Abstract
The crystal structures of channels and transporters reveal the chemical nature of ion-binding sites and, thereby, constrain mechanistic models for their transport processes. However, these structures, in and of themselves, do not reveal equilibrium selectivity or transport preferences, which can be discerned only from various functional assays. In this Review, I explore the relationship between cation transport protein structures, equilibrium binding measurements, and ion transport selectivity. The primary focus is on K+-selective channels and nonselective cation channels because they have been extensively studied both functionally and structurally, but the principles discussed are relevant to other transport proteins and molecules.
Ion transport proteins, including channels, exchangers, and pumps, facilitate the selective transfer of ions across biological membranes (Hille, 2001). Ion channels transport their substrates by electrodiffusion at rates that approach 108 s−1, whereas exchangers and pumps are orders of magnitude slower because of the obligatory coupling of the transport event to a conformational change. Crystal structures of transport proteins reveal the arrangement of atoms that directly interact with transported ions and are used to develop detailed models for the structural basis for selectivity. The mechanisms by which these proteins achieve transport selectivity are defined by their three-dimensional structure and can depend on both kinetic and thermodynamic factors. But inferring function from structures can be tricky, in part because transport molecules often adopt slightly different conformations depending on the nature of the bound ligand. For example, the small molecules valinomycin and nonactin readily yield crystal structures bound to Na+ or K+ ions with coordination chemistry typical for the bound ions (Kilbourn et al., 1967; Dobler and Phizackerley, 1974; Neupertlaves and Dobler, 1975; Steinrauf et al., 1982). By simply comparing these structures, one might conclude that these small molecules are nonselective. However, transport assays, equilibrium ion-binding assays, and computational studies reveal a substantial K+ selectivity for these small molecules highlighting the importance of integrating information from a three-dimensional structure with experimentally determined ion selectivity measurements (Moore and Pressman, 1964; Graven et al., 1966; Izatt et al., 1985; Eisenman and Alvarez, 1992).
Caution must also be exercised when comparing equilibrium properties of transport proteins that move ions at vastly different rates, as equilibrium properties do not always predict transport selectivity. In the case of exchangers and pumps, hereafter referred to as “transporters,” the active sites tend to be buried in the middle of the protein and accessible to only one side of the membrane at a time. The obligatory conformational change required to transport the ion (and any other substrate) limits the transport rate (Dutzler et al., 2002; Yamashita et al., 2005; Boudker et al., 2007; Weyand et al., 2008; Gao et al., 2009; Ressl et al., 2009; Liao et al., 2012; Nyblom et al., 2013; Zhou et al., 2014). Therefore, transporter selectivity could be determined by the equilibrium-binding preference for ions alone, which can be experimentally determined with a binding assay and rationalized from co-structures of the transporter with ion (Boudker et al., 2007; Picollo et al., 2009; Reyes et al., 2013).
Ion channels, on the other hand, feature an ion conduction pathway that spans the protein in at least one conformation (called the open conformation) and contains a series of selective ion-binding sites (Hille, 2001; Jiang et al., 2002; Zimmermann and Dutzler, 2011; Baconguis and Gouaux, 2012). Although ions moving by electrodiffusion through the channel encounter energy barriers, these barriers are rapidly surmounted with thermal fluctuations, and ion channels can sometimes conduct at rates that are near diffusion limited. So, unlike transporters where transport is slow relative to the time it takes to equilibrate a binding site, channels can transport ions more rapidly than the ion-binding sites equilibrate.
Ion channel selectivity in a lipid membrane is typically quantified using one of two operational definitions (Blatz and Magleby, 1984; Eisenman et al., 1986; Heginbotham and MacKinnon, 1993; Hille, 2001). One is the ratio of ion conductances for different ions, e.g., Na+ versus K+ ions. Conductance measures the rate of ionic throughput and provides information as to the largest barrier(s) the ions encounter as they pass through the channel. The second is the permeability ratio measured by the value of the reversal potential when gradients of two ions are present that compete for transit through the channel. It is important to note that conductance ratios and permeability ratios measure distinctly different channel properties that are not easily related to one another, or to experimentally determined equilibrium selectivity, which relates to the inherent preference of a channel for an ion. Importantly, several recent studies have shown that the transport selectivity of ion channels can differ from the equilibrium selectivity derived from various experimental methods (Liu and Lockless, 2013; Sauer et al., 2013). These equilibrium properties facilitate the direct correlation between structures and measures of ion selectivity, and are an additional parameter that must be considered in models to explain ion channel selectivity.
Below, I discuss functional, structural, and computational studies of K+ channels and some nonselective channels, with the goal of connecting their equilibrium ion-binding properties to their structures and selective ion conduction.
Structures of k+-selective channels
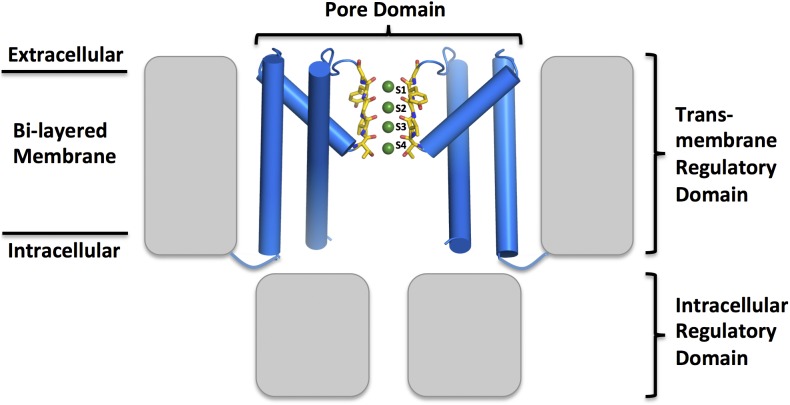
The diversity of available K+ channel crystal structures has facilitated our understanding of the molecular details of ion channel function (Doyle et al., 1998; Morais-Cabral et al., 2001; Zhou et al., 2001; Jiang et al., 2002, 2003; Kuo et al., 2003; Long et al., 2005, 2007; Nishida et al., 2007; Tao et al., 2009; Uysal et al., 2009; Clarke et al., 2010; Mari et al., 2011; Whorton and MacKinnon, 2011; Brohawn et al., 2012; Miller and Long, 2012). These channels can be roughly divided into two functional/structural units: the transmembrane pore that conducts ions, and the regulatory domains that determine if the channel is open or closed (Fig. 1) (Hille, 2001). The regulatory domains include transmembrane voltage sensors, Ca2+-binding domains, PAS (Per, ARNT, Sim) domains, Kir cytoplasmic domains, and cyclic nucleotide domains, which are tethered to the cytoplasmic portion of the transmembrane pore domain. The eight transmembrane helices of the pore domain form an inverted teepee, with each of the four subunits contributing two helices. In the open ion-conducting conformation, the intracellular region of the helices is splayed open to reveal a water-filled cavity that can accommodate a hydrated ion. Both electrophysiology and crystallography reveal a modest five- to sevenfold equilibrium selectivity for K+ over Na+ in the cavity, which may result from the different hydration radius of the two ions and the space available in the cavity (Neyton and Miller, 1988; Zhou and MacKinnon, 2004).
Figure 1.
Structure of a generic tetrameric cation channel. The pore domain (in blue) is found in all tetrameric cation channels. This region of the channel selectively transports ions across the membrane and integrates regulatory signals through transmembrane and intracellular regulatory domains that can be attached to the N or C terminus of the channel (in gray). In the K+ channel pore shown here, the selectivity filter (in yellow) is composed of discrete ion-binding sites indicated with the green spheres. Other tetrameric cation channels (Na+, Ca2+, and nonselective) have the same overall architecture but with a different selectivity filter.
