Fission yeast has been used to elucidate mechanisms of heterochromatin assembly and maintenance. Other epigenetic phenomena, including nucleosome remodeling and nuclear organization, are also being explored in this species.
Abstract
This article discusses the advances made in epigenetic research using the model organism fission yeast Schizosaccharomyces pombe. S. pombe has been used for epigenetic research since the discovery of position effect variegation (PEV). This is a phenomenon in which a transgene inserted within heterochromatin is variably expressed, but can be stably inherited in subsequent cell generations. PEV occurs at centromeres, telomeres, ribosomal DNA (rDNA) loci, and mating-type regions of S. pombe chromosomes. Heterochromatin assembly in these regions requires enzymes that modify histones and the RNA interference (RNAi) machinery. One of the key histone-modifying enzymes is the lysine methyltransferase Clr4, which methylates histone H3 on lysine 9 (H3K9), a classic hallmark of heterochromatin. The kinetochore is assembled on specialized chromatin in which histone H3 is replaced by the variant CENP-A. Studies in fission yeast have contributed to our understanding of the establishment and maintenance of CENP-A chromatin and the epigenetic activation and inactivation of centromeres.
OVERVIEW
This article discusses the advances made in epigenetic regulation using the fission yeast Schizosaccharomyces pombe as a model organism. S. pombe has been exploited for the investigation of epigenetically regulated processes since the discovery of position effect variegation (PEV), originally observed in Drosophila. This is a phenomenon in which a transgene is variably expressed, but often the expression pattern is then stably inherited in subsequent cell generations. This variable gene expression, however, occurs only when the transgene integrates in or near heterochromatic regions of the genome. The multitude of studies exploiting PEV or gene silencing in fission yeast have led to great progress not only in elucidating the mechanisms of heterochromatin assembly and maintenance, but understanding how heterochromatin creates a repressive environment. Because many of the mechanisms used to create repressive chromatin environments are common to higher eukaryotes including humans, S. pombe has provided a good model organism for epigenetic research.
Heterochromatin is found mainly at centromeres, telomeres, ribosomal DNA (rDNA) loci, and the mating-type regions of S. pombe chromosomes. Heterochromatin assembly requires the RNA interference (RNAi) machinery and enzymes that modify histones. One of the key modifying enzymes is the histone lysine (K) methyltransferase (HKMT or KMT) Clr4, which methylates histone H3 on lysine 9 (H3K9), now considered a hallmark of heterochromatin.
Centromeres consist of two types of chromatin: the pericentric heterochromatin regions and CENP-A (also referred to as CenH3 or Cnp1 in S. pombe)—containing chromatin. Pericentric heterochromatin plays a crucial function in mitotic and meiotic cell divisions, providing genome stability and sister-chromatid cohesion, partly by supporting the establishment of adjacent centromeric CENP-A-containing chromatin. CENP-A chromatin is required for the formation of a protein structure called the kinetochore, which interacts with spindle-microtubules to segregate chromosomes in mitosis and meiosis. CENP-A is a centromere-specific histone H3 variant, which replaces the canonical H3 histone in centromeric nucleosomes, and is targeted to centromeres by the combined action of chaperone proteins and loading factors. Heterochromatin in the mating-type region is essential for regulating gene expression, which directs the sexual life cycle of S. pombe. It provides a silencing mechanism to prevent the expression of inappropriate mating-type information in a haploid cell, yet allows for mating-type switching at the appropriate time. Heterochromatin at telomeres plays an important function in maintaining the integrity of linear chromosomes. Heterochromatin at rDNA repeats may suppress recombination, thus maintaining genome integrity.
In addition to heterochromatin research, S. pombe is used to study several other aspects of epigenetic control such as chromatin remodeling and nuclear organization. The relatively small size of its genome allows for convenient genomic analysis, especially when coupled to the simplicity of working with a unicellular organism in which one can exploit both the haploid and diploid phases of the life cycle. S. pombe has also become an important model organism for understanding how epigenetic mechanisms operate on a genome-wide scale, referred to as epigenomics.
1. SCHIZOSACCHAROMYCES POMBE LIFE CYCLE
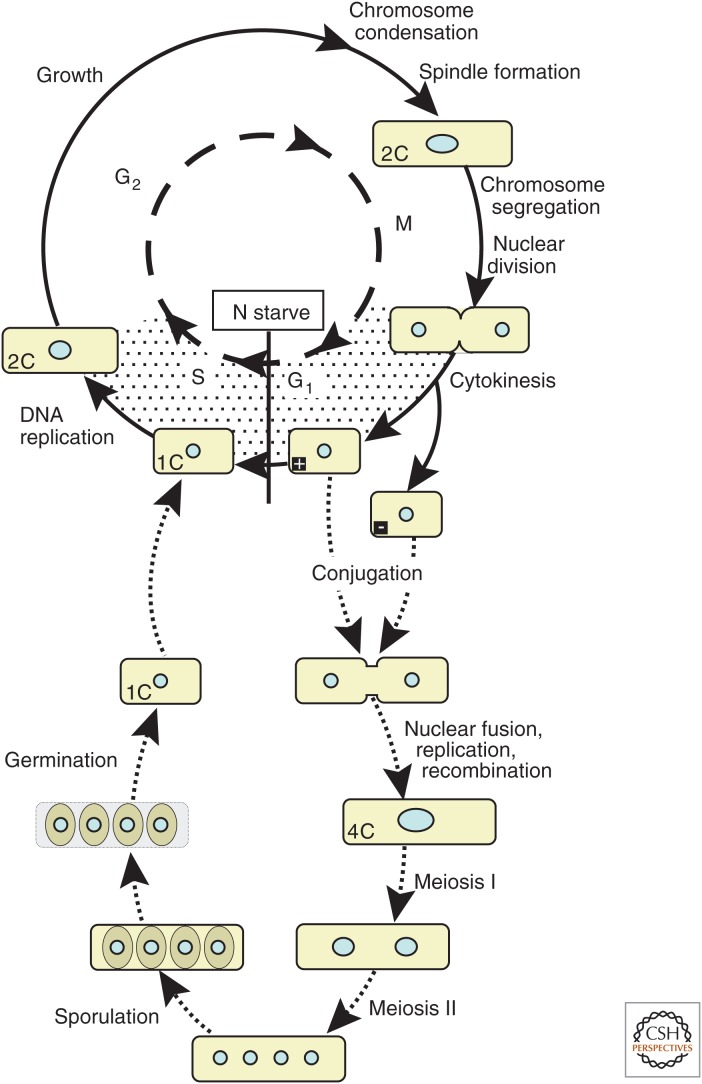
Fission yeast, Schizosaccharomyces pombe, is found in the fermentations involved in the production of beer in subtropical regions; “pombe” is the Swahili word for beer. S. pombe is primarily a haploid (1N) unicellular organism. In nutrient-rich media, wild-type cells undergo a mitotic division approximately every 2 h. However, a variety of conditions, or conditional mutants, can be used to block cells at distinct stages of the cell cycle or synchronize cell cultures in G1/S, G2, or at metaphase. This is particularly useful because the G1 phase is very short in normally growing cultures and cells pass almost immediately into S phase following cytokinesis; the major portion of the cell cycle is spent in G2 (Fig. 1) (Egel 2004).
Figure 1.
Life cycle of the fission yeast, S. pombe. Fission yeast has a short G1 taking less than 10% of the cell cycle (stippled area is expanded to aid representation). In rich medium, G1 cells proceed into S phase followed by a long G2 (∼70% of the cell cycle), mitosis, and cytokinesis. When starved of nitrogen, cells of opposite mating-type (+ and −) conjugate, after which nuclei fuse in a process known as karyogamy. Premeiotic replication and recombination allows meiosis I and II to proceed, resulting in four haploid nuclei that are separated into four spores in an ascus. Provision of rich medium allows germination of spores and resumption of the vegetative cell cycle.
S. pombe has two mating types, named plus (+) and minus (−), and, like Saccharomyces cerevisiae, can switch between opposite mating types. Mating types are equivalent to dimorphic sexes in higher eukaryotes, although they are haploid. The information for both mating types resides in the genome as epigenetically regulated silent cassettes: The (+) information cassette is found at the mat2-P locus and the (−) information cassette at the mat3-M locus. These silent loci provide the genetic template for mating-type identity, but mating type itself is determined by the information (+ or −) that resides at the active mat1 locus. Switching of information at the active mat1 locus occurs by recombination between one of the silent loci (mat2-P or mat3-M) and the mat1 locus according to a strict pattern (Egel 2004). When starved of nitrogen, cells stop dividing and arrest in G1, which promotes the sexual phase of the life cycle through conjugation of pairs of “+” and “−” cells to form diploid zygotes (Fig. 1). After mating and nuclear fusion, premeiotic replication occurs (increasing DNA content from 2N to 4N). This is followed by the pairing and recombination of homologous chromosomes, and ends with the reductional meiosis I division and equational meiosis II division. This produces four separate haploid nuclei (1N) that become encapsulated into spores enclosed in an ascus. The subsequent provision of a rich nutrient source allows germination and resumption of vegetative growth and mitotic cell division.
Nonswitching derivatives have been isolated or constructed in which all cells are either “+” or “−” mating type. This facilitates controlled mating between strains of distinct genotypes. Although S. pombe is normally haploid, it is possible to select for diploid strains. Such diploid cells can then divide by vegetative mitotic growth until starved of nitrogen when they, too, undergo meiosis and form “azygotic asci.”
2. SCREENS TO IDENTIFY HETEROCHROMATIN COMPONENTS
The type of chromatin surrounding a gene can strongly influence its expression. This was originally shown in the fruit fly, Drosophila melanogaster, in which chromosomal rearrangements that move the white gene close to centromeric heterochromatin led to its variable expression in eye facets causing variegation in eye coloration (see Fig. 1 in Elgin and Reuter 2013). Studies in S. pombe showed this through the variable silencing of a reporter gene when inserted into different centromeric regions. Scientists have subsequently studied silent chromatin (i.e., heterochromatin) as well as active chromatin (euchromatin) by exploiting PEV of reporter or translocated genes. In fact, in S. pombe, this approach has been fundamental in identifying many of the molecular components that define heterochromatin structures at centromeres and other regions of the genome.
The first genetic screens to identify modifiers of PEV were performed by selecting for mutants that allow expression of the normally silent mat2-P and mat3-M cassettes. Mutants were identified because of aberrant mating patterns or when selecting for expression of a ura4+ transgene inserted in the silent mating-type region (Thon and Klar 1992; Ekwall and Ruusala 1994; Thon et al. 1994). These screens identified a set of genes called cryptic loci regulators (i.e., the clr1+, clr2+, clr3+, clr4+), as well as the rik1+ and swi6+ genes (for the gene products see Table 1). These proteins are the functional counterparts of the silent information regulator (Sir) proteins found in S. cerevisiae (described in Grunstein and Gasser 2013). The link between heterochromatin formation occurring at centromeres and in the mat2-mat3 region became clear when several of the Clr proteins were shown to be required for silencing at centromeres as well as the mating-type loci in S. pombe (Allshire et al. 1995).
Table 1.
Gene products identified in S. pombe forward PEV screens
| Gene product | Molecular function | Reference(s) |
|---|---|---|
| Clr1 (Cryptic loci regulator 1) | Zinc ion binding protein, part of the SHREC complex | Thon and Klar 1992 |
| Clr2 (Cryptic loci regulator 2) | Transcription-silencing protein Clr2; part of the SHREC complex | Ekwall and Ruusala 1994; Thon et al. 1994 |
| Clr3 (Cryptic loci regulator 3) | Histone deacetylase class II (H3-K14-specific); part of the SHREC complex | Ekwall and Ruusala 1994; Thon et al. 1994; Grewal et al. 1998 |
| Clr4 (Cryptic loci regulator 4) | Histone methyltransferase KMT activity (H3-K9-specific) | Ekwall and Ruusala 1994; Thon et al. 1994 |
| Rik1 | DNA-binding protein; part of the CLRC ubiquitin ligase complex | Egel et al. 1989; Ekwall and Ruusala 1994 |
| Swi6 | Chromo domain/shadow protein, heterochromatin protein 1 homolog | Ekwall and Ruusala 1994; Lorentz et al. 1994 |
| Clr6 (Cryptic loci regulator 6) | Histone deacetylase class I | Grewal et al. 1998 |
| Ago1 was identified as Csp9 (centromere: suppressor of position effect 9) | Argonaute protein, part of the RITS complex | Ekwall et al. 1999; Volpe et al. 2003 |
| Rpb7 was identified as Csp3 (centromere: suppressor of position effect 3) | DNA-directed RNA polymerase II subunit | Ekwall et al. 1999; Djupedal et al. 2005 |
| Rpb2 | DNA-directed RNA polymerase II subunit | Kato et al. 2005 |
| Epe1 (Enhanced position effect 1) | jmjC domain protein | Ayoub et al. 2003 |
| Cwf10 was identified as Csp4 | Splicing factor | Bayne et al. 2008; Ekwall et al. 1999 |
| Prp39 was identified as Csp5 | Splicing factor | Bayne et al. 2008; Ekwall et al. 1999 |
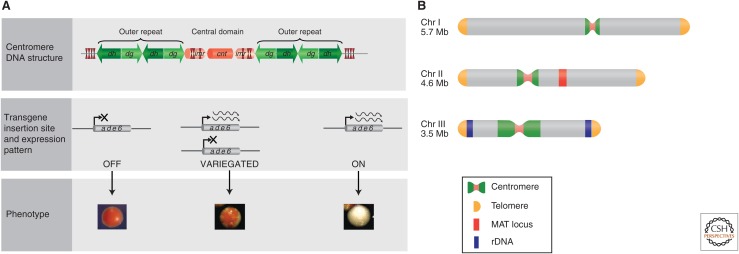
To understand how the PEV screens have helped in identifying components involved in centromeric heterochromatin, it is useful to know the nature of the underlying DNA sequence at centromeres. At the DNA level, centromere regions in fission yeast are composed of outer repeats (subdivided into elements known as dg and dh), flanking a central domain. The central domain includes the innermost repeats (imr), and a central core (cnt; Fig. 2A). The three centromeres—cen1, cen2, and cen3—occupy ∼40, ∼60, and ∼120 kb on chromosomes I, II, and III, respectively (Fig. 2B; reviewed in Egel 2004; Pidoux and Allshire 2004). The repetitive nature of fission yeast centromere DNA resembles the larger, more complex repeated structures associated with many metazoan centromeres, but they are more amenable to manipulation. Because repetitive DNA frequently correlates with the presence of heterochromatin in other eukaryotes (see also Martienssen and Moazed 2014), this suggested that fission yeast centromeres might have heterochromatic properties such as the ability to hinder gene expression. Reporter gene silencing has been monitored in fission yeast by phenotypic assays similar to those used in S. cerevisiae. For example, when the ura4+ reporter is silenced, colonies can form on counterselective media containing 5-fluoroorotic acid. An alternative assay monitors the silencing of the ade6+ reporter, which results in red (repressed) rather than white (expressed) colony color (Fig. 2A). Thus, placement of a normally expressed gene, such as ura4+ or ade6+ within the centromere (as defined by the outer repeat and central domain) results in transcriptional silencing. Compared to the outer repeats, silencing of ade6+ within the central domain is comparatively unstable, resulting in variegated colonies (i.e., either red, white, or red–white sectored colonies). Moreover, just 1 kb distal to the outer repeats no silencing occurs (Allshire et al. 1994, 1995), indicating that transcriptional repression is confined to the centromere.
