Significance
We developed an efficient and accurate computational framework, RABIT (regression analysis with background integration), and comprehensively integrated public transcription factor (TF)-binding profiles with TCGA tumor-profiling datasets in 18 cancer types. To systematically search for cancer-associated TFs, RABIT controls the effect of tumor-confounding factors on transcriptional regulation, such as copy number alteration, DNA methylation, and TF somatic mutation. Our predicted TF regulatory activity in tumors is highly consistent with the knowledge from cancer gene databases and reveals many previously unidentified cancer-associated TFs. We also analyzed RNA-binding protein regulation in cancer and demonstrated that RABIT is a general platform for predicting oncogenic gene expression regulators.
Keywords: regulatory inference, tumor profiling, transcription factor, RNA-binding protein
Abstract
Despite the rapid accumulation of tumor-profiling data and transcription factor (TF) ChIP-seq profiles, efforts integrating TF binding with the tumor-profiling data to understand how TFs regulate tumor gene expression are still limited. To systematically search for cancer-associated TFs, we comprehensively integrated 686 ENCODE ChIP-seq profiles representing 150 TFs with 7484 TCGA tumor data in 18 cancer types. For efficient and accurate inference on gene regulatory rules across a large number and variety of datasets, we developed an algorithm, RABIT (regression analysis with background integration). In each tumor sample, RABIT tests whether the TF target genes from ChIP-seq show strong differential regulation after controlling for background effect from copy number alteration and DNA methylation. When multiple ChIP-seq profiles are available for a TF, RABIT prioritizes the most relevant ChIP-seq profile in each tumor. In each cancer type, RABIT further tests whether the TF expression and somatic mutation variations are correlated with differential expression patterns of its target genes across tumors. Our predicted TF impact on tumor gene expression is highly consistent with the knowledge from cancer-related gene databases and reveals many previously unidentified aspects of transcriptional regulation in tumor progression. We also applied RABIT on RNA-binding protein motifs and found that some alternative splicing factors could affect tumor-specific gene expression by binding to target gene 3′UTR regions. Thus, RABIT (rabit.dfci.harvard.edu) is a general platform for predicting the oncogenic role of gene expression regulators.
Tumorigenesis is a multistep process requiring alterations in gene expression programs (1, 2). Transcription factors (TFs) are instrumental in driving these gene expression programs, and misregulation of these TFs can result in the acquisition of tumor-related properties (3). For example, E2F1 is overexpressed in many cancer types and promotes tumor proliferation by regulating expression of genes involved in cell differentiation, metabolism, and development (4). As another example, FOXM1 plays an important role in promoting cell proliferation and cell cycle progression through transcriptional activation of many G2/M-specific genes. Increased FOXM1 gene expression was detected in numerous cancer types, and FOXM1 is a promising therapeutic target for cancer treatment (5). TFs also play critical roles in inducing the tumor microenvironment for metastasis. For example, SNAI1/2, TWIST, and ZEB1/2 orchestrate the expression of genes involved in cell polarity, cell–cell contact, cytoskeleton structure, and extracellular matrix degradation. The joint effect of these TFs promotes cancer cell motility and invasion in the metastatic process (2, 6).
With the rapid development of high-throughput technologies, large amounts of datasets have been generated for regulatory proteins. For example, the ENCODE project generated 689 ChIP-seq TF-binding profiles (7, 8). Additionally, several studies have profiled the recognition motifs for hundreds of TFs, which could be integrated together to elucidate the genome-wide regulatory network (9). Meanwhile, the TCGA project generated datasets for over 18 cancer types, which include gene expression, copy number alteration (CNA), DNA methylation, and somatic mutation profiles (10). All of these resources provided a rich base for cancer integrative analysis (11, 12).
Despite the rapid growth of genomic data, the knowledge on how gene expression programs in tumors are controlled by TFs is still limited. As one challenge, the experimental condition of public ChIP-seq data, such as stem cell line, may not match the physiological condition of a specific cancer type. Even though analysis can be done between ChIP-seq data and cancer type with similar conditions (13), it remains to be seen how to use most public ChIP-seq profiles across diverse cancer types. Meanwhile, the cancer genome is highly unstable, and the gene expression change could arise from CNAs not under the direct effect of TF regulation (14). To overcome these difficulties and search for TFs driving tumor-specific gene expression patterns, we developed an integration framework, RABIT (regression analysis with background integration). We also applied RABIT to RNA-binding protein (RBP) recognition motifs to predict cancer-associated RBPs, demonstrating its potential as a general platform for finding expression regulators in cancers.
Results
Landscape of Transcriptional Regulation in Cancer.