The cavity ion is dehydrated as it enters the channel’s selectivity filter, the portion of the channel responsible for its high selectivity. The selectivity filter (sites S1–S4) is 12 Å long and decorated with evenly spaced oxygen atoms contributed from the backbone carbonyls and hydroxyls from threonine amino acids (Fig. 2). This arrangement of oxygen atoms is found in all K+-selective channels studied to date, suggesting a strong evolutionary constraint on this region of the channel (Morais-Cabral et al., 2001; Zhou et al., 2001; Jiang et al., 2003; Kuo et al., 2003; Long et al., 2007; Tao et al., 2009; Clarke et al., 2010; Brohawn et al., 2012; Miller and Long, 2012). The distribution of electron density in the selectivity filter differs depending on whether the structure was solved in the presence of K+ ions, Na+ ions, or a mixture of ions, indicating that the K+ and Na+ ions interact with the channel differently within this region. In both the Streptomyces lividans KcsA channel and the Methanobacterium thermoautotrophicum MthK channel, electron density corresponding to K+ ions (or water molecules) is observed at the center of the four oxygen cages of sites S1–S4; anomalous difference maps show that at high K+ concentrations, the ions are distributed near equally across the four binding sites, each in a square anti-prismatic configuration (Zhou et al., 2001; Zhou and MacKinnon, 2003; Ye et al., 2010).
Figure 2.
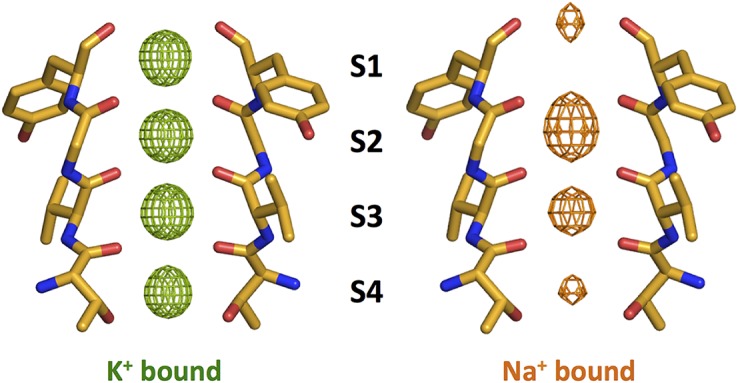
Comparison of ion distribution in MthK K+ channel. (Left) The selectivity filter of MthK with K+ ions (Protein Data Bank [PDB] accession no. 3LDC). An Fo–Fc ion omit map (4σ) shows the electron density in the filter. (Right) The selectivity filter of MthK with Na+ ions (PDB accession no. 3LDE). An Fo–Fc ion omit map (4σ) shows the electron density in the filter. Structures are from Ye et al. (2010).
When K+ ions are replaced with Na+ ions, the electron density in the selectivity filter is different. In the KcsA channel, the selectivity filter undergoes a conformational change in the presence of Na+ ions that significantly alters the positions of the carbonyl oxygen atoms, and electron density is primarily observed in sites S1 and S4 (Zhou et al., 2001; Zhou and MacKinnon, 2003). This Na+-containing filter, however, is in a constricted conformation that is unable to conduct ions. In contrast, the position of the protein atoms in the MthK channel is virtually identical in the presence of either K+ or Na+ ions (Fig. 2) (Ye et al., 2010). The electron density observed in the filter likely corresponds to a mixture of Na+ ions and water molecules, with density observed in the plane of the oxygen atoms, in the center of an oxygen cage or in between these configurations. Unlike with K+ ions, where anomalous difference maps can be used to estimate the contribution of K+ ions to the electron densities, it is not possible to experimentally determine the contribution from Na+ ions. Instead, the location of Na+ ions must be inferred from coordination preferences and other indirect methods. Because MthK conducts Na+ ions in the absence of K+, its selectivity filter in the presence of Na+ ions is likely to represent the conformation of the channel during Na+ conduction.
Although some K+ channels conduct Na+ ions at appreciable rates (>10 pS), they interact with the channel differently than K+ ions. The physical–chemical properties of channel–ion interactions have been extensively discussed elsewhere, including in a series of reviews in this journal, and will not be discussed in detail here (Gouaux and Mackinnon, 2005; Alam and Jiang, 2011; Dixit and Asthagiri, 2011; Nimigean and Allen, 2011; Roux et al., 2011; Varma et al., 2011; Horn et al., 2014). In brief, these interactions include field strength, charge–dipole interactions, and ion coordination number, which, together with forces controlling the position of the atoms directly interacting with the ions, determine the free energy landscape within the filter. The contributions from all of these properties can be captured in the equilibrium-binding preference of the ion for the channel. In the simplest scenario, where K+ or Na+ ions are transferred from a solvent to a site in the channel, the selectivity is quantified as:
where ΔΔG < 0 is K+ selective, ΔΔG > 0 is Na+ selective, and ΔΔG = 0 is nonselective.
K+ channels have an equilibrium preference for K+ ions over Na+ ions in their selectivity filter. Electrophysiology experiments using Ba2+ to block several channels demonstrated a >1,000-fold equilibrium preference for K+ ions in the extracellular “lock-in” sites with K+ affinities of ∼20 µM (Neyton and Miller, 1988; Piasta et al., 2011). In KcsA, MthK, and NaK2K channels, low concentrations of K+ outcompete high concentrations of Na+ ions for binding to the filter, based on x-ray crystallography (Morais-Cabral et al., 2001; Zhou et al., 2001; Zhou and MacKinnon, 2003; Thompson et al., 2009; Sauer et al., 2013). This qualitative result demonstrates that the selective ion-binding sites are indeed within the selectivity filter. Titrations in both solution and solid-state nuclear magnetic resonance shows a K+-dependent change in chemical shift of atoms in the selectivity filter (Bhate et al., 2010; Imai et al., 2010). Free energy calculations and atomistic molecular dynamics (MD) simulations also demonstrate an equilibrium preference for K+ ions, even when Na+ ions are not restricted to the center of observed K+ sites (Luzhkov and Aqvist, 2001; Noskov and Roux, 2006; Varma and Rempe, 2007; Egwolf and Roux, 2010; Kim and Allen, 2011). And finally, equilibrium ion binding to several K+ channels was measured using isothermal titration calorimetry (ITC) (Lockless et al., 2007; Liu et al., 2012; Liu and Lockless, 2013). The observed K+ affinities are in the range of 10–100 µM, which is consistent with electrophysiology measurements of the koff rate of 105 s−1 for the last ion leaving the filter of Shaker channel (if one assumes the diffusion-limited kon rate of 109 M−1 s−1; KD = koff /kon of ∼10 µM) (Baukrowitz and Yellen, 1996).
Several mechanistic explanations for the equilibrium selectivity have been proposed based on MD simulations and free energy calculations. On the one hand, Roux and colleagues proposed from MD simulations that K+ selectivity arises from a field strength match between the partial negative charge of the carbonyl oxygen atoms and the charge density of K+ ions, which is different from that of the smaller Na+ ions (Roux et al., 2011). This conclusion was based on both a model ion-binding site mimic of site S2 of KcsA, where coordinating dipoles are free to move in a defined area of the structure, and on simulations of the multi-ion free energy landscape of a K+ channel, where a greater selectivity for carbonyl oxygen atoms versus water molecules is observed (Noskov et al., 2004; Egwolf and Roux, 2010). Rempe and colleagues, on the other hand, argued that chemistry is important, but not the driving force in selectivity of K+ channels. Using free energy calculations with density functional theory, they found selectivity arises from an environment that imposes constraints on ion-binding sites (Varma et al., 2008, 2011). This topological control emerges from intramolecular hydrogen bonds within the ion-binding molecule to constrain the number of oxygen atoms that coordinate the ions, leading to over-coordination. Further experiments are necessary to parse the role of these two mechanistic hypotheses.
Although the primary molecular determinants of equilibrium selectivity are not yet resolved, chemists in the guest–host field and well-studied ion-binding proteins highlight an easy-to-calculate parameter that often correlates with the equilibrium preference for a cation: the size of the cage formed by the contacting oxygen atoms and the size of the dehydrated ion (Dietrich, 1985; Harding, 2002). This does not mean that the structure is rigid—as a rigid structure would not be able to enclose an ion—but that the energy minimum of the system is an oxygen-lined site that matches the size of the “site” created by the water molecules around a hydrated ion. Although this is a correlative parameter, not a mechanistic principle, a simple explanation is that the oxygen atoms in the ion-binding site mimic the hydration of water without the entropic penalty associated with ordering water molecules around an ion. Although the exact affinities vary depending on the channel and technique used to measure ion affinities, all examined K+ channels have equilibrium preference for K+ ions over Na+ ions, consistent with their preference to conduct K+ ions.
Nonselective tetrameric cation channels
Nonselective tetrameric cation channels including hyperpolarization-activated cyclic nucleotide–gated (HCN) and cyclic nucleotide–gated (CNG) channels show a low preference for K+/Na+ during ion conduction, as measured by reversal potential (Hille, 2001; Craven and Zagotta, 2006). While we await structures of bona fide members of these families, it is informative to compare the structural, transport, and equilibrium properties of the nonselective Bacillus cereus NaK and Vibrio parahaemolyticus TrkH channels with those of K+ channels. NaK is a homotetramer with an overall architecture similar to that of K+ and nonselective channels (Shi et al., 2006; Alam and Jiang, 2009a). From the channel’s cavity, ions enter oxygen-lined sites similar to sites S3 and S4 in K+ channels (Fig. 3 A). Upon leaving site S3, they encounter an expanded portion of the filter, with coordination by carbonyl oxygen atoms from site S3 and water molecules in what would be site S2 in K+ channels. An overlay of the crystal structures showed that the protein portion of the channel’s selectivity filter is the same in either Na+ or K+ ions. The ion-binding sites S3 and S4 in NaK are the same size as those found in the selectivity filter of K+ channels and K+-selective small molecules, suggesting a transport and equilibrium-binding preference for K+ ions. However, a single-channel experiment demonstrates that the channel nonselectively conducts both K+ and Na+ ions, hinting that the transport properties of a channel cannot be gleaned from the size of the oxygen-lined cavities and ions in the selectivity filter (Derebe et al., 2011a). A free energy–perturbation MD study attributes the lack of transport selectivity of WT NaK channel to differences in hydration of bound ions in the pore compared with K+ channels (Noskov and Roux, 2007). The same study also revealed a slight equilibrium preference for K+ ions at site S3 and a slight Na+ preference at site S4. A different study points out that K+ ions tend to reside in the center of the oxygen cage, whereas Na+ ions prefer the plane of carbonyl oxygen atoms, which is also seen in KcsA K+ channels (Shen and Guo, 2009; Kim and Allen, 2011). An experimental determination of NaK channel selectivity at equilibrium has not been published to compare with these predictions.
Figure 3.
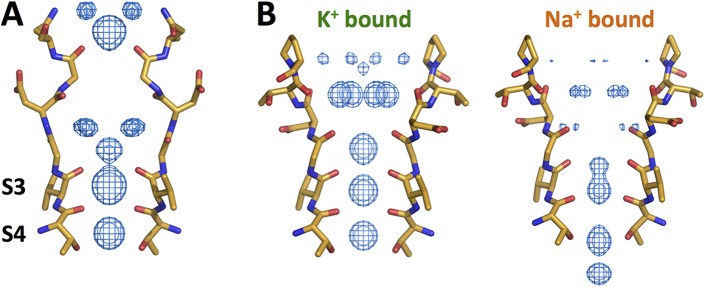
The selectivity filter of the nonselective NaK and NaK2CNG-D channels. An Fo–Fc ion omit map (5σ) shows the electron density in the filter. (A) The WT NaK channel in the presence of K+ ions (PDB accession no. 3E8H). (B) Comparison of electron density in nonselective NaK2CNG-D channel. (Left) The selectivity filter of NaK2CNG-D with bound K+ ions (PDB accession no. 3K03). (Right) The selectivity filter of NaK2CNG-D with bound Na+ ions (PDB accession no. 3K04). Structures are from Alam and Jiang (2009b) and Derebe et al. (2011a).
NaK channels can be converted into CNG-like channels that are nonselective during ion conduction and are blocked by divalent cations by replacing their filter with the eukaryotic CNG channel’s selectivity filter sequence (Derebe et al., 2011a,b). Structures of these CNG-like channels (called NaK2CNG-X, where X is D, E, or N depending on the amino acid at position 66) show that both K+ and Na+ are able to bind within the selectivity filter (Fig. 3 B). As with K+ channels, K+ ions prefer to bind in the oxygen cage, whereas Na+ ions are found in various positions throughout the filter. Potentials of mean force calculations showed that NaK2CNG-D is not selective for K+ or Na+ at equilibrium, potentially explaining the lack of transport selectivity (Wang et al., 2014). However, recent ITC and x-ray crystallography experiments revealed that the NaK2CNG-D and other mutant NaK channels have a large preference for K+ to Na+ ions at equilibrium (Liu and Lockless, 2013; Sauer et al., 2013).
Aside from the different number of oxygen-lined sites, CNG and K+ channels are remarkably similar but have different permeation preferences. Indeed, the nonselective NaK channel can be transformed into a K+-selective channel by swapping in the selectivity filter sequence of a K+ channel (TVGYGD), demonstrating that NaK protein is a reasonable system in which to study both K+ and nonselective ion conduction (Derebe et al., 2011a). This so-called NaK2K channel exhibits K+-selective ion conduction and a selectivity filter that is identical to the conductive conformations of all K+ channels determined to date. Not surprisingly, NaK2K also has an equilibrium preference for K+ ions, as demonstrated by x-ray crystallography and ITC (Liu and Lockless, 2013; Sauer et al., 2013). The primary structural difference between the nonselective CNG channel and the K+-selective NaK2K channel is that NaK2K has four sites, as is typical of K+ channels, whereas NaK2CNG-X channels have three sites.
The straightforward transformation of NaK channels to K+-like and CNG-like channels could partially explain pre-structure experiments in which deleting two amino acids from the Shaker K+ channels to mimic the sequence of CNG channels led to nonselective ion conduction and divalent block (Heginbotham et al., 1992). These CNG-converting mutations likely destroyed the mechanism whereby K+ channels translate their equilibrium preference into selective ion conduction (discussed in the next section), but, like NaK2CNG-D, select K+ ions at equilibrium. However, the number of observed sites in crystal structures does not always correlate with the permeation selectivity. For example, NaK2K(Y55F) has four binding sites but is nonselective during ion conduction (Sauer et al., 2011). A mutation in MthK to eliminate half of the oxygen atoms comprising site S4 renders the channel nonselective, but the equivalent mutation in Shaker retains the ability to selectively conduct K+ ions (Heginbotham et al., 1994; Derebe et al., 2011a). The lack of correlation between permeation selectivity and the number of binding sites in the queue suggests that additional parameters are needed to explain transport selectivity. Indeed, mutations of amino acids one shell away from the selectivity filter can alter the conduction, selectivity, and inactivation properties of channels with identical selectivity filter sequences, demonstrating that all information is not contained in the direct ion-binding ligands (Starkus et al., 1997; Cordero-Morales et al., 2006, 2011; Sauer et al., 2011; McCoy and Nimigean, 2012). In the future, it will be important to address the roles amino acids not directly contacting ions/substrates have in determining a transport protein’s selectivity.
Recent crystal structures of TrkH and KtrB suggest that the lack of correlation between selective ion transport and equilibrium binding is not an artifact of using engineered NaK mutant channels (Cao et al., 2011, 2013; Vieira-Pires et al., 2013). TrkH and its homologues were originally described as K+ transporters based on the observation that deleting them slowed or eliminated bacterial growth at low K+ concentrations (Dosch et al., 1991; Schlösser et al., 1995). Recent structural and functional data unambiguously show that TrkH is a nonselective channel (Cao et al., 2011, 2013). Unlike K+ channels and the NaK channel, the selectivity filter is formed from four nonidentical sequences that create ion-binding sites reminiscent of sites S3 and S4 in K+ channels and NaK channels; the resolutions of the available structures are too low to reliably determine the size of the binding sites, but the similar overall architecture to that of K+ channels and NaK channels suggests a K+ preference. The distantly related KtrB from Bacillus subtilis is remarkably similar in structure to TrkH, suggesting that KtrB is also a nonselective channel (Vieira-Pires et al., 2013). These data demonstrate that an equilibrium ion preference is not sufficient to explain ion transport selectivity in some channels, further calling into doubt our ability to predict transport properties from structures alone.