Figure 2.
Distinct outer repeat heterochromatin and central kinetochore domains at fission yeast centromeres. (A, top) Representation of a fission yeast centromere. The central domain (pink, kinetochore) is composed of imr and cnt elements, the outer repeats contain transcribed dg and dh repeats (green, heterochromatin). All three centromeres have a similar overall arrangement; however, the number of outer repeats differs: cen1 (40 kb) has two, cen2 (65 kb) has three, and cen3 (110 kb) has approximately 13. Clusters of transfer RNA (tRNA) genes (double arrowheads) occur in the imr region and at the extremities of all three centromeres. (Middle) Schematically shows transcription patterns of marker genes placed within the outer repeats, central domain, or beyond the centromere. (Bottom) Images showing the phenotype of S. pombe colonies of ade6+ transgenics inserted at various sites within the centromere. Cells expressing ade6+ from a transgene inserted in sequences outside the centromere form white colonies. When ade6+ is inserted at sites within the outer repeats, expression is silenced and red colonies are formed. Expression of ade6+ from the central domain is typically variegated, resulting in red, white, and sectored colonies. (B) A schematic representation of S. pombe chromosomes. The three chromosomes are depicted showing the four main regions of heterochromatin: centromere, telomere, mat2/3, and rDNA regions.
3. DIFFERENT TYPES OF HETEROCHROMATIN
Apart from centromeres, other repressive regions of the genome can silence transgene expression, notably the mating-type loci (mat2-mat3), the rDNA region, and telomeres (reviewed in Egel 2004). Together, these constitute the four regions of heterochromatin in S. pombe (Fig. 2B). With the possible exception of the rDNA region, it turns out that each region of silent chromatin has an essential function. At centromeres, heterochromatin ensures normal chromosome segregation (Allshire et al. 1995; Ekwall et al. 1995), whereas at the mating-type loci it facilitates and regulates mating-type switching (reviewed in Egel 2004). Silent chromatin formed adjacent to telomeres (Nimmo et al. 1994; Kanoh et al. 2005) plays a role in meiotic chromosome segregation (Nimmo et al. 1998), whereas the function of heterochromatin in the rDNA region of S. pombe has not been established yet (Thon and Verhein-Hansen 2000; Cam et al. 2005). It could be involved in maintaining rDNA stability by preventing recombination between rDNA repeat sequences just like in S. cerevisiae (described in Grunstein and Gasser 2013).
The mechanisms that S. pombe uses to achieve heterochromatic silencing distinguish it from S. cerevisiae, making it more akin to Neurospora crassa, plants, and metazoa. So, although S. pombe and S. cerevisiae have silent chromatin at telomeres, the mating-type loci, and rDNA regions, only S. pombe shows silencing at centromeres. Silencing is mediated by distinct histone-modifying activities (in particular, histone H3K9 methylation), RNAi proteins, the RNA polymerase (RNA Pol) II machinery and homologs of heterochromatin protein 1 (HP1; see Table 1). These mechanisms are conserved in higher eukaryotes. Interestingly S. pombe, in contrast to N. crassa and many other eukaryotes, appears to lack any detectable DNA methylation (Wilkinson et al. 1995), a common mechanism used for silencing chromatin in higher eukaryotes (see Li and Zhang 2014; Pikaard and Mittelsten Sheid 2014); thus, silencing in fission yeast is mediated primarily by chromatin modifications and the RNAi machinery.
4. S. POMBE CENTROMERES: A PARADIGM FOR STUDYING HETEROCHROMATIN
Heterochromatin formation at centromeres is a complex process involving chromatin factors, noncoding RNA, RNA Pol II, and its associated proteins. It is also strictly controlled by chromatin boundaries (see Sec. 4.5), which prevent the spreading of heterochromatin. It is important to note that chromatin boundaries need to be maintained throughout the cell cycle. This section discusses these factors and processes, which together control the formation of heterochromatin.
4.1. Chromatin Factors
In chromatin, the amino-terminal tails of histones H3 and H4 are subject to a range of posttranslational modifications, which generally correlate with active or repressed states (summarized in Allis et al. 2014). The centromeric heterochromatin domain at outer repeats in fission yeast contains di- and trimethylated histone H3 at lysine 9 (H3K9me2 and H3K9me3; Nakayama et al. 2001; Yamada et al. 2005). In most eukaryotic organisms, H3K9me2 and H3K9me3 are characteristic of silent heterochromatin. Because H3K9 methylation extends over a long stretch of heterochromatin, how does the process of heterochromatinization occur? Studies indicate it predominantly spreads through the concerted action of histone deacetylases (HDACs), the Clr4 histone lysine methyltransferase (KMT/HMTase), and the HP1 homolog, Swi6.
HDACs such as Clr3, Clr6, and Sir2 deacetylate histone H3 at various lysine residues, which contribute to generating repressive heterochromatin (discussed in Shankaranarayana 2003; Wiren et al. 2005; Seto and Yoshida 2014). More recent studies, dissecting where, when, and how the different enzymes work, have identified that Clr3 operates as part of a complex called SHREC, which helps to deacetylate chromatin (Fig. 3) (Sugiyama et al. 2007). Clr4 is the key KMT/HKMT that methylates H3K9 over the centromeric outer repeats. Clr4 functions as part of a complex called CLRC, which, through methylation of H3K9, creates a specific binding site recognized by factors containing a chromodomain motif. Interestingly, Clr4 contains a chromodomain; thus, it not only methylates H3K9, but also binds H3K9me2/3, likely effecting adjacent H3K9 methylation through its catalytic domain in a sequential fashion (Zhang et al. 2008).
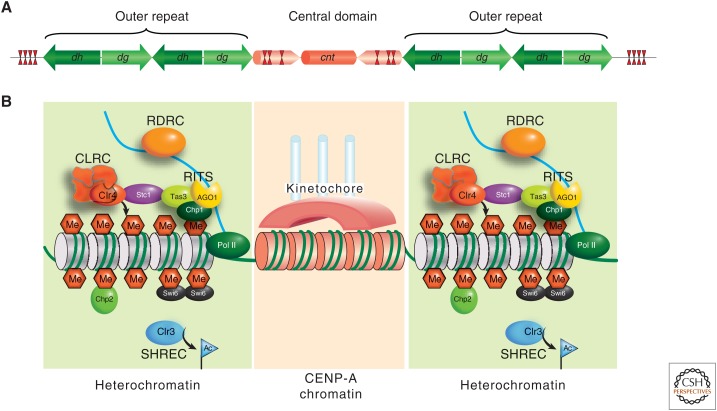
Figure 3.
Centromeric chromatin domains in S. pombe. (A) A schematic representation of the symmetrical DNA arrangement in S. pombe centromeres. (B) Heterochromatin: outer repeats are packaged in nucleosomes, which are methylated on lysine 9 of histone H3 (H3K9me) by Clr4 as part of the CLRC complex. This allows the binding of chromodomain proteins Chp1 (a component of the RNAi RITS complex), Chp2, and Swi6. Collectively, these and other factors, including the SHREC complex-containing Clr3 histone deacetylase activity and the RDRC complex, act to assemble and propagate heterochromatin. Central “kinetochore” chromatin: CENP-A is found in the central domain where it probably replaces the majority of histone H3 to form specialized nucleosomes (coral colored). In addition to CENP-A, several proteins assemble at the central domain chromatin to form the inner and outer kinetochore multiprotein structures (coral arc). See Figure 6 for a description of the kinetochore.
Swi6 is a chromodomain-containing HP1 homolog known to bind methylated H3K9 and contribute to the formation of silent chromatin over the outer repeats (Fig. 3) (Bannister et al. 2001; Nakayama et al. 2001). In addition to the chromodomain, Swi6 has another functional domain called the chromoshadow domain, which causes self-dimerization (Cowieson et al. 2000). This suggests an important role for nucleosome-bridging in heterochromatin assembly (Canzio et al. 2011), presumably altering nucleosome repeat length in heterochromatin compared to euchromatin (Lantermann et al. 2010). Reporter gene insertions at the outer repeats (e.g., the ura4+ gene) are notably enriched in H3K9me2 and Swi6 proteins, stressing how important Clr4-mediated H3K9 methylation and Swi6 heterochromatin spreading is not only within the centromere outer repeats but also into neighboring interposing sequences (Cowieson et al. 2000; Nakayama et al. 2001).
Chp1, another chromodomain protein, also associates with outer repeat chromatin at centromeres by binding methylated H3K9, as does Swi6. Chp1 is a member of the RNA-induced transcriptional silencing or RITS complex—another player in the heterochromatin-forming process. The RITS complex is part of the RNAi machinery, which is also required for the complete methylation of histone H3K9 over the outer repeats and inserted reporter genes (Partridge et al. 2002; Motamedi et al. 2004; Sadaie et al. 2004). The contribution of RNAi to heterochromatin formation is further explained in Section 4.2 and in Martienssen and Moazed (2014).
Altogether there are four chromodomain proteins in S. pombe (Chp1, Chp2, Swi6, and Clr4). Two of these, Clr4 and Chp1, have reduced affinities for methylated H3K9 when H3K4 is acetylated by the histone lysine acetyltransferase (HAT or sometimes abbreviated to KAT) enzyme, Mst1. This suggests that H3K4 methylation acts like a switch: After DNA replication, Clr4/Chp1 occupancy at H3K9me2/3 is promoted on newly deposited histones where H3K4 is unacetylated. On Mst1 acetylation of H3K4, occupancy switches to favor Chp2/Swi6 as the cell cycle progresses and heterochromatin is fully reassembled after DNA replication (Xhemalce and Kouzarides 2010). The concept of heterochromatin maintenance throughout the cell cycle is expanded in Section 4.4.
However, chromodomains are not the only interaction surface between these HP1-like proteins with chromosomes. Interestingly, Swi6 was recently shown to have a strong RNA-binding activity via a hinge region in the protein, and this RNA-binding domain has a specific function in the destruction of transcripts originating from heterochromatin regions (described in Sec. 4.2). Mutations in the hinge domain lead to reduced silencing of such transcripts, but the integrity of heterochromatin is not affected, indicating that Swi6 performs an effector function downstream from methylated H3K9 (Keller et al. 2012). Thus, chromatin factors including histone modifiers and HP1-like proteins cooperate to specify heterochromatin in S. pombe.
4.2. RNAi and the RNA Pol II Machinery in Heterochromatin Assembly
RNAi is an important mechanism contributing to heterochromatin formation in S. pombe. The phenomenon of RNAi was first discovered in Caenorhabditis elegans in which the expression of double-stranded RNA (dsRNA) abolished the expression of a homologous gene. It soon became apparent that this form of RNAi is related to the process of transcriptional gene silencing (TGS) described in plants (see Pikaard and Mittelsten Scheid 2014) and quelling in N. crassa (Aramayo and Selker 2013). These are processes of silencing that occur when a region is transcriptionally active, and transcripts that generate regions of dsRNA (e.g., through the self-annealing of inverted repeats) can be processed into small RNA fragments (termed small RNA biogenesis). These small RNAs are taken up by effector complexes and can trigger silent chromatin via targeting activities, which cause DNA methylation and histone modification (see Figs. 1 and 2 of Martienssen and Moazed 2014). This process of silencing appears to be in operation from S. pombe to plants and metazoans, including mammals.
Studies of the components of the RNAi machinery in S. pombe have led to significant advances in our understanding of RNAi-mediated chromatin modification and silencing. Mutants in the RNAi machinery in S. pombe result in reduced H3K9me2 and loss of silencing over the outer repeats of centromeres (Volpe et al. 2002). Surprisingly at the time, these RNAi mutants revealed overlapping noncoding RNA (ncRNA) transcripts of a discrete size, originating from centromeric outer repeats. These ncRNAs were homologous to naturally occurring small dsRNAs called small interfering RNAs (siRNAs; ∼21 nt) that had been isolated and sequenced from S. pombe (Reinhart and Bartel 2002). We now know that these long noncoding double-stranded centromere repeat transcripts are cleaved by the Dicer (Dcr1) enzyme to generate siRNAs. These siRNAs then act to guide the RNAi machinery to homologous transcripts.
One of the initial questions in trying to elucidate how TGS worked in S. pombe was “Which RNA polymerase was responsible for the noncoding transcription of centromere repeats?” Mutation of either subunit of RNA Pol II (Rpb2 and Rpb7) results in defective centromere silencing (Djupedal et al. 2005; Kato et al. 2005) although these mutations display very different phenotypes. The rpb7-1 mutant shows reduced levels of centromere repeat transcription, resulting in less ncRNA and, consequently, less siRNA production and a loss of silent chromatin. This implies that RNA Pol II is required for the transcription of centromere repeats, which then provides the primary substrate for RNAi. In contrast, centromeric transcripts in the rpb2-m203 mutant are produced but not processed into siRNA, and H3K9 methylation at centromeres is reduced. These studies indicate that RNAi not only requires an RNA Pol II transcript but that, like other RNA-processing events, the production of centromeric siRNA may be coupled to transcription by interactions between the RNAi machinery, chromatin, histone-modifying enzymes, and RNA Pol II.
The RNAi machinery in S. pombe is complex and not yet fully understood. In addition to transcription of noncoding centromeric outer repeats by RNA Pol II and the processing of transcripts into siRNAs by Dcr1, the key activities of the RNAi machinery involve two complexes: RITS and RDRC (RNA-directed RNA polymerase complex; summarized in Fig. 4 of Martienssen and Moazed 2014). The RITS complex incorporates siRNAs to then direct it to centromere outer repeats via sequence recognition as well as H3K9me2/3 recognition through the Chp1 chromodomain (see Sec. 4.3). RDRC is recruited to amplify the process of TGS by generating more long double-stranded ncRNAs through the action of Rdp1 (RNA-directed RNA polymerase 1). Rdp1 transcribes from transcripts primed with siRNAs presented by the RITS complex. The chromatin-modifying machineries that execute chromatin changes include the CLRC (for Clr4-Rik1-Cul4 complex) and the SHREC complex. CLRC is recruited by the RITS complex via the Stc1 protein (Bayne et al. 2010; see Sec. 4.4). Once recruited, Clr4 methylates H3K9 over the outer repeats and this allows it to bind directly to H3K9me2/3 via its chromodomain. The HP1 homologs Swi6 and Chp2 play further roles in establishing and maintaining heterochromatin.
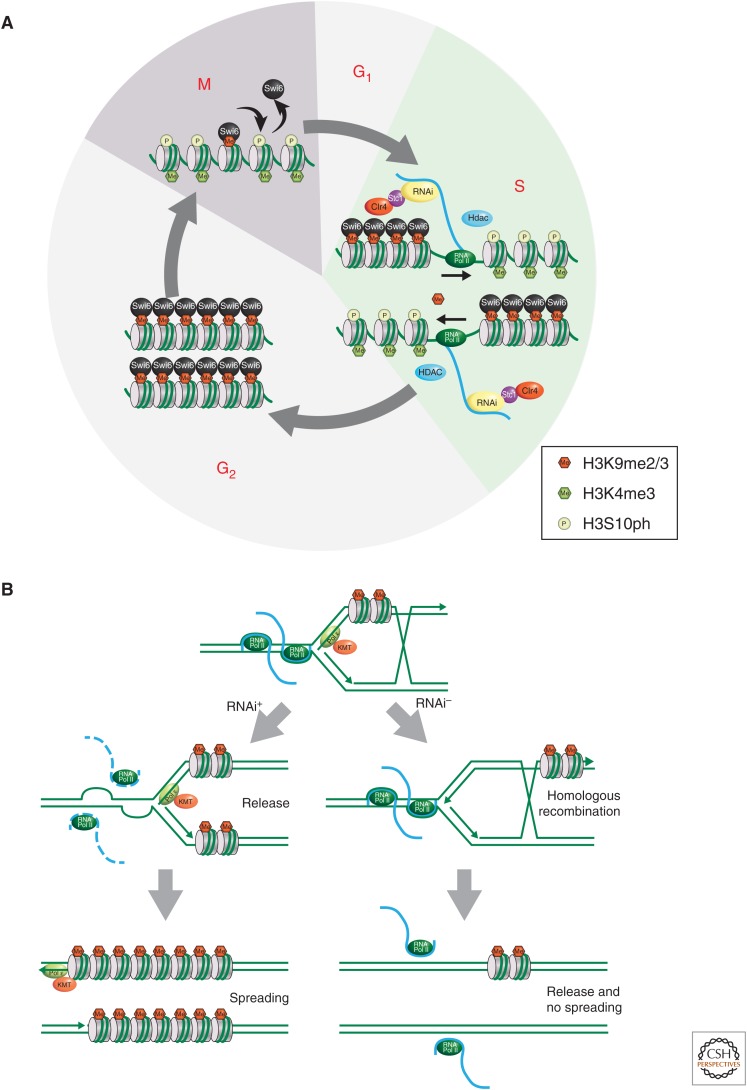
Figure 4.
Cell-cycle regulation of centromere heterochromatin assembly. (A) Heterochromatin located at chromosomal centromeres becomes differentially methylated and phosphorylated on histones throughout the cell cycle as indicated. These modifications control the binding of the heterochromatin protein Swi6. During mitosis Swi6 is displaced by H3S10 phosphorylation. Swi6 binding is reestablished during subsequent DNA replication (S phase) when a more accessible chromatin structure permits RNA Pol II to transcribe centromeric DNA. This, in turn, recruits the RNAi machinery to direct H3K9me methylation. (B) Replication-coupled RNAi model (Li et al. 2011). This figure illustrates an alternative model for how RNAi works at centromeres. Here, RNAi serves to release RNA Pol II from chromatin to avoid collision with DNA replication machinery during S phase. See text for further details. (A, Adapted from Djupedal and Ekwall 2008.)
Initiation of transcription, transcriptional elongation, and transcript processing are as important for heterochromatin assembly as they are for euchromatic gene expression. Several associated factors and activities are important for these different steps of the RNA Pol II transcription cycle, in addition to RNA Pol II itself. FACT, an RNA Pol II-associated chromatin assembly factor, and Spt6, another RNA Pol II-associated protein both colocalize to pericentric repeats. Spt6 is specifically required for facilitating trimethylation of H3K9, Swi6 binding, siRNA production, and recruitment of the HDAC enzyme Clr3 (Kiely et al. 2011). Mutations in the FACT component, Pob3, have a similar phenotype to spt6 implicating FACT in the same processes (Lejeune et al. 2007).
Interestingly, mutations in chromatin-modifying activities or RNA-processing factors have been shown to suppress the need for Dcr1. For example, loss of Mst2 activity, a H3K14-specific acetyltransferase, completely suppresses dcr1 mutants; that is, it eliminates the need for the RNAi machinery in heterochromatin maintenance, but not in the establishment of new heterochromatin (Reddy et al. 2011). This suggests that an important role of the RNAi directed heterochromatin process is to prevent Mst2 activity, which might interfere with CLRC recruitment.
Another example of bypassing the RNAi pathway is by knocking out the gene encoding Mlo3, involved in mRNP biogenesis and RNA quality control. This also suppresses the need for Dcr1 (Reyes-Turcu et al. 2011). It was suggested that in mlo3 ago1 knockout cells there is an aberrant accumulation of centromeric transcripts, which results in the recruitment of the CLRC complex via Rik1 (Hong et al. 2005; Horn et al. 2005). In wild-type cells, CLRC acts downstream from the RNAi pathway and is required for heterochromatin silencing by recruiting and promoting the activity of the Clr4 enzyme. Thus, this RNAi-independent recruitment mechanism of CLRC allows the induction of heterochromatin assembly at repeat sequences in mlo3 ago1 knockout cells (Reyes-Turcu et al. 2011).
Another RNA Pol II linked event is splicing of pre-mRNA by the spliceosome. There is not a general requirement for the spliceosome or the splicing process in heterochromatin formation, although some specific splicing factors are required for siRNA production (Bayne et al. 2008; Bernard et al. 2010). It remains to be determined how exactly these splicing factors contribute to RNAi, but their physical interaction with the RDRC suggests a direct role in RNAi. Findings in C. elegans and Cryptococcus neoformans corroborate the idea that splicing can be linked to RNAi. Clearly, heterochromatin formation is a complex process involving the RNA Pol II machinery, RNAi, splicing factors, and chromatin-modifying machinery.
4.3. Establishment of Heterochromatin
Detailed analysis of the genetic requirements for the initial establishment and the subsequent maintenance of heterochromatin indicate that these steps are distinct. Two models have been suggested to explain how heterochromatin is established over repeat sequences.
The first model suggests that RITS-associated siRNA may home in on nascent transcripts as they emerge from RNA Pol II engaged with the homologous locus. Consistent with this notion, artificial tethering of the RITS subunit Tas3 to the mRNA of the normally active ura4+ locus can initiate heterochromatin formation over the ura4+ gene (Buhler et al. 2006). But where do the initial siRNAs that trigger heterochromatin formation come from? The production of spurious small “primal” RNA by RNA degradation has been suggested to start off the heterochromatin establishment process (Halic and Moazed 2010). Alternatively, RNA Pol II transcripts could fold into self-structured double-stranded regions that are cleaved by Dcr1 to initiate heterochromatin formation (Djupedal et al. 2009; see Fig. 2 of Martienssen and Moazed 2014). Once recognition has taken place, the RITS-siRNA complex might be stabilized on these transcripts resulting in the recruitment of chromatin-modifying activities such as Clr4 as part of the CLRC complex (see Fig. 4 of Martienssen and Moazed 2014). Consistent with this idea, artificial tethering of Clr4 can completely bypass the need for RNAi during synthetic heterochromatin assembly (Kagansky et al. 2009).
A surprising result was that centromeric siRNAs were lost in cells lacking Clr4 activity (Noma et al. 2004; Hong et al. 2005). It is possible that the absence of Clr4 affects siRNA production by destabilizing associations between various components at the interface between transcription, RNAi, and chromatin modification (Motamedi et al. 2004). In some repeat regions, Clr4 can trigger RNAi independently of H3K9 methylation (Gerace et al. 2010) making it possible that Clr4 has additional targets for its HKMTase activity apart from histone H3, which might be important for siRNA amplification from repeat sequences.
A second model proposes that H3K9 methylation may be required to allow the generation of siRNAs in cis via the action of various RNA-processing activities (e.g., Rdp1) on primary centromeric transcripts (Noma et al. 2004; Sugiyama et al. 2005). Experiments with mutants of the RITS component Tas3, in which Tas3 is separated from the complex (so that Tas3 still interacts with Chp1 binding to H3K9me but not with Ago1), suggest that H3K9me is required before the production of small RNAs (Partridge et al. 2007). However, experiments showing that cells with H3K9 substitutions still produce small RNA argue against this model (Djupedal et al. 2009; Gerace et al. 2010). Clearly, more research is needed to understand what essential initiating steps versus reinforcing steps are required to generate heterochromatin.
4.4. Maintenance of Heterochromatin
Once heterochromatin is established, it must be maintained through cell division. The heterochromatin protein Swi6 is released from chromatin during mitosis because of the phosphorylation of the adjacent H3S10 residue (Fig. 4A) (Chen et al. 2008). Therefore, it is essential that Swi6 binding and the heterochromatic state are reestablished in S phase, when chromatin is replicated. Transcription of noncoding repeat sequences plays a crucial role in this maintenance mechanism, as it does for the establishment phase. The implication of RNA Pol II in heterochromatin assembly was initially thought to be a paradox because condensed heterochromatin was considered refractory to RNA polymerase activity. However, it appears that RNA Pol II is associated with the repeat sequences mainly during S phase of the cell cycle (Chen et al. 2008; Kloc et al. 2008). The decondensation of heterochromatin that occurs in this short window of time during DNA replication, before new heterochromatin is made, leads to a transient burst of RNA Pol II activity. This burst of transcription is thought to result in a boost of siRNA production leading to recruitment of heterochromatin assembly factors (Fig. 4A). As described in Section 4.1, other chromatin factors (i.e., HDACs) are also recruited at this stage to allow for reassembly of heterochromatin. A popular model is that siRNAs guide the RITS and RDRC complexes (i.e., RNAi machinery) to the chromosomes via the nascent noncoding RNA, providing a platform for the RNAi machinery (Motamedi et al. 2004). Interestingly, the LIM domain protein, Stc1, interacts both with the RITS Ago1 protein and CLRC; tethering of Stc1 is sufficient for heterochromatin assembly (Bayne et al. 2010). Thus, Stc1 seems to constitute the missing link between chromatin modification and the RNAi pathway, providing an explanation to why RNAi is needed to target H3K9 methylation.
In an alternative but not mutually exclusive model, RNAi plays a role in transcriptional termination leading to the avoidance of collisions between DNA polymerase and RNA Pol II (Fig. 4B) (Zaratiegui et al. 2011). In the absence of RNAi, experiments showed that such collisions cause stalled DNA replication forks that are repaired by homologous recombination (Zaratiegui et al. 2011). In this model, the HKMT activity is recruited via association with DNA polymerase epsilon interacting with the CLRC complex, linking the heterochromatin assembly process to lagging strand synthesis of DNA (Li et al. 2011). Thus, there is evidence for two distinct ways of CLRC recruitment to chromosomes in S. pombe. Clearly, further experiments are needed to determine their relative importance in the maintenance of heterochromatin.
4.5. Chromatin Boundaries: The Antisilencing Activity of Epe1 and Cullin Ligases
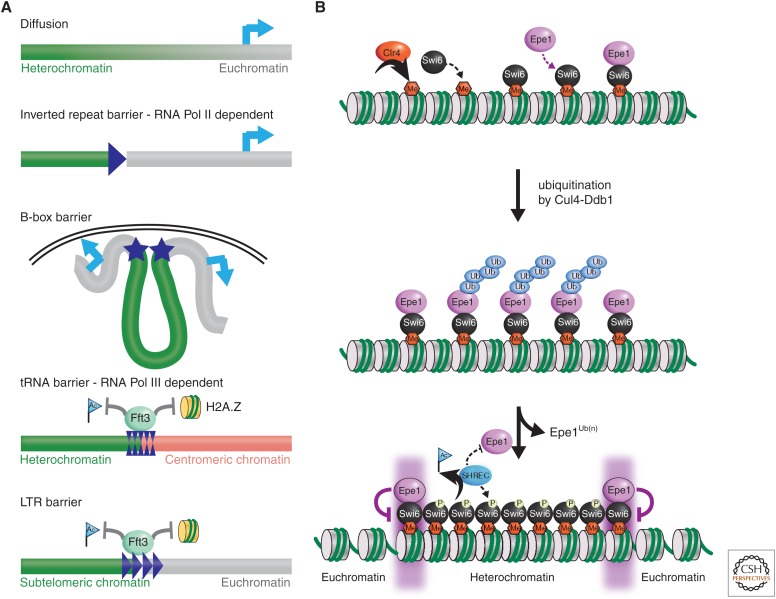
A key question is how distinct boundaries are set and maintained between heterochromatin and euchromatin. There are several types of heterochromatin boundary elements in S. pombe: diffuse boundaries, RNA Pol II inverted repeat (IR) barriers, B-box barriers, RNA Pol III-dependent tRNA barriers, and RNA Pol III-independent LTR barriers (long terminal repeat sequences derived from retrotransposons) elements (Fig. 5A) (Scott et al. 2007; Stralfors et al. 2011). Several tRNA genes reside between the outer repeats and the central kinetochore domain (double arrowheads in Fig. 2A). These have been shown to act as a barrier preventing heterochromatin from encroaching into the central domain (Scott et al. 2006).
Figure 5.
Chromatin boundaries and the boundary mechanism involving Epe1 (for enhancement of position effect) in S. pombe. (A) A schematic representation of different types of boundary elements in S. pombe. (B) The mechanism of boundary function by Epe1. Epe1 associates with Swi6, however, when the antisilencing factor is ubiquitinated by the Cul4-Ddb1 ligase and degraded in the heterochromatin region to allow for heterochromatin assembly. However, at boundaries, Epe1 is somehow protected from degradation, thus restricting the spreading of heterochromatin. Phosphorylation of Swi6 contributes to the dissociation of Epe1 at heterochromatin, while promoting the association with the HDAC complex SHREC in maintaining histone hypoacetylation. (A, Adapted from Scott et al. 2007.)
One of the best-studied boundary factors, Epe1, was first identified through a mutation that enhances heterochromatin formation (Ayoub et al. 2003). Epe1 interacts with Swi6 and somehow restricts H3K9 methylation to within heterochromatic regions. It also reduces siRNA production and promotes RNA Pol II occupancy in heterochromatin (Zofall and Grewal 2006; Isaac et al. 2007; Trewick et al. 2007), thereby limiting heterochromatin assembly beyond (IR) boundaries in the mat2-mat3 region and pericentric regions of chromosomes 1 and 3 (Trewick et al. 2007; Braun et al. 2011). Based on its function in restricting heterochromatin domains from spreading into euchromatin, Epe1 has been termed an antisilencing factor. How, then, is the function of Epe1 restricted to chromatin boundary regions? It is thought to uniformly associate with heterochromatin through association with Swi6; however, the ubiquitinating action of the cullin-dependent ligase Cul4-Ddb1 degrades Epe1 in heterochromatin so that it becomes restricted to the heterochromatin/euchromatin boundary regions (Fig. 5B) (Braun et al. 2011). What prevents the ubiquitin ligase from acting on Epe1 at boundaries is currently an open question.
Determining and maintaining a particular chromatin structure is clearly a dynamic process involving the interaction and competition of many factors. Epe1 operates in the context of many other chromatin factors, and research is beginning to show how it interacts or competes with these other proteins and complexes. Epe1 counteracts HDAC activity by competing with the SHREC complex (containing the Clr3 HDAC, SNF2 remodeling factor, Mit1, and telomere end protection protein, Ccq1) for binding to heterochromatin (Sugiyama et al. 2007). Interestingly, phosphorylation of Swi6 by casein kinase 2 (CK2) promotes SHREC binding and heterochromatin spreading, which leads to reduced Epe1 levels (Shimada et al. 2009). Reduced phosphorylation of Swi6 leads to the opposite situation, with less SHREC and more Epe1, and hence reduced spreading of heterochromatin. Exactly why SHREC requires CK2-dependent phosphorylation of Swi6 to localize at the heterochromatic domain is not yet known.
The Fun30 remodeling factor Fft3, an ATPase helicase of the SNF2 family, also functions at boundaries (see Becker and Workman 2013 for details of how the SNF2 enzymes work). Fft3 is localized to the centromeric tRNA and subtelomeric LTR boundary regions, where it prevents euchromatin formation in the centromeric domain and subtelomeric regions (Stralfors et al. 2011). Thus, Fft3 performs a new type of boundary function at tRNAs, which does not limit the spreading of heterochromatin, but prevents euchromatin marks such as H3/H4 acetylation or the histone variant H2A.Z from invading the insulated regions. Fft3 works similarly at subtelomeric LTR boundaries preventing the formation of euchromatin in these regions (Buchanan et al. 2009). It is currently not known how chromatin remodeling at these boundaries leads to this type of insulator function, but one possibility is that this function involves changes in higher-order chromatin structure and nuclear organization (see Sec. 8 for a discussion of nuclear organization in S. pombe).
5. S. POMBE CENTRAL DOMAIN CHROMATIN AND KINETOCHORES
The central domain is the site of kinetochore assembly and its chromatin is very distinct from the flanking outer repeat heterochromatin. The kinetochore is the protein structure that forms at centromeres to facilitate the separation of sister chromatids along spindle fibers during M phase. The variegated silencing of reporter genes in the ∼10-kb central domain of cen1 was indicative of a different chromatin configuration, which is essentially independent of Clr4 and therefore does not involve methylation of H3K9 (Fig. 3). The distinct chromatin composition was initially shown by micrococcal nuclease analysis (see Elgin and Reuter 2013 for an explanation of MNase digestion), which revealed a smear in contrast to the regular 150-bp ladder characteristic of flanking outer repeat chromatin (Polizzi and Clarke 1991; Takahashi et al. 1992). This distinct pattern differentiated central domain chromatin from both heterochromatin and euchromatin, reflecting its assembly into distinctive chromatin and the assembly of the kinetochore over it. Later research in various eukaryotes showed that all active centromere chromatin examined contained a histone H3-like protein known as CENP-A (or CenH3), which is critical for specifying the site of kinetochore assembly (Fig. 6).
Figure 6.
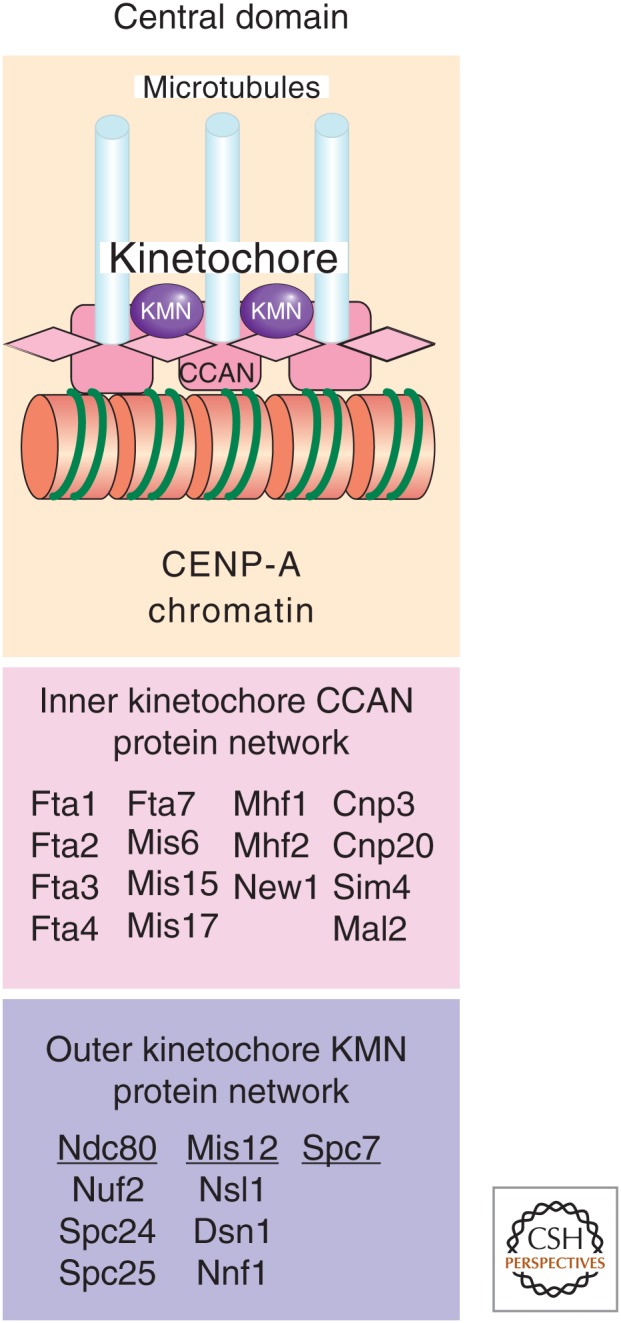
Centromeric chromatin and kinetochores in S. pombe. The central domain of S. pombe centromeres is composed of CENP-A-containing chromatin and a multiprotein network that makes up the kinetochore. The inner constitutive centromere-associated network (CCAN) and outer KMN kinetochore protein networks are depicted and the different protein components listed below. Kinetochore assembly within the central domain mediates attachment to microtubules on spindle formation and chromosome segregation.
5.1. The Central Domain Contains CENP-A
In the central core domain of fission yeast centromeres, the majority of histone H3 is replaced by the variant CENP-A ortholog known as Cnp1 (Takahashi et al. 2000). If CENP-ACnp1 chromatin structure is disrupted (as in the case of kinetochore mutants), then the specific smeared micrococcal nuclease digestion pattern reverts to a pattern more typical of bulk chromatin (i.e., a nucleosomal ladder) without affecting adjacent heterochromatin or gene silencing in the outer repeats (Pidoux et al. 2003; Hayashi et al. 2004). Furthermore, the fact that CENP-ACnp1 and other kinetochore proteins only associate with the central domain shows that the central kinetochore domain is structurally complex and functionally distinct from outer repeat silent heterochromatin (Fig. 6).
It has been estimated, based on Cnp1-GFP intensity, that there are 680 CENP-ACnp1 molecules per cell in S. pombe (Coffman et al. 2011). Quantitation using single-molecule microscopy, however, concluded that there are 72–82 molecules of CENP-ACnp1 per centromere cluster (containing all three centromeres) in G2 cells. Estimates based on ChIP-seq (chromatin immunoprecipitation coupled to DNA sequencing) measurements yielded between 19 and 24 CENP-A nucleosomes per centromere; hence, a total of 64 molecules per haploid cell (Lando et al. 2012). These results indicate that there are ∼10–20 CENP-ACnp1 particles at each centromere and CENP-A is deposited in mid to late G2 phase of the cell cycle.
Although not all known kinetochore domain proteins have been tested, thus far it appears that silencing within the central domain is a result of the steric hindrance caused from the assembly of an intact kinetochore. The inner kinetochore consists of the CCAN totaling 15 proteins. This includes the S. pombe homologs of several CENP proteins (Fig. 6) (reviewed in Perpelescu and Fukagawa 2011). Outside of CCAN, we find the “outer kinetochore” KMN complex, which is made up of another conserved network of proteins including the Ndc80 and Mis12 protein subcomplexes and Spc7 (reviewed in Buttrick and Millar 2011). Together, these protein networks bind centromeric CENP-A chromatin, helping to form stable kinetochore-microtubule attachments. This large complex of proteins presumably restricts access of RNA Pol II to reporter genes placed within this region and thereby impedes their transcription. In conditional temperature-sensitive kinetochore mutants, the kinetochore integrity is clearly only partially functional at the growth permissive temperature and this allows for increased transcription of reporter genes. An advantage of this is that a normally silent reporter gene that becomes active has been used to assay for defects in central core chromatin, leading to the identification of novel kinetochore proteins (Pidoux et al. 2003).
5.2. Epigenetic Inheritance of the Functional Centromere State
Evidence from many organisms has contributed to the hypothesis that CENP-A is the epigenetic mark that specifies centromere identity. Recent observations in Drosophila indicate that CENP-A chromatin is able to self-propagate. Studies in fission yeast have contributed to our understanding of CENP-A maintenance as well as the establishment of this unique chromatin state. In addition, elegant studies on formation of neocentromeres and inactivation of dicentric chromosomes highlight the importance of S. pombe in epigenetic research.
An interesting epigenetic phenomenon has been described with respect to the assembly of functional centromeres in fission yeast using plasmids containing minimal regions for centromere function. Plasmids that retain only part of an outer repeat and most of the central domain can assemble a functional centromere, albeit inefficiently. Once this functional centromere state has been established, however, it can be propagated through many mitotic divisions and even through meiosis (Steiner and Clarke 1994; Ngan and Clarke 1997). One interpretation is that the outer repeats provide an environment that is favorable for kinetochore assembly (Pidoux and Allshire 2005), but once assembled, CENP-ACnp1 chromatin and thus the kinetochore is propagated at this position by a templating mechanism that may be coupled to replication. It is possible that heterochromatin somehow induces or aids the deposition of CENP-ACnp1 in the central domain (Fig. 7A) such that only one block of heterochromatin is sufficient for kinetochore assembly. In fact, heterochromatin and RNAi are required for de novo assembly of CENP-A chromatin on naked DNA introduced into cells (Folco et al. 2008). Further evidence for the epigenetic nature of centromeric chromatin in S. pombe came from studies on neocentromeres (Ishii et al. 2008). Neocentromeres can be formed in subtelomeric regions, lacking any centromeric DNA sequences, on induced deletion of the centromere. This suggests that the formation of neocentromeres in the vicinity of subtelomeric heterochromatin probably reflects a key role for heterochromatin in centromere establishment (Castillo et al. 2013). Another illustration of epigenetic regulation in S. pombe is provided by the way that cells deal with dicentric chromosomes. One centromere in a dicentric chromosome can become inactivated because of disassembly of the kinetochore and the CENP-A chromatin and assembly of heterochromatin over the central domain (Sato et al. 2012). Even in the absence of heterochromatin components, centromeric shutdown can occur by the assembly of a hypoacetylated chromatin state. In addition, inactivated centromeres can become reactivated in certain situations. These observations serve to show the plasticity and epigenetic nature of centromeric chromatin; the same DNA can be assembled into a variety of different chromatin states, which have different functions.
Figure 7.
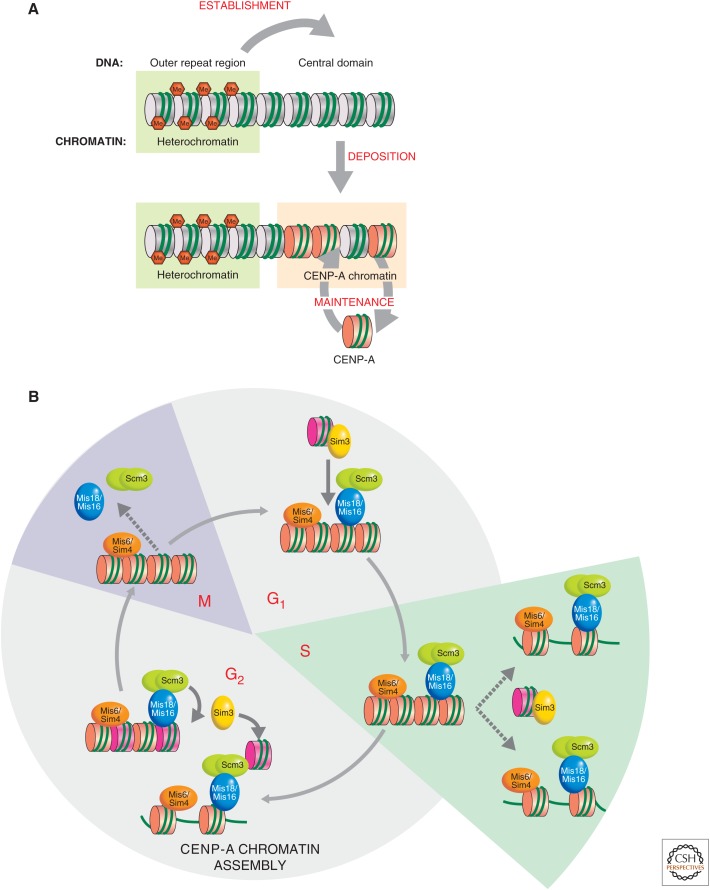
CENP-A chromatin establishment and propagation through the cell cycle. (A) Central domain DNA alone is unable to establish a functional centromere; outer repeats are required. Loss of heterochromatin from established centromeres does not affect CENP-Acnp1 or kinetochore maintenance in the central domain. This suggests that heterochromatin may, in some way, initially direct the site of CENP-Acnp1 chromatin and thus kinetochore assembly. (B) Cell-cycle dependency of CENP-ACnp1 recruitment in S. pombe. In S. pombe, centromeric DNA is replicated and existing CENP-ACnp1 is diluted by nucleosome segregation to sister chromatids during S phase. Recruitment of new CENP-A occurs during the G2 phase, indicated by the pink nucleosomes. The Sim3 histone chaperone interacts with new CENP-ACnp1 and delivers it to the centromere, where it is received by Scm3 and assembled into nucleosomes by unknown factors and mechanisms. Nucleosome gaps could be filled or H3 nucleosomes could be replaced. Scm3 is shown as a dimer interacting directly with Mis18. Scm3 recruitment at centromeres requires the Sim4/Mis6 and Mis16/Mis18 complexes. Mis16/Mis18 and Scm3 are removed from centromeres during mitosis and reassociate in G1. (B, Adapted from Mellone et al. 2009.)
Once CENP-ACnp1 chromatin is established, heterochromatin is not required to propagate it in subsequent cell divisions. What is the mechanism of CENP-A propagation through the cell cycle? Unlike in Drosophila and mammals in which loading occurs in G1, CENP-ACnp1 deposition in S. pombe occurs mainly during the G2 phase of the cell cycle (Fig. 7B) (Lando et al. 2012). Kinetochore proteins themselves govern the localization and assembly of CENP-ACnp1 specifically within the central domain (Goshima et al. 1999; Takahashi et al. 2000; Pidoux et al. 2003). Several loading factors and chaperones for CENP-ACnp1 have been identified. Mis16, an RpAp46/RpAp48-like protein, and Mis18/KNL are conserved centromeric proteins required for CENP-A loading and histone deacetylation in the central core region (Hayashi et al. 2004). They dissociate from centromeres, from early mitosis (prophase) until mid-anaphase, but remain associated for the rest of the cell cycle (Fujita et al. 2007). Sim3, a NASP-like protein, binds CENP-ACnp1 and acts as a chaperone to deliver it to centromeres (Dunleavy et al. 2007). Scm3 (ortholog of HJURP) is a CENP-ACnp1 chaperone required for assembly of CENP-A chromatin, and it dissociates from centromeres in mitosis similar to Mis16 and Mis18 (Pidoux et al. 2009; Williams et al. 2009). It is currently unknown what restricts CENP-ACnp1 replenishment to the G2 phase of the cell cycle in S. pombe.
In addition to the chaperones and loading factors, SNF2 family members of chromatin-remodeling factors are also required for CENP-ACnp1 loading. The SNF2 remodeler Hrp1 (a Chd1 ortholog) is localized to centromeric chromatin where it is required to maintain high CENP-ACnp1 levels (Walfridsson et al. 2005). Unstable noncoding transcripts that originate from the centromeric central core chromatin have been recently described in S. pombe (Choi et al. 2011). During the process of noncoding transcription in this region, nucleosome disassembly and reassembly reactions must occur, and it is conceivable that Hrp1 acts in these transcription-coupled chromatin remodeling and assembly activities to evict H3 and maintain CENP-ACnp1 at centromeres. Consistent with this, loss of factors required for the reassembly and stabilization of H3 nucleosomes behind RNA Pol II, such as FACT and Clr6-complex II, promotes CENP-A assembly in place of histone H3 in noncentromeric regions of the genome (Choi et al. 2012). Moreover, some subunits of the Mediator complex are required for CENP-ACnp1 localization, and the reduced levels of CENP-ACnp1 in Mediator mutants can be suppressed by RNA Pol II inhibitors (Carlsten et al. 2012). This finding suggests that proper levels of centromeric transcription are critical to ensure inheritance of the functional centromere state.
Thus, initial establishment of CENP-ACnp1 chromatin in S. pombe requires flanking heterochromatin. The subsequent maintenance of CENP-A chromatin is independent of heterochromatin, but depends on several loading factors and chaperones to facilitate G2 stage–specific assembly of CENP-A chromatin. The chromatin-remodeling factor Hrp1 and appropriate levels of noncoding transcription help to maintain high CENP-ACnp1 levels at centromeres.
5.3. Additional Chromatin Modifications Required for Mediating Sister-Centromere Cohesion, Condensation, and Normal Chromosome Segregation
How do outer repeat heterochromatin and CENP-ACnp1 kinetochore chromatin affect the overall function of chromosome segregation? Experiments with naked plasmid DNA constructs have shown that outer repeats contribute to the assembly of functional centromeres (through unknown mechanisms), which allow these plasmids to segregate on mitotic and meiotic spindles. However, as already mentioned, neither the outer repeats nor the central domain alone are sufficient to assemble a functional centromere (Clarke and Baum 1990; Takahashi et al. 1992; Baum et al. 1994; Ngan and Clarke 1997; Pidoux and Allshire 2004).
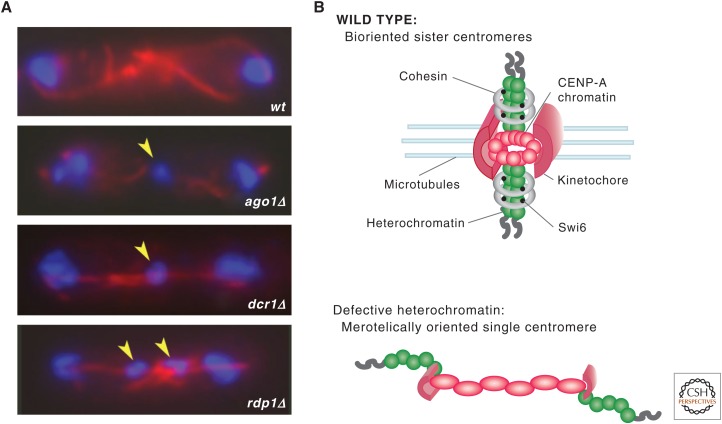
Mutants that cause loss of silencing at the outer repeats (i.e., those defective in Clr4, RNAi components, or Swi6) have elevated rates of mitotic chromosome loss and a high incidence of lagging chromosomes on late anaphase spindles (Fig. 8A) (Allshire et al. 1995; Ekwall et al. 1996; Bernard et al. 2001; Nonaka et al. 2002; Hall et al. 2003; Volpe et al. 2003). Additionally, cells lacking Swi6 are defective in cohesion at centromeres, but retain cohesion along the chromosome arms (Bernard et al. 2001; Nonaka et al. 2002). One function of Swi6 is to recruit cohesin to outer repeat chromatin to mediate tight physical cohesion between sister centromeres. This ensures that sister kinetochores are tightly held together during the formation of a properly bioriented spindle in which sister kinetochores attach to microtubule fibers emanating from opposite spindle poles (Fig. 8B). Thus, one function of heterochromatin at centromeres is to mediate cohesion.
Figure 8.
Defective heterochromatin leads to abnormal centromere structures. (A) Cells lacking RNAi or heterochromatin components display elevated rates of chromosome loss and lagging chromosomes (indicated by yellow arrows) on late anaphase spindles. Chromosomal DNA is stained by DAPI (blue) and mitotic spindle microtubules are labeled for immunofluorescence (IF) (red). (B) A schematic three-dimensional figure of a normal centromere illustrates the outer heterochromatin regions (green circles) decorated with Swi6 (black circles), which recruits cohesin to ensure sister-chromatid cohesion. The central domain consists of CENP-A-containing chromatin (red circles), associated with opposing kinetochores on each sister chromatid. Lagging chromosomes in cells with defective heterochromatin may be the result of disorganized kinetochores such that one centromere may attach to microtubules from opposite poles. Such merotelic orientation could persist into anaphase, in which the breakage of attachment with one pole or the other would lead to random segregation and result in chromosome loss/gain events.
Cohesin is also strongly associated with telomeres and the mat2-mat3 region (Bernard et al. 2001; Nonaka et al. 2002) and it is involved in the formation of subtelomeric heterochromatin (Dheur et al. 2011). In addition, cohesin is recruited to silent chromatin formed on a ura4+ gene in response to an adjacent ectopic centromere repeat, underscoring the link between heterochromatin and cohesion (Partridge et al. 2002). Thus, the recruitment of cohesin seems to be a general property of Swi6-associated silent chromatin. How Swi6 chromatin brings about cohesin recruitment is not known, but Swi6 does interact with the cohesin subunit Psc3 (Nonaka et al. 2002). In addition, Dfp1, the regulatory subunit of the conserved kinase Hsk1 (Cdc7), interacts with Swi6 and is required to recruit cohesin to centromeres (Bailis et al. 2003). Cohesion at vertebrate centromeres from prophase to anaphase onset also depends on a protein known as Shugoshin. In S. pombe, Shugoshin associates with the centromere on phosphorylation of histone H2A at S121 by Bub1 to ensure proper chromosome segregation in mitosis and meiosis (Kawashima et al. 2010). Thus, cohesin and associated factors are not only required at centromeres, but along the length of chromosomes to mediate chromosome condensation and segregation functions during M phase.
The histone H2A variant H2A.Z was initially identified as a factor that when mutated led to defects in chromosome segregation (Carr et al. 1994). Histone H2A and its variant H2A.Z play important roles in chromosome segregation by acting as chromatin receptors for condensin recruitment both at kinetochores and along the entire chromosome to shape mitotic chromosomes during cell division. Recruitment of condensin to chromatin via H2A and H2A.Z only occurs in its phosphorylated state and is a cell-cycle regulated process (Tada et al. 2011). Acetylation of H2A.Z by the HAT enzyme Mst1 is also important for proper structure and separation of anaphase chromosomes (Kim et al. 2009). H2A.Z itself is not normally present at centromeres, but is required for expression of the centromere protein CENP-CCnp3 (Fig. 6), and perturbs centromere function when mislocalized to centromeres in mutant conditions (Ahmed et al. 2007; Buchanan et al. 2009; Hou et al. 2010). Histone H2B is also implicated in centromere function, probably through ubiquitination, because mutations near the ubiquitination site cause anaphase defects and changes in centromeric chromatin structure (Maruyama et al. 2006). Thus, all core histones, together with the variants CENP-ACnp1 (a histone H3 variant) and H2A.Z, are involved in orchestrating proper centromeric and chromosome-wide chromatin structure in S. pombe important for sister-centromere cohesion, condensation, and normal chromosome segregation.
