Significance
Microbes hold promise as an inflammatory bowel disease (IBD) therapy. Lactococcus lactis, which has not been appreciated as a beneficial microbe, attenuated colitis in three preclinical mouse IBD models. Neither colonization nor an intact bacterium throughout the colon per se was required. Rather, host lysozyme-mediated lysis in an inflamed colon led to L. lactis’s release of its superoxide dismutase, which was necessary for its colitis-attenuating and oxidative stress-reducing activity. Overall, these findings unveil a mechanism by which a bacterium offers benefits to the host but requires the host for targeted release of this beneficial activity. Furthermore, because L. lactis is generally regarded as safe, it represents an opportunity for rapid bench-to-bedside testing in IBD.
Keywords: Lactococcus lactis, oxidative stress, lysozyme, colitis, probiotics
Abstract
Beneficial microbes that target molecules and pathways, such as oxidative stress, which can negatively affect both host and microbiota, may hold promise as an inflammatory bowel disease therapy. Prior work showed that a five-strain fermented milk product (FMP) improved colitis in T-bet−/− Rag2−/− mice. By varying the number of strains used in the FMP, we found that Lactococcus lactis I-1631 was sufficient to ameliorate colitis. Using comparative genomic analyses, we identified genes unique to L. lactis I-1631 involved in oxygen respiration. Respiration of oxygen results in reactive oxygen species (ROS) generation. Also, ROS are produced at high levels during intestinal inflammation and cause tissue damage. L. lactis I-1631 possesses genes encoding enzymes that detoxify ROS, such as superoxide dismutase (SodA). Thus, we hypothesized that lactococcal SodA played a role in attenuating colitis. Inactivation of the sodA gene abolished L. lactis I-1631’s beneficial effect in the T-bet−/− Rag2−/− model. Similar effects were obtained in two additional colonic inflammation models, Il10−/− mice and dextran sulfate sodium-treated mice. Efforts to understand how a lipophobic superoxide anion (O2−) can be detoxified by cytoplasmic lactoccocal SodA led to the finding that host antimicrobial-mediated lysis is a prerequisite for SodA release and SodA’s extracytoplasmic O2− scavenging. L. lactis I-1631 may represent a promising vehicle to deliver antioxidant, colitis-attenuating SodA to the inflamed intestinal mucosa, and host antimicrobials may play a critical role in mediating SodA’s bioaccessibility.
Inflammatory bowel disease (IBD) pathophysiology is driven by both host genetic mutations and the gut microbiota. Immune dysregulation in IBD can result from deficiencies in acute inflammatory response pathways (1) or impaired counterregulation of immune responsiveness (2). Host production of reactive oxygen species (ROS) is an evolutionarily conserved response to microbes (3). However, chronic and excessive ROS up-regulate host inflammatory pathways (4, 5) and result in oxidative stress. Chronic intestinal inflammation and oxidative stress affect not only the host but also the microbiota. Oxidative stress within the lumen is a fitness challenge for gut anaerobic bacteria. IBD patient fecal microbiomes reflect a pattern of response to oxidative stress with enrichments in genes for sulfate transport and cysteine and glutathione metabolism (6). In IBD, oxidative stress contributes to chronic inflammation and dysbiosis, and modulating oxidative stress may help to restore intestinal homeostasis.
Beneficial microbes hold promise for IBD inflammation and dysbiosis (7). However, human clinical trials have shown mixed results (8, 9) because of variations in microbes under study and patient heterogeneity. Preclinical studies that use model systems that recapitulate key features of the human disease are needed to elucidate the mechanism of action of beneficial microbes on hosts and their microbiota. Such information facilitates clinical trial design by identifying patients with the host and microbial features most likely to benefit from the bioactivity of a beneficial microbe. Identifying microbes that target molecules and pathways such as oxidative stress—which negatively affects both host and microbiota—affords opportunities for new IBD therapies.
Building on prior studies examining how a five-strain fermented milk product (FMP) affected the gut microbiome in a preclinical model of colitis and human subjects (10–12), herein we focused on how individual bacterial strains in the FMP affected host response in several preclinical colitis models. One of the five strains, Lactococcus lactis subsp. lactis CNCM I-1631 (L. lactis I-1631), reduced gut oxidative stress and attenuated colitis in three mouse models of colonic inflammation: BALB/c T-bet−/− Rag2−/− mice, BALB/c Il10−/− mice, and BALB/c wild-type mice treated with dextran sulfate sodium (DSS), a colitogenic mucosal disruptant. The colitis-attenuating activity of L. lactis I-1631 was dependent on L. lactis I-1631 superoxide dismutase A (SodA), which reduced colonic epithelial ROS. Our data also support that host factors, increased at sites of inflammation, facilitated targeted delivery of L. lactis I-1631 effects. Specifically, lysis of L. lactis I-1631 by the host peptidoglycan hydrolase and antimicrobial lysozyme-1 appeared to mediate L. lactis I-1631 SodA release and reduction in host oxidative stress.
Results
An L. lactis FMP Attenuates Colitis in T-bet−/− Rag2−/−, Il10−/−, and DSS-Treated Mice.
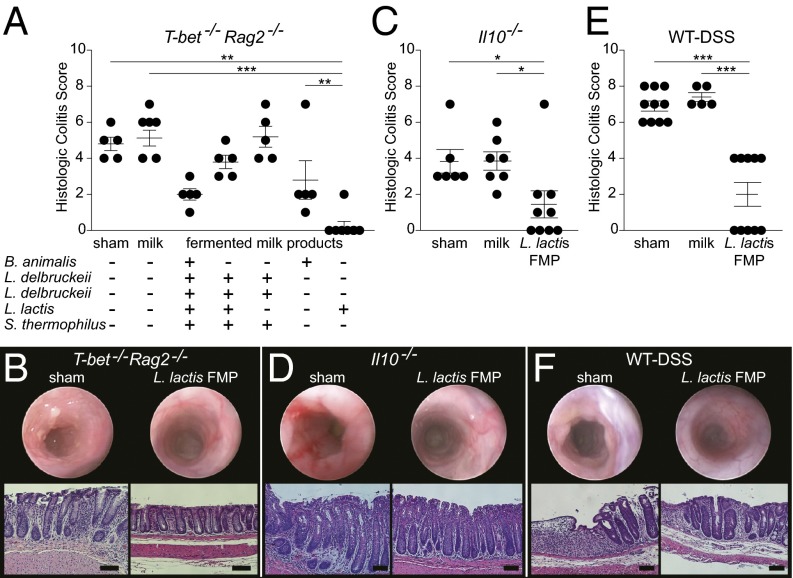
Unraveling the specific bioactivities that beneficial microbes afford to a host is challenging. Prior work suggested that a five-strain FMP attenuated colitis in an innate immune model of colitis (10) and furnished the opportunity to determine the contributions of these strains to attenuating colitis. We began by varying the number of strains present in the five-strain FMP fed to T-bet−/− Rag2−/− mice (13). Two single-strain FMPs prepared with either Bifidobacterium animalis subsp. lactis I-2494 (B. animalis I-2494) or L. lactis I-1631 ameliorated colitis to a level comparable to the five-strain FMP (Fig. 1A). This was not unexpected for B. animalis I-2494, because this probiotic species has antiinflammatory activities in various animal models (14, 15). However, in our experiments, the L. lactis I-1631 FMP was highly effective in reducing colitis (Fig. 1A) and, heretofore, L. lactis has not been widely recognized as a beneficial microbe.
Fig. 1.
L. lactis FMP attenuates colitis in T-bet−/− Rag2−/−, Il10−/−, and DSS-treated BALB/c wild-type mice. (A) Histologic colitis scores from T-bet−/− Rag2−/− mice treated as labeled. Symbols represent data from individual mice from three experiments. (B) Endoscopic distal colon images (Upper) and H&E section photomicrographs from distal colons (Lower) of T-bet−/− Rag2−/− mice treated as labeled. (C) Histologic colitis scores from Il10−/− mice, treated as labeled. Symbols represent data from individual mice from three experiments. (D) Endoscopic distal colon images and H&E section photomicrographs from distal colons of Il10−/− mice treated as labeled. (E) Histologic colitis scores from DSS-exposed mice treated as labeled. Symbols represent data from individual mice from three experiments. (F) Endoscopic distal colon images and H&E section photomicrographs from distal colons of DSS-treated wild-type mice treated as labeled. Error bars indicate mean ± SEM; Kruskal–Wallis test with post hoc Dunn’s comparison test. *P < 0.05, **P < 0.01, and ***P < 0.001. (Scale bars, 100 μm.)
Using endoscopic imaging of T-bet−/− Rag2−/− mice fed the L. lactis I-1631 FMP, we observed a marked reduction in colonic inflammation versus sham-handled mice, which were fed an equal volume of water (Fig. 1B, representative distal colon endoscopic images). With subsequent evaluation using microscopy, L. lactis I-1631 FMP-fed mice showed no evidence of colitis in contrast to sham-handled mice (Fig. 1B, representative micrographs).
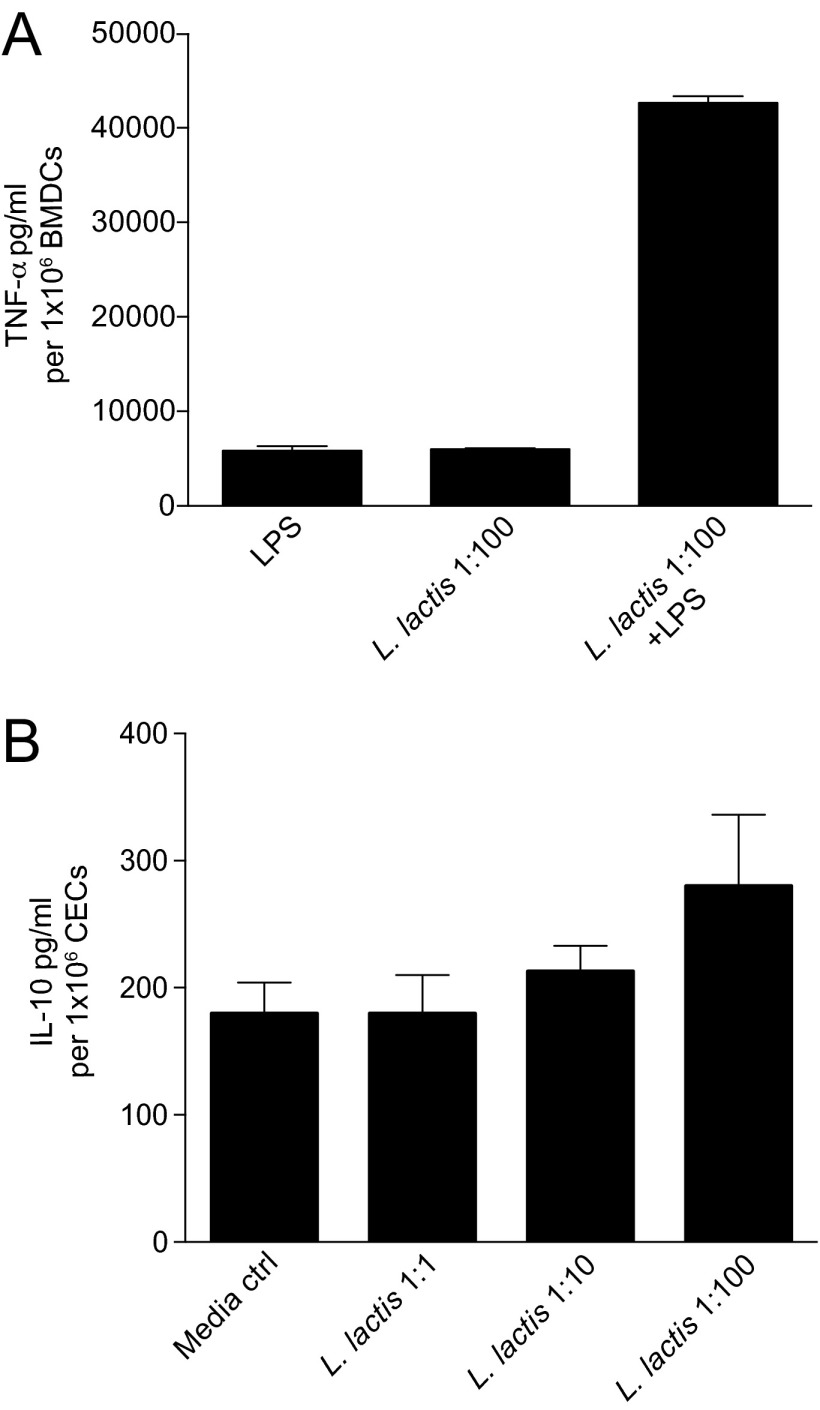
L. lactis I-1631 is a bacterium used in the dairy industry, because it imparts desirable properties to food. There is limited knowledge that it possesses antiinflammatory bioactivities. In in vitro assays used to screen beneficial microbes, such as attenuation of LPS-induced TNF-α production or amplification of IL-10 production (16, 17), L. lactis I-1631 did not exhibit these activities (Fig. S1). To mitigate concerns that this strain’s effects were exclusive to the T-bet−/− Rag2−/− model and to evaluate whether its effects would extend to other intestinal inflammation models, we tested the L. lactis I-1631 FMP in Il10−/− mice and wild-type mice treated with DSS. L. lactis I-1631 FMP attenuated colitis in Il10−/− (Fig. 1C) and DSS-treated wild-type mice (Fig. 1E and Table S1) compared with controls. Endoscopic imaging of the distal colons from Il10−/− (Fig. 1D) and DSS-treated mice (Fig. 1F) demonstrated a marked reduction in colonic inflammation in L. lactis I-1631 FMP-fed mice compared with sham-treated mice (Fig. 1 D and F).