Role for a queue of ions
The conversion between K+-selective and nonselective channels by changing the number of binding sites suggests that the queue of ion-binding sites is critical for transport selectivity. Crystal structures of both K+-selective and nonselective tetrameric cation channels reveal queues of nearly identical ion-binding sites that prefer K+ ions to Na+ ions. The discrepancy between the selectivity of K+-selective and nonselective channels is at least partially resolved if we consider the queue of ion-binding sites in the selectivity filter instead of the properties of individual sites.
Several lines of evidence support the notion that the queue of K+-selective ion-binding sites is the relevant structural and functional unit in the K+ channel’s selectivity filter. First, the selectivity filter of KcsA undergoes ion-dependent conformational change; when K+ ions are replaced with Na+ ions, the filter rearranges to a constricted conformation that primarily retains sites S1 and S4 (Zhou et al., 2001). Second, the distributions of ions (K+, Rb+, Cs+, and Tl+) can vary from each other and can vary in different channels, suggesting a different energy profile along the whole selectivity filter, not simply each site individually (Morais-Cabral et al., 2001; Zhou and MacKinnon, 2003; Lam et al., 2014). Third, mutations that alter direct interactions with the ions can have different outcomes; a threonine-to-alanine mutation at site S4 of MthK K+ channel leads to loss of K+ selectivity, whereas the same mutation in Shaker K+ channel does not (Heginbotham et al., 1994; Derebe et al., 2011a). Fourth, chemical modification of the backbone alters the distribution of Rb+ ions throughout the filter, not just near the modified site (Valiyaveetil et al., 2006; Matulef et al., 2013). Finally, the location of the channel blocker Ba2+ depends on whether Na+ or K+ is present and can differ in different channels (Lockless et al., 2007; Guo et al., 2014; Lam et al., 2014). Collectively, these data argue that the selectivity filter functions as a unit that binds a queue of ions and water, not as a series of independent sites.
Qualitative equilibrium properties of a queue can be inferred from studies of small, single-site K+-selective molecules. Structures of K+-selective cryptands, crown ethers, and antibiotics reveal that (a) ion-binding sites are similar in size to the ion’s hydration radius, and (b) they can deform to bind less preferred ion. These molecules bind K+ ions in a cage of oxygen atoms ∼2.8 Å in diameter, but can deform to bind to the smaller Na+ ion with its optimal 2.4-Å diameter (Fig. 4) (Kilbourn et al., 1967; Dobler and Phizackerley, 1974; Neupertlaves and Dobler, 1975; Kauffmann et al., 1976; Steinrauf et al., 1982; Izatt et al., 1985; Cram and Ho, 1986; Chekhlov, 2005a,c). Because many molecules change their shape depending on the bound ligand, it’s difficult to predict which ion would be optimally bound to a particular ion-binding site without knowing the equilibrium preference in advance. However, one might expect that if an ion-binding molecule does not change its shape in the presence of different ions, the size of the ion-binding site can be readily calculated from either structure. Additionally, if a structure is solved in different mixtures of ions, the most preferred ion would outcompete the less preferred ion to establish the size of the ion-binding molecule.
Figure 4.
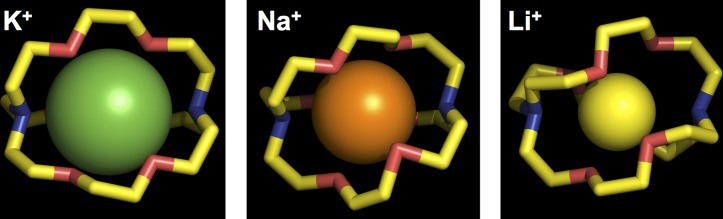
Crystal structures of K+-selective [2,2,2]cryptand. (Left) K+ ion is green with an average O–K+ distance of 2.8 Å. (Middle) Na+ ion is orange with an average O–Na+ distance of 2.6 Å. (Right) Li+ ion is yellow with an average O–Na+ distance of 2.4 Å. Structures are from Chekhlov (2003, 2005a,b).
In the case of the K+-selective [2.2.2]cryptand, the distance between the Na+ ion and oxygen atoms averages 2.6 Å, which one would infer is not optimally bound (Chekhlov, 2005b) (Fig. 4). The non-optimal size of the ion-binding cavity may explain the lower affinity of the molecule for Na+ ions (Kauffmann et al., 1976). The inability of this [2.2.2]cryptand to deform to accommodate the smaller ion undoubtedly contributes to its high K+ selectivity. To adapt to the smaller Na+ ion, the oxygen atoms will either move in to reduce the size of the cage (causing strain), or the Na+ ion will need to bind in a different location than the center of the cage. The smaller Na+-selective [2.2.1]cryptand, with its slightly smaller cavity, prefers Na+ to K+ but is also able to bind both ions (Kauffmann et al., 1976; Cram and Ho, 1986). Thus, selectivity is the potential to bind an ion at the energy minimum, whose effects can be propagated to other parts of the structure or solvent. Either way, this disrupts the binding potential for a K+ ion that prefers to bind in the center of the cage with its own optimal coordination distance.
The selectivity filter is the equivalent of an enzyme-active site, so it is critical to consider its behavior as a connected unit in much the same way that an enzyme-active site is more than the sum of its parts. In both K+-selective and nonselective mutant NaK channels, Na+ and K+ ions bind to different sites across their selectivity filters (Derebe et al., 2011a,b). The occupancy of the sites is two to three K+ ions based on flux ratio measurements in several channels and in KcsA by comparing the electron density in K+ to the electron-dense thallium, whose occupancy is more easily obtained (Hodgkin and Keynes, 1955; Begenisich and De Weer, 1980; Spalding et al., 1981; Vestergaard-Bogind et al., 1985; Zhou and MacKinnon, 2003). Even with these constraints, there are a large number of possible models that could explain the selectivity and ion conduction of the channels. However, structures aid in eliminating possible models by highlighting a few reasonable assumptions. First, K+ ions bind in the center of the oxygen cage, in sites whose size is ideal for K+ ions. Second, Na+ ions can bind in the filter but do not necessarily occupy the same location as K+ ions (Zhou et al., 2001; Ye et al., 2010). This is likely because the selectivity filter does not provide optimal coordination along the entire length. Third, in mixtures of K+ and Na+, the electron density corresponding to K+ ions tends to be in the center of the oxygen cage (Zhou and MacKinnon, 2003; Ye et al., 2010; Sauer et al., 2013). This is consistent with the ITC observation that K+ has a higher affinity for the channels than Na+ and would indicate that the channel is “built” to bind queues of K+ ions over Na+ ions (Lockless et al., 2007; Liu et al., 2012; Liu and Lockless, 2013). Because K+ ions bind with higher affinity than Na+ ions, it is likely that K+ sets the “register” of the queue such that K+ ions will reside in the center of the oxygen cages at optimal coordination distances, whereas Na+ ions will be relegated to suboptimal positions.
In multi-ion queues, ions bound to selective sites influence one another’s conduction properties, which can lead to the well-known anomalous mole fraction effect (Hille and Schwarz, 1978; Almers and McCleskey, 1984; Hess and Tsien, 1984; Korn and Ikeda, 1995). In a K+ channel, the premise is that a K+ ion binds with high affinity when there is only one ion in the filter. The binding of a second ion, either Na+ or K+, will weaken the affinity of the queue of ions. If a Na+ ion is bound, the mixed ion queue encounters a barrier for the Na+ ion to move further into the filter, such that the queue is more likely to reset back to where it came from than allow the Na+ ion to transit further through the filter. The origin of the barrier is not clear, but several nonexclusive models have been proposed. A barrier could result from K+ ions residing longer in a site than Na+ ions, creating a physical barrier whereby the Na+ ion is unable to advance to an already occupied site (Hille and Schwarz, 1978; Korn and Ikeda, 1995). Intriguingly, a similar principle has been proposed to explain the nuclear pore complex’s selectivity for nuclear transport factor proteins (Zilman, 2009; Zilman et al., 2010). Or, the carbonyl oxygen atoms themselves could present a selective barrier to Na+ ion movement relative to K+ ions (Thompson et al., 2009). Alternatively, the location of the K+ ions in the center of the oxygen cage could force a Na+ ion to bind in a suboptimal position in its site, creating an environment where the ion is more likely to dissociate back than advance to the next site.
An extension of this idea is that destroying the multi-ion mechanism could lead to nonselective ion conduction. The ion selectivity and conduction properties of single- and multi-ion queues were widely discussed, long before structures were available (Bezanilla and Armstrong, 1972; Hille and Schwarz, 1978). In single-ion queues, the largest barrier encountered by each ion determines ion selectivity measured by relative conductances, whereas the rate of ion binding determines selectivity when measured by reversal potential. In essence, the ions move independently of one another, and their properties do not necessarily correlate with the equilibrium preferences of the channel. A discordance between the equilibrium and ion conduction selectivity was recently observed with mutant NaK channels that nonselectively conduct K+ or Na+ ions, yet retain their overall equilibrium selectivity for K+ ions (Liu and Lockless, 2013; Sauer et al., 2013). This suggests that these nonselective channels have a broken multi-ion mechanism, which might also explain how mutations in the KCNJ K+ channel and the GIRK2 channel and mutations to convert Shaker into a CNG-like channel resulted in nonselective conduction (Heginbotham et al., 1992; Slesinger et al., 1996; Choi et al., 2011). In each case, the equilibrium selectivity is likely preserved, but the transport selectivity is not. Perhaps the multi-ion mechanism is used to ensure equilibrium-based transport, as it is difficult to maintain selective barriers for similarly sized ions like Na+ and K+. An equilibrium-based transport mechanism also ensures that the desired ion is transported even if the local concentration of either ion changes significantly.
Multi-ion queues can also facilitate high ion conduction rates. Both K+ and nonselective channels are able to transport ions at rates approaching the diffusion limit (Hille, 2001). In other words, an ion is bound to the channel for ∼10 ns. This means that the channel must not bind the ions too tightly, as transport would be limited by the rate at which they can leave the channel. This is a particular problem for K+ channels, as the ion must reside in the channel long enough to discriminate between the similarly sized K+ and Na+ ions. The equilibrium ion-binding measurements in both K+ and nonselective channels reveal a K+ binding affinity in the mid-micromolar range, suggesting a koff rate of 105 s−1, which is too slow (Neyton and Miller, 1988; Baukrowitz and Yellen, 1996; Piasta et al., 2011; Liu et al., 2012; Liu and Lockless, 2013). Therefore, K+ ions must bind with lower affinity during ion conduction. One possible resolution to this conundrum is if the binding of multiple ions lowers the affinity of all ions in the queue, then selectivity is retained even at low affinities. The low millimolar-range affinities are outside of the experimental range for most equilibrium techniques but would be necessary to account for the high ion conduction rates.
A different problem exists with NaK2CNG-D channels, which are selective for K+ at equilibrium but nonselective during ion conduction. If K+ ions bind in the 10-µM range but Na+ ions bind in the 10-mM range, why do both ions go through the channel equally well in bi-ionic conditions (Derebe et al., 2011a)? One might expect that the K+ ion would block ion conduction because its high affinity would result in long residence times in the channel, unless its affinity is weakened from multiple ions interacting in the filter during ion conduction. This would then suggest that NaK2CNG-D is behaving similarly to K+ channels, but without selective ion conduction. What then are the differences between NaK2CNG-D and K+ channels that lead to their divergent ion conduction selectivities?
Conclusion
The equilibrium ion-binding properties of ion channels and transporters can be difficult to discern from crystal structures alone, as proteins often adopt different lowest energy states depending on the ions bound. In cases where transport is slow, their inherent ion-binding preferences can be used to infer their transport preferences. However, in cases where transport is fast, the transport selectivity can hide their equilibrium preferences by accentuating the kinetics of ions hopping through a channel over its inherent ion-binding preferences. Thus, depending on the arrangement of ion-binding sites in a channel’s selectivity filter, one can achieve either selective or nonselective ion transport.
The equilibrium K+ selectivity of some nonselective channels suggests a potential mechanism whereby they could evolve into a fast K+-selective channel. K+ channels and nonselective channels like CNG and HCN are related to one another in both sequence and structure, suggesting an evolutionary link between them. Swap experiments show that only a few mutations separate a nonselective channel from a K+-selective channel. One might imagine an evolutionary path between these channels in which the equilibrium preference for a K+ ion in a nonselective channel evolves into a K+-selective channel through these few mutations to create the selective ion queue. Alternatively, a slow single-ion channel with an equilibrium and transport preference for K+ ions could be transformed into a fast multi-ion channel through mutations that create a queue of K+-selective ion-binding sites, as is seen in most K+ channels studied to date.
In the case of multi-ion selectivity filters, such as those found in K+ channels, the selectivity filter can be viewed as the active site that interacts with different queues of ions and water molecules. At least three properties emerge from multi-ion queues: (1) high conductance by reducing the affinity of multiple bound ions versus single ions; (2) high selectivity by allowing disfavored ions time to dissociate back into solution; and, consequently, (3) robust selectivity in an environment where ion concentrations can change. For transporters and carriers, the equilibrium preference and slow transport naturally create robust selectivity. In all these cases, equilibrium-based ion selectivity is achieved by slowing transport enough so that the disfavored ion is able to dissociate back into solution before transport takes place.
Acknowledgments
I thank David Dawson for stimulating conversations and critical comments on the manuscript, and members of the Lockless laboratory for helpful discussions.
The Welch Foundation (grant A-1742) and Texas A&M Startup Funds support our work on ion channel selectivity.
The author declares no competing financial interests.
Elizabeth M. Adler served as editor.
Footnotes
Abbreviations used in this paper:
- ITC
- isothermal titration calorimetry
- MD
- molecular dynamics
References
- Alam A., and Jiang Y.. 2009a. High-resolution structure of the open NaK channel. Nat. Struct. Mol. Biol. 16:30–34. 10.1038/nsmb.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam A., and Jiang Y.. 2009b. Structural analysis of ion selectivity in the NaK channel. Nat. Struct. Mol. Biol. 16:35–41. 10.1038/nsmb.1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam A., and Jiang Y.. 2011. Structural studies of ion selectivity in tetrameric cation channels. J. Gen. Physiol. 137:397–403. 10.1085/jgp.201010546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almers W., and McCleskey E.W.. 1984. Non-selective conductance in calcium channels of frog muscle: calcium selectivity in a single-file pore. J. Physiol. 353:585–608. 10.1113/jphysiol.1984.sp015352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baconguis I., and Gouaux E.. 2012. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature. 489:400–405. 10.1038/nature11375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baukrowitz T., and Yellen G.. 1996. Use-dependent blockers and exit rate of the last ion from the multi-ion pore of a K+ channel. Science. 271:653–656. 10.1126/science.271.5249.653 [DOI] [PubMed] [Google Scholar]
- Begenisich T., and De Weer P.. 1980. Potassium flux ratio in voltage-clamped squid giant axons. J. Gen. Physiol. 76:83–98. 10.1085/jgp.76.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F., and Armstrong C.M.. 1972. Negative conductance caused by entry of sodium and cesium ions into the potassium channels of squid axons. J. Gen. Physiol. 60:588–608. 10.1085/jgp.60.5.588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhate M.P., Wylie B.J., Tian L., and McDermott A.E.. 2010. Conformational dynamics in the selectivity filter of KcsA in response to potassium ion concentration. J. Mol. Biol. 401:155–166. 10.1016/j.jmb.2010.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatz A.L., and Magleby K.L.. 1984. Ion conductance and selectivity of single calcium-activated potassium channels in cultured rat muscle. J. Gen. Physiol. 84:1–23. 10.1085/jgp.84.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudker O., Ryan R.M., Yernool D., Shimamoto K., and Gouaux E.. 2007. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 445:387–393. 10.1038/nature05455 [DOI] [PubMed] [Google Scholar]
- Brohawn S.G., del Mármol J., and MacKinnon R.. 2012. Crystal structure of the human K2P TRAAK, a lipid- and mechano-sensitive K+ ion channel. Science. 335:436–441. 10.1126/science.1213808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Jin X., Huang H., Derebe M.G., Levin E.J., Kabaleeswaran V., Pan Y., Punta M., Love J., Weng J., et al. . 2011. Crystal structure of a potassium ion transporter, TrkH. Nature. 471:336–340. 10.1038/nature09731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Pan Y., Huang H., Jin X., Levin E.J., Kloss B., and Zhou M.. 2013. Gating of the TrkH ion channel by its associated RCK protein TrkA. Nature. 496:317–322. 10.1038/nature12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekhlov A.N. 2003. Crystal structure of (2.2.2-cryptand)lithium perchlorate. Russ. J. Coord. Chem. 29:828–832. 10.1023/B:RUCO.0000008393.57920.7d [DOI] [Google Scholar]
- Chekhlov A.N. 2005a. Synthesis and crystal structure of (2.2.2-cryptand)potassium bicarbonate trihydrate. Russ. J. Inorg. Chem. 50:1556–1560. [Google Scholar]
- Chekhlov A.N. 2005b. Synthesis and crystal structure of (2.2.2-cryptand)sodium nitrate. Russ. J. Inorg. Chem. 50:418–422. [Google Scholar]
- Chekhlov A.N. 2005c. Synthesis and crystal structure of aqua(18-crown-6)(triphenylphosphine oxide)potassium bromide. Russ. J. Inorg. Chem. 50:888–893. [Google Scholar]
- Choi M., Scholl U.I., Yue P., Björklund P., Zhao B., Nelson-Williams C., Ji W., Cho Y., Patel A., Men C.J., et al. . 2011. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 331:768–772. 10.1126/science.1198785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke O.B., Caputo A.T., Hill A.P., Vandenberg J.I., Smith B.J., and Gulbis J.M.. 2010. Domain reorientation and rotation of an intracellular assembly regulate conduction in Kir potassium channels. Cell. 141:1018–1029. 10.1016/j.cell.2010.05.003 [DOI] [PubMed] [Google Scholar]
- Cordero-Morales J.F., Cuello L.G., Zhao Y., Jogini V., Cortes D.M., Roux B., and Perozo E.. 2006. Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 13:311–318. 10.1038/nsmb1069 [DOI] [PubMed] [Google Scholar]
- Cordero-Morales J.F., Jogini V., Chakrapani S., and Perozo E.. 2011. A multipoint hydrogen-bond network underlying KcsA C-type inactivation. Biophys. J. 100:2387–2393. 10.1016/j.bpj.2011.01.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram D.J., and Ho S.P.. 1986. Host-guest complexation. 39. Cryptahemispherands are highly selective and strongly binding hosts for alkali metal ions. J. Am. Chem. Soc. 108:2998–3005. 10.1021/ja00271a032 [DOI] [PubMed] [Google Scholar]
- Craven K.B., and Zagotta W.N.. 2006. CNG and HCN channels: Two peas, one pod. Annu. Rev. Physiol. 68:375–401. 10.1146/annurev.physiol.68.040104.134728 [DOI] [PubMed] [Google Scholar]
- Derebe M.G., Sauer D.B., Zeng W., Alam A., Shi N., and Jiang Y.. 2011a. Tuning the ion selectivity of tetrameric cation channels by changing the number of ion binding sites. Proc. Natl. Acad. Sci. USA. 108:598–602. 10.1073/pnas.1013636108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derebe M.G., Zeng W., Li Y., Alam A., and Jiang Y.. 2011b. Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore. Proc. Natl. Acad. Sci. USA. 108:592–597. 10.1073/pnas.1013643108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich B. 1985. Coordination chemistry of alkali and alkaline-earth cations with macrocyclic ligands. J. Chem. Educ. 62:954–964. 10.1021/ed062p954 [DOI] [Google Scholar]
- Dixit P.D., and Asthagiri D.. 2011. Perspectives on: Ion selectivity: Thermodynamics of ion selectivity in the KcsA K+ channel. J. Gen. Physiol. 137:427–433. 10.1085/jgp.201010533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobler M., and Phizackerley R.P.. 1974. The crystal structure of the NaNCS complex of nonactin. Helv. Chim. Acta. 57:664–674. 10.1002/hlca.19740570319 [DOI] [PubMed] [Google Scholar]
- Dosch D.C., Helmer G.L., Sutton S.H., Salvacion F.F., and Epstein W.. 1991. Genetic analysis of potassium transport loci in Escherichia coli: Evidence for three constitutive systems mediating uptake potassium. J. Bacteriol. 173:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., and MacKinnon R.. 1998. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 280:69–77. 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Dutzler R., Campbell E.B., Cadene M., Chait B.T., and MacKinnon R.. 2002. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature. 415:287–294. 10.1038/415287a [DOI] [PubMed] [Google Scholar]
- Egwolf B., and Roux B.. 2010. Ion selectivity of the KcsA channel: A perspective from multi-ion free energy landscapes. J. Mol. Biol. 401:831–842. 10.1016/j.jmb.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman G., and Alvarez O.. 1992. Ionic selectivity of proteins: Lessons from molecular dynamics simulations of valinomycin. Biomembrane Structure and Function: The State of the Art. Gaber B.P. and Easwaran K.R.K., Adenine Press, Schenectady, NY: 321–351. [Google Scholar]
- Eisenman G., Latorre R., and Miller C.. 1986. Multi-ion conduction and selectivity in the high-conductance Ca++-activated K+ channel from skeletal muscle. Biophys. J. 50:1025–1034. 10.1016/S0006-3495(86)83546-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Lu F., Zhou L., Dang S., Sun L., Li X., Wang J., and Shi Y.. 2009. Structure and mechanism of an amino acid antiporter. Science. 324:1565–1568. 10.1126/science.1173654 [DOI] [PubMed] [Google Scholar]
- Gouaux E., and Mackinnon R.. 2005. Principles of selective ion transport in channels and pumps. Science. 310:1461–1465. 10.1126/science.1113666 [DOI] [PubMed] [Google Scholar]
- Graven S.N., Lardy H.A., and Rutter A.. 1966. Antibiotics as tools for metabolic studies. VI. Damped oscillatory swelling of mitochondria induced by nonactin, monactin, dinactin, and trinactin. Biochemistry. 5:1735–1742. 10.1021/bi00869a041 [DOI] [PubMed] [Google Scholar]