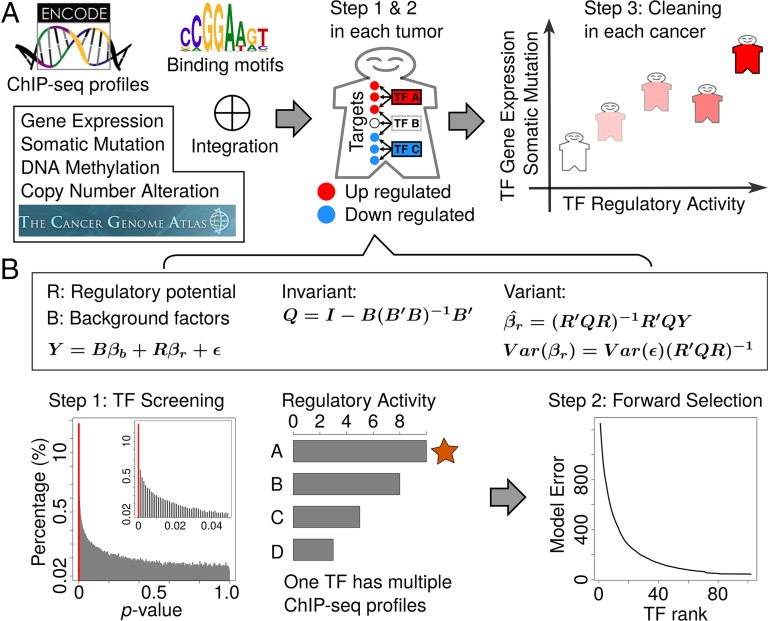
To systematically search for TFs that drive tumor-specific gene expression patterns, we developed an integration framework, RABIT (rabit.dfci.harvard.edu). As a key distinction from other algorithms integrating gene expression and TF ChIP-seq data (15–18), RABIT better captures the properties of cancer cells, such as CNA and DNA methylation, that shape tumor gene expression independently from TF regulation. Additionally, somatic mutations of the TF-coding region can perturb transcriptional regulation. As another difficulty of transcriptional regulation analysis in cancer, most public ChIP-seq datasets were generated under experimental conditions distinct from those in cancers. To model and control the above confounding factors, RABIT uses three steps to identify TFs that drive tumor-specific gene expression patterns at both the individual tumor level and the whole cancer-type level (Fig. 1). In step 1, RABIT screens for TFs that significantly affect the gene expression patterns in each tumor and select the most relevant ChIP-seq profile if multiple profiles exists for the same TF. In step 2, RABIT further selected a subset of TFs among those screened in step 1 to achieve an optimized model error. In step 3, RABIT investigates how well the public ChIP-seq profiles can capture the active TF targets in each cancer type and clean up insignificant TFs.
Fig. 1.
Search transcription factors driving tumor-specific gene expression patterns. (A) The input to RABIT framework includes TF ChIP-seq profiles, recognition motifs, and tumor-profiling datasets. RABIT uses three steps to identify TFs that drive tumor-specific gene expression patterns at both the individual tumor level and the whole cancer-type level. In steps 1 and 2, for each tumor sample, RABIT tests whether the TF target genes are significantly up-regulated or down-regulated compared with the normal controls. In step 3, for each cancer type, RABIT tests whether the TF gene expression and somatic mutation are correlated with the scores of TF regulatory activity on target genes across all tumors and cleans up TFs with poor correlation. (B) In step 1, the efficient Frisch–Waugh–Lovell method of linear regression is applied to test the impact of TFs on target gene regulation after controlling for background factors. A set of TFs with significant regulatory activity is screened. If one TF has several ChIP-seq profiles from different conditions, RABIT only keeps the profile that gives the largest statistical effect of regulatory activity on target genes. In step 2, RABIT further selects a subset of TFs among those screened in step 1 by stepwise forward selection to achieve an optimized model error.
We first collected 686 ChIP-seq profiles from the ENCODE project, representing 150 TFs and 90 cell types (7). For a given TF ChIP-seq dataset, candidate target genes are identified by weighting the number of binding sites by their distance to the transcription start site (TSS) of each gene, using the BETA method we developed (19). Then, in each tumor sample, RABIT tests whether the putative target genes of a TF show significant differential expression compared with the normal controls (step 1 in Fig. 1). To correct for the influence of gene CNA, promoter DNA methylation, promoter CpG content, and promoter degree (total number of ChIP-seq peaks near the gene TSS) on gene expression (SI Appendix, Fig. S1), RABIT uses multivariate linear regression (Table 1 and SI Appendix, Fig. S2A). Because this linear regression needs to be conducted against many ChIP-seq profiles and in a large collection of tumor samples, RABIT uses the efficient Frisch–Waugh–Lovell (FWL) method for regression (20). FWL separates factors that are invariant in each tumor, such as CNA and promoter degree, and only regresses against each variable ChIP-seq profile to speed up the calculation (Fig. 1B).
Table 1.
Multivariate linear regression for TF regulatory activity
| Covariate | Coefficient | SE | t value | P value |
| Promoter degree | 0.0013 | 0.0003 | 4.99 | 6.14e-07 |
| CpG content | 0.1823 | 0.0384 | 4.75 | 2.05e-06 |
| Gene CNA | 0.7281 | 0.0211 | 34.55 | 7.92e-252 |
| Promoter methylation | −0.6742 | 0.0744 | −9.07 | 1.37e-19 |
| Regulatory potential | 0.6359 | 0.0417 | 15.24 | 4.79e-52 |
Using linear regression, the effect of TF regulatory potential on target gene expression is evaluated after controlling for background effects of promoter degree, promoter CpG content, gene CNA, and promoter methylation. In this example, the ENCODE MYC ChIP-seq in the MCF-7 cell line is analyzed together with TCGA data of breast tumor TCGA-AO-A03P-01A. The significance of the regression coefficient is evaluated by the t test, and the TF regulatory activity score is defined as the t value (regression coefficient/SE) of the TF regulatory potential, which is shown in the bold text row.
After running the regression in each tumor, a regulatory activity score can be defined for each TF, which is the t value of the linear regression coefficient t test (coefficient/SE). When one TF has several ChIP-seq profiles from different cell lines and conditions, RABIT only keeps the ChIP-seq profile that gives the largest absolute value of regulatory activity score on target genes (Fig. 1B). For example, among 20 ENCODE MYC ChIP-seq profiles, the MCF7 cell profile is selected for most TCGA breast tumors (SI Appendix, Fig. S3). To achieve an optimized model error in predicting tumor gene expression patterns, RABIT further applies stepwise forward selection to find a subset of TFs among those screened previously (step 2 in Fig. 1B).
After modeling the TF activity on individual tumor level, RABIT investigates how well the public ChIP-seq profiles used can capture the active TF targets in each cancer type (step 3 in Fig. 1). RABIT tests whether the TF gene expression and somatic mutations are correlated with the differential expression of TF target genes across all tumors in one cancer type. As an example, the GATA3 expression level is positively associated with its target gene differential expression across breast tumors, and the presence of GATA3 somatic mutation is negatively associated with its target gene differential expression (Table 2 and SI Appendix, Fig. S2B). Those TFs with insignificant cross-tumor correlation are removed from the results.
Table 2.
Multivariate linear regression for TF regulatory activity
| Covariate | Coefficient | SE | t value | P value |
| TF gene expression | 0.9444 | 0.0480 | 19.66 | 4.95e-64 |
| TF somatic mutation | −1.4455 | 0.2485 | −5.82 | 1.08e-08 |
In each cancer type, the number of nonsynonymous mutation on the TF-coding region is counted in each tumor and evaluated together with TF gene expression against the response variable of the TF regulatory activity scores (t values in Table 1) across tumors by linear regression. GATA3 analysis with TCGA breast tumors is used as example here.
With the RABIT framework, we integrated 686 ENCODE ChIP-seq profiles with 7484 TCGA tumor profiles over 18 cancer types (Fig. 2 and SI Appendix, Table S1) (7, 10). The impact of TFs on tumor gene expression predicted by our framework is highly consistent with previous knowledge. For example, RABIT predicted the target genes of MYC to be significantly up-regulated in numerous cancers (star in Fig. 2), consistent with the known role of MYC as an oncogenic TF (21). FOXM1 also up-regulates its target genes in many cancer types (star in Fig. 2) and could become a potential therapeutic target for many cancer types (5). For MYC and FOXM1, they are clustered together with several other TFs by their regulatory similarity across cancer types (cluster 1 in SI Appendix, Fig. S4). The target genes of TFs in this cluster are preferentially up-regulated in most cancer types, which indicates these TFs as pervasive oncogenic regulators. Another example, RAD21, a member of the cohesion complex with important roles in chromosome maintenance (22), has its target genes repressed in breast cancer (BRCA) (Fig. 2 and SI Appendix, Fig. S5). One recent study reported a high level of RAD21 expression to be indicative of poor prognosis and resistance to chemotherapy in breast cancers (23), which is consistent with our findings. We found RAD21 is clustered with a set of TFs, whose target genes are generally repressed in most cancer types (cluster 3 in SI Appendix, Fig. S4).
Fig. 2.
The landscape of transcriptional regulation in cancer. RABIT calculates the percentage of tumors with TF targets differentially regulated in each cancer type. The upper red triangle represents the percentage of tumors with target genes up-regulated, and the lower blue triangle represents the percentage down-regulated. Only TFs with targets differentially regulated in greater than 50% of tumors in more than two cancer types are shown. The cancer name is displayed by TCGA abbreviation with the platform used for gene expression profiling.
Besides capturing knowledge from previous studies, our analysis also predicted putative TF functions in cancer. For example, the target genes of SPI1 are significantly up-regulated in Glioblastoma (GBM) and kidney renal clear cell carcinoma (KIRC), as predicted by RABIT (Fig. 2). SPI1 is clustered with several other TFs, featured by significant up-regulation of target genes in both GBM and KIRC (cluster 4 in SI Appendix, Fig. S4). We found that high SPI1 expression is associated with poor patient survival in GBM and KIRC (SