrakis 1953 |
| Lymphocyte (s) | 9.0 | 4.1 | n, a | 2115.1 | 6.2 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 4.1 | n, a | 1278.2 | 6.2 | Petrakis 1953 |
| Lymphocyte (s) | 6.8 | 4.1 | n, a | 1217.0 | 6.3 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 4.2 | n, a | 950.4 | 6.4 | Petrakis 1953 |
| Lymphocyte (s) | 6.5 | 4.2 | n, a | 1113.7 | 6.4 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 4.2 | n, a | 1286.0 | 6.4 | Petrakis 1953 |
| Lymphocyte (s) | 7.6 | 4.2 | n, a | 1512.1 | 6.4 | Petrakis 1953 |
| Lymphocyte (s) | 6.6 | 4.2 | n, a | 1142.7 | 6.4 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 4.3 | n, a | 1299.0 | 6.5 | Petrakis 1953 |
| Lymphocyte (s) | 7.6 | 4.3 | n, a | 1503.6 | 6.5 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 4.3 | n, a | 950.4 | 6.5 | Petrakis 1953 |
| Lymphocyte (s) | 6.9 | 4.3 | n, a | 1252.5 | 6.5 | Petrakis 1953 |
| Lymphocyte (s) | 8.3 | 4.3 | n, a | 1818.2 | 6.6 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 4.3 | n, a | 966.9 | 6.6 | Petrakis 1953 |
| Lymphocyte (s) | 6.6 | 4.3 | n, a | 1150.1 | 6.6 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 4.3 | n, a | 1286.0 | 6.6 | Petrakis 1953 |
| Lymphocyte (s) | 7.1 | 4.3 | n, a | 1325.1 | 6.6 | Petrakis 1953 |
| Lymphocyte (s) | 7.2 | 4.3 | n, a | 1356.6 | 6.6 | Petrakis 1953 |
| Lymphocyte (s) | 7.5 | 4.4 | n, a | 1485.0 | 6.6 | Petrakis 1953 |
| Lymphocyte (s) | 6.5 | 4.4 | n, a | 1106.4 | 6.7 | Petrakis 1953 |
| Lymphocyte (s) | 7.6 | 4.4 | n, a | 1503.6 | 6.7 | Petrakis 1953 |
| Lymphocyte (s) | 7.4 | 4.4 | n, a | 1435.4 | 6.7 | Petrakis 1953 |
| Lymphocyte (s) | 6.3 | 4.4 | n, a | 1056.6 | 6.7 | Petrakis 1953 |
| Lymphocyte (s) | 6.6 | 4.4 | n, a | 1135.6 | 6.7 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 4.4 | n, a | 966.9 | 6.7 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 4.4 | n, a | 1299.0 | 6.8 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 4.5 | n, a | 1666.4 | 6.8 | Petrakis 1953 |
| Lymphocyte (s) | 9.0 | 4.5 | n, a | 2095.0 | 6.8 | Petrakis 1953 |
| Lymphocyte (s) | 5.5 | 4.5 | n, a | 811.7 | 6.8 | Petrakis 1953 |
| Lymphocyte (s) | 6.2 | 4.5 | n, a | 1014.6 | 6.8 | Petrakis 1953 |
| Lymphocyte (s) | 6.8 | 4.5 | n, a | 1221.9 | 6.8 | Petrakis 1953 |
| Lymphocyte (s) | 7.6 | 4.5 | n, a | 1503.6 | 6.8 | Petrakis 1953 |
| Lymphocyte (s) | 5.8 | 4.5 | n, a | 897.3 | 6.9 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 4.6 | n, a | 1666.4 | 6.9 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 4.6 | n, a | 1299.0 | 6.9 | Petrakis 1953 |
| Lymphocyte (s) | 7.4 | 4.6 | n, a | 1453.6 | 7.0 | Petrakis 1953 |
| Lymphocyte (s) | 7.8 | 4.6 | n, a | 1588.3 | 7.0 | Petrakis 1953 |
| Lymphocyte (s) | 9.0 | 4.6 | n, a | 2105.0 | 7.0 | Petrakis 1953 |
| Lymphocyte (s) | 8.9 | 4.6 | n, a | 2075.5 | 7.0 | Petrakis 1953 |
| Lymphocyte (s) | 6.1 | 4.6 | n, a | 1000.9 | 7.0 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 4.6 | n, a | 1286.0 | 7.0 | Petrakis 1953 |
| Lymphocyte (s) | 6.3 | 4.6 | n, a | 1035.5 | 7.1 | Petrakis 1953 |
| Lymphocyte (s) | 6.6 | 4.6 | n, a | 1150.1 | 7.1 | Petrakis 1953 |
| Lymphocyte (s) | 7.6 | 4.6 | n, a | 1503.6 | 7.1 | Petrakis 1953 |
| Lymphocyte (s) | 6.9 | 4.7 | n, a | 1239.8 | 7.1 | Petrakis 1953 |
| Lymphocyte (s) | 7.8 | 4.7 | n, a | 1605.5 | 7.1 | Petrakis 1953 |
| Lymphocyte (s) | 9.0 | 4.7 | n, a | 2085.5 | 7.1 | Petrakis 1953 |
| Lymphocyte (s) | 7.8 | 4.7 | n, a | 1595.5 | 7.1 | Petrakis 1953 |
| Lymphocyte (s) | 6.2 | 4.7 | n, a | 1007.9 | 7.2 | Petrakis 1953 |
| Lymphocyte (s) | 7.5 | 4.7 | n, a | 1478.5 | 7.2 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 4.7 | n, a | 1666.4 | 7.2 | Petrakis 1953 |
| Lymphocyte (s) | 6.5 | 4.7 | n, a | 1132.6 | 7.2 | Petrakis 1953 |
| Lymphocyte (s) | 6.8 | 4.7 | n, a | 1209.7 | 7.2 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 4.7 | n, a | 1286.0 | 7.2 | Petrakis 1953 |
| Lymphocyte (s) | 7.4 | 4.7 | n, a | 1437.3 | 7.2 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 4.7 | n, a | 1307.0 | 7.2 | Petrakis 1953 |
| Lymphocyte (s) | 7.4 | 4.8 | n, a | 1429.3 | 7.3 | Petrakis 1953 |
| Lymphocyte (s) | 5.8 | 4.8 | n, a | 894.0 | 7.3 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 4.8 | n, a | 1675.4 | 7.3 | Petrakis 1953 |
| Lymphocyte (s) | 7.6 | 4.8 | n, a | 1520.3 | 7.3 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 4.8 | n, a | 1275.9 | 7.4 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 4.9 | n, a | 1291.5 | 7.4 | Petrakis 1953 |
| Lymphocyte (s) | 7.2 | 4.9 | n, a | 1346.3 | 7.4 | Petrakis 1953 |
| Lymphocyte (s) | 7.4 | 4.9 | n, a | 1437.3 | 7.4 | Petrakis 1953 |
| Lymphocyte (s) | 7.5 | 4.9 | n, a | 1470.4 | 7.4 | Petrakis 1953 |
| Lymphocyte (s) | 6.7 | 4.9 | n, a | 1194.4 | 7.5 | Petrakis 1953 |
| Lymphocyte (s) | 9.0 | 4.9 | n, a | 2095.0 | 7.5 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 4.9 | n, a | 953.5 | 7.5 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 4.9 | n, a | 1666.4 | 7.5 | Petrakis 1953 |
| Lymphocyte (s) | 7.1 | 5.0 | n, a | 1314.9 | 7.6 | Petrakis 1953 |
| Lymphocyte (s) | 8.1 | 5.0 | n, a | 1719.8 | 7.6 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 5.0 | n, a | 1666.4 | 7.7 | Petrakis 1953 |
| Lymphocyte (s) | 7.9 | 5.1 | n, a | 1614.3 | 7.7 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 5.1 | n, a | 966.9 | 7.7 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 5.1 | n, a | 1283.6 | 7.7 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 5.1 | n, a | 1293.6 | 7.8 | Petrakis 1953 |
| Lymphocyte (s) | 8.4 | 5.1 | n, a | 1846.4 | 7.8 | Petrakis 1953 |
| Lymphocyte (s) | 7.7 | 5.1 | n, a | 1571.3 | 7.8 | Petrakis 1953 |
| Lymphocyte (s) | 6.6 | 5.1 | n, a | 1150.1 | 7.8 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 5.1 | n, a | 1657.9 | 7.8 | Petrakis 1953 |
| Lymphocyte (s) | 8.1 | 5.1 | n, a | 1711.1 | 7.8 | Petrakis 1953 |
| Lymphocyte (s) | 6.8 | 5.2 | n, a | 1224.7 | 7.9 | Petrakis 1953 |
| Lymphocyte (s) | 7.5 | 5.2 | n, a | 1495.0 | 7.9 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 5.2 | n, a | 1684.4 | 7.9 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 5.2 | n, a | 970.6 | 7.9 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 5.3 | n, a | 1691.8 | 8.0 | Petrakis 1953 |
| Lymphocyte (s) | 6.0 | 5.3 | n, a | 940.2 | 8.1 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 5.3 | n, a | 1684.4 | 8.1 | Petrakis 1953 |
| Lymphocyte (s) | 8.1 | 5.3 | n, a | 1719.8 | 8.1 | Petrakis 1953 |
| Lymphocyte (s) | 5.6 | 5.4 | n, a | 843.0 | 8.1 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 5.4 | n, a | 1307.0 | 8.2 | Petrakis 1953 |
| Lymphocyte (s) | 9.8 | 5.4 | n, a | 2486.4 | 8.2 | Petrakis 1953 |
| Lymphocyte (s) | 9.0 | 5.4 | n, a | 2085.5 | 8.2 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 5.4 | n, a | 1674.1 | 8.2 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 5.5 | n, a | 1666.4 | 8.3 | Petrakis 1953 |
| Lymphocyte (s) | 7.1 | 5.5 | n, a | 1314.9 | 8.4 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 5.5 | n, a | 1691.8 | 8.4 | Petrakis 1953 |
| Lymphocyte (s) | 7.3 | 5.5 | n, a | 1388.8 | 8.4 | Petrakis 1953 |
| Lymphocyte (s) | 8.6 | 5.6 | n, a | 1912.5 | 8.5 | Petrakis 1953 |
| Lymphocyte (s) | 9.0 | 5.6 | n, a | 2095.0 | 8.5 | Petrakis 1953 |
| Lymphocyte (s) | 7.5 | 5.6 | n, a | 1487.0 | 8.5 | Petrakis 1953 |
| Lymphocyte (s) | 7.2 | 5.6 | n, a | 1356.6 | 8.6 | Petrakis 1953 |
| Lymphocyte (s) | 6.1 | 5.7 | n, a | 977.2 | 8.7 | Petrakis 1953 |
| Lymphocyte (s) | 6.8 | 5.7 | n, a | 1229.6 | 8.7 | Petrakis 1953 |
| Lymphocyte (s) | 7.6 | 5.7 | n, a | 1512.1 | 8.7 | Petrakis 1953 |
| Lymphocyte (s) | 8.4 | 5.7 | n, a | 1856.2 | 8.7 | Petrakis 1953 |
| Lymphocyte (s) | 5.6 | 5.8 | n, a | 824.1 | 8.8 | Petrakis 1953 |
| Lymphocyte (s) | 7.1 | 5.8 | n, a | 1309.1 | 8.8 | Petrakis 1953 |
| Lymphocyte (s) | 9.2 | 5.9 | n, a | 2215.0 | 9.0 | Petrakis 1953 |
| Lymphocyte (s) | 6.3 | 6.0 | n, a | 1049.4 | 9.1 | Petrakis 1953 |
| Lymphocyte (s) | 7.8 | 6.0 | n, a | 1597.1 | 9.1 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 6.0 | n, a | 1675.4 | 9.1 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 6.1 | n, a | 1684.4 | 9.3 | Petrakis 1953 |
| Lymphocyte (s) | 9.0 | 6.1 | n, a | 2095.0 | 9.4 | Petrakis 1953 |
| Lymphocyte (s) | 7.0 | 6.2 | n, a | 1307.0 | 9.5 | Petrakis 1953 |
| Lymphocyte (s) | 6.9 | 6.3 | n, a | 1267.8 | 9.5 | Petrakis 1953 |
| Lymphocyte (s) | 9.2 | 6.3 | n, a | 2205.2 | 9.6 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 6.4 | n, a | 1684.4 | 9.7 | Petrakis 1953 |
| Lymphocyte (s) | 9.2 | 6.4 | n, a | 2205.2 | 9.8 | Petrakis 1953 |
| Lymphocyte (s) | 9.2 | 6.5 | n, a | 2215.0 | 9.9 | Petrakis 1953 |
| Lymphocyte (s) | 9.1 | 6.6 | n, a | 2134.8 | 10.0 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 6.6 | n, a | 1666.4 | 10.1 | Petrakis 1953 |
| Lymphocyte (s) | 8.0 | 6.7 | n, a | 1666.4 | 10.2 | Petrakis 1953 |
| Lymphocyte (s) | 6.6 | 6.8 | n, a | 1162.1 | 10.4 | Petrakis 1953 |
| Lymphocyte (s) | 9.0 | 6.9 | n, a | 2124.7 | 10.5 | Petrakis 1953 |
| Lymphocyte (s) | 9.1 | 6.9 | n, a | 2164.8 | 10.5 | Petrakis 1953 |
| Lymphocyte (s) | 9.1 | 7.0 | n, a | 2144.5 | 10.7 | Petrakis 1953 |
| Lymphocyte (s) | 11.1 | 7.4 | n, a | 3158.4 | 11.3 | Petrakis 1953 |
| Lymphocyte (s) | 12.1 | 8.5 | n, a | 3762.4 | 13.0 | Petrakis 1953 |
| Lymphocyte polymorph (m) | 25.1 | 17.9 | cc, a | 849.2 | 6.5 | Bedi and Goldstein 1976 |
| Lymphocyte polymorph (m) | 22.8 | 18.0 | cc, a | 773.8 | 6.5 | Bedi and Goldstein 1976 |
| Lymphocyte polymorph (m) | 21.4 | 18.0 | cc, a | 728.5 | 6.5 | Bedi and Goldstein 1976 |
| Lymphocyte polymorph (m) | 20.0 | 18.1 | cc, a | 683.1 | 6.6 | Bedi and Goldstein 1976 |
| Lymphocyte polymorph (m) | 24.4 | 18.2 | cc, a | 826.6 | 6.6 | Bedi and Goldstein 1976 |
| Lymphocyte polymorph (m) | 21.3 | 18.2 | cc, a | 724.8 | 6.6 | Bedi and Goldstein 1976 |
| Lymphocyte polymorph (m) | 27.1 | 18.2 | cc, a | 916.9 | 6.6 | Bedi and Goldstein 1976 |
| Lymphocyte polymorph (m) | 22.9 | 18.5 | cc, a | 777.6 | 6.7 | Bedi and Goldstein 1976 |
| Lymphocyte polymorph (m) | 25.7 | 18.9 | cc, a | 871.8 | 6.9 | Bedi and Goldstein 1976 |
| Lymphocyte polymorph (m) | 28.0 | 19.0 | cc, a | 946.9 | 6.9 | Bedi and Goldstein 1976 |
| Lymphocyte polymorph (m) | 26.2 | 19.0 | cc, a | 886.8 | 6.9 | Bedi and Goldstein 1976 |
| Megakaryocyte (m) | 15.5 | 3.9 | d, p | 1949.8 | 13.7 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 15.3 | 4.4 | d, p | 1875.3 | 15.4 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 16.1 | 4.4 | d, p | 2352.1 | 15.4 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 16.5 | 4.4 | d, p | 2185.1 | 15.4 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 16.3 | 4.6 | d, p | 2267.6 | 16.1 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 16.8 | 5.3 | d, p | 2482.7 | 18.6 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 21.3 | 7.0 | d, p | 5059.8 | 24.5 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 20.8 | 7.3 | d, p | 4711.8 | 25.6 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 22.4 | 8.6 | d, p | 5884.9 | 30.1 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 27.5 | 10.4 | d, p | 10889.2 | 36.4 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 27.6 | 10.6 | d, p | 11008.4 | 37.1 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 30.2 | 10.9 | d, p | 14421.8 | 38.2 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 30.6 | 12.6 | d, p | 15002.5 | 44.1 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 32.3 | 12.8 | d, p | 17644.4 | 44.8 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 33.3 | 14.1 | d, p | 19334.4 | 49.4 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 33.8 | 14.5 | d, p | 20218.5 | 50.8 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 33.8 | 16.1 | d, p | 20218.5 | 56.4 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 34.2 | 16.8 | d, p | 20944.8 | 58.8 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 34.1 | 16.9 | d, p | 20761.6 | 59.2 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 34.4 | 17.2 | d, p | 21314.4 | 60.2 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 34.4 | 17.2 | d, p | 21314.4 | 60.2 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 35.0 | 17.7 | d, p | 22449.3 | 62.0 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 36.0 | 17.7 | d, p | 24429.0 | 62.0 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 35.4 | 18.6 | d, p | 23227.8 | 65.1 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 35.4 | 19.3 | d, p | 23227.8 | 67.6 | Ishibashi et al. 1986 |
| Megakaryocyte (m) | 36.4 | 19.6 | d, p | 25252.4 | 68.6 | Ishibashi et al. 1986 |
| Monocyte (s) | 21.4 | 19.3 | cc, a | 728.5 | 7.0 | Bedi and Goldstein 1976 |
| Monocyte (s) | 32.1 | 19.9 | cc, a | 1081.8 | 7.2 | Bedi and Goldstein 1976 |
| Monocyte (s) | 26.3 | 20.0 | cc, a | 890.6 | 7.3 | Bedi and Goldstein 1976 |
| Monocyte (s) | 30.8 | 20.1 | cc, a | 1036.9 | 7.3 | Bedi and Goldstein 1976 |
| Monocyte (s) | 30.6 | 20.2 | cc, a | 1033.1 | 7.3 | Bedi and Goldstein 1976 |
| Monocyte (s) | 30.9 | 20.3 | cc, a | 1040.7 | 7.4 | Bedi and Goldstein 1976 |
| Monocyte (s) | 46.2 | 21.5 | cc, a | 1543.3 | 7.8 | Bedi and Goldstein 1976 |
| Primary spermatocyte (m) | 37.6 | 2.6 | cc, a | 1184.8 | 7.6 | Leuchtenberger et al. 1956 |
| Primary spermatocyte (m) | 28.1 | 3.3 | cc, a | 980.5 | 9.7 | Leuchtenberger et al. 1956 |
| Primary spermatocyte (m) | 59.3 | 3.3 | cc, a | 1593.2 | 9.7 | Leuchtenberger et al. 1956 |
| Primary spermatocyte (m) | 42.1 | 3.9 | cc, a | 1275.2 | 11.3 | Leuchtenberger et al. 1956 |
| Primary spermatocyte (m) | 69.4 | 4.6 | cc, a | 1764.7 | 13.3 | Leuchtenberger et al. 1956 |
| Primary spermatocyte (m) | 45.1 | 4.6 | cc, a | 1333.5 | 13.4 | Leuchtenberger et al. 1956 |
| Primary spermatocyte (m) | 48.6 | 5.1 | cc, a | 1399.9 | 14.7 | Leuchtenberger et al. 1956 |
| Primary spermatocyte (m) | 48.2 | 5.4 | cc, a | 1392.4 | 15.8 | Leuchtenberger et al. 1956 |
| Secondary spermatocyte (m) | 14.2 | 1.5 | cc, a | 629.2 | 4.2 | Leuchtenberger et al. 1956 |
| Secondary spermatocyte (m) | 13.1 | 1.9 | cc, a | 597.0 | 5.4 | Leuchtenberger et al. 1956 |
| Secondary spermatocyte (m) | 17.6 | 1.9 | cc, a | 723.4 | 5.5 | Leuchtenberger et al. 1956 |
| Secondary spermatocyte (m) | 19.9 | 2.3 | cc, a | 783.5 | 6.8 | Leuchtenberger et al. 1956 |
| Secondary spermatocyte (m) | 33.4 | 2.6 | cc, a | 1097.0 | 7.7 | Leuchtenberger et al. 1956 |
| Secondary spermatocyte (m) | 25.9 | 2.7 | cc, a | 929.9 | 8.0 | Leuchtenberger et al. 1956 |
| Secondary spermatocyte (m) | 20.6 | 2.9 | cc, a | 801.3 | 8.4 | Leuchtenberger et al. 1956 |
| Secondary spermatocyte (m) | 26.2 | 3.0 | cc, a | 936.9 | 8.7 | Leuchtenberger et al. 1956 |
| Spermatid (m) | 5.2 | 0.6 | cc, a | 327.5 | 1.8 | Leuchtenberger et al. 1956 |
| Spermatid (m) | 8.7 | 1.0 | cc, a | 457.6 | 2.9 | Leuchtenberger et al. 1956 |
| Spermatid (m) | 5.3 | 1.0 | cc, a | 331.6 | 3.0 | Leuchtenberger et al. 1956 |
| Spermatid (m) | 8.0 | 1.1 | cc, a | 433.3 | 3.2 | Leuchtenberger et al. 1956 |
| Spermatid (m) | 8.7 | 1.4 | cc, a | 457.6 | 4.0 | Leuchtenberger et al. 1956 |
| Spermatid (m) | 9.0 | 1.5 | cc, a | 467.8 | 4.3 | Leuchtenberger et al. 1956 |
| Spermatid (m) | 9.5 | 1.5 | cc, a | 484.5 | 4.4 | Leuchtenberger et al. 1956 |
| Spermatid (m) | 6.5 | 1.5 | cc, a | 378.6 | 4.5 | Leuchtenberger et al. 1956 |
| Spermatogonium (m) | 39.3 | 1.9 | cc, a | 1219.4 | 5.6 | Leuchtenberger et al. 1956 |
| Spermatogonium (m) | 23.4 | 2.3 | cc, a | 870.5 | 6.7 | Leuchtenberger et al. 1956 |
| Spermatogonium (m) | 19.7 | 2.6 | cc, a | 778.4 | 7.4 | Leuchtenberger et al. 1956 |
| Spermatogonium (m) | 38.6 | 2.8 | cc, a | 1205.2 | 8.1 | Leuchtenberger et al. 1956 |
| Spermatogonium (m) | 51.1 | 3.4 | cc, a | 1446.3 | 10.0 | Leuchtenberger et al. 1956 |
| Spermatogonium (m) | 26.7 | 3.6 | cc, a | 948.4 | 10.5 | Leuchtenberger et al. 1956 |
| Spermatogonium (m) | 35.9 | 3.6 | cc, a | 1149.7 | 10.5 | Leuchtenberger et al. 1956 |
| Spermatogonium (m) | 51.9 | 3.8 | cc, a | 1461.0 | 11.0 | Leuchtenberger et al. 1956 |
| Spermatozoon (m) | 11.7 | 6.4 | c, a | 404.5 | 3.1 | Royere et al. 1988 |
| Spermatozoon (m) | 11.9 | 6.7 | cc, a | 410.9 | 3.3 | Royere et al. 1988 |
| Spermatozoon (m) | 11.1 | 6.7 | cc, a | 384.9 | 3.3 | Royere et al. 1988 |
| Spermatozoon (m) | 11.0 | 7.0 | cc, a | 380.5 | 3.4 | Royere et al. 1988 |
| Spermatozoon (m) | 11.6 | 7.0 | cc, a | 400.8 | 3.5 | Royere et al. 1988 |
| Spermatozoon (m) | 12.0 | 7.3 | cc, a | 414.2 | 3.6 | Royere et al. 1988 |
| Spermatozoon (m) | 11.5 | 7.4 | cc, a | 398.4 | 3.6 | Royere et al. 1988 |
| Spermatozoon (m) | 11.4 | 7.5 | cc, a | 394.0 | 3.7 | Royere et al. 1988 |
| Spermatozoon (m) | 13.0 | 7.7 | cc, a | 447.9 | 3.8 | Royere et al. 1988 |
| Spermatozoon (m) | 11.7 | 7.9 | cc, a | 405.1 | 3.9 | Royere et al. 1988 |
| Syncytiotrophoblast (m) | 81.0 | 1.8 | cc, p | 769.0 | 6.4 | Galton 1962 |
| Syncytiotrophoblast (m) | 72.0 | 1.9 | cc, p | 712.3 | 6.7 | Galton 1962 |
| Syncytiotrophoblast (m) | 79.0 | 2.2 | cc, p | 756.6 | 7.7 | Galton 1962 |
| Syncytiotrophoblast (m) | 77.0 | 2.2 | cc, p | 744.1 | 7.8 | Galton 1962 |
| Villous stroma (m) | 82.0 | 2.1 | cc, p | 775.2 | 7.4 | Galton 1962 |
| Villous stroma (m) | 108.0 | 2.1 | cc, p | 927.1 | 7.5 | Galton 1962 |
| Villous stroma (m) | 96.0 | 2.4 | cc, p | 858.8 | 8.5 | Galton 1962 |
| Villous stroma (m) | 102.0 | 2.5 | cc, p | 893.3 | 8.7 | Galton 1962 |
Data from 19 cell types used in analyses shown in Figures 1 and 2. The original (i.e., reported) values for cell/nuclear size and DNA content are listed in columns 2 and 3, and the original units in which these values were expressed are listed in column 4. For analyses, all of the measures of cell/nuclear size were converted to units of cell volume (μ3) shown in column 5, and measures of DNA content were converted to picograms of DNA shown in column 6. Letters in parentheses following the listing of cell type in column 1 denote whether the value is a measurement of a single cell (s) or the mean of a population of cells (m). The first notation used in column 4 indicates whether cell/nuclear size measurements were originally reported as cell diameter (d), nuclear cross-sectional area in calibrated arbitrary units (c), nuclear cross-sectional area in μ2 (cc), nuclear diameter in μ (n), or cell volume in μ3 (v). The second notation in column 4 indicates whether DNA content measurements were reported in units of ploidy level (p), arbitrary absorbance units (a), or picograms of DNA (pg). Cell types are reported based on cell-type designations given in original studies.
Computation of mean cell volumes: Studies reported cell or nuclear size using one of four measurements: cell volume, cell diameter/radius, nuclear cross-sectional area, or nuclear diameter/radius. Measurements of cell diameter/radius were converted to cell volume assuming cells were spherical. Measurements of nuclear cross-sectional area and diameter/radius were converted to nuclear volume assuming nuclei were spherical and that nuclear cross-sectional areas were circular. All estimates of nuclear volume were then converted to cell volume using a previously reported relationship between cell and nuclear volume of human cells (Swanson et al. 1991): cell volume = 44.2 (nuclear volume)0.65 as indicated in the text. In one case (Klinger and Schwarzacher 1960), measurements of nuclear area were given in calibrated arbitrary units. In this case, we converted back from arbitrary units to units of squared micrometers before computing nuclear radius (in micrometers).
Computation of mean DNA content: DNA content measurements were originally reported in one of three ways: as picograms of DNA, as ploidy level, or as arbitrary absorbance units of DNA. To convert ploidy level to pg DNA, we multiplied by the typical haploid DNA content of human cells (3.5 pg [Gregory 2012]). To convert arbitrary units to pg DNA, we determined the mean arbitrary unit value of cells that the original investigators of each study designated as diploid, and then assumed this corresponded to 2 × 3.5 = 7 pg of DNA to convert values from arbitrary units to pg DNA. Thus, each of the seven studies in which this conversion was performed had a unique conversion factor: Bedi and Goldstein 1976 (2.75 arbitrary units/pg DNA), Klinger and Schwarzacher 1960 (6.9 arbitrary units/pg DNA), Leuchtenberger et al. 1956 (0.34 arbitrary units/pg DNA), Meek and Harbison 1967 (2.1 arbitrary units/pg DNA), Petrakis 1953 (0.66 arbitrary units/pg DNA), Royere et al. 1988 (2.03 arbitrary units/pg DNA), and Swartz 1956 (0.37 pg). This conversion is possible because the Feulgen staining method results in a linear relationship between light absorbance and DNA content as discussed in the text.
Averaging individal cell measurements: For cases in which individual cell measurements were reported, we averaged all cell volume and DNA content measurements to yield a single population mean for each cell type within each study. Mean cell volumes of these cell types were 1574 µ3 (amnion epithelial cells [Klinger and Schwarzacher 1960]), 2936 µ3 (hepatocytes [Meek and Harbison 1967]), 1441 µ3 (lymphocytes [Petrakis 1953]), 764 µ3 (lymphocytes [Bedi and Goldstein 1976]), 817 µ3 (lymphocyte polymorphs [Bedi and Goldstein 1976]), and 1051 µ3 (monocytes [Bedi and Goldstein 1976]). Corresponding mean DNA contents were 12, 19.9, 7.1, 6.8, 6.7, and 7.3 pg, respectively. These means were used in the analyses presented in the text.
In cases in which the relative content of DNA was originally expressed in “arbitrary units” in the original studies, we converted these measures to pg DNA given that the Feulgen staining method results in a linear relationship between light absorbance and DNA content (Biesterfeld et al. 2011). In each case, a specific conversion factor was used for each cell type based on the observed relationship between light absorbance and DNA content in the particular study (see Table 1). This was done to avoid any bias associated with differences in DNA compaction level or uptake of stain across cell types. The conversions used for each cell type are given in Table 1.
Finally, in many studies considered here, only the population means of cell size and DNA content were reported. Thus, to remain consistent, for those studies that reported individual cell measurements, we calculated mean values and used these in the analyses. So, all points shown in the figures for humans represent means of DNA content and cell size from cell populations. In Table 1, we give all values for cell size and DNA content reported in the original sources, both population mean values and individual cell values. In the majority of cases, cell volumes were not directly reported, so we estimated these volumes from measures of nuclear size based on a published relationship between nuclear volume and cell volume across human cell types (see Table 1 for further details of methodology and data) (Swanson et al. 1991).
We evaluated the relationship between mean nuclear DNA content (pg) and cell volume (µ3) using a linear mixed model of the form log10 (DNA) = a + b log10 (cell volume) + C + D + E, where a and b are regression coefficients, C and D represent random effects of cell type (random slope and intercept, respectively), and E represents residual errors (Pinheiro and Bates 2000). This statistical model accounts for within-group autocorrelation, allowing us to include cells from multiple cell types in a single analysis. To determine whether DNA content was correlated with cell volume within cell types, we used ordinary least-squares regression. We verified normality of residuals using quantile–quantile plots.
Results
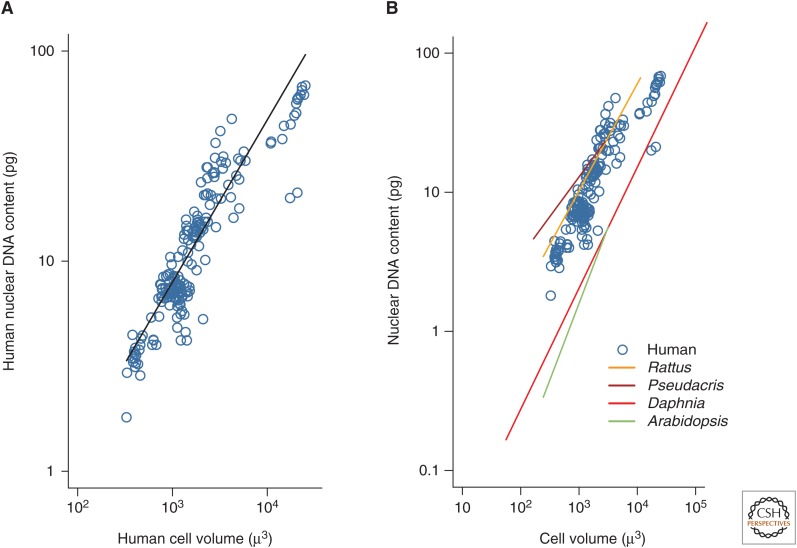
Our analyses of these data indicate that the amount of DNA in human cells varies systematically with cell size among cell types, such that larger cells contain substantially more DNA (Fig. 1A) (mean log10 [DNA] = −1.4 + 0.77 log10 [cell volume], r2 = 0.82, F1, 175 = 36.6, P < 1 × 10−9). Figure 1A shows that the mean DNA content for cell populations varies from 1.8 to 68.6 pg across cell types ranging from haploid sperm to polyploid megakaryocytes. For 17 of the 19 cell types shown in this relationship, cell volume was calculated based on some measure of nucleus size. Without converting to cell volume, we still observed a significant, positive relationship between nuclear volume and DNA content among these cell types (log10 [DNA, pg] = −0.25 + 0.54 log10 [nuclear volume, µ3], P = 3.8 × 10−10). For the two remaining cell types in which we had direct measures of cell diameter (cardiomyocytes and megakaryocytes), we also observed a significant positive relationship between DNA content and cell volume (log10 [DNA] = −0.71 + 0.52 log10 [cell volume], P = 1.0 × 10−10).
Figure 1.
Mean DNA content for cell populations. (A) Mean nuclear DNA content versus cell volume for healthy human cell populations from 19 cell types (log10 [DNA]) = −1.3 + 0.74 log10 [cell volume], r2 = 0.81; data in Table 1). The characteristic diploid human cell contains 7 pg DNA. (B) Relationship between DNA content and cell size in humans (blue) in comparison to previously reported relationships in the rat (Rattus norvegicus, log10 [DNA] = −1.3 + 0.77 log10 [cell volume] [Heizer 1955]), frog (Pseudacris obscura, log10 [DNA] = −0.6 + 0.57 log10 [cell volume] [Bachmann et al. 1966]), crustacean (Daphnia pulex, log10 [DNA] = −2.3 + 0.87 log10 [cell volume] [Beaton and Hebert 1989]), and plant (Arabidopsis thalania, log10 [DNA] = −3.1 + 1.1 log10 [cell volume] [Jovtchev et al. 2006]). Lines were fitted to data for nonhumans using ordinary least-squares regression. The human data shown here are the same as those shown in A.
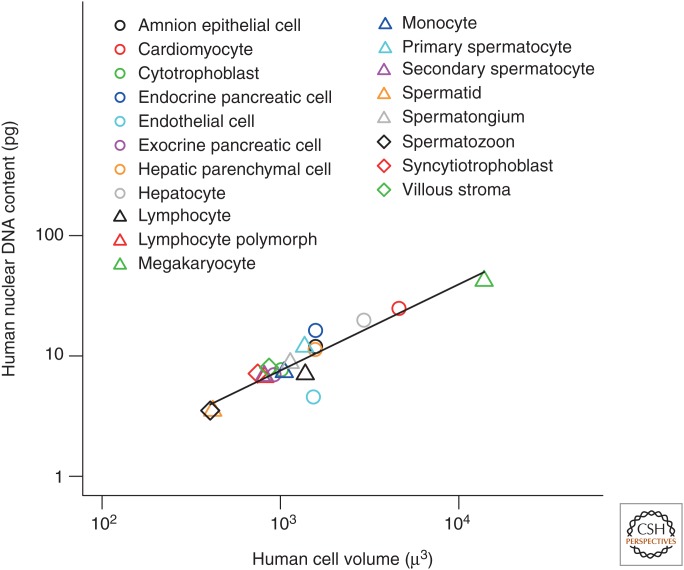
When DNA content and cell volume were averaged over all cells for each cell type, we observed a similar relationship between mean DNA content and cell volume (Fig. 2) (mean log10 [DNA] = −1.3 + 0.72 log10 [cell volume], r2 = 0.82, F1,17 = 86.8, P = 4.3 × 10−8). Note, however, that the range in DNA content of individual cells from these populations was even greater, as one would expect. At one extreme, the largest megakaryocytes contained 448 pg of DNA (see Royere et al. 1988 in Table 1). This indicates that these cells have likely undergone six genome duplication events from their diploid state.
Figure 2.
Relationship between mean nuclear DNA content and mean cell size in diploid and polyploid human cells by cell type for data shown in Figure 1. Each point represents mean cell size and DNA content from all cell populations of a given cell type.
Moreover, within cell types, the same correlations between nuclear DNA content and cell volume were observed in five of the six cases in which mean cell volume varied by a factor of 5 or more (megakaryocytes, hepatocytes, lymphocytes, hepatic parenchymal cells, and amnion epithelial cells). In these five cases, DNA content was positively correlated with cell volume (ordinary least-squares [OLS] regression, all P ≤ 1.6 × 10−9). In the case of cardiomyocytes, however, we did not observe a statistically significant correlation (P = 0.1).
RELATIONSHIP OF NUCLEAR DNA CONTENT TO CELL SIZE: HUMANS COMPARED WITH OTHER SPECIES
Methodology
To determine whether the relationships between cell size and DNA content were similar in humans and in other species, we used data from the few studies available on intraspecific variation in cell size and nuclear DNA content (Heizer 1955; Bachmann et al. 1966; Beaton and Hebert 1989; Jovtchev et al. 2006). Studies of nonhuman cells typically reported nuclear size instead of cell size. To convert nuclear volume to cell volume, we used the formula log10 (cell volume) = 0.88 + log10 (nuclear volume), which we obtained by fitting a linear function to log10-transformed cell and nuclear size measurements from vertebrate (Olmo 1983) and plant (Price et al. 1973) cells from a diverse assortment of species using ordinary least squares.
Results
The relationship between nuclear DNA content and cell volume in humans is similar to that previously reported for other species. DNA content is similarly correlated with cell volume in the rat Rattus norvegicus, the frog Pseudacris obscura, the crustacean Daphnia pulex, and the plant Arabidopsis thalania (Fig. 1B). These results indicate that concomitant changes in cell volume and nuclear DNA content may occur across very different cell types and species, although this has only rarely been investigated.
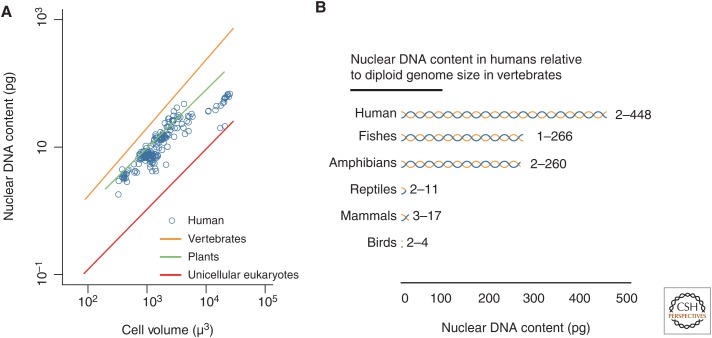
The relationship between nuclear DNA content and cell volume in humans also appears similar to the relationships observed between diploid genome size and cell volume in plants (Price et al. 1973), vertebrate animals (Olmo 1983), and unicellular eukaryotes (Fig. 3A) (Shuter et al. 1983). Interestingly, the range of nuclear DNA content in human cells is comparable to the range of diploid genome sizes across all vertebrates (Fig. 3B). However, one must be cautious in interpreting similarities and differences among these interspecific relationships given possible differences in methodology, possible differences in the relationship between nuclear volume and cell volume among taxa and, in some cases, a fairly limited sampling of species from these groups (e.g., unicellular eukaryotes).
Figure 3.
Relationship between nuclear DNA content and cell volume in humans. (A) Relationship between nuclear DNA content and cell size in diploid and polyploid human cells (blue) in comparison to previously reported relationships for diploid cells of vertebrates (log10 [DNA] = −1.91 + 1.07 log10 [cell volume]), unicellular eukaryotes (log10 [DNA] = −2.81 + 0.945 log10 [cell volume]), and angiosperms (log10 [DNA] = −1.75 + 0.912 log10 [cell volume]). Lines were fitted to previously reported relationships using ordinary least-squares regression. The range of unicellular eukaryote cell and genome sizes is truncated to clearly show data for other groups. Note that the human data shown in this figure are the same as those shown in Figure 1A. (B) Range of nuclear DNA content in individual human cells in comparison to ranges of diploid genome sizes within vertebrate groups. DNA content was rounded to the nearest whole number.
CONCLUSIONS
Our analysis of previously published data raises questions regarding the prevalence of endoreplication in human cells and its relationship to cell size. It remains to be seen whether the relationship presented here extends across the hundreds of different cell types in the human body. Our common assumption of DNA constancy across somatic cells in humans and other species has not been adequately investigated. The number of animal species for which we have measured the amount of DNA in different cell types of different sizes is restricted to only a handful of model organisms. Endoreplication may be a common and/or functional trait in the somatic cells of humans and other animals (Lee et al. 2009; Ullah et al. 2009; Anatskaya and Vinogradov 2011; De Veylder et al. 2011), as it has long been viewed in plants. Some have argued that perhaps this process serves to provide extra gene copies as an insurance policy against DNA damage, or that larger cells simply require greater levels of protein synthesis for maintenance and production (Lee et al. 2009; De Veylder et al. 2011). Certainly, in some human cell types (e.g., megakaryocytes), polyploid nuclei are considered normal and functional. And in other cell types not considered here (e.g., muscle cells), multinucleated cells are common and functional. The relationship between cell size, nuclear DNA content, and gene expression within and across cell types is only beginning to be explored (Marguerat and Bähler 2012), but clearly genomes are much more dynamic than was previously thought (Parfrey et al. 2008).
We have moved beyond the view that genomes are immutable blueprints. Most of the cells shown in Figure 1 are presumably postmitotic, but many show quantities of nuclear DNA well beyond the expected value of about 7 pg.
The notion that human cells may have quite different genetic constitutions depending on their size would have profound implications for our understanding of cell structure and function.
The data shown here suggest that perhaps the quantity of nuclear DNA content in human cells is best viewed as a distribution of values that reflects cell size distributions, rather than as a single value. That is, most cells may be of a similar size and contain something close to the characteristic amount of nuclear DNA, but a relatively small number of cells may be large with high amounts of DNA.
Changes in the DNA content of somatic cells may occur by endoreplication of individual genes or the entire genome (i.e., polyploidization). Often, it is assumed that genome duplication via endoreplication is responsible for any increases in nuclear DNA content beyond the typical diploid quantity. Of the cell types shown here, it is simply not possible to generalize the extent to which this may be the case because most data are averages of cell populations. However, for the two cell types for which we have data on the individual cells of a cell population (amnion epithelial cells and hepatocytes), a closer look at the data provides some clues (see Table 1). In these data, the increases in DNA content with cell size appear to cluster around values expected from endopolyploidization (e.g., 7, 14, 28, etc.), but there is considerably more variation than one would expect simply based on error. Thus, the data suggest DNA content has increased as a result of some combination of partial and complete genome duplication in these two cell types.
However, the results shown here do not address the question of whether genome size controls cell size, or cell size controls genome size. This has long been debated in the literature, and has yet to be resolved. It is our hope that our comparisons of DNA–cell size relationships within species to genome size–cell size relationships across species may help point to a more general understanding. We were surprised by the qualitatively similar relationships between the two. They highlight the fact that we have much to learn about the various mechanisms that lead to increases in cellular DNA content, and how such increases affect gene expression (Therman et al. 1983; Marguerat and Bähler 2012). Again, here, broadly comparative approaches may be insightful. In particular, we suggest that it may be useful to relate relevant structural features (genome size and cell size) and functional features (gene expression and polyploidization) to the life span, replication rate, and metabolic rate of cells.
ACKNOWLEDGMENTS
We thank the population biology group at University of Florida for feedback.
REFERENCES
*Reference is also in this collection.