- Guo R., Zeng W., Cui H., Chen L., and Ye S.. 2014. Ionic interactions of Ba2+ blockades in the MthK K+ channel. J. Gen. Physiol. 144:193–200. 10.1085/jgp.201411192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding M.M. 2002. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr. D Biol. Crystallogr. 58:872–874. 10.1107/S0907444902003712 [DOI] [PubMed] [Google Scholar]
- Heginbotham L., and MacKinnon R.. 1993. Conduction properties of the cloned Shaker K+ channel. Biophys. J. 65:2089–2096. 10.1016/S0006-3495(93)81244-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heginbotham L., Abramson T., and MacKinnon R.. 1992. A functional connection between the pores of distantly related ion channels as revealed by mutant K+ channels. Science. 258:1152–1155. 10.1126/science.1279807 [DOI] [PubMed] [Google Scholar]
- Heginbotham L., Lu Z., Abramson T., and MacKinnon R.. 1994. Mutations in the K+ channel signature sequence. Biophys. J. 66:1061–1067. 10.1016/S0006-3495(94)80887-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess P., and Tsien R.W.. 1984. Mechanism of ion permeation through calcium channels. Nature. 309:453–456. 10.1038/309453a0 [DOI] [PubMed] [Google Scholar]
- Hille B. 2001. Ion Channels of Excitable Membranes. Third edition Sinauer Associates, Sunderland, MA: 814 pp. [Google Scholar]
- Hille B., and Schwarz W.. 1978. Potassium channels as multi-ion single-file pores. J. Gen. Physiol. 72:409–442. 10.1085/jgp.72.4.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin A.L., and Keynes R.D.. 1955. The potassium permeability of a giant nerve fibre. J. Physiol. 128:61–88. 10.1113/jphysiol.1955.sp005291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn R., Roux B., and Åqvist J.. 2014. Permeation redux: Thermodynamics and kinetics of ion movement through potassium channels. Biophys. J. 106:1859–1863. 10.1016/j.bpj.2014.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Osawa M., Takeuchi K., and Shimada I.. 2010. Structural basis underlying the dual gate properties of KcsA. Proc. Natl. Acad. Sci. USA. 107:6216–6221. 10.1073/pnas.0911270107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izatt R.M., Bradshaw J.S., Nielsen S.A., Lamb J.D., and Christensen J.J.. 1985. Thermodynamic and kinetic data for cation macrocycle interaction. Chem. Rev. 85:271–339. 10.1021/cr00068a003 [DOI] [Google Scholar]
- Jiang Y., Lee A., Chen J., Cadene M., Chait B.T., and MacKinnon R.. 2002. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. 10.1038/417515a [DOI] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B.T., and MacKinnon R.. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41. 10.1038/nature01580 [DOI] [PubMed] [Google Scholar]
- Kauffmann E., Lehn J.M., and Sauvage J.P.. 1976. Enthalpy and entropy of formation of alkali and alkaline-earth macrobicyclic cryptate complexes. Helv. Chim. Acta. 59:1099–1111. 10.1002/hlca.19760590414 [DOI] [Google Scholar]
- Kilbourn B.T., Dunitz J.D., Pioda L.A.R., and Simon W.. 1967. Structure of the K+ complex with nonactin, a macrotetrolide antibiotic possessing highly specific K+ transport properties. J. Mol. Biol. 30:559–563. 10.1016/0022-2836(67)90370-1 [DOI] [PubMed] [Google Scholar]
- Kim I., and Allen T.W.. 2011. On the selective ion binding hypothesis for potassium channels. Proc. Natl. Acad. Sci. USA. 108:17963–17968. 10.1073/pnas.1110735108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn S.J., and Ikeda S.R.. 1995. Permeation selectivity by competition in a delayed rectifier potassium channel. Science. 269:410–412. 10.1126/science.7618108 [DOI] [PubMed] [Google Scholar]
- Kuo A., Gulbis J.M., Antcliff J.F., Rahman T., Lowe E.D., Zimmer J., Cuthbertson J., Ashcroft F.M., Ezaki T., and Doyle D.A.. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 300:1922–1926. 10.1126/science.1085028 [DOI] [PubMed] [Google Scholar]
- Lam Y.L., Zeng W., Sauer D.B., and Jiang Y.. 2014. The conserved potassium channel filter can have distinct ion binding profiles: Structural analysis of rubidium, cesium, and barium binding in NaK2K. J. Gen. Physiol. 144:181–192. 10.1085/jgp.201411191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Li H., Zeng W., Sauer D.B., Belmares R., and Jiang Y.. 2012. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science. 335:686–690. 10.1126/science.1215759 [DOI] [PubMed] [Google Scholar]
- Liu S., and Lockless S.W.. 2013. Equilibrium selectivity alone does not create K+-selective ion conduction in K+ channels. Nat. Commun. 4:2746. [DOI] [PubMed] [Google Scholar]
- Liu S., Bian X., and Lockless S.W.. 2012. Preferential binding of K+ ions in the selectivity filter at equilibrium explains high selectivity of K+ channels. J. Gen. Physiol. 140:671–679. 10.1085/jgp.201210855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless S.W., Zhou M., and MacKinnon R.. 2007. Structural and thermodynamic properties of selective ion binding in a K+ channel. PLoS Biol. 5:e121 10.1371/journal.pbio.0050121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., and Mackinnon R.. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903. 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- Long S.B., Tao X., Campbell E.B., and MacKinnon R.. 2007. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 450:376–382. 10.1038/nature06265 [DOI] [PubMed] [Google Scholar]
- Luzhkov V.B., and Aqvist J.. 2001. K+/Na+ selectivity of the KcsA potassium channel from microscopic free energy perturbation calculations. Biochim. Biophys. Acta. 1548:194–202. 10.1016/S0167-4838(01)00213-8 [DOI] [PubMed] [Google Scholar]
- Mari S.A., Pessoa J., Altieri S., Hensen U., Thomas L., Morais-Cabral J.H., and Müller D.J.. 2011. Gating of the MlotiK1 potassium channel involves large rearrangements of the cyclic nucleotide-binding domains. Proc. Natl. Acad. Sci. USA. 108:20802–20807. 10.1073/pnas.1111149108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulef K., Komarov A.G., Costantino C.A., and Valiyaveetil F.I.. 2013. Using protein backbone mutagenesis to dissect the link between ion occupancy and C-type inactivation in K+ channels. Proc. Natl. Acad. Sci. USA. 110:17886–17891. 10.1073/pnas.1314356110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy J.G., and Nimigean C.M.. 2012. Structural correlates of selectivity and inactivation in potassium channels. Biochim. Biophys. Acta. 1818:272–285. 10.1016/j.bbamem.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.N., and Long S.B.. 2012. Crystal structure of the human two-pore domain potassium channel K2P1. Science. 335:432–436. 10.1126/science.1213274 [DOI] [PubMed] [Google Scholar]
- Moore C., and Pressman B.C.. 1964. Mechanism of action of valinomycin on mitochondria. Biochem. Biophys. Res. Commun. 15:562–567. 10.1016/0006-291X(64)90505-4 [DOI] [Google Scholar]
- Morais-Cabral J.H., Zhou Y., and MacKinnon R.. 2001. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature. 414:37–42. 10.1038/35102000 [DOI] [PubMed] [Google Scholar]
- Neupert-Laves K., and Dobler M.. 1975. The crystal structure of a K+ complex of valinomycin. Helv. Chim. Acta. 58:432–442. 10.1002/hlca.19750580212 [DOI] [PubMed] [Google Scholar]
- Neyton J., and Miller C.. 1988. Discrete Ba2+ block as a probe of ion occupancy and pore structure in the high-conductance Ca2+-activated K+ channel. J. Gen. Physiol. 92:569–586. 10.1085/jgp.92.5.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimigean C.M., and Allen T.W.. 2011. Origins of ion selectivity in potassium channels from the perspective of channel block. J. Gen. Physiol. 137:405–413. 10.1085/jgp.201010551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida M., Cadene M., Chait B.T., and MacKinnon R.. 2007. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 26:4005–4015. 10.1038/sj.emboj.7601828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov S.Y., and Roux B.. 2006. Ion selectivity in potassium channels. Biophys. Chem. 124:279–291. 10.1016/j.bpc.2006.05.033 [DOI] [PubMed] [Google Scholar]
- Noskov S.Y., and Roux B.. 2007. Importance of hydration and dynamics on the selectivity of the KcsA and NaK channels. J. Gen. Physiol. 129:135–143. 10.1085/jgp.200609633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov S.Y., Bernèche S., and Roux B.. 2004. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 431:830–834. 10.1038/nature02943 [DOI] [PubMed] [Google Scholar]