Fig. S1.
L. lactis I-1631 does not exhibit typical activities of beneficial microbes. (A) TNF-α levels in isolated mouse bone marrow-derived dendritic cells treated with LPS and/or cocultured with L. lactis I-1631 at a ratio of 1:100 bacterial to dendritic cells. Bars represent mean ± SEM of three independent experiments. (B) IL-10 levels in isolated mouse primary colonic epithelial cells cocultured with L. lactis I-1631 at various ratios of bacterial to epithelial cells. Bars represent mean ± SEM of three independent experiments.
Table S1.
Disease activity index for WT-DSS mice
| FMP fed | Disease activity index*,† |
| Sham | 3.57 ± 0.19 |
| Milk | 3.80 ± 0.13 |
| L. lactis | 0.83 ± 0.28 |
| (P = 0.0010 vs. sham)† | |
| (P = 0.0021 vs. milk) |
Disease activity index = (combined score of weight loss, stool consistency, and bleeding)/3; mean score ± SEM.
P values were determined using the Kruskal–Wallis test with post hoc Dunn’s comparison test.
L. lactis Ameliorates Colitis in a SodA-Dependent Manner.
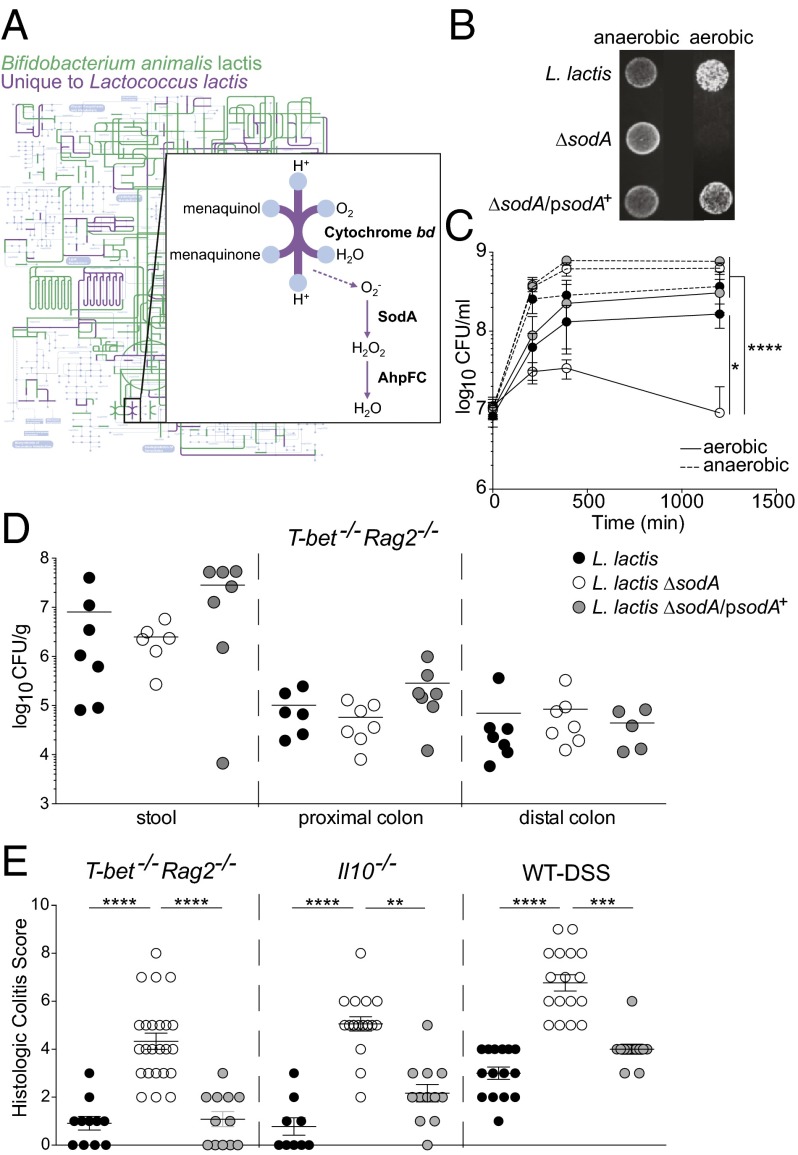
Given the phylogenetic and metabolic differences between B. animalis I-2494 and L. lactis I-1631, we hypothesized that their effects on intestinal inflammation derived from distinct mechanisms. We explored the metabolic differences distinguishing these two species by projecting on the KEGG (Kyoto Encyclopedia of Genes and Genomes) metabolic map the functions ascribed to the clusters of orthologous groups (COGs) of proteins encoded by the B. animalis I-2494 genome. Predicted functions present in L. lactis I-1631 but absent from B. animalis I-2494 were also projected on the same map (Fig. 2A). Out of the 792 nonredundant COGs assigned to B. animalis I-2494, 639 (∼80%) were shared with L. lactis I-1631. Out of the 1,230 nonredundant COGs assigned to the L. lactis I-1631 genome, 591 COGs were not detected in B. animalis I-2494. Consideration of L. lactis I-1631’s unique COGs led us to pathways and functions involved in oxygen utilization/respiration (e.g., cytochrome bd and menaquinone pathways) and oxygen radical detoxification (e.g., SodA) (Fig. 2A). SodA converts the superoxide anion (O2−) into hydrogen peroxide.
Fig. 2.
L. lactis I-1631 ameliorates colitis in a SodA-dependent manner. (A) iPath projection of KEGG metabolic pathways for predicted functions of B. animalis I-2494 (green) and predicted functions present in L. lactis I-1631 and absent from B. animalis I-2494 (purple). The KEGG global map (gray) is in the background layer. (Inset) Details and adaptation of the oxidative phosphorylation pathway present in L. lactis I-1631 and absent from B. animalis I-2494, with superoxide generation indicated by the dashed arrow. (B) Photograph of L. lactis I-1631, the ΔsodA mutant, and the ΔsodA/psodA+ strain grown on plates under anaerobic and aerobic conditions. (C) Growth curves of L. lactis I-1631, the ΔsodA mutant, and the ΔsodA/psodA+ strain grown in milk under anaerobic and aerobic conditions. Data represent mean ± SD; two-way ANOVA with post hoc Bonferroni’s multiple comparison test (MCT). (D) L. lactis I-1631, ΔsodA mutant, and ΔsodA/psodA+ strain enumerations in the stool and large intestine of T-bet−/− Rag2−/− mice. Symbols represent data from individual mice. (E) Histologic colitis scores from T-bet−/− Rag2−/−, Il10−/−, and DSS-exposed mice treated as labeled. Symbols represent data from individual mice from three experiments. Error bars indicate mean ± SEM; Kruskal–Wallis test with post hoc Dunn’s comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To test whether L. lactis I-1631’s capacity to detoxify superoxide contributes to its ability to reduce colitis in mice, we generated an isogenic deletion mutant lacking sodA. As expected, the sodA deletion (ΔsodA) mutant displayed compromised growth kinetics in vitro when cultivated aerobically on solid medium or in milk (Fig. 2B). The complemented strain (ΔsodA/psodA+) was able to restore aerobic growth under both conditions (Fig. 2 B and C). This growth defect did not affect survival of the ∆sodA mutant in vivo in T-bet−/− Rag2−/− mice fed single-strain FMPs, using L. lactis ΔsodA versus L. lactis I-1631 or L. lactis ΔsodA/psodA+, by comparing culturable counts of these strains plated from stool and proximal and distal colon homogenates (Fig. 2D). Thus, we were able to test whether these single-strain FMPs attenuated inflammation in T-bet−/− Rag2−/−, Il10−/−, and DSS-exposed wild-type mice. Loss of sodA abrogated the colitis-attenuating effects of L. lactis I-1631, whereas the fermented milk with the complemented strain restored the inflammation-dampening effects in all three models (Fig. 2E and Tables S2 and S3).
Table S2.
Polymorphonuclear cell-infiltration scores
| Mouse genotype | FMP fed | PMN score*,† |
| T-bet−/− Rag2−/− | L. lactis | 0.44 ± 0.13 |
| L. lactis ∆sodA | 1.59 ± 0.12 | |
| (P < 0.0001 vs. L. lactis) | ||
| (P = 0.4022 vs. L. lactis ∆sodA/psodA+) | ||
| L. lactis ∆sodA/psodA+ | 0.67 ± 0.33 | |
| Il10−/− | L. lactis | 0.67 ± 0.33 |
| L. lactis ∆sodA | 2.50 ± 0.29 | |
| (P = 0.1040 vs. L. lactis) | ||
| (P = 0.0148 vs. L. lactis ∆sodA/psodA+) | ||
| L. lactis ∆sodA/psodA+ | 0.50 ± 0.22 | |
| WT-DSS | L. lactis | 0.71 ± 0.17 |
| L. lactis ∆sodA | 2.29 ± 0.18 | |
| (P = 0.0008 vs. L. lactis) | ||
| (P = 0.0778 vs. L. lactis ∆sodA/psodA+) | ||
| L. lactis ∆sodA/psodA+ | 1.00 ± 0.00 |
Neutrophilic inflammation was scored as follows: 0, absent; 1, mild (scattered lamina propria neutrophils only); 2, moderate (cryptitis, crypt abscesses); 3, severe (sheetlike or submucosal neutrophils); mean score ± SEM.
P values were determined using the Kruskal–Wallis test with post hoc Dunn’s comparison test.
Table S3.
Disease activity index for WT-DSS mice
| FMP fed | Disease activity index*,† |
| L. lactis | 2.27 ± 0.34 |
| L. lactis ∆sodA | 3.53 ± 0.15 |
| (P = 0.0093 vs. L. lactis) | |
| (P = 0.0018 vs. L. lactis ∆sodA/psodA+) | |
| L. lactis ∆sodA/psodA+ | 1.57 ± 0.32 |
Disease activity index = (combined score of weight loss, stool consistency, and bleeding)/3; mean score ± SEM.
P values were determined using the Kruskal–Wallis test with post hoc Dunn’s comparison test.