- *.Amodeo AA, Skothein JM. 2015. Cell size control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anatskaya OV, Vinogradov AE. 2011. Somatic polyploidy promotes cell function under stress and energy depletion: Evidence from tissue-specific mammal transcriptome. Funct Integr Genomic 10: 433–446. [DOI] [PubMed] [Google Scholar]
- Bachmann K, Goin OB, Goid CJ. 1966. Hylid frogs: Polyploid classes of DNA in liver nuclei. Science 154: 650–651. [DOI] [PubMed] [Google Scholar]
- Beaton MJ, Hebert PDN. 1989. Miniature genomes and endopolyploidy in cladoceran crustaceans. Genome 32: 1048–1053. [Google Scholar]
- Bedi KS, Goldstein DJ. 1976. Apparent anomalies in nuclear Feulgen-DNA contents. Role of systematic microdensitometric errors. J Cell Biol 71: 68–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesterfeld S, Gerres K, Fischer-Wein G, Bocking A. 1994. Polyploidy in non-neoplastic tissues. J Clin Pathol 47: 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesterfeld S, Beckers S, Del Carmen Villa Cadenas M, Schramm M. 2011. Feulgen staining remains the gold standard for precise DNA image cytometry. Anticancer Res 31: 53–58. [PubMed] [Google Scholar]
- Chieco P, Derenzini M. 1999. The Feulgen reaction 75 years on. Histochem Cell Biol 111: 345–358. [DOI] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A. 2011. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 16: 624–634. [DOI] [PubMed] [Google Scholar]
- Dorman A, Graham D, Curran B, Henry K, Leader M. 1990. Ploidy of smooth muscle tumours: Retrospective image analysis study of formalin fixed, paraffin wax embedded tissue. J Clin Pathol 43: 465–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrie MG, Swartz FJ. 1974. Diploid, tetraploid, and octaploid β cells in the islets of Langerhans of the normal human pancreas. Diabetes 23: 583–588. [DOI] [PubMed] [Google Scholar]
- Feulgen R, Rosenbeck HC. 1942. Manual of histological demonstration technique. Butterworth, London. [Google Scholar]
- Galton M. 1962. DNA content of placental nuclei. J Cell Biol 13: 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. 2005. The evolution of the genome, pp. 1–768. Academic, New York. [Google Scholar]
- Gregory TR. 2012. Animal genome size database, www.genomesize.com. [Google Scholar]
- Hancock V, Martin JF, Lelchuk R. 2008. The relationship between human megakaryocyte nuclear DNA content and gene expression. Brit J Haematol 85: 692–697. [DOI] [PubMed] [Google Scholar]
- Heizer P. 1955. Desoxyribose nucleic acid (DNA) content and size of rat liver nuclei during thioac etamide intoxication and recovery. Chromosoma 7: 281–327. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Ruggeri ZM, Harker LA, Burstein SA. 1986. Separation of human megakaryocytes by state of differentiation on continuous gradients of Percoll: Size and ploidy analysis of cells identified by monoclonal antibody to glycoprotein IIb/IIIa. Blood 67: 1286–1292. [PubMed] [Google Scholar]
- Jovtchev G, Schubert V, Barrow M, Schubert I. 2006. Nuclear DNA content and nuclear and cell volumes are positively correlated in angiosperms. Cytogenet Genome Res 114: 77–82. [DOI] [PubMed] [Google Scholar]
- Klinger HP, Schwarzacher HG. 1960. The sex chromatin and heterochromatic bodies in human diploid and polyploid nuclei. J Biophys Biochem Cytol 8: 345–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ. 2009. Endoreplication: Polyploidy with purpose. Genes Dev 23: 2461–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuchtenberger C, Leuchtenberger R, Schrader F, Weir DR. 1956. Reduced amounts of desoxyribose nucleic acid in testicular germ cells of infertile men with active spermatogenesis. Lab Invest 5: 422–440. [PubMed] [Google Scholar]
- Marguerat S, Bähler J. 2012. Coordinating genome expression with cell size. Trends Genet 28: 560–565. [DOI] [PubMed] [Google Scholar]
- Meek ES, Harbison JFA. 1967. Nuclear area and deoxyribonucleic acid content in human liver cell nuclei. J Anat 101: 487–489. [PMC free article] [PubMed] [Google Scholar]
- Mirsky AE, Ris H. 1949. Variable and constant components of chromosomes. Nature 163: 666–667. [DOI] [PubMed] [Google Scholar]
- *.Niklas KJ. 2015. A phyletic perspective on cell growth. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmo E. 1983. Nucleotype and cell size in vertebrates: A review. Basic Appl Histochem 27: 227–256. [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJG, Katz LA. 2008. The dynamic nature of eukaryotic genomes. Mol Biol Evol 25: 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrakis NL. 1953. Microspectrophotometric estimation of the desoxyribonucleic acid (DNA) content of individual normal and leukemic human lymphocytes. Blood 8: 905–915. [PubMed] [Google Scholar]
- Pinheiro J, Bates D. 2000. Mixed-effects models in S and S-PLUS. Springer, New York. [Google Scholar]
- Price HJ, Sparrow AH, Nauman AF. 1973. Correlations between nuclear volume, cell volume and DNA content in meristematic cells of herbaceous angiosperms. Cell Mol Life Sci 29: 1028–1029. [Google Scholar]
- Royere D, Hamamah S, Nicolle JC, Barthelemy C, Lansac J. 1988. Freezing and thawing alter chromatin stability of ejaculated human spermatozoa: Fluorescence acridine orange staining and Feulgen-DNA cytophotometric studies. Gamete Res 21: 51–57. [DOI] [PubMed] [Google Scholar]
- Shuter BJ, Thomas JE, Taylor WD, Zimmerman AM. 1983. Phenotypic correlates of genomic DNA content in unicellular eukaryotes and other cells. Am Nat 122: 26–44. [Google Scholar]
- Swanson JA, Lee M, Knapp PE. 1991. Cellular dimensions affecting the nucleocytoplasmic volume ratio. J Cell Biol 115: 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz FJ. 1956. The development in the human liver of multiple desoxyribose nucleic acid (DNA) classes and their relationship to the age of the individual. Chromosoma 8: 53–72. [DOI] [PubMed] [Google Scholar]
- Szarski H. 1976. Cell size and nuclear DNA content in vertebrates. Int Rev Cytol 44: 93–111. [DOI] [PubMed] [Google Scholar]
- Therman E, Sarto GE, Stubblefield PA. 1983. Endomitosis: A reappraisal. Human Genet 63: 13–18. [DOI] [PubMed] [Google Scholar]
- Ullah Z, Lee CY, Lilly MA, DePamphilis ML. 2009. Developmentally programmed endoreduplication in animals. Cell Cycle 8: 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vliegen HW, Bruschke VG, Van der Laarse A. 1990. Different response of cellular DNA content to cardiac hypertrophy in human and rat heart myocytes. Comp Biochem Physiol 95A: 109–114. [DOI] [PubMed] [Google Scholar]
- Winkelmann M, Pfitzer P, Schneider W. 1987. Significance of polyploidy in megakaryocytes and other cells in health and tumor disease. Klin Wochenschr 65: 1115–1131. [DOI] [PubMed] [Google Scholar]
- Wohlschlaeger J, Levkau B, Brockhoff G, Schmitz KJ, von Winterfeld M, Takeda A, Takeda N, Stypmann J, Vahlhaus C, Schmid C, et al. 2010. Hemodynamic support by left ventricular assist devices reduces cardiomyocyte DNA content in the failing human heart. Circulation 121: 989–996. [DOI] [PubMed] [Google Scholar]