- Nyblom M., Poulsen H., Gourdon P., Reinhard L., Andersson M., Lindahl E., Fedosova N., and Nissen P.. 2013. Crystal structure of Na+, K+-ATPase in the Na+-bound state. Science. 342:123–127. 10.1126/science.1243352 [DOI] [PubMed] [Google Scholar]
- Piasta K.N., Theobald D.L., and Miller C.. 2011. Potassium-selective block of barium permeation through single KcsA channels. J. Gen. Physiol. 138:421–436. 10.1085/jgp.201110684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picollo A., Malvezzi M., Houtman J.C., and Accardi A.. 2009. Basis of substrate binding and conservation of selectivity in the CLC family of channels and transporters. Nat. Struct. Mol. Biol. 16:1294–1301. 10.1038/nsmb.1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressl S., Terwisscha van Scheltinga A.C., Vonrhein C., Ott V., and Ziegler C.. 2009. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature. 458:47–52. 10.1038/nature07819 [DOI] [PubMed] [Google Scholar]
- Reyes N., Oh S., and Boudker O.. 2013. Binding thermodynamics of a glutamate transporter homolog. Nat. Struct. Mol. Biol. 20:634–640. 10.1038/nsmb.2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B., Bernèche S., Egwolf B., Lev B., Noskov S.Y., Rowley C.N., and Yu H.. 2011. Perspectives on: Ion selectivity: Ion selectivity in channels and transporters. J. Gen. Physiol. 137:415–426. 10.1085/jgp.201010577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer D.B., Zeng W., Raghunathan S., and Jiang Y.. 2011. Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation. Proc. Natl. Acad. Sci. USA. 108:16634–16639. 10.1073/pnas.1111688108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer D.B., Zeng W., Canty J., Lam Y., and Jiang Y.. 2013. Sodium and potassium competition in potassium-selective and non-selective channels. Nat. Commun. 4:2721 10.1038/ncomms3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlösser A., Meldorf M., Stumpe S., Bakker E.P., and Epstein W.. 1995. TrkH and its homolog, TrkG, determine the specificity and kinetics of cation transport by the Trk system of Escherichia coli. J. Bacteriol. 177:1908–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R., and Guo W.. 2009. Ion binding properties and structure stability of the NaK channel. Biochim. Biophys. Acta. 1788:1024–1032. 10.1016/j.bbamem.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Shi N., Ye S., Alam A., Chen L., and Jiang Y.. 2006. Atomic structure of a Na+- and K+-conducting channel. Nature. 440:570–574. 10.1038/nature04508 [DOI] [PubMed] [Google Scholar]
- Slesinger P.A., Patil N., Liao Y.J., Jan Y.N., Jan L.Y., and Cox D.R.. 1996. Functional effects of the mouse weaver mutation on G protein-gated inwardly rectifying K+ channels. Neuron. 16:321–331. 10.1016/S0896-6273(00)80050-1 [DOI] [PubMed] [Google Scholar]
- Spalding B.C., Senyk O., Swift J.G., and Horowicz P.. 1981. Unidirectional flux ratio for potassium ions in depolarized frog skeletal muscle. Am. J. Physiol. 241:C68–C75. [DOI] [PubMed] [Google Scholar]
- Starkus J.G., Kuschel L., Rayner M.D., and Heinemann S.H.. 1997. Ion conduction through C-type inactivated Shaker channels. J. Gen. Physiol. 110:539–550. 10.1085/jgp.110.5.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinrauf L.K., Hamilton J.A., and Sabesan M.N.. 1982. Crystal structure of valinomycin-sodium picrate. Anion effects on valinomycin-cation complexes. J. Am. Chem. Soc. 104:4085–4091. 10.1021/ja00379a008 [DOI] [Google Scholar]
- Tao X., Avalos J.L., Chen J., and MacKinnon R.. 2009. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 Å resolution. Science. 326:1668–1674. 10.1126/science.1180310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.N., Kim I., Panosian T.D., Iverson T.M., Allen T.W., and Nimigean C.M.. 2009. Mechanism of potassium-channel selectivity revealed by Na+ and Li+ binding sites within the KcsA pore. Nat. Struct. Mol. Biol. 16:1317–1324. 10.1038/nsmb.1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uysal S., Vásquez V., Tereshko V., Esaki K., Fellouse F.A., Sidhu S.S., Koide S., Perozo E., and Kossiakoff A.. 2009. Crystal structure of full-length KcsA in its closed conformation. Proc. Natl. Acad. Sci. USA. 106:6644–6649. 10.1073/pnas.0810663106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiyaveetil F.I., Sekedat M., MacKinnon R., and Muir T.W.. 2006. Structural and functional consequences of an amide-to-ester substitution in the selectivity filter of a potassium channel. J. Am. Chem. Soc. 128:11591–11599. 10.1021/ja0631955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S., and Rempe S.B.. 2007. Tuning ion coordination architectures to enable selective partitioning. Biophys. J. 93:1093–1099. 10.1529/biophysj.107.107482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S., Sabo D., and Rempe S.B.. 2008. K+/Na+ selectivity in K channels and valinomycin: Over-coordination versus cavity-size constraints. J. Mol. Biol. 376:13–22. 10.1016/j.jmb.2007.11.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S., Rogers D.M., Pratt L.R., and Rempe S.B.. 2011. Perspectives on: Ion selectivity: Design principles for K+ selectivity in membrane transport. J. Gen. Physiol. 137:479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard-Bogind B., Stampe P., and Christophersen P.. 1985. Single-file diffusion through the Ca2+-activated K+ channel of human red cells. J. Membr. Biol. 88:67–75. 10.1007/BF01871214 [DOI] [PubMed] [Google Scholar]
- Vieira-Pires R.S., Szollosi A., and Morais-Cabral J.H.. 2013. The structure of the KtrAB potassium transporter. Nature. 496:323–328. 10.1038/nature12055 [DOI] [PubMed] [Google Scholar]
- Wang Y., Chamberlin A.C., and Noskov S.Y.. 2014. Molecular strategies to achieve selective conductance in NaK channel variants. J. Phys. Chem. B. 118:2041–2049. [DOI] [PubMed] [Google Scholar]
- Weyand S., Shimamura T., Yajima S., Suzuki S., Mirza O., Krusong K., Carpenter E.P., Rutherford N.G., Hadden J.M., O’Reilly J., et al. . 2008. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 322:709–713. 10.1126/science.1164440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton M.R., and MacKinnon R.. 2011. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 147:199–208. 10.1016/j.cell.2011.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Singh S.K., Kawate T., Jin Y., and Gouaux E.. 2005. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 437:215–223. 10.1038/nature03978 [DOI] [PubMed] [Google Scholar]
- Ye S., Li Y., and Jiang Y.. 2010. Novel insights into K+ selectivity from high-resolution structures of an open K+ channel pore. Nat. Struct. Mol. Biol. 17:1019–1023. 10.1038/nsmb.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Levin E.J., Pan Y., McCoy J.G., Sharma R., Kloss B., Bruni R., Quick M., and Zhou M.. 2014. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature. 505:569–573. 10.1038/nature12811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., and MacKinnon R.. 2003. The occupancy of ions in the K+ selectivity filter: Charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates. J. Mol. Biol. 333:965–975. 10.1016/j.jmb.2003.09.022 [DOI] [PubMed] [Google Scholar]
- Zhou Y., and MacKinnon R.. 2004. Ion binding affinity in the cavity of the KcsA potassium channel. Biochemistry. 43:4978–4982. 10.1021/bi049876z [DOI] [PubMed] [Google Scholar]
- Zhou Y., Morais-Cabral J.H., Kaufman A., and MacKinnon R.. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 414:43–48. 10.1038/35102009 [DOI] [PubMed] [Google Scholar]
- Zilman A. 2009. Effects of multiple occupancy and interparticle interactions on selective transport through narrow channels: Theory versus experiment. Biophys. J. 96:1235–1248. 10.1016/j.bpj.2008.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilman A., Di Talia S., Jovanovic-Talisman T., Chait B.T., Rout M.P., and Magnasco M.O.. 2010. Enhancement of transport selectivity through nano-channels by non-specific competition. PLOS Comput. Biol. 6:e1000804 10.1371/journal.pcbi.1000804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann I., and Dutzler R.. 2011. Ligand activation of the prokaryotic pentameric ligand-gated ion channel ELIC. PLoS Biol. 9:e1001101 10.1371/journal.pbio.1001101 [DOI] [PMC free article] [PubMed] [Google Scholar]