L. lactis I-1631 Reduces Primary Colonic Epithelial Cell Superoxide Levels.
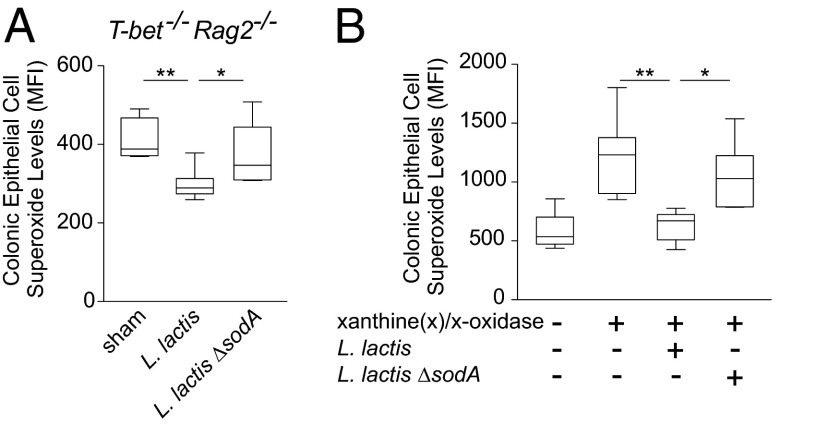
Oxidative stress occurs when there is an imbalance between ROS production and scavenging. To determine whether L. lactis I-1631 was lowering oxidative stress in vivo, we measured superoxide levels in colonic epithelial cells (CECs) isolated from T-bet−/− Rag2−/− mice that were fed L. lactis I-1631 fermented milk or sham-handled. Levels of colonic epithelial superoxide radicals were lowest in mice that received the L. lactis I-1631 FMP compared with controls (Fig. 3A).
Fig. 3.
L. lactis I-1631 reduces primary CEC superoxide levels in vivo and in vitro. (A) Superoxide levels in CECs isolated from T-bet−/− Rag2−/− mice and stained with DHE. T-bet−/− Rag2−/− mice were treated as labeled. Box and whiskers plot; data reflect samples from four experiments; unpaired t test. (B) Superoxide levels in CECs isolated from wild-type mice and stained with DHE. Cells were stimulated with xanthine and xanthine oxidase or unstimulated. Additional treatment with L. lactis I-1631 or L. lactis ΔsodA culture lysates is as labeled. Box and whiskers plot; data reflect six experiments. One-way ANOVA with post hoc Bonferroni’s MCT. *P < 0.05 and **P < 0.01.
To evaluate the ability of L. lactis I-1631 to lower colonic epithelial superoxide radicals, we established an in vitro assay that used primary CECs and stimulation of ROS production using xanthine and xanthine oxidase. Lysates from L. lactis I-1631 lowered superoxide levels in stimulated CECs in a SodA-dependent manner (Fig. 3B).
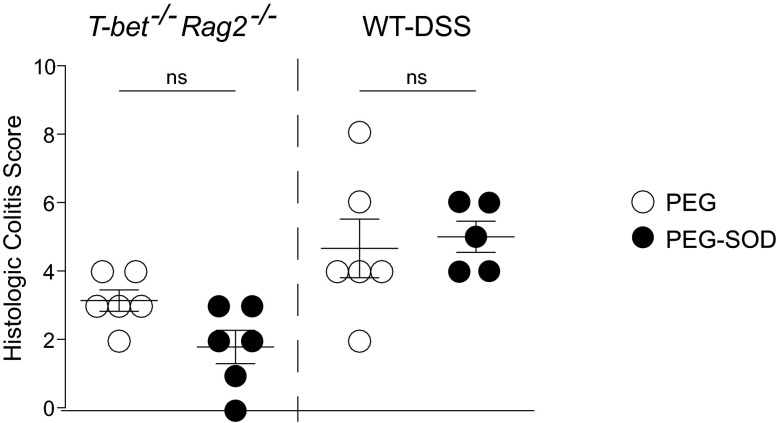
The antiinflammatory activity of SOD has been explored for several inflammatory diseases; however, efficacy has been inconsistent, attributable to its short half-life secondary to rapid hydrolysis and proteolysis. Chemical modification with polyethylene glycol (PEG) can improve its stability (18). However, PEG is an osmotic laxative, which is problematic from a symptom perspective in preclinical models of intestinal inflammation and IBD patients. Aware of this caveat, we undertook a pilot experiment using DSS wild-type and T-bet−/− Rag2−/− mice fed either PEG or PEG coupled to superoxide dismutase. The differences in colitis scores were not statistically significant between the experimental and control groups (Fig. S2), and all of the mice had very loose stools. These data support the concept that how SodA is delivered to a host may affect SodA’s ability to attenuate colitis.
Fig. S2.
Delivery vehicle of SodA influences its effectiveness in reducing colitis. The graph shows histologic colitis scores (y axis) from T-bet−/− Rag2−/− mice and DSS-treated BALB/c wild-type mice that were fed PEG or PEG coupled to superoxide dismutase at a dose of 13 mg⋅kg−1⋅d−1. Symbols represent data from individual mice from three independent experiments. Error bars indicate mean ± SEM; Mann–Whitney test. ns, not significant.
Lysozyme-Mediated Lysis of L. lactis I-1631 Is Required for Colitis Attenuation.
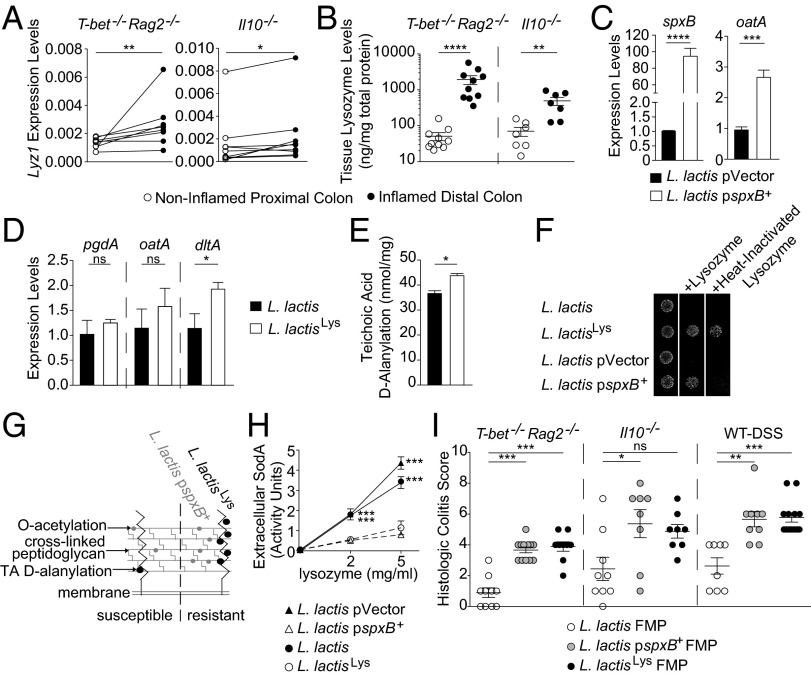
Although our data supported that the colitis-attenuating effects of L. lactis I-1631 were SodA-dependent (Fig. 2) and that L. lactis I-1631 lowered oxidative stress in the colonic mucosa and superoxide levels within colonic epithelial cells (Fig. 3), it was unclear how lactococcal SodA enzyme would encounter host-generated O2−, because lactococcal SodA is predicted to be cytoplasmic by PSORT (www.psort.org). Because L. lactis lacks a secretion system that would enable targeted delivery of SodA to host cells, we hypothesized that lactococcal lysis was a required step for SodA release into the intestinal lumen. Host-derived peptidoglycan hydrolases (PGHs), such as lysozyme-1 (encoded by lyz1) or PGLYRP-2, could contribute to L. lactis I-1631 lysis within the intestine (19). Expression levels of lyz1 were detected in colonic tissue and significantly elevated in inflamed versus healthy colonic mucosa from T-bet−/− Rag2−/− mice (Fig. 4A), consistent with prior findings in mouse models of colitis and IBD patients (20, 21). However, expression levels of pglyrp-2 were below the level of detection. Protein-based determinations of colonic tissue lysozyme-1 levels from T-bet−/− Rag2−/− and Il10−/− mice confirmed that lysozyme-1 levels were markedly elevated in inflamed versus noninflamed colonic tissues (Fig. 4B). Collectively, these data support that colonic inflammation may contribute to lactococcal lysis, providing a mechanism by which SodA could reach the host.
Fig. 4.
Lysozyme-mediated lysis of L. lactis I-1631 is required for colitis attenuation. (A) Colonic tissue lyz1 expression levels from T-bet−/− Rag2−/− and Il10−/− mice. Wilcoxon matched-pairs test (T-bet−/− Rag2−/−; non-Gaussian data distribution); paired t test (Il10−/−). (B) Colonic tissue lysozyme-1 protein levels from T-bet−/− Rag2−/− and Il10−/− mice. Symbols represent data from individual mice from three experiments. Mean ± SEM; Mann–Whitney test. (C and D) mRNA expression levels of genes contributing to lactococcal lysozyme resistance. Mean ± SEM of three experiments; unpaired t test. (E) d-alanylation of teichoic acids in the lysozyme-resistant mutant L. lactisLys and its parent. Mean ± SEM of three experiments; unpaired t test. (F) Lysozyme-resistant L. lactis I-1631 mutants grown on media with lysozyme or heat-inactivated lysozyme. (G) Model of the lactococcal peptidoglycan layer with two lysozyme resistance mechanisms. (H) Extracellular SodA levels of strains grown in lysozyme. Mean ± SEM of three experiments; two-way ANOVA with post hoc Bonferroni’s MCT. (I) Histologic colitis scores from T-bet−/− Rag2−/−, Il10−/−, and DSS-exposed mice treated as labeled. Symbols represent data from individual mice from three experiments. Mean ± SEM; Kruskal–Wallis test with post hoc Dunn’s comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant.
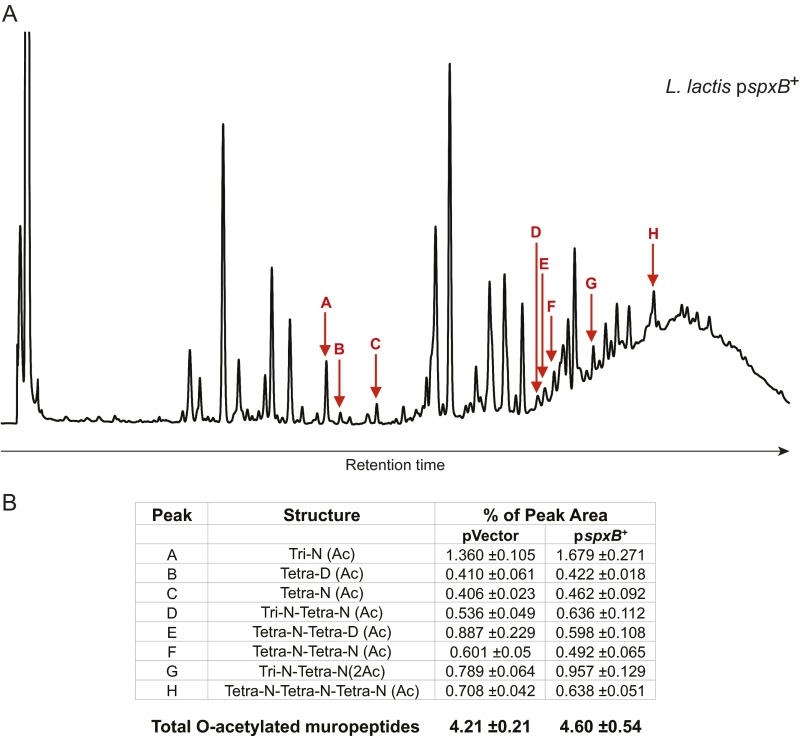
To further interrogate this hypothesis, we questioned whether impeding L. lactis’s lysis would impair SodA release and diminish L. lactis I-1631’s colitis-attenuating effect. O-acetylation is a peptidoglycan modification that confers lysozyme resistance to gram-positive bacteria (19). In the L. lactis MG1363 strain, the regulator SpxB up-regulates the expression of oatA, the gene encoding the O-acetyltransferase responsible for peptidoglycan O-acetylation, and spxB overexpression leads to lysozyme resistance (19). We transformed L. lactis I-1631 with a multiple-copy plasmid containing the spxB gene from L. lactis MG1363 (pspxB+) or the empty plasmid (pVector). As expected, the pspxB+-transformed strain was resistant to lysozyme compared with the pVector-carrying strain (Fig. 4F). We confirmed that expression of spxB and oatA was increased (20- and 2-fold, respectively) in the L. lactis I-1631 pspxB+ strain, similar to what was observed with L. lactis MG1363 (Fig. 4C) (19). In line with these transcription results, the L. lactis pspxB+ strain had a higher percentage of O-acetylated muropeptides compared with the pVector control strain (4.60 ± 0.54% and 4.21 ± 0.21%; Fig. S3). In agreement with a previous report, we observed that a modest difference in PG O-acetylation has a strong effect on lysozyme susceptibility (19).
Fig. S3.
Peptidoglycan structure analysis of L. lactis pspxB+. (A) Representative HPLC profile of muropeptides obtained by mutanolysin digestion of peptidoglycan extracted from L. lactis pspxB+. Red arrows indicate O-acetylated muropeptides. (B) Table listing O-acetylated muropeptides and their respective percent abundances in L. lactis pspxB+ and the plasmid vector control strain. Ac, acetylation; D, d-Asp; Disaccharide, GlcNAc-MurNAc; iGln, α-amidated isoGlu (or γGlu); N, d-Asn; Tetra, disaccharide tetrapeptide (l-Ala-d-iGln-l-Lys-d-Ala); Tri, disaccharide tripeptide (l-Ala-d-iGln-l-Lys). The total percentage of O-acetylated muropeptides was calculated as follows: percentage = Σ monomers (Ac) + 1/2 Σ dimers (Ac) + Σ dimers (2Ac) + 1/3 Σ trimers (Ac). Data represent mean ± SD of three independent culture and media preparations.
We also isolated a spontaneous lysozyme-resistant mutant derived from L. lactis I-1631 (L. lactisLys) and characterized its lysozyme-resistant phenotype. Three genes contribute to L. lactis lysozyme resistance (oatA, pgdA, dltA) (19, 22). Of those, only dltA showed a statistically significant increase in expression (∼1.8-fold) in L. lactisLys compared with L. lactis I-1631 (Fig. 4D). The dltA gene belongs to the dltABCD operon, which encodes the machinery for teichoic acid (TA) d-alanylation (22). We quantified the TA d-alanylation and observed an average 9.3% increase of TA d-alanylation in L. lactisLys compared with L. lactis I-1631 (Fig. 4E). Increasing TA d-alanylation decreases the negative charge of the cell wall, conferring resistance to cationic antimicrobial peptides (CAMPs) (22). Lysozyme exerts its antimicrobial effects via its PGH and/or CAMP activities (23). Heat inactivation of lysozyme abolishes its enzymatic activity whereas conserving its CAMP potential (24). Using heat-inactivated lysozyme, we found that L. lactisLys is resistant to the CAMP activity of lysozyme, consistent with its increased TA d-alanylation (Fig. 4F). In contrast, resistance of L. lactis pspxB+ to lysozyme was lost with heat activation compared with its isogenic control (Fig. 4F), suggesting that its lysozyme resistance was dependent on lysozyme’s hydrolytic activity, consistent with its up-regulation of the O-acetylation pathway (Fig. S3). Thus, lysozyme resistance of these strains was dependent upon two distinct mechanisms (Fig. 4G).
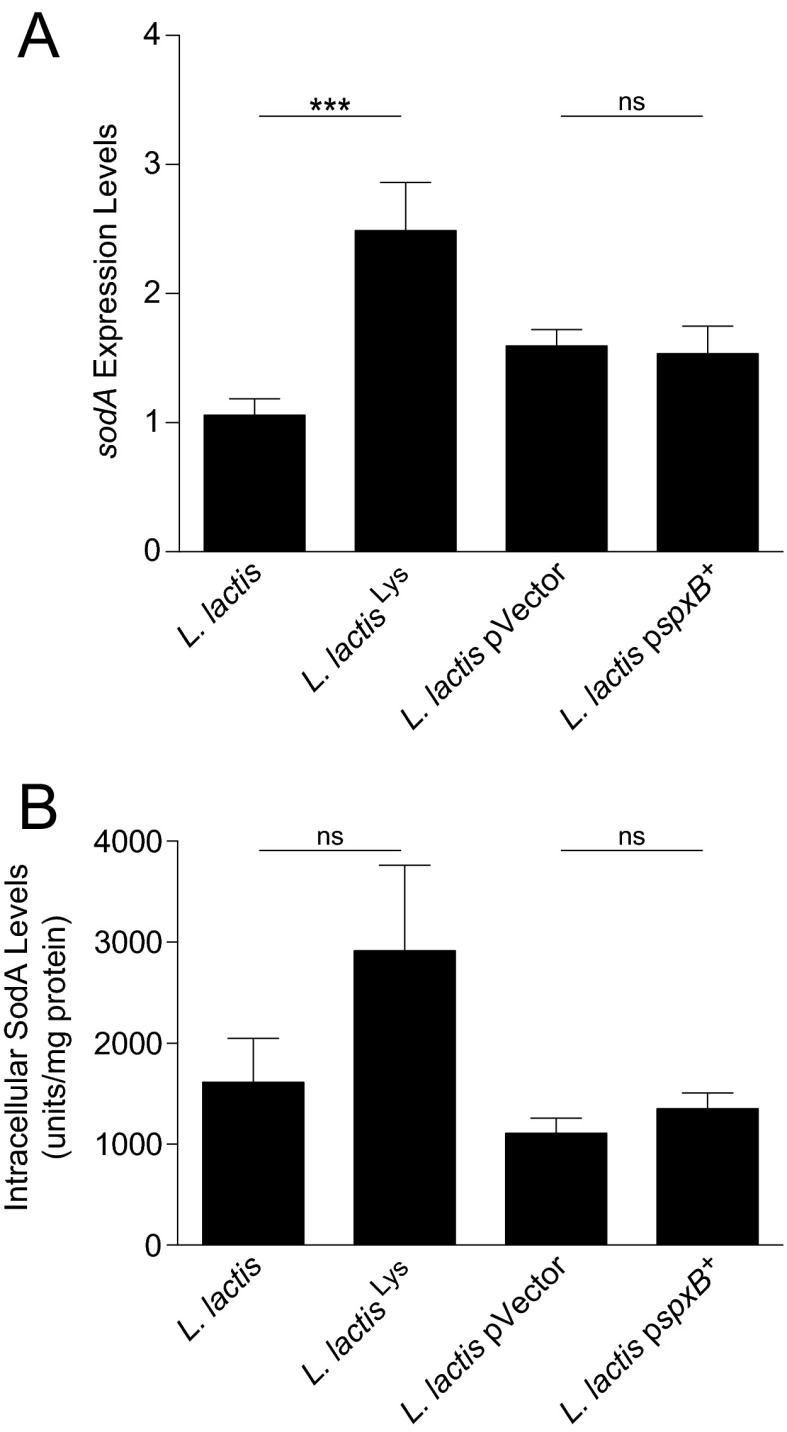
To test our hypothesis of lysozyme-mediated lactococcal SodA release, we measured extracellular SodA release with lysozyme exposure in vitro. SodA release was markedly decreased in both lysozyme-resistant strains (Fig. 4H). To ensure that the observed phenotype was not due to altered sodA expression, we measured sodA expression levels (Fig. S4A) and intracellular SodA levels (Fig. S4B) in the lysozyme-resistant mutants and found that they were equal or higher compared with their respective parental strains. We tested FMPs of the mutants compared with L. lactis I-1631 in the three models of intestinal inflammation and observed that the lysozyme-resistant strains were less effective in reducing colitis than the lysozyme-sensitive wild-type strain (Fig. 4I) and empty vector control [T-bet−/− Rag2−/−: n = 11; 3 ± 0.3 (mean colitis score ± SEM); DSS wild type: n = 8; 4 ± 0.5]. These data support that lysozyme-mediated lysis is a critical step for L. lactis I-1631’s colitis attenuation.
Fig. S4.
SodA levels in the lysozyme-resistant mutants. (A) Relative mRNA expression levels of sodA in lysozyme-resistant mutants. Bars represent mean ± SEM of three independent experiments. ***P < 0.001; unpaired t test. ns, not significant. (B) Intracellular SodA levels in lysozyme-resistant mutants. Bars represent mean ± SEM of three independent experiments; unpaired t test.
Discussion
We identify a bacterium that exhibits a host-beneficial activity facilitated by a host factor. We found that an L. lactis strain, which was not appreciated as a beneficial microbe and that naturally produces superoxide dismutase, attenuates colitis in three different mouse models and lowers colonic epithelial oxidative stress. Neither colonization nor an intact bacterium throughout the colon per se is required. Rather, lysozyme-mediated lysis at inflamed colonic sites contributes to L. lactis’s release of its cytoplasmic SodA, which is necessary for L. lactis’s colitis-attenuating activity. Collectively, our results suggest that a bacterium used in FMPs may have an activity that ameliorates intestinal inflammation and that targeted delivery of this beneficial activity need not be synthetically engineered.
Probiotic strains have been explored as a tool to combat oxidative stress. In vitro experiments suggest that L. lactis has antioxidant properties (25). Two studies expressed L. lactis catalase and SOD in Lactobacillus casei BL23. The catalase-expressing L. casei strain was protective against trinitrobenzenesulfonic acid (TNBS)-induced colitis in rats (26), and the L. casei sodA-expressing strain reduced intestinal inflammation in DSS-exposed mice (27). Also, a recombinant L. lactis strain expressing lactococcal SOD from another strain improved TNBS-induced colitis in rats (28). In another study, a Lactobacillus gasseri strain expressing Streptococcus thermophilus MnSOD reduced inflammation in Il10−/− mice (29). Our studies, in contrast to the aforementioned, tested the nonengineered L. lactis I-1631 in three genetically distinct colitis models and used a sodA deletion mutant and its complement to demonstrate that SodA was necessary and sufficient to reduce colitis.
Our experiments support that L. lactis I-1631 susceptibility to lysozyme-mediated lysis is required to reduce inflammation. Peptidoglycan hydrolases, degrading the peptidoglycan of bacterial cell walls, may be derived from the host or bacteria. Lysozyme-1 is a host-produced peptidoglycan hydrolase. Whereas many studies have focused on lysozyme production by small intestinal Paneth cells, lysozyme-1 levels are increased in the inflamed gastrointestinal tracts and feces of IBD patients versus healthy controls (20, 30). In the mouse models of chronic colitis used in this study, lysozyme-1 was elevated at sites of colonic inflammation. lyz1 encodes an epithelial-expressed lysozyme, which is what we examined, as opposed to Lyz2, which is produced by lamina propria myeloid cells. The use of lysozyme-resistant L. lactis I-1631 mutants in three models of colitis enabled us to test the hypothesis that lysozyme-mediated lysis was a key step in the targeted release of SodA from L. lactis I-1631 to inflamed regions of the colon.
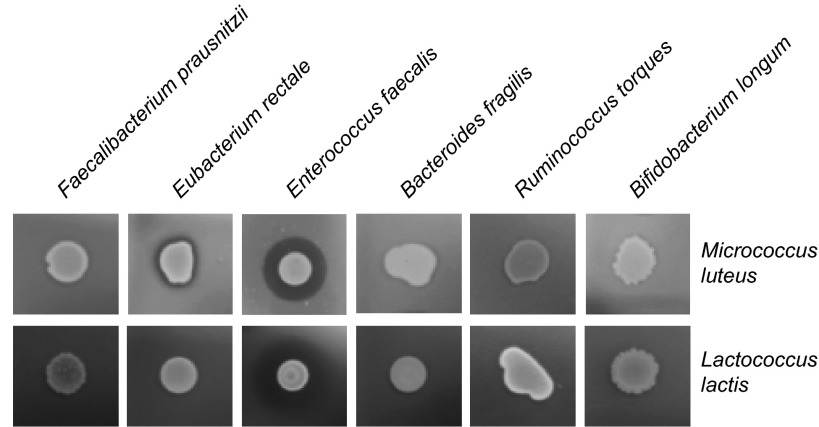
Bacteria, including members of the gut microbiota, may be another source of peptidoglycan hydrolytic activity required for L. lactis I-1631 lysis. Bacterial growth and division require peptidoglycan hydrolysis so that bacteria can change shape, polarity, and their growth dynamics (31, 32). Such PGH can be released into bacterial culture media and can even affect cell-wall rigidity of other species, as shown previously (31). We tested the ability of members of a “healthy” human gut microbiota (Faecalibacterium prausnitzii, Eubacterium rectale, Enterococcus faecalis, Bacteroides fragilis, Ruminococcus torques, and Bifidobacterium longum) to produce PGH using the conventional Micrococcus luteus assay. E. rectale and E. faecalis digested M. luteus cell wall, and E. faecalis digested L. lactis I-1631 cell wall (Fig. S5). Thus, PGH produced by human gut microbiota members could participate in L. lactis I-1631 lysis. Some PGHs are associated with prophages that can be induced by inflammatory conditions, suggesting that colitis may tune resident gut microbes’ PGH activity and potentially lyse L. lactis I-1631 (33, 34). However, the relative contribution and importance of bacterial compared with host lysozyme-1 remains to be elucidated and represents a topic for future investigation.