6. HETEROCHROMATIN FORMATION AT OTHER SILENT REGIONS
Many of the molecular mechanisms controlling heterochromatin formation at centromeres also operate at other silenced chromatin regions such as the mating-type locus, telomeres, and rDNA (Fig. 2A). However, there are also additional pathways that are specific for these distinct regions; these are discussed below.
6.1. Assembly of Silent Chromatin at cenH Regions
At the DNA sequence level, most heterochromatic regions contain centromere homologous (cenH) DNA regions that are capable of assembling Clr4-dependent silent heterochromatin. cenH sequences were originally defined at the outer repeat regions of centromeres, but cenH DNA sequences can also be found at the silent mating-type loci (mat2-mat3; Noma et al. 2001) and at a subtelomeric region adjacent to the terminal telomeric repeats (consensus TTACAGG) synthesized by telomerase (Nimmo et al. 1994; Allshire et al. 1995; Nimmo et al. 1998; Kanoh et al. 2005). Seven of the 20-kb long cenH region that resides between mat2 and mat3 shares a high degree of sequence similarity with the centromeric outer repeats (Grewal and Klar 1997). In addition, a 0.5-kb DNA sequence located within the telomere-associated sequences (TASs) that occupy up to 40 kb proximal to the telomeres of chromosomes I and II shares >84% identity with cenH (Kanoh et al. 2005). This suggested that cenH repeats might act in cis to affect silent heterochromatin assembly.
Indeed, ectopic centromeric outer repeats (dg) or related cenH DNA sequences are sufficient to silence adjacent marker genes in a Clr4-dpendent manner (Ayoub et al. 2000; Partridge et al. 2002; Volpe et al. 2003). As already discussed, transcription of centromeric outer repeats (and cenH sequences) and the processing of resulting dsRNA by Dcr1 and the RNAi machinery provides a mechanism by which Clr4 is recruited to trigger the assembly of silent chromatin (Volpe et al. 2002).
6.2. Mechanisms of Heterochromatin Formation at the Mating-Type Loci
At the mating-type loci, two distinct mechanisms are involved in the establishment and maintenance of heterochromatin. This was revealed by experiments involving treatment of cells with trichostatin A, an HDAC inhibitor, in which derepression of silencing could be achieved only in RNAi mutant backgrounds (Hall et al. 2002). In addition to RNAi, an RNAi-independent silencing mechanism based on the DNA-binding proteins Atf1 and Pcr1 (which bind near cenH) and the HDAC Clr6 operates at the silent mating-type loci to maintain heterochromatin (Hall et al. 2002; Jia et al. 2004). RNAi acts to establish silent chromatin; however, its maintenance is achieved by the Atf1- and Pcr1-based silencing system that can maintain heterochromatin even in the absence of RNAi (Jia et al. 2004; Kim et al. 2004). Consistent with this notion, cells lacking components of the RNAi machinery and Atf1/Pcr1 show a complete loss of silent chromatin at the mat2-mat3 region.
6.3. Mechanisms of Heterochromatin Formation at Telomeres
Overlapping mechanisms of silencing also function at telomeres. Terminal repeats are bound by the telomere repeat binding protein Taz1, which, in turn, recruits the Clr4 HKMT and thus Swi6 to form heterochromatin. However, RNAi also acts via the cenH region within the TAS elements to form an extended region of silent chromatin at telomeres (Nimmo et al. 1994; Allshire et al. 1995; Kanoh et al. 2005). Interestingly, heterochromatin formation contributes to the protection of chromosome ends even in the absence of telomerase, which typically acts to maintain telomere length. In a recent study, a subset of cells that survive telomerase ablation were found to maintain the integrity of chromosome ends through a mechanism of continual amplification and rearrangement of heterochromatic sequences, which, in turn, induces the recruitment of telomere end-protection factors Pot1 and Ccq1 (a component of SHREC). These strains were consequently named HAATI mutants (for heterochromatin amplification-mediated and telomerase-independent; Jain et al. 2010). Swi6-GFP localization in HAATI cells and the requirement of Clr4 for HAATI indicates that the amplified regions are indeed heterochromatic. Moreover, maintenance of linear chromosomes in HAATI cells depends on the Ccq1 component of the SHREC complex, which contains HDAC activity and plays a role in heterochromatin spreading (see Sec. 4.1). Thus, Ccq1 provides a molecular link between telomeric end-protecting protein complexes and histone-modifying activities involved in heterochromatin assembly.
6.4. Facultative Heterochromatin
In addition to the larger constitutive heterochromatin regions found at centromeres, the mating-type region, rDNA, and telomeres, there are also so-called facultative heterochromatin regions in S. pombe (Zofall et al. 2012). These heterochromatic “islands” are present in vegetative cells and contain clusters of genes that are expressed and, hence, euchromatic in meiotic cells. Epe1 and RNA degradation factors are important for the regulation of the meiotically induced genes in these islands. RNA degradation factors maintain silencing of the islands during vegetative growth, but Epe1 is needed to disassemble the heterochromatic islands during meiosis to allow for expression of the meiotic genes in response to nutritional signals. Some meiotically induced genes also cluster in the subtelomeric regions (Mata et al. 2002), and one such meiotic cluster resides in the tel1L (left subtelomere of chromosome 1) region. This cluster is hypoacetylated in mitotic cells and repression of the genes requires the HDAC Clr3, indicative of its facultative heterochromatin structure (Wiren et al. 2005).
Another type of transient heterochromatin is found in intergenic regions between some convergent gene pairs in S. pombe (Gullerova and Proudfoot 2008). This heterochromatin apparently depends on convergent transcription in the G1 phase of the cell cycle, leading to local RNAi-dependent H3K9 methylation, which recruits Swi6 and cohesin. siRNA homologous to such intergenic regions have, so far, eluded detection. The accumulation of cohesin by interaction with Swi6 in these regions aids transcription termination in the G2 phase at sites upstream of the termination sites used in G1, so that alternative 3′ mRNA ends are generated. Interestingly, many of the genes that encode the RNAi machinery components are themselves autoregulated by this process (Gullerova et al. 2011).
7. NUCLEOSOME REMODELING IN S. POMBE
Chromatin structure and function are not solely determined by histone modifications and mechanisms that facilitate this. Chromatin remodeling is also an important component in the regulation of chromatin structure and function. Although we have already alluded to a few chromatin remodeling proteins in our discussion of S. pombe heterochromatin formation and propagation, this section elaborates on the status of research in this field. Another important aspect that governs chromatin structure is nuclear architecture, which is the topic of Section 8.
SNF2 nucleosome-remodeling enzymes play key roles in epigenetic processes, such as the regulation of transcription and the loading or exchange of histones because of their ability to alter the positions of nucleosomes in the genome (Becker and Workman 2013). Homo sapiens has 53 SNF2 enzymes, whereas S. pombe has only 20 (Flaus et al. 2006). The CHD-type SNF2 enzyme, Mit1, is part of the SHREC complex discussed in Section 4.5 and more extensively in Martienssen and Moazed (2014). It is required for silencing at the mating-type region, telomeres, and pericentric heterochromatin in S. pombe, but not at the 5′ end of genes (Sugiyama et al. 2007; Pointner et al. 2012).
The Chd1-like SNF2 remodelers, Hrp1 and Hrp3, are involved in silencing at pericentric heterochromatin and at the mating-type region. It is plausible that the role of Hrp1 at centromeres in CENP-A loading is mechanistically related to its ability, in conjunction with the Nap1 chaperone, to evict H3 at many gene promoters and its role in nucleosome assembly (Walfridsson et al. 2007; Pointner et al. 2012). Recently, Hrp1 and Hrp3 were implicated in the genome-wide suppression of cryptic transcription in gene-coding regions (Hennig et al. 2012; Pointner et al. 2012; Shim et al. 2012). The hrp1 and hrp3 mutants show reduced nucleosome arrays in coding regions, which allows for cryptic initiation of transcription by RNA Pol II. A mechanistic explanation for this phenomenon is the demonstration of a prominent nucleosome spacing activity for Hrp1 and Hrp3 in vitro (Pointner et al. 2012). Thus, Hrp1 and Hrp3 remodelers seem to have distinct activities that, at intergenic regions, favor disassembly versus coding regions, in which they favor the assembly and spacing of nucleosomes. The action of Hrp1 in intergenic regions is strongly correlated to topoisomerases I and II (Durand-Dubief et al. 2010). Topoisomerases I and II are generally required to maintain nucleosome-free regions both at gene promoters and in 3′ intergenic regions, most likely by removing negative supercoils that stabilize nucleosomes (Durand-Dubief et al. 2007, 2011). It is likely that this function of topoisomerases stimulates the nucleosome disassembly function of SNF2-remodeling enzymes including Hrp1. It is possible that the absence of Top I and II from coding regions alters the outcome of the remodeling process in favor of the spacing activity. Another example of SNF2 chromatin remodelers implicated in gene silencing is the Swr1 complex that replaces H2A with H2A.Z (Buchanan et al. 2009; see Fig. 9 of Henikoff and Smith 2014). Thus, although the research on SNF2 enzymes in S. pombe is still in its budding stages, it is already clear that these enzymes play key roles in epigenetic regulating chromatin structure and control.
Figure 9.
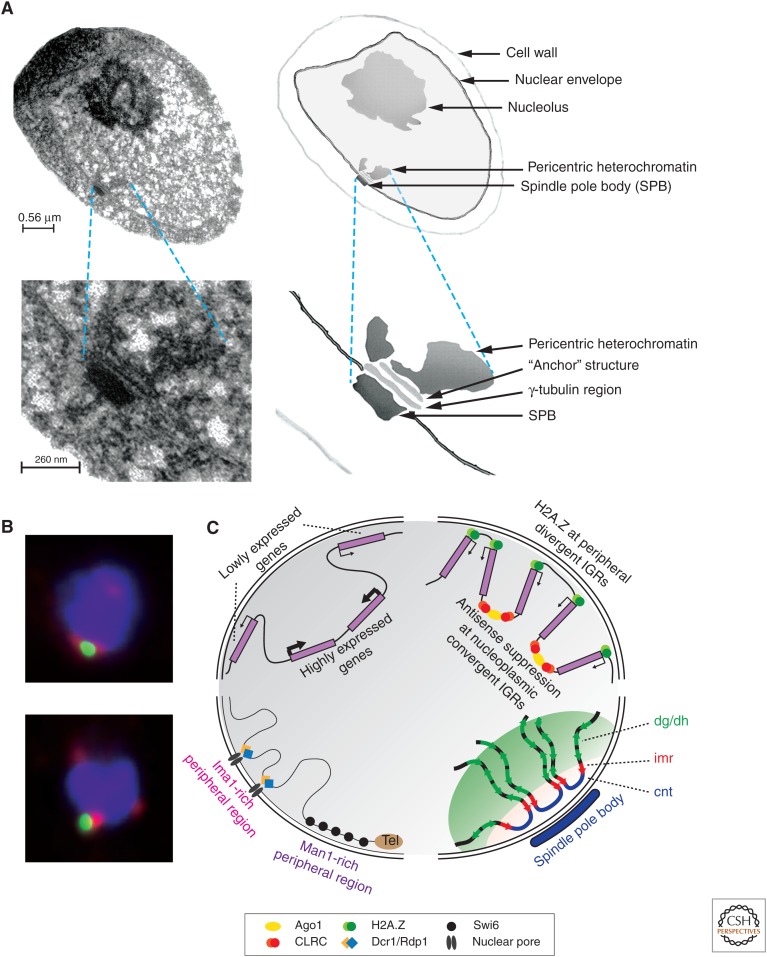
Nuclear organization in S. pombe. (A) Electron microscopy analysis of an S. pombe nucleus. (Top) Micrograph of a cross-section through a high-pressure fixed and Lowicryl-embedded interphase S. pombe cell. The cellular structures are indicated: cell wall, nuclear envelope, nucleolus, heterochromatin region, and SPB. (Bottom) A higher magnification of the same nucleus. The nuclear structures indicated are SPB, γ-tubulin region, anchor structure, and the centromeric heterochromatin. (B) Two interphase nuclei with heterochromatin (centromeres, telomeres, and the silent mat2-mat3 loci) decorated by red fluorescent immunolocalization of Swi6, and kinetochore chromatin (centromeres only) decorated by green fluorescent immunolocalization of CENP-ACnp1. The red signals, not in close proximity to green, represent telomeres or the mat2-mat3 loci. All centromeres are clustered at the nuclear periphery adjacent to the SPB. (C) A model for chromatin organization at the fission yeast nuclear periphery. (Top) Genes with low expression levels tend to associate with the nuclear periphery, whereas highly expressed genes tend to reside in the nuclear interior. (Top) Localization of divergent intergenic regions and H2A.Z at the nuclear envelope may present a mechanism for anchoring the promoters of convergent gene pairs at the periphery. (Bottom left) Differential localization of Ima1, nuclear pores, and Man1. The inner membrane proteins Ima1 and Man1 are not equally distributed at the nuclear periphery, but rather occupy distinct areas that interact with different chromosomal regions. The subtelomeric chromatin is associated with Man1-rich peripheral regions in which Swi6 is also located. Ima1 is colocalized with Dcr1 and Rdp1 at nuclear pores. (Bottom right) Organization of centromeric DNA at the SPB. Central domain cnt and imr regions are localized closer to the SPB than the heterochromatic dg and dh repeats. The two centromeric domains are shaded in colors symbolizing the different IF localization of Swi6 (dg-dh repeats) and CENP-ACnp1 (imr/cnt regions). (A, Reprinted from Kniola et al. 2001; C, bottom right, Adapted from Takahashi et al. 1992.)
8. NUCLEAR ORGANIZATION IN S. POMBE
Although the role of nuclear architecture in epigenetic control is well established in mammalian systems (see Dekker and Misteli 2014), our knowledge in S. pombe is largely descriptive at present. The S. pombe interphase nucleus is spherical and consists of a chromatin-rich and an RNA-rich part, the latter of which contains the nucleolus (Fig. 9A). All heterochromatin is peripheral in the nucleus. The three centromeres are in fact attached to the nuclear periphery, adjacent to the spindle pole body (SPB) throughout interphase, but pericentric heterochromatin can be cytologically distinguished from centromeric chromatin by IF microscopy; kinetochores and centromeric CENP-ACnp1 chromatin are visualized as being surrounded by a layer of pericentric heterochromatin (Fig. 9B,C) (Kniola et al. 2001). The mating-type region can also be found localized near the nuclear envelope in close proximity with the SPB (Alfredsson-Timmins et al. 2007). The telomeres, however, although found at the nuclear periphery, are not fixed in location, unlike the centromeres or mating-type regions.
Interestingly, specific transcriptionally active regions, such as tRNA and 5S rRNA genes transcribed by RNA Pol III, cluster and colocalize with centromeres (Iwasaki et al. 2010). It is hypothesized that this association with centromeres is mediated through interaction of the condensin complex with the RNA Pol III transcription machinery. Another interesting feature of nuclear organization is the distribution of retrotransposon elements such as Tf2 known to be silenced by HDACs; these elements cluster as discrete nuclear structures at the nuclear periphery in a CENP-B-dependent fashion, presumed to be a mechanism of host genome surveillance and defense (Cam et al. 2008). There is also evidence for the existence of chromosome territories in S. pombe (Molnar and Kleckner 2008; Tanizawa et al. 2010; concept illustrated in Fig. 1 of Dekker and Misteli 2014). How these observations are interconnected and relate to nuclear function requires further investigation; however, the application of other methodology has begun to address these questions.
Studies using the DamID approach have shown how the nuclear envelope and nuclear pores are involved in chromosomal organization (Fig. 9C). In DamID, a protein with a known nuclear location is fused to DNA adenine methylase (Dam). When chromatin or DNA associates with the Dam-fusion protein, contacted regions of the genome can be mapped based on the presence of adenine methylation added by the Dam fusion in vivo. DamID thus allows transient associations to be detected. Using the nuclear envelope proteins Man1 and Ima1 as DamID fusion partners, repressed genes were frequently found to localize to the periphery (Steglich et al. 2012). This verifies IF data that show that Ima1 specifically binds to heterochromatic regions. However, Ima1 is dispensable for the tethering of centromeric DNA to the SPB region of the nuclear periphery (King et al. 2008; Hiraoka et al. 2011). Rather, the Ima1-interacting loci are enriched with the RNAi components, Dcr1 and Rdp1. The Ima1 protein is mainly localized around nuclear pores (Fig. 9C) where Dcr1 is involved in an RNA degradation mechanism that contributes to keeping stress-induced genes repressed (Woolcock et al. 2012). Man1 target loci are mostly within subtelomeric regions and bound by the heterochromatin protein Swi6 (Steglich et al. 2012). Aberrant expression of Man1, in contrast to Ima1, leads to the delocalization of telomeres from the nuclear periphery (Gonzalez et al. 2012). This implies that Man1 has a function in anchoring chromosomal regions to the inner membrane.
Little is known about the changes in nuclear organization that occur during gene activation in S. pombe. Clusters of inducible genes such as the meiosis-induced genes found at the subtelomere 1L region (Mata et al. 2002) are spatially subject to repression by the HDAC enzyme Clr3 in mitotic cells—that is, their peripheral localization requires Clr3 activity (Hansen et al. 2005; Wiren et al. 2005). Other meiotic genes are posttranscriptionally repressed by an RNA elimination machinery that also directs assembly of heterochromatin islands during vegetative growth (see also Sec. 6.4). To allow for expression of the meiotic genes, some RNA degrading factors are inactivated in meiotic cells by the formation of a nuclear dot structure containing the Mei2 protein (Harigaya et al. 2006; Zofall et al. 2012). Evidence also suggests that nitrogen starvation can induce movement of stress-induced genes away from the nuclear membrane when a nitrogen-repressed gene cluster becomes activated (Alfredsson-Timmins et al. 2009). Thus, these studies show that the S. pombe nucleus is highly ordered and drastic changes in the nuclear organization seem to accommodate gene activation and repression. Chromosomes occupy distinct territories and are anchored via specific interactions with inner membrane proteins, nuclear pores, and the SPB. Chromatin-modifying activities seem to play key roles in the dynamics of the higher-order organization of the S. pombe nucleus.
9. GENOMICS AND EPIGENOMICS IN S. POMBE
Historically, the study of chromatin and epigenetics has used techniques such as chromatin immunoprecipitation (ChIP) and IF that have focused necessarily either on particular loci or regions of the genome (ChIP) or provided low-resolution results (IF). Because S. pombe is such a tractable model system to study, great progress has nonetheless been made in characterizing regions of heterochromatin in this way. The entire DNA sequence, in itself, has been useful in studying the evolution of the underlying DNA sequences surrounding heterochromatic regions and has provided a foundation for the extensive epigenetic studies and facilitated the accurate mapping of histone modifications and proteins that associate with particular genomic regions. Recent epigenomic mapping and characterization studies have significantly accelerated progress by coupling our knowledge from the complete sequencing of S. pombe with the advent of high-throughput technology.
The DNA sequence has revealed a compact genome, 13.8 Mb in size, with only 4824 protein-coding genes (Wood et al. 2002). The small genome size of S. pombe makes it a convenient and inexpensive model organism for genomics and epigenomics (see Fig. 19 of Allis et al. 2014 for a comparison). The first epigenome maps produced in S. pombe were generated using spotted microarrays. Technology developments, including high-resolution tiling microarrays and parallel sequencing, have allowed even more comprehensive mapping of large and small RNA and nucleosome positions. Genome-wide nucleosome mapping in S. pombe revealed a surprisingly short linker length and positioning mechanisms that are distinct from those of S. cerevisiae (Lantermann et al. 2010). Examples of enzymatic functions subjected to epigenomic analysis in S. pombe have included HDAC, KAT and SNF2-remodeling enzymes, and the RNAi machinery (Wiren et al. 2005; Durand-Dubief et al. 2007; Nicolas et al. 2007; Walfridsson et al. 2007; Johnsson et al. 2009; Garcia et al. 2010; Halic and Moazed 2010; Hogan et al. 2010; Woolcock et al. 2011).
By comparing the S. pombe genome with the related fission yeasts, Schizosaccharomyces octosporus and Schizosaccharomyces japonicus, interesting insights into the evolution of centromeres, transposable elements, and RNAi were gained (Rhind et al. 2011). Centromeric DNA regions of all three fission yeasts consist of symmetric arrangements of repeat sequences and transposons, or transposon-like sequences. In S. japonicus, small RNAs are produced from transposon sequences, whereas in S. pombe, the majority of the small RNAs are produced from centromeric repeats indicating that the RNAi machinery has evolved from an ancestral role in transposon silencing to its role in S. pombe heterochromatin assembly. Genome-wide approaches have also recently been developed to study nuclear organization by combining chromosome conformation capture with parallel sequencing (Tanizawa et al. 2010), and Dam-ID with inner membrane proteins (Steglich et al. 2012) and nuclear pore proteins (Woolcock et al. 2012). These approaches are clearly paving the way for a holistic molecular understanding of epigenetics and cell biology in S. pombe.
10. CONCLUDING REMARKS
The fungus S. pombe has emerged as a very powerful system to discover and elucidate epigenetic phenomena. S. pombe epigenetics research began with the studies of mating type and centromeric silencing (i.e., the PEV phenomena). This led the community of epigenetic researchers to focus on heterochromatin assembly and centromere identity. Currently, new epigenomics tools are allowing us to explore other aspects of epigenetics in S. pombe including nucleosome remodeling and nuclear organization. Exciting discoveries related to epigenetic regulation will continue to be made in this excellent model organism as we strive to understand in detail the underlying mechanisms.
ACKNOWLEDGMENTS
We thank Monika Lachner for excellent help with figures and commenting on the manuscript. R.C.A thanks Alison L. Pidoux, Lakxmi Subramanian, and Sharon A. White for comments. Research in the Allshire laboratory is supported by the Wellcome Trust [095021], [065061], and EC FP7 “EpiGeneSys” Network of Excellence [HEALTH-F4-2010-257082], along with core funding from the Wellcome Trust to the Wellcome Trust Centre for Cell Biology [092076]. R.C.A. is a Wellcome Trust Principal Fellow. K.E. thanks Babett Steglich for proofreading and Figure 9C, and Jenna Persson for commenting on the manuscript. Research in the Ekwall laboratory is supported by grants from the Swedish Cancer Society and the Swedish Research Council (VR).
REFERENCES
*Reference is also in this collection.
- Ahmed S, Dul B, Qiu X, Walworth NC. 2007. Msc1 acts through histone H2A.Z to promote chromosome stability in Schizosaccharomyces pombe. Genetics 177: 1487–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfredsson-Timmins J, Henningson F, Bjerling P. 2007. The Clr4 methyltransferase determines the subnuclear localization of the mating-type region in fission yeast. J Cell Sci 120: 1935–1943. [DOI] [PubMed] [Google Scholar]
- Alfredsson-Timmins J, Kristell C, Henningson F, Lyckman S, Bjerling P. 2009. Reorganization of chromatin is an early response to nitrogen starvation in Schizosaccharomyces pombe. Chromosoma 118: 99–112. [DOI] [PubMed] [Google Scholar]
- *.Allis CD, Jenuwein T, Reinberg D. 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739. [DOI] [Google Scholar]
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G. 1994. Position effect variegation at fission yeast centromeres. Cell 76: 157–169. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9: 218–233. [DOI] [PubMed] [Google Scholar]
- *.Aramayo R, Selker EU. 2013. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb Perspect Biol 5: a017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Goldshmidt I, Lyakhovetsky R, Cohen A. 2000. A fission yeast repression element cooperates with centromere-like sequences and defines a mat silent domain boundary. Genetics 156: 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Noma K, Isaac S, Kahan T, Grewal SI, Cohen A. 2003. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol Cell Biol 23: 4356–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis JM, Bernard P, Antonelli R, Allshire RC, Forsburg SL. 2003. Hsk1-Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat Cell Biol 5: 1111–1116. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. [DOI] [PubMed] [Google Scholar]
- Baum M, Ngan VK, Clarke L. 1994. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol Biol Cell 5: 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, Portoso M, Kagansky A, Kos-Braun IC, Urano T, Ekwall K, Alves F, Rappsilber J, Allshire RC. 2008. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science 322: 602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, et al. 2010. Stc1: A critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140: 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Becker PB, Workman JL. 2013. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol 5: a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542. [DOI] [PubMed] [Google Scholar]
- Bernard P, Drogat J, Dheur S, Genier S, Javerzat JP. 2010. Splicing factor Spf30 assists exosome-mediated gene silencing in fission yeast. Mol Cell Biol 30: 1145–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Garcia JF, Rowley M, Rougemaille M, Shankar S, Madhani HD. 2011. The Cul4-Ddb1(Cdt)2 ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 144: 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan L, Durand-Dubief M, Roguev A, Sakalar C, Wilhelm B, Stralfors A, Shevchenko A, Aasland R, Ekwall K, Francis Stewart A. 2009. The Schizosaccharomyces pombe JmjC-protein, Msc1, prevents H2A.Z localization in centromeric and subtelomeric chromatin domains. PLoS Genet 5: e1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. 2006. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125: 873–886. [DOI] [PubMed] [Google Scholar]
- Buttrick GJ, Millar JB. 2011. Ringing the changes: Emerging roles for DASH at the kinetochore-microtubule interface. Chromosome Res 19: 393–407. [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819. [DOI] [PubMed] [Google Scholar]
- Cam HP, Noma K, Ebina H, Levin HL, Grewal SI. 2008. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 451: 431–436. [DOI] [PubMed] [Google Scholar]
- Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B. 2011. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell 41: 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsten JO, Szilagyi Z, Liu B, Lopez MD, Szaszi E, Djupedal I, Nystrom T, Ekwall K, Gustafsson CM, Zhu X. 2012. Mediator promotes CENP-A incorporation at fission yeast centromeres. Mol Cell Biol 32: 4035–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr AM, Dorrington SM, Hindley J, Phear GA, Aves SJ, Nurse P. 1994. Analysis of a histone H2A variant from fission yeast: Evidence for a role in chromosome stability. Mol Gen Genet 245: 628–635. [DOI] [PubMed] [Google Scholar]
- Castillo AG, Pidoux AL, Catania S, Durand-Dubief M, Choi ES, Hamilton G, Ekwall K, Allshire RC. 2013. Telomeric repeats facilitate CENP-A(Cnp1) incorporation via telomere binding proteins. PLoS One 8: e69673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. 2008. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737. [DOI] [PubMed] [Google Scholar]
- Choi ES, Strålfors A, Castillo AG, Durand-Dubief M, Ekwall K, Allshire RC. 2011. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem 286: 23600–23607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi ES, Strålfors A, Catania S, Castillo AG, Svensson JP, Pidoux AL, Ekwall K, Allshire RC. 2012. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-ACnp1 in fission yeast. PLoS Genet 8: e1002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Baum MP. 1990. Functional analysis of a centromere from fission yeast: A role for centromere-specific repeated DNA sequences. Mol Cell Biol 10: 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Wu P, Parthun MR, Wu JQ. 2011. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol 195: 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. 2000. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol 10: 517–525. [DOI] [PubMed] [Google Scholar]
- *.Dekker J, Misteli T. 2014. Long-range chromatin interactions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheur S, Saupe SJ, Genier S, Vazquez S, Javerzat JP. 2011. Role for cohesin in the formation of a heterochromatic domain at fission yeast subtelomeres. Mol Cell Biol 31: 1088–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Ekwall K. 2008. Molecular biology. The paradox of silent heterochromatin. Science 320: 624–625. [DOI] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. 2005. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 19: 2301–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Kos-Braun IC, Mosher RA, Soderholm N, Simmer F, Hardcastle TJ, Fender A, Heidrich N, Kagansky A, Bayne E, et al. 2009. Analysis of small RNA in fission yeast; centromeric siRNAs are potentially generated through a structured RNA. EMBO J 28: 3832–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Pidoux AL, Monet M, Bonilla C, Richardson W, Hamilton GL, Ekwall K, McLaughlin PJ, Allshire RC. 2007. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol Cell 28: 1029–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M, Sinha I, Fagerstrom-Billai F, Bonilla C, Wright A, Grunstein M, Ekwall K. 2007. Specific functions for the fission yeast Sirtuins Hst2 and Hst4 in gene regulation and retrotransposon silencing. EMBO J 26: 2477–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M, Persson J, Norman U, Hartsuiker E, Ekwall K. 2010. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J 29: 2126–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M, Svensson JP, Persson J, Ekwall K. 2011. Topoisomerases, chromatin and transcription termination. Transcription 2: 66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R. 2004. The molecular biology of Schizosaccharomyces pombe: Genetics, genomics and beyond. Springer, Berlin. [Google Scholar]
- Egel R, Willer M, Nielsen O. 1989. Unblocking of meiotic crossing-over between the silent mating-type cassettes of fission yeast, conditioned by the recessive, pleiotropic mutant rik1. Curr Genet 15: 407–410. [Google Scholar]
- Ekwall K, Ruusala T. 1994. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics 136: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R. 1995. The chromodomain protein Swi6: A key component at fission yeast centromeres. Science 269: 1429–1431. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R. 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 109: 2637–2648. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Cranston G, Allshire RC. 1999. Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics 153: 1153–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Elgin SCR, Reuter G. 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 5: a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T. 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34: 2887–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC. 2008. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. 2007. Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β, and M18BP1. Dev Cell 12: 17–30. [DOI] [PubMed] [Google Scholar]
- Garcia JF, Dumesic PA, Hartley PD, El-Samad H, Madhani HD. 2010. Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev 24: 1758–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace EL, Halic M, Moazed D. 2010. The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol Cell 39: 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Y, Saito A, Sazer S. 2012. Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina. Nucleus 3: 60–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. 1999. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev 13: 1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Klar AJ. 1997. A recombinationally repressed region between mat2 and mat3 loci shares homology with centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146: 1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Bonaduce MJ, Klar AJ. 1998. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150: 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Grunstein M, Gasser SM. 2013. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb Perspect Biol 5: a017491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ. 2008. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132: 983–995. [DOI] [PubMed] [Google Scholar]
- Gullerova M, Moazed D, Proudfoot NJ. 2011. Autoregulation of convergent RNAi genes in fission yeast. Genes Dev 25: 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D. 2010. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140: 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237. [DOI] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI. 2003. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci 100: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KR, Burns G, Mata J, Volpe TA, Martienssen RA, Bahler J, Thon G. 2005. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol Cell Biol 25: 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. 2006. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442: 45–50. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729. [DOI] [PubMed] [Google Scholar]
- *.Henikoff S, Smith MM. 2014. Histone variants and epigenetics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig BP, Bendrin K, Zhou Y, Fischer T. 2012. Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription. EMBO Rep 13: 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Maekawa H, Asakawa H, Chikashige Y, Kojidani T, Osakada H, Matsuda A, Haraguchi T. 2011. Inner nuclear membrane protein Ima1 is dispensable for intranuclear positioning of centromeres. Genes Cells 16: 1000–1011. [DOI] [PubMed] [Google Scholar]
- Hogan CJ, Aligianni S, Durand-Dubief M, Persson J, Will WR, Webster J, Wheeler L, Mathews CK, Elderkin S, Oxley D, et al. 2010. Fission yeast Iec1-Ino80-mediated nucleosome eviction regulates nucleotide and phosphate metabolism. Mol Cell Biol 30: 657–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EJ, Villen J, Gerace EL, Gygi S, Moazed D. 2005. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol 2: 106–111. [DOI] [PubMed] [Google Scholar]
- Horn PJ, Bastie JN, Peterson CL. 2005. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev 19: 1705–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Wang Y, Kallgren SP, Thompson J, Yates JR III, Jia S. 2010. Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast. J Biol Chem 285: 1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac S, Walfridsson J, Zohar T, Lazar D, Kahan T, Ekwall K, Cohen A. 2007. Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics 175: 1549–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K. 2008. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321: 1088–1091. [DOI] [PubMed] [Google Scholar]
- Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K. 2010. Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell 21: 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Hebden AK, Nakamura TM, Miller KM, Cooper JP. 2010. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature 467: 223–227. [DOI] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI. 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976. [DOI] [PubMed] [Google Scholar]
- Johnsson A, Durand-Dubief M, Xue-Franzen Y, Ronnerblad M, Ekwall K, Wright A. 2009. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep 10: 1009–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC. 2009. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324: 1716–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F. 2005. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15: 1808–1819. [DOI] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. 2005. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309: 467–469. [DOI] [PubMed] [Google Scholar]
- Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327: 172–177. [DOI] [PubMed] [Google Scholar]
- Keller C, Adaixo R, Stunnenberg R, Woolcock KJ, Hiller S, Buhler M. 2012. HP1Swi6 mediates the recognition and destruction of heterochromatic RNA transcripts. Mol Cell 47: 215–227. [DOI] [PubMed] [Google Scholar]
- Kiely CM, Marguerat S, Garcia JF, Madhani HD, Bahler J, Winston F. 2011. Spt6 is required for heterochromatic silencing in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol 31: 4193–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Choi ES, Shin JA, Jang YK, Park SD. 2004. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. J Biol Chem 279: 42850–42859. [DOI] [PubMed] [Google Scholar]
- Kim HS, Vanoosthuyse V, Fillingham J, Roguev A, Watt S, Kislinger T, Treyer A, Carpenter LR, Bennett CS, Emili A, et al. 2009. An acetylated form of histone H2A.Z regulates chromosome architecture in Schizosaccharomyces pombe. Nat Struct Mol Biol 16: 1286–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Drivas TG, Blobel G. 2008. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell 134: 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. 2008. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol 18: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniola B, O”Toole E, McIntosh JR, Mellone B, Allshire R, Mengarelli S, Hultenby K, Ekwall K. 2001. The domain structure of centromeres is conserved from fission yeast to humans. Mol Biol Cell 12: 2767–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D, Endesfelder U, Berger H, Subramanian L, Dunne PD, McColl J, Klenerman D, Carr AM, Sauer M, Allshire RC, et al. 2012. Quantitative single-molecule microscopy reveals that CENP-ACnp1 deposition occurs during G2 in fission yeast. Open Biol 2: 120078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. 2010. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 17: 251–257. [DOI] [PubMed] [Google Scholar]
- Lejeune E, Bortfeld M, White SA, Pidoux AL, Ekwall K, Allshire RC, Ladurner AG. 2007. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr Biol 17: 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Li E, Zhang Y. 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 6: a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Martienssen R, Cande WZ. 2011. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature 475: 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentz A, Ostermann K, Fleck O, Schmidt H. 1994. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene 143: 139–143. [DOI] [PubMed] [Google Scholar]
- *.Martienssen R, Moazed D. 2014. RNAi and heterochromatin assembly. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Nakamura T, Hayashi T, Yanagida M. 2006. Histone H2B mutations in inner region affect ubiquitination, centromere function, silencing and chromosome segregation. EMBO J 25: 2420–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bahler J. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147. [DOI] [PubMed] [Google Scholar]
- Mellone BG, Zhang W, Karpen GH. 2009. Frodos found: Behold the CENP-A “ring” bearers. Cell 137: 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Kleckner N. 2008. Examination of interchromosomal interactions in vegetatively growing diploid Schizosaccharomyces pombe cells by Cre/loxP site-specific recombination. Genetics 178: 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113. [DOI] [PubMed] [Google Scholar]
- Ngan VK, Clarke L. 1997. The centromere enhancer mediates centromere activation in Schizosaccharomyces pombe. Mol Cell Biol 17: 3305–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI. 2007. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol 14: 372–380. [DOI] [PubMed] [Google Scholar]
- Nimmo ER, Cranston G, Allshire RC. 1994. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J 13: 3801–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC. 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392: 825–828. [DOI] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293: 1150–1155. [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI. 2004. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36: 1174–1180. [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4: 89–93. [DOI] [PubMed] [Google Scholar]
- Partridge JF, DeBeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ. 2007. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell 26: 593–602. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC. 2002. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol 12: 1652–1660. [DOI] [PubMed] [Google Scholar]
- Perpelescu M, Fukagawa T. 2011. The ABCs of CENPs. Chromosoma 120: 425–446. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC. 2004. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res 12: 521–534. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC. 2005. The role of heterochromatin in centromere function. Philos Trans R Soc Lond B Biol Sci 360: 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Richardson W, Allshire RC. 2003. Sim4: A novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J Cell Biol 161: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. 2009. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell 33: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pikaard CS, Mittelsten Sheid O. 2014. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointner J, Persson J, Prasad P, Norman-Axelsson U, Stralfors A, Khorosjutina O, Krietenstein N, Svensson JP, Ekwall K, Korber P. 2012. CHD1 remodelers regulate nucleosome spacing in vitro and align nucleosomal arrays over gene coding regions in S. pombe. EMBO J 31: 4388–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzi C, Clarke L. 1991. The chromatin structure of centromeres from fission yeast: Differentiation of the central core that correlates with function. J Cell Biol 112: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BD, Wang Y, Niu L, Higuchi EC, Marguerat SB, Bahler J, Smith GR, Jia S. 2011. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev 25: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI. 2011. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol 18: 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J. 2004. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J 23: 3825–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Masuda F, Takayama Y, Takahashi K, Saitoh S. 2012. Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr Biol 22: 658–667. [DOI] [PubMed] [Google Scholar]
- Scott KC, Merrett SL, Willard HF. 2006. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol 16: 119–129. [DOI] [PubMed] [Google Scholar]
- Scott KC, White CV, Willard HF. 2007. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS One 2: e1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Seto E, Yoshida M. 2014. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6: a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI. 2003. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol 13: 1240–1246. [DOI] [PubMed] [Google Scholar]
- Shim YS, Choi Y, Kang K, Cho K, Oh S, Lee J, Grewal SI, Lee D. 2012. Hrp3 controls nucleosome positioning to suppress non-coding transcription in eu- and heterochromatin. EMBO J 31: 4375–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Dohke K, Sadaie M, Shinmyozu K, Nakayama J, Urano T, Murakami Y. 2009. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev 23: 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich B, Filion G, van Steensel B, Ekwall K. 2012. The inner nuclear membrane proteins Man1 and Ima1 link to two different types of chromatin at the nuclear periphery in S. pombe. Nucleus 3: 77–87. [DOI] [PubMed] [Google Scholar]
- Steiner NC, Clarke L. 1994. A novel epigenetic effect can alter centromere function in fission yeast. Cell 79: 865–874. [DOI] [PubMed] [Google Scholar]
- Stralfors A, Walfridsson J, Bhuiyan H, Ekwall K. 2011. The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet 7: e1001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. 2005. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci 102: 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504. [DOI] [PubMed] [Google Scholar]
- Tada K, Susumu H, Sakuno T, Watanabe Y. 2011. Condensin association with histone H2A shapes mitotic chromosomes. Nature 474: 477–483. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215–2219. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. 1992. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 3: 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K. 2010. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res 38: 8164–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Klar AJ. 1992. The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics 131: 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Cohen A, Klar AJ. 1994. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics 138: 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Verhein-Hansen J. 2000. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155: 551–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. 2007. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J 26: 4670–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC. 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res 11: 137–146. [DOI] [PubMed] [Google Scholar]
- Walfridsson J, Bjerling P, Thalen M, Yoo EJ, Park SD, Ekwall K. 2005. The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucleic Acids Res 33: 2868–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. 2007. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J 26: 2868–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CR, Bartlett R, Nurse P, Bird AP. 1995. The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res 23: 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. 2009. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell 33: 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiren M, Silverstein RA, Sinha I, Walfridsson J, Lee HM, Laurenson P, Pillus L, Robyr D, Grunstein M, Ekwall K. 2005. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J 24: 2906–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Woolcock KJ, Gaidatzis D, Punga T, Buhler M. 2011. Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nat Struct Mol Biol 18: 94–99. [DOI] [PubMed] [Google Scholar]
- Woolcock KJ, Stunnenberg R, Gaidatzis D, Hotz HR, Emmerth S, Barraud P, Buhler M. 2012. RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes Dev 26: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xhemalce B, Kouzarides T. 2010. A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev 24: 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. 2005. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell 20: 173–185. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marin L, Chang AY, Goto D, et al. 2011. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature 479: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI. 2008. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388. [DOI] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. 2006. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell 22: 681–692. [DOI] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI. 2012. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335: 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]