Fig. S5.
Peptidoglycan hydrolase activity of select and representative members of the gut microbiota. (Upper) Heat-inactivated M. luteus. (Lower) Heat-inactivated L. lactis. Overnight cultures from the strains listed above the panels were added on top of the agar, and clearing of the agar indicates lysis.
Both bacterial and host cells have evolved several strategies to lyse bacteria. Some bacteria can kill or lyse each other via bacteriocin-mediated killing or type VI secretion systems. Both epithelial and myeloid cell subsets also possess a range of antimicrobial molecules, including c-type lectins, defensins, and cathelicidins. Because myeloid cells contribute to the inflammatory infiltrates in colitis, they represent a source for antimicrobials that may potentially lyse L. lactis. In this study, however, we focused on lysozyme-1, not lysozyme-2, which we did not detect in the inflamed colon and which is produced by myeloid cells.
We questioned whether L. lactis I-1631 and its lysis were necessary to deliver SOD to the colon and investigated delivering SOD to mice without bacteria. The protein was PEGylated, which is required for its stability (18). Delivering superoxide dismutase in this way could help standardize dosage and efficacy and appears a more direct approach than using bacteria. However, PEG–SOD did not improve colitis to the same degree as SodA delivered by L. lactis I-1631, and caused diarrhea and weight loss. Bacterial delivery, besides ensuring that the active compound is released directly at the site of inflammation, may afford additional benefits. The use of bacteria that produce, lyse, and release SOD may circumvent issues of oxidative damage caused by the accumulation of intermediate metabolites, such as hydrogen peroxide, observed in a study that examined using lecithinized SOD in DSS-exposed mice (35). L. lactis I-1631 also possesses alkyl hydroperoxide reductase (AhpC), which is predicted to reduce hydrogen peroxide and is produced during superoxide reduction, obviating the need for enzymes from other sources to detoxify such intermediate metabolites. For these reasons, L. lactis I-1631 is appealing.
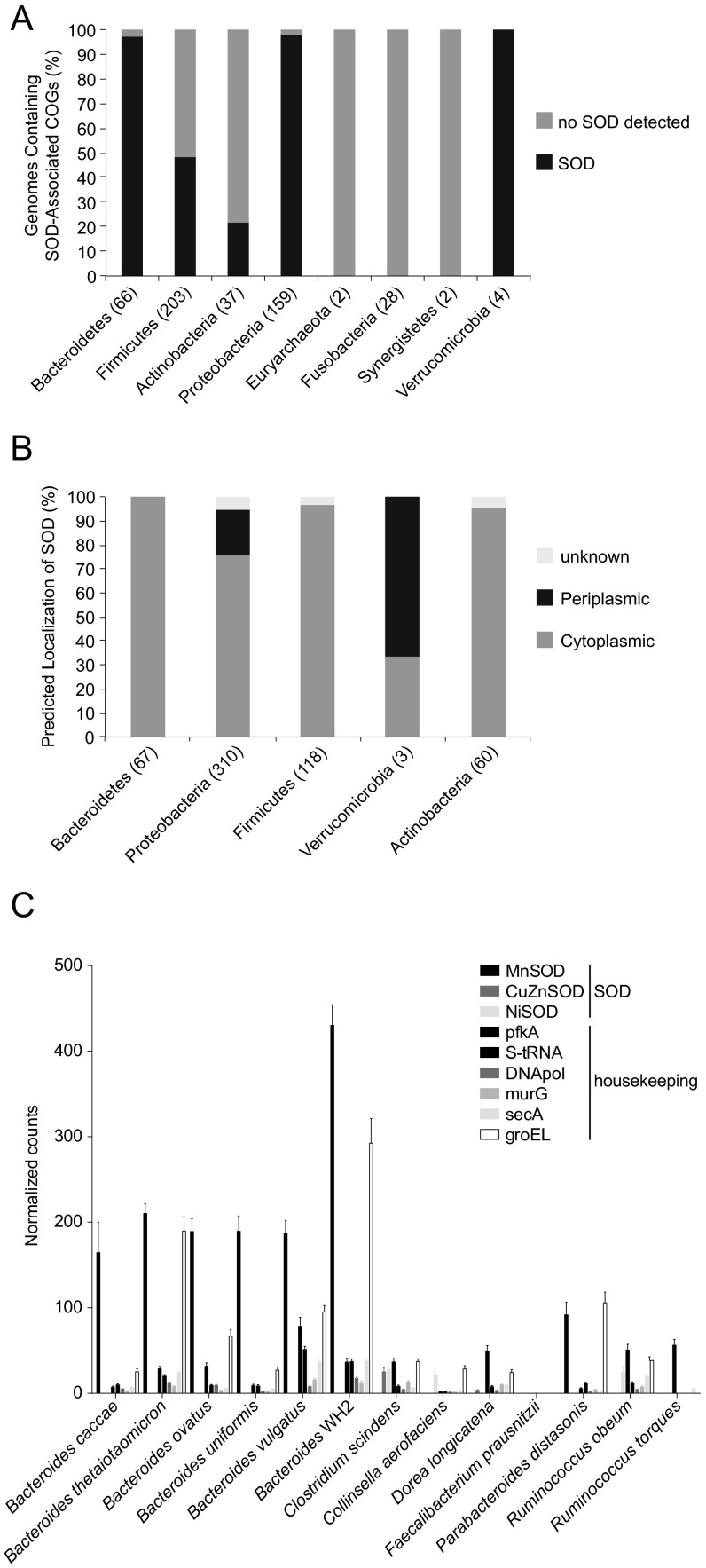
However, we wondered whether members of the human gut microbiota had SOD genomic potential similar to L. lactis I-1631. We searched the genomes of 497 strains, sequenced under the Human Microbiome Project, for the presence of SOD activities, as well as four Akkermansia sp. available from the National Center for Biotechnology Information. Of 501 genomes, we found 330 with predicted SOD function (Fig. S6A). Predicted SOD function was absent from Archaea and from Fusobacteria and Synergistetes. Bacteroidetes, Proteobacteria, and Verrumicrobia were predominantly composed of species encoding at least one SOD enzyme. For Firmicutes and Actinobacteria, the distribution was more genus- and species-specific.
Fig. S6.
SOD genomic potential and expression in representative gut microbiomes. (A) Percentage of genomes containing SOD-associated clusters of orthologous groups. Data are grouped along the x axis by their phylogenetic assignment at the phylum level. In parentheses are the numbers of genomes examined per phylum. (B) Putative localization of predicted SOD proteins. Data are grouped along the x axis by phylum. In parentheses are the numbers of genes examined per phylum. (C) Bars depict the proportion of normalized read counts assignable to the annotation (legend) out of all read counts. Data represent RNAseq profiles of five mice colonized with a 20-member consortium across four time points. Counts were normalized to reads per kilobase gene length per million mapped reads. Bars represent mean number of normalized read counts ± SEM.
Because SOD localization appears critical for its bioavailability, we predicted the localization of the SOD proteins using PSORT. We found that SOD was not predicted to be secreted extracellularly but was either cytoplasmic or periplasmic in the case of bacteria with an outer membrane (Fig. S6B). This observation supports our hypothesis that lysis of SOD-carrying bacteria is key to scavenging host-produced ROS. We also investigated the gut microbiome’s ability to express SOD-encoding genes by interrogating metatranscriptomics data obtained from gnotobiotic mice colonized with a 20-strain consortium (11). SOD was highly expressed in Bacteroidetes species compared with other members of the consortium (Fig. S6C), suggesting that Bacteroidetes might be a source of bacterial SOD in vivo.
Using bacteria as vehicles to deliver biologically active molecules is a promising approach for the treatment of many diseases. Our experiments support that the nonengineered L. lactis I-1631 strain reduces colitis in three murine models, does so in a SodA-dependent fashion, and requires host lysozyme to facilitate delivery. Our study challenges the perception that beneficial microbes need to remain viable throughout the gastrointestinal tract to confer their health benefit. Our findings also delineate a mechanism by which beneficial microbial strains offer up benefits to the host and highlight how specific characteristics of the host may be necessary to promote a bacterium’s beneficial activity.
Materials and Methods
Growth Conditions and Bacterial Strains.
For bacterial strains and plasmids used in this study, see Table S4. For routine cultures, L. lactis I-1631 strains were grown at 37 °C under aerobic conditions in M17 broth (BD Biosciences) supplemented with 0.5% lactose (Sigma-Aldrich), M17L media. For all other strains and culture conditions, see SI Materials and Methods.
Table S4.
Strains and plasmids
| Strain/plasmid | Description | Growth conditions | Source |
| L. lactis I-1631 | L. lactis subsp. lactis CNCM I-1631 (L. lactis I-1631) | 37 °C | Danone Nutricia Research |
| L. lactis ΔsodA | Deletion mutant of sodA gene derived from L. lactis I-1631 | 37 °C, anaerobic | This study |
| L. lactisLys | Spontaneous lysozyme-resistant, derived from L. lactis I-1631 | 37 °C, lysozyme (1 mg/mL) | This study |
| L. lactis ΔsodA/psodA+ | L. lactis I-1631 ΔsodA in cis complemented by the plasmid psodA | 37 °C, erythromycin (2.5 µg/mL) | This study |
| L. lactis pspxB+ | L. lactis I-1631 overexpressing spxB gene carried on the pVES3910 | 37 °C, chloramphenicol (2.5 µg/mL) | This study |
| L. lactis pVector | L. lactis I-1631 containing pVE3916 | 37 °C, chloramphenicol (2.5 µg/mL) | This study |
| Bacteroides fragilis DSM 9669 | 37 °C, anaerobic | DSMZ | |
| E. coli repA | E. coli TG1 supEhsd_5 thi_(lac-proAB)F[traD36_ proAB_ lac1qlacZ_M15] _ repA | 37 °C | (41) |
| Bifidobacterium animalis subsp. lactis DN-173 010 | 37 °C, anaerobic | (10) | |
| Bifidobacterium longum Bll96 | 37 °C, anaerobic | Danone Nutricia Research | |
| Enterococcus faecalis Enfz2 | 37 °C, anaerobic | Danone Nutricia Research | |
| Eubacterium rectale DSM 17629 | 37 °C, anaerobic | DSMZ | |
| Faecalibacterium prausnitzii DSM17677 | 37 °C, anaerobic | DSMZ | |
| Lactobacillus delbrueckii subsp. bulgaricus DN-100 130 | 37 °C | (10) | |
| Lactobacillus delbrueckii subsp. bulgaricus DN-100 182 | 37 °C | (10) | |
| Ruminococcus torques ATCC 27756 | 37 °C, anaerobic | DSMZ | |
| Streptococcus thermophiles DN-001 171 | 37 °C | (10) | |
| pVector | Multiple-copy plasmid with a broad host range replicon pWV01 (pVE3916) | (19) | |
| pspxB+ | Derivative of pVE3916 encoding spxB from L. lactis MG1363 (pVES3910) | (19) | |
| pORI280 | repA-negative lacZ+ derivative of pWV01 | (40) | |
| psodA | Derivative of pORI280 carrying a functional sodA gene | This study | |
| pG+h9 | Thermosensitive replicative plasmid, erythromycin-resistant | (36) | |
| pG+hCm | Thermosensitive replicative plasmid, chloramphenicol-resistant | (36) | |
DSMZ, German Collection of Microorganisms and Cell Cultures.
Study Products.
FMPs were prepared before each experiment. The five-strain FMP, described previously (10), was provided by Danone Nutricia Research. For details, see SI Materials and Methods.
Animal Husbandry.
Specified pathogen-free BALB/c wild-type, BALB/c T-bet−/− Rag2−/− (13), and BALB/c Il10−/− mice were weaned and randomized into experimental cages between postnatal days 21 and 25. Mice were fed standard mouse chow [PicoLab Mouse Diet 20 (5058); LabDiet] and housed in the barrier facility at the Harvard School of Public Health under a 12-h light cycle. Animal experiments were approved and conducted in accordance with the Harvard Medical School Standing Committee on Animals and National Institutes of Health guidelines. For details, see SI Materials and Methods.
Histology.
Sections were examined and the degree of colitis was scored as previously described (13). Image acquisition was performed on a Nikon Eclipse Ni-U, equipped with a Nikon digital sight Fi2 color camera using NIS-Elements basic research software.
Mouse Colonoscopy.
Rigid colonoscopy was performed on mice using a miniendoscope (Karl Storz). For details, see SI Materials and Methods.
Bioinformatics.
Genes of B. animalis subsp. lactis I-2494 and L. lactis subsp. lactis CNCM I-1631 were organized into COGs using the CD-Search Tool (www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi). Retrieved COGs were projected on the global map of KEGG metabolism using iPath v1 software (pathways.embl.de).
Gene Inactivation and Complementation.
The sodA gene was deleted by using the pG+h9 gene replacement system (36). The ΔsodA mutant was in cis complemented by the chromosomal insertion of psodA. For details, see SI Materials and Methods.
Enumeration of L. lactis in the Digestive Tract.
Absolute enumeration of L. lactis I-1631 strains was conducted by administering FMP (150 mg) to BALB/c T-bet−/− Rag2−/− mice 4–5 h before stool and/or tissue collection. For details, see SI Materials and Methods.
Colonic Epithelial Cell Isolation.
For procedural details, see SI Materials and Methods.
Dihydroethidium Assay.
CECs were isolated and incubated with dihydroethidium (DHE) (3 µM; Calbiochem) in RPMI (Corning) for 20 min at 37 °C, followed by staining and flow cytometry. For details and bacterial lysate coincubation experiments, see SI Materials and Methods.
Quantitative RT-PCR.
Quantitative (q) real-time PCR was performed using a Stratagene Mx3005P instrument (Agilent Technologies). For details, see SI Materials and Methods. All primers used for expression analysis are listed in Table S5.
Table S5.
Primers
| Gene target, locus ID; gene name | Primer | Sequence, 5′-3′ | Method | Source |
| LCRE1631_00740; spxB (chromosome) | spxB1-K-F | TCGGAATCAAAAGCACCCAA | RT-qPCR | This study |
| spxB1-K-R | TGCTCCGTCGAAATTTGATCC | |||
| LLCRE1631_00572; oatA | oatA-1-F | TTTGTGGGCGGGATTACGTT | RT-qPCR | This study |
| oatA-1-R | CCTTGCGAAATTTGCCACCA | |||
| LLCRE1631_02065; pgdA | pgdA-2-F | AGATGCGGTGCCACAGATTA | RT-qPCR | This study |
| pgdA-2-R | GCCTTAGCTCCCCCATCTTG | |||
| LLCRE1631_00726; dltA | dltA-1-F | ATTGTACTTGCCGAGCGTGA | RT-qPCR | This study |
| dltA-1-R | TCCGCTCAAATGGTGTGTGA | |||
| LLCRE1631_01647; sodA | sodA-F | GCTTCGTCAAAGAAAGGTTCAAGC | RT-qPCR | This study |
| sodA-R | CGAATGTCTTCTGGAATTGC | |||
| LLCRE1631_02229; tuf2 | Tuf F2 | GAACGCGACACTGACAAACC | RT-qPCR | This study |
| Tuf R2 | AGCAACTGTACCACGACCAG | |||
| llmg_1155; spxB (plasmid) | spxB1-P-F | TTGATGACCGACGCTTGCT | RT-qPCR | This study |
| spxB1-P-R | TAACGAAGGCGCCAAGGAAT | |||
| LLCRE1631_01647; sodA | OFF2685F | TAAAGGAAAGCCTCATTCATCATTAGTAATTTCTCCTTTAAATCCATG | Mutagenesis | This study |
| OFF2688R* | TCAATACAGCGCAAGCTTTGCCAATGGACATCTTAAAAGGAGAAATT | Mutagenesis | This study | |
| OFF2686R | CATCTTAAAAGGAGAAATTACTAATGATGAATGAGGCTTTCCTTTA | Mutagenesis | This study | |
| OFF2687F* | GTAACCCATTTGTGCCGATAAAGCTTGTCCAAGCAAA | Mutagenesis and complementation | This study | |
| OFF2689 | TGGTTTCAGTGAAGTTCGTA | Complementation | This study | |
| Lyz1 | mLyz1-F2 | GAGACCGAAGCACCGACTATG | RT-qPCR | MGH PrimerBank |
| mLyz1-R2 | CGGTTTTGACATTGTGTTCGC |
MGH, Massachusetts General Hospital.
HindIII restriction sites are in bold.
ELISA.
Samples were processed using the Mouse Lysozyme (LZM) ELISA Kit (MyBioSource). Total protein levels were determined by the BCA assay (Pierce). Lysozyme levels were normalized to the total protein content of the respective samples. For details, see SI Materials and Methods.
Spontaneous Mutant Selection.
For selection of lysozyme-resistant L. lactis clones, see SI Materials and Methods.
d-Alanylation of Teichoic Acids.
d-Ala was quantified by HPLC. For details, see SI Materials and Methods.
Superoxide Dismutase A Activity Assay.
The SodA activity assay was adapted from ref. 37. For details, see SI Materials and Methods.
Statistical Analysis.
All statistical analysis was performed using Prism 5 (GraphPad).
SI Materials and Methods
Growth Conditions and Bacterial Strains.
Erythromycin (2.5 µg/mL; Sigma-Aldrich) and chloramphenicol (2.5 µg/mL; Corning) were added as indicated. The ΔsodA mutant strain was grown in an anaerobic hood (Coy Laboratory Products). For growth curve kinetics, overnight cultures were grown in M17L in the presence of antibiotics and diluted 1:100 into prewarmed milk (as described for FMP production) without antibiotic. Cultures were grown for 20 h at 37 °C either aerobically with agitation or in an anaerobic hood. Colony-forming units (CFUs) per milliliter were determined at 0, 3.5, 6.5, and 20 h of incubation by plating dilutions on M17L agar plates. Lysozyme-containing M17L agar (M17LA) plates were prepared by adding 1 mg/mL chicken egg white lysozyme (Sigma-Aldrich) to melted (∼50 °C) M17L media supplemented with 15 g/mL Bacto Agar (BD Biosciences) and immediately poured into Petri dishes. When specified as heat-inactivated, lysozyme was incubated for 2 h at 80 °C before use in the lysozyme test and 2 mg/mL was added to the melted agar medium.
Study Products.
Ultra-high-temperature pasteurized skim and whole milks (Parmalat) were mixed for final values of 3.6% (wt/vol) protein and 3% (wt/vol) fat. After pasteurization, this nonfermented milk was reserved as a milk control or inoculated with 1% overnight bacterial culture (in either M17L or milk) for fermentation at 37 °C. After fermentation (when pH 4.8–4.9, ∼16 h), bacterial counts were measured and only FMPs with a minimum bacterial count of 108 CFU/mL were used as study products. For both the five-strain FMP and single-strain FMPs, 150 mg (∼107 CFUs) was orally instilled daily and ∼100 mg per mouse was provided for ad libitum consumption in each cage. Bacterial counts (CFU/mL) for FMPs were obtained before each experiment.
Animal Husbandry.
T-bet−/− Rag2−/− mice were fed FMPs or milk or sham-handled and fed an equal volume of water from postnatal day (p) 28 ± 5 d old until p56 ± 5 d old, at which point they were killed. Il10−/− mice underwent the same 28-d feeding intervention starting at p52 ± 5 d until p80 ± 5 d old (age- and sex-matched within each experiment), followed by sacrifice. Wild-type BALB/c mice from p28 ± 5 d were fed with study product (detailed above), milk, or water control for 10 d, with 3% (wt/vol) dextran sulfate sodium salt (DSS) (Affymetrix) added to the drinking water from experimental day 3 through 7. Body weights were monitored every 3 d. The same feeding regimens were followed for mice fed PEG (5,000 molecular weight) or PEG (5,000 molecular weight) coupled to bovine superoxide dismutase (Sigma-Aldrich) at a dose of 13 mg⋅kg−1⋅d−1. Dosage of SOD was based on a prior study (38). For all three models, mice were killed on the day the feeding ended and colons were harvested, fixed, and embedded, and slides were generated for histologic analysis.
Histology.
The degree of polymorphonuclear cell infiltration was scored as follows: 0, absent; 1, mild (scattered lamina propria neutrophils only); 2, moderate (cryptitis, crypt abscesses); or 3, severe (sheetlike or submucosal neutrophils).
Mouse Colonoscopy.
Mice were anesthetized with isoflurane (Butler Schein) (5% for induction, 2% for maintenance). Once anesthetized, a rigid colonoscope (Karl Storz) was introduced and images were obtained. Postprocedure mice were observed for signs of distress. Typical time for anesthesia and procedure was 10 min per mouse.
Disease Activity Index.
The disease activity index was determined according to Murthy et al. (39) by scoring changes in weight, hemoccult positivity or gross bleeding, and stool consistency (combined score/3).
Gene Inactivation and Complementation.
For sodA deletion, ∼500-bp DNA fragments located upstream and downstream of the sodA-encoding region were amplified with the primer sets OFF2685F–OFF2688R and OFF2686R–OFF2687F. The two fragments were fused together using an overlap extension PCR. The final PCR product was cloned into the restriction site HindIII of the pG+h9. The procedure of integration and excision was performed as previously described (36). psodA is a lactococcal nonreplicative plasmid obtained by cloning sodA, amplified with the primers OFF2687F and OFF2689, into the HindIII/EcoRV restriction sites of the pORI280 plasmid (40). psodA was propagated in Escherichia coli TG1 repA (41). psodA was cotransformed into L. lactis I-1631 ∆sodA with the temperature-sensitive helper plasmid pG+hCm (36), which provides repA in trans for conditional replication of the psodA. The resulting erythromycin- and chloramphenicol-resistant transformants were reisolated and submitted to a temperature shift (38 °C) to select for single-crossover recombination events resulting from the insertion of psodA into the chromosome. Erythromycin-resistant and chloramphenicol-sensitive colonies were tested for chromosomal integration of psodA.
Enumeration of L. lactis in the Digestive Tract.
Stool samples were collected in PBS (900 µL), weighed, and 10-fold serially diluted. For colonic tissue-associated L. lactis I-1631 counts, mice were killed; colons were harvested, gently cleaned with sterilized PBS, equally divided into two parts (i.e., proximal and distal colons), collected into 2 mL of PBS, weighed, and homogenized with the TissueRuptor (Qiagen). One hundred microliters of the dilutions was plated on L. lactis I-1631-specific solid media [M17L supplemented with 15 g/L Bacto Agar and noninhibitory concentrations of rifampicin (1 µg/mL; Sigma-Aldrich) and polymyxin B (5 µg/mL; Sigma-Aldrich)]. At these concentrations, rifampicin and polymyxin B inhibit growth of endogenous intestinal microbes while permitting growth of L. lactis I-1631 and mutant derivatives. For the antibiotic-resistant strains, the indicated antibiotics were added to the L. lactis-selective media. After a 24-h incubation at 37 °C, CFUs were counted and bacterial abundance was determined as CFU/g of wet stool or colonic tissue.
Bone Marrow-Derived Dendritic Cell Isolation and Coculture.
Mouse bone marrow-derived dendritic cells (BMDCs) were generated as previously described (13) and purified using anti-mouse CD11c-coupled magnetic beads. L. lactis I-1631 was cocultured with BMDCs at a ratio of 1:100 bacterial to dendritic cells in the presence or absence of LPS in RPMI with FBS, 10 mM Hepes, 2 mM l-glutamine, and 0.35% (vol/vol) β-mercapto-EtOH in a 24-well plate for 24 h at 37 °C in a cell-culture incubator at 5% CO2. LPS (100 ng/mL) was added to the wells to induce TNF-α production. After 24 h the supernatants were collected and TNF-α levels were determined using ELISA.
Colonic Epithelial Cell Isolation and Coculture.
Colons were opened and contents were removed by rinsing with Dulbecco’s PBS (DPBS; Corning), and EDTA-based stripping of the epithelium was used for epithelial cell isolation. After cleansing, colons were incubated twice with 5 mM EDTA in DPBS for 15 min at 37 °C followed by vigorous shaking for 15 s. Supernatants were removed after each incubation, pooled, pelleted, and washed with DPBS. Pellets were incubated for 20 min at 37 °C in RPMI (Corning) containing 5% fetal bovine calf serum, dispase (50 µg/mL; StemCell Technologies), and DNase I (25 µg/mL; Roche). Cells were then passed through 100- and 70-µm cell strainers and washed with 5 mM EDTA in PBS to generate single-cell suspensions.
For coculture, CECs were washed twice with cold DPBS, quantified by hemocytometer, and resuspended in RPMI with 5% (vol/vol) FBS, 10 mM Hepes, 2 mM l-glutamine, and 0.35% (vol/vol) β-mercapto-EtOH at 1 × 107 cells per mL in a 24-well plate. L. lactis I-1631 was cocultured with CECs at a ratio of 1:1, 1:10, or 1:100 bacterial to epithelial cells for 24 h at 37 °C in a cell-culture incubator at 5% CO2. After 24 h the supernatants were collected and IL-10 levels were determined using ELISA.
ELISA.
Cytokines were measured in tissue-culture supernatants. TNF-α levels were determined using Mouse TNF ELISA Set II (BD OptEIA) according to the manufacturer’s instructions. IL-10 levels were determined using the Mouse IL-10 ELISA Set (BD OptEIA) according to the manufacturer’s instructions. Total supernatant protein concentration was measured and used to calculate the cytokine concentration in pg/mg.
For tissue lysozyme levels, the proximal and distal colons from age- and sex-matched BALB/c T-bet−/− Rag2−/− and BALB/c Il10−/− mice were collected in PBS, homogenized using the TissueRuptor (Qiagen), and frozen at −80 °C. Two freeze–thaw cycles were performed to break open the cells. Samples were processed using the Mouse Lysozyme (LZM) ELISA Kit (MyBioSource) according to the manufacturer’s instructions. Total protein levels were determined by the BCA assay (Pierce). Lysozyme levels were normalized to the total protein content of the respective samples.
Dihydroethidium Assay.
Cells were stained for 20 min at 4 °C with purified anti-mouse CD16/32 (clone 93), Pacific Blue-labeled anti-CD45 (clone 30-F11), PE-Cy7–labeled anti-CD326 (clone G8.8), Pacific Blue-labeled anti-rat IgG2b (clone RTK4530), or PE-Cy7–labeled anti-rat IgG2a (clone RTK2758). All antibodies were obtained from BioLegend. Flow cytometry using a BD LSR II (BD Biosciences) was performed on the stained cells, and data were analyzed with FACSDiva software 6.1.3 (BD Biosciences) and FlowJo 10.0.5 software (TreeStar). Of the many reagents that can be used to measure ROS, dihydroethidium is highly desirable, because it is specific in its detection of superoxide radicals, fixable, and retained well by cells, and can be quickly measured in a population of cells using flow cytometry (42).
For bacterial lysate coincubation experiments, CECs from WT BALB/c mice were obtained as described above, stimulated for 60 min at 37 °C with xanthine (100 mM; Sigma-Aldrich)/xanthine oxidase (30 mU per 1 × 106 ECs; Sigma-Aldrich), and simultaneously treated with bacterial lysates from L. lactis I-1631 (108 CFUs) or its mutant derivatives (108 CFUs). Bacterial lysates were obtained by treating overnight cultures of L. lactis I-1631 (∼108 CFU/mL) or L. lactis I-1631 ΔsodA (∼108 CFU/mL) with lysozyme (5 mg/mL; Sigma-Aldrich) in the presence of cOmplete Protease Inhibitor mixture (one tablet per 10 mL; Roche) for 60 min at 37 °C, centrifuging at 10,000 × g for 5 min, and collecting the supernatant. After coincubation, CECs were stained with DHE, followed by staining with cell-surface markers and quantification by flow cytometry. Data were analyzed using FACSDiva software (BD Biosciences) and FlowJo 10.0.5 (TreeStar).
RT-qPCR.
For lysozyme-1 expression levels, proximal and distal colons from BALB/c T-bet−/− Rag2−/− mice were flash-frozen, weighed, and homogenized in Qiazol (Qiagen) using the TissueRuptor (Qiagen). RNA was isolated using Qiazol reagent and Phase Lock Gel tubes (5 Prime) followed by chloroform extraction. DNase treatments were performed on all of the RNA samples using the Ambion DNA-free Kit and following the manufacturer’s instructions for rigorous DNA removal. Samples were quantified using a NanoPhotometer Pearl (Denville), and total RNA was reverse-transcribed using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative real-time PCR was performed using the KAPA Biosystems SYBR Fast Universal qPCR Kit. Actin was used as the housekeeping gene to normalize samples. Primers are listed in Table S5.
For bacterial mRNA expression levels, bacteria in exponential growth phase (50 mL) were collected when the OD600 reached 0.5 and subjected to centrifugation at 4,800 × g for 1 min, and then the bacterial pellets were flash-frozen and stored at −80 °C. Once thawed, bacterial pellets were resuspended in Qiazol (Qiagen), 0.3 g of 0.1-mm zirconia/silica beads (BioSpec Products) were added, and cells were lysed using a BioSpec Mini-Beadbeater at maximum speed for 1.5 min at 4 °C. RNA was extracted according to Qiagen-recommended procedures. DNase treatment, cDNA synthesis, and quantitative real-time PCR were performed as described above. The L. lactis housekeeping gene tuf2 was used for normalization. For spxB gene expression, primer sets (spxB1-K and spxB1-P) were used to quantify mRNA originating from either the chromosomal or plasmid spxB genes, respectively. Relative mRNA abundance was calculated using the chromosomal gene expression level as a reference. All primers used for expression analysis are listed in Table S5.
Spontaneous Mutant Selection.
Overnight cultures of L. lactis I-1631 were spread onto M17L agar plates supplemented with 2 mg/mL chicken egg white lysozyme (Sigma-Aldrich). After a 24-h incubation at 37 °C, ∼20 randomly selected spontaneous resistant colonies were streaked on M17LA and isolated as single colonies. Clones with no growth defect compared with the parent strain and with a stable resistant phenotype were used for experiments herein.
d-Alanylation of Teichoic Acids.
Overnight cultures of the different strains were harvested by centrifugation at 4,800 × g for 10 min at 4 °C, washed twice with 20 mM ammonium acetate buffer (pH 4.7), and resuspended in the same buffer with an adjusted volume to reach OD600 150. Cells were heat-inactivated at 100 °C for 10 min and then freeze-dried. d-alanine was released from bacterial cells by alkaline hydrolysis with 0.1 N NaOH for 1 h at 37 °C as reported previously (43). After neutralization with 0.1 N HCl, the cells were removed and the supernatant was lyophilized. d-Ala was quantified by HPLC after derivatization with Marfey’s reagent (1-fluoro-2,4-dinitrophenyl-5-l-alanine amide; Sigma-Aldrich) as described previously (22) by comparison with a standard curve with d-Ala derivatives in the range of 50–1,000 pmol.
Peptidoglycan Structure Analysis.
PG was extracted from exponentially growing cultures (OD600 0.75) of the different L. lactis strains and mutants as described previously (44). PG was then hydrolyzed with mutanolysin, and the resulting soluble muropeptides were reduced and separated by RP-HPLC with an Agilent UHPLC 1290 system using an ammonium phosphate buffer and methanol linear gradient as described previously (45). The eluted muropeptides were detected by UV absorbance at 206 nm. Muropeptides were identified according to their retention time by comparison with a reference chromatogram or according to their mass determined by MALDI-TOF mass spectrometry (45). The different muropeptides were quantified by integration of the peaks on the chromatogram. The amount of each muropeptide was expressed as the ratio of the peak area over the sum of all of the peak areas. The percentage of acetylated muropeptides was calculated as the sum of the amounts of all of the identified acetylated muropeptides.
Superoxide Dismutase Activity Assay.
Overnight cultures of L. lactis I-1631 were centrifuged for 5 min at 20,000 × g at room temperature. Culture volumes were adjusted to OD600 1. Bacterial pellets were washed twice with cold PBS, resuspended in 1 mL of PBS, and exposed to 0, 2, or 5 mg of chicken egg white lysozyme (Sigma-Aldrich) at 37 °C. After a 30-min incubation, bacteria were pelleted by centrifugation at 20,000 × g at 4 °C for 5 min. Supernatant was collected as the extracellular fraction for the SOD activity assay. For intracellular SodA activity, 2 mL PBS-washed overnight-cultured bacteria was resuspended in PBS (1 mL), and 150 mg of zirconia/silica beads (0.1 mm; BioSpec Products) was added followed by 2 min of bead beating at 4 °C with a Mini-Beadbeater (BioSpec Products) set at maximum speed. Total protein was measured with a Bradford Reagent Kit (Sigma-Aldrich) following the manufacturer’s recommendations. For the SodA activity assay, 40 µL of the test sample was added to 80 µL of a 50 mM Tris⋅cacodylate (pH 8.5) solution containing 1 mM EDTA and 5 µL 2.6 mM HCl-dissolved pyrogallol (Sigma-Aldrich). The autooxidation kinetics of the pyrogallol were followed by measuring the OD at 405 nm every 15 s for 10 min. Serial dilutions of bovine superoxide dismutase (Sigma-Aldrich) were used as a standard. SodA activity was normalized to the bacterial OD. SodA activity was normalized to the bacterial OD or the total protein content in the case of extracellular or intracellular SodA activities, respectively.
Growth of Representative Gut Bacteria.
Bacteroides fragilis DSM 9669, Bifidobacterium longum Bll96 (Danone Strain Collection), Enterococcus faecalis Enfz2 (Danone Strain Collection), Eubacterium rectale DSM 17629, Faecalibacterium prausnitzii DSM 17677, and Ruminococcus torques ATCC 27756 were grown in Brain Heart Infusion (BHI) medium (Becton Dickinson) supplemented with 5 g/L yeast extract (Becton Dickinson), 5 mg/L hemin (Calbiochem), 1 g/L cellobioase (Sigma-Aldrich), 1 g/L maltose (Sigma-Aldrich), and 0.5 g/L cysteine (Sigma-Aldrich). Cultures were incubated in an anaerobic chamber with the following gas mixture: 80% N2, 10% CO2, 10% H2.
Peptidoglycan Hydrolase Activity.
The ability of select, representative members of the human gut microbiota (see strains listed in Growth of Representative Gut Bacteria) to produce peptidoglycan hydrolases was assessed by testing their capacity to lyse heat-killed Micrococcus lysodeikticus (M. luteus) or L. lactis CNCM I-1631 as previously described (46). Heat-inactivated M. luteus was obtained from Sigma-Aldrich. Heat-inactivated L. lactis I-1631 was generated by incubating stationary-phase cultures at 100 °C for 60 min. Heat-inactivated bacteria were added at a final concentration of 0.04% (wt/wt) to melted BHI-based media containing 15 g/L agar. Seven microliters of overnight cultures of the representative gut microbiota strains, grown in BHI, was spotted onto the surface of BHI agar and the plates were incubated anaerobically at 37 °C for 5 or 10 d for the M. luteus- or L. lactis-containing plates, respectively.
Phylogenetic Distribution of Superoxide Dismutase Genomic Potential.
Genomes of 497 human gut microbiome strains sequenced by the Human Microbiome Project (hmpdacc.org/catalog/) were searched for the presence of predicted superoxide dismutase enzymes (i.e., COG0605 and COG2032). Four genomes available for Akkermansia species were manually searched for the presence of superoxide dismutases by using BlastP (blast.ncbi.nlm.nih.gov/Blast.cgi) and using the L. lactis SodA protein (GenBank accession no. AAA85266.1) as a query sequence. In total, 330 proteins were retrieved. Subcellular localization of the predicted SOD was determined using PSORTb v3.0.2 software (www.psort.org/psortb/), adjusting the parameters as follows: gram-positive bacteria without an outer membrane for Firmicutes (except Negativicutes) and Actinobacteria, gram-positive bacteria for Negativicutes, and gram-negative bacteria for Bacteroidetes, Proteobacteria, and Verrucomicrobia.
SOD Expression in a Published Metatranscriptomic Analysis of Gnotobiotic Mice Associated with a Defined Consortium.
Fecal RNA-sequencing (RNAseq) profiles of mice (n = 5) colonized with a defined mixture of 15 human gut-derived commensals and 5 strains isolated from a fermented milk product were obtained from the McNulty et al. study (11). Only transcripts assigned to select housekeeping genes [pfkA: 6-phosphofructokinase (KEGG orthology K00850); S-tRNA: seryl-tRNA synthetase (K01875); DNApol: DNA polymerase III, alpha-subunit (K02337); murG: UDP-N-acetylglucosamine:LPS N-acetylglucosamine transferase (K02563); secA: preprotein translocase subunit SecA (ATPase, RNA helicase) (K03070); groEL: chaperonin GroEL (K04077)] as well as superoxide dismutase genes [MnSOD: superoxide dismutase, Fe/Mn family (EC:1.15.1.1) (K04564); CuZnSOD: Cu/Zn superoxide dismutase (EC:1.15.1.1) (K04565); NiSOD: superoxide dismutase (EC:1.15.1.1) (K00518)] and obtained at 14-, 15-, 35-, and 36-wk time points were used for subsequent analysis. Counts were normalized to reads per kilobase gene length per million mapped reads.
Supplementary Material
Acknowledgments
We thank members of the W.S.G. laboratory for discussions and advice. This study was supported by a grant from Danone Nutricia Research, R01CA154426 (National Cancer Institute), K08AI078942 (National Institute of Allergy and Infectious Diseases), a Burroughs Wellcome Career in Medical Sciences Award, and a Searle Scholars Award.
Footnotes
Conflict of interest statement: P.V., G.Q., P.G., C.B., M.D., and J.v.H.V. are employees of and hold equity in Danone Nutricia Research. This study was supported by a grant from Danone Nutricia Research, R01CA154426 (National Cancer Institute), K08AI078942 (National Institute of Allergy and Infectious Diseases), a Burroughs Wellcome Career in Medical Sciences Award, and a Searle Scholars Award.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501897112/-/DCSupplemental.
References
- 1.Uhlig HH, et al. 2014. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology 147(5):990–1007.e3.
- 2.Packey CD, Sartor RB. Interplay of commensal and pathogenic bacteria, genetic mutations, and immunoregulatory defects in the pathogenesis of inflammatory bowel diseases. J Intern Med. 2008;263(6):597–606. doi: 10.1111/j.1365-2796.2008.01962.x. [DOI] [PubMed] [Google Scholar]
- 3.Neish AS. Redox signaling mediated by the gut microbiota. Free Radic Res. 2013;47(11):950–957. doi: 10.3109/10715762.2013.833331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christman JW, Blackwell TS, Juurlink BH. Redox regulation of nuclear factor kappa B: Therapeutic potential for attenuating inflammatory responses. Brain Pathol. 2000;10(1):153–162. doi: 10.1111/j.1750-3639.2000.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474(7351):298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 6.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shanahan F. Probiotics in inflammatory bowel disease—Therapeutic rationale and role. Adv Drug Deliv Rev. 2004;56(6):809–818. doi: 10.1016/j.addr.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Bibiloni R, et al. VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol. 2005;100(7):1539–1546. doi: 10.1111/j.1572-0241.2005.41794.x. [DOI] [PubMed] [Google Scholar]
- 9.Bourreille A, et al. FLORABEST Study Group Saccharomyces boulardii does not prevent relapse of Crohn’s disease. Clin Gastroenterol Hepatol. 2013;11(8):982–987. doi: 10.1016/j.cgh.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 10.Veiga P, et al. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc Natl Acad Sci USA. 2010;107(42):18132–18137. doi: 10.1073/pnas.1011737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNulty NP, et al. 2011. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 3(106):106ra106.
- 12.Veiga P, et al. Changes of the human gut microbiome induced by a fermented milk product. Sci Rep. 2014;4:6328. doi: 10.1038/srep06328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131(1):33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philippe D, et al. Bifidobacterium lactis attenuates onset of inflammation in a murine model of colitis. World J Gastroenterol. 2011;17(4):459–469. doi: 10.3748/wjg.v17.i4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SW, et al. Bifidobacterium lactis inhibits NF-kappaB in intestinal epithelial cells and prevents acute colitis and colitis-associated colon cancer in mice. Inflamm Bowel Dis. 2010;16(9):1514–1525. doi: 10.1002/ibd.21262. [DOI] [PubMed] [Google Scholar]
- 16.Helwig U, et al. Lactobacilli, bifidobacteria and E. coli nissle induce pro- and anti-inflammatory cytokines in peripheral blood mononuclear cells. World J Gastroenterol. 2006;12(37):5978–5986. doi: 10.3748/wjg.v12.i37.5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drakes M, Blanchard T, Czinn S. Bacterial probiotic modulation of dendritic cells. Infect Immun. 2004;72(6):3299–3309. doi: 10.1128/IAI.72.6.3299-3309.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pyatak PS, Abuchowski A, Davis FF. Preparation of a polyethylene glycol: Superoxide dismutase adduct, and an examination of its blood circulation life and anti-inflammatory activity. Res Commun Chem Pathol Pharmacol. 1980;29(1):113–127. [PubMed] [Google Scholar]
- 19.Veiga P, et al. SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J Biol Chem. 2007;282(27):19342–19354. doi: 10.1074/jbc.M611308200. [DOI] [PubMed] [Google Scholar]
- 20.Fahlgren A, Hammarström S, Danielsson A, Hammarström M-L. Increased expression of antimicrobial peptides and lysozyme in colonic epithelial cells of patients with ulcerative colitis. Clin Exp Immunol. 2003;131(1):90–101. doi: 10.1046/j.1365-2249.2003.02035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saha S, et al. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-γ. Cell Host Microbe. 2010;8(2):147–162. doi: 10.1016/j.chom.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giaouris E, Briandet R, Meyrand M, Courtin P, Chapot-Chartier M-P. Variations in the degree of D-alanylation of teichoic acids in Lactococcus lactis alter resistance to cationic antimicrobials but have no effect on bacterial surface hydrophobicity and charge. Appl Environ Microbiol. 2008;74(15):4764–4767. doi: 10.1128/AEM.00078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibrahim HR, Thomas U, Pellegrini A. A helix-loop-helix peptide at the upper lip of the active site cleft of lysozyme confers potent antimicrobial activity with membrane permeabilization action. J Biol Chem. 2001;276(47):43767–43774. doi: 10.1074/jbc.M106317200. [DOI] [PubMed] [Google Scholar]
- 24.Ibrahim HR, et al. Partially unfolded lysozyme at neutral pH agglutinates and kills gram-negative and gram-positive bacteria through membrane damage mechanism. J Agric Food Chem. 1996;44(12):3799–3806. [Google Scholar]
- 25.Amaretti A, et al. Antioxidant properties of potentially probiotic bacteria: In vitro and in vivo activities. Appl Microbiol Biotechnol. 2013;97(2):809–817. doi: 10.1007/s00253-012-4241-7. [DOI] [PubMed] [Google Scholar]
- 26.LeBlanc JG, et al. Use of superoxide dismutase and catalase producing lactic acid bacteria in TNBS induced Crohn’s disease in mice. J Biotechnol. 2011;151(3):287–293. doi: 10.1016/j.jbiotec.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Watterlot L, et al. Intragastric administration of a superoxide dismutase-producing recombinant Lactobacillus casei BL23 strain attenuates DSS colitis in mice. Int J Food Microbiol. 2010;144(1):35–41. doi: 10.1016/j.ijfoodmicro.2010.03.037. [DOI] [PubMed] [Google Scholar]
- 28.Han W, et al. Improvement of an experimental colitis in rats by lactic acid bacteria producing superoxide dismutase. Inflamm Bowel Dis. 2006;12(11):1044–1052. doi: 10.1097/01.mib.0000235101.09231.9e. [DOI] [PubMed] [Google Scholar]
- 29.Carroll IM, et al. Anti-inflammatory properties of Lactobacillus gasseri expressing manganese superoxide dismutase using the interleukin 10-deficient mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2007;293(4):G729–G738. doi: 10.1152/ajpgi.00132.2007. [DOI] [PubMed] [Google Scholar]
- 30.van der Sluys Veer A, et al. Fecal lysozyme in assessment of disease activity in inflammatory bowel disease. Dig Dis Sci. 1998;43(3):590–595. doi: 10.1023/a:1018823426917. [DOI] [PubMed] [Google Scholar]
- 31.Frirdich E, Gaynor EC. Peptidoglycan hydrolases, bacterial shape, and pathogenesis. Curr Opin Microbiol. 2013;16(6):767–778. doi: 10.1016/j.mib.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Smith TJ, Blackman SA, Foster SJ. Autolysins of Bacillus subtilis: Multiple enzymes with multiple functions. Microbiology. 2000;146(Pt 2):249–262. doi: 10.1099/00221287-146-2-249. [DOI] [PubMed] [Google Scholar]
- 33.Banks DJ, Lei B, Musser JM. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect Immun. 2003;71(12):7079–7086. doi: 10.1128/IAI.71.12.7079-7086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeMarini DM, Lawrence BK. Prophage induction by DNA topoisomerase II poisons and reactive-oxygen species: Role of DNA breaks. Mutat Res. 1992;267(1):1–17. doi: 10.1016/0027-5107(92)90106-c. [DOI] [PubMed] [Google Scholar]
- 35.Ishihara T, et al. Therapeutic effect of lecithinized superoxide dismutase against colitis. J Pharmacol Exp Ther. 2009;328(1):152–164. doi: 10.1124/jpet.108.144451. [DOI] [PubMed] [Google Scholar]
- 36.Biswas I, Gruss A, Ehrlich SD, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175(11):3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nandi A, Chatterjee IB. Assay of superoxide dismutase activity in animal tissues. J Biosci. 1988;13(3):305–315. [Google Scholar]
- 38.Seguí J, et al. Superoxide dismutase ameliorates TNBS-induced colitis by reducing oxidative stress, adhesion molecule expression, and leukocyte recruitment into the inflamed intestine. J Leukoc Biol. 2004;76(3):537–544. doi: 10.1189/jlb.0304196. [DOI] [PubMed] [Google Scholar]
- 39.Murthy SNS, et al. Treatment of dextran sulfate sodium-induced murine colitis by intracolonic cyclosporin. Dig Dis Sci. 1993;38(9):1722–1734. doi: 10.1007/BF01303184. [DOI] [PubMed] [Google Scholar]
- 40.Leenhouts K, et al. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet. 1996;253(1-2):217–224. doi: 10.1007/s004380050315. [DOI] [PubMed] [Google Scholar]
- 41.Garault P, Letort C, Juillard V, Monnet V. Branched-chain amino acid biosynthesis is essential for optimal growth of Streptococcus thermophilus in milk. Appl Environ Microbiol. 2000;66(12):5128–5133. doi: 10.1128/aem.66.12.5128-5133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Owusu-Ansah E, Yavari A, Banerjee U. 2008. A protocol for in vivo detection of reactive oxygen species. Protocol Exchange, 10.1038/nprot.2008.23.
- 43.Kovács M, et al. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J Bacteriol. 2006;188(16):5797–5805. doi: 10.1128/JB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyrand M, et al. Peptidoglycan N-acetylglucosamine deacetylation decreases autolysis in Lactococcus lactis. Microbiology. 2007;153(Pt 10):3275–3285. doi: 10.1099/mic.0.2007/005835-0. [DOI] [PubMed] [Google Scholar]
- 45.Courtin P, et al. Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an L,D-carboxypeptidase involved in peptidoglycan maturation. J Bacteriol. 2006;188(14):5293–5298. doi: 10.1128/JB.00285-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fontana R, et al. Paradoxical response of Enterococcus faecalis to the bactericidal activity of penicillin is associated with reduced activity of one autolysin. Antimicrob Agents Chemother. 1990;34(2):314–320. doi: 10.1128/aac.34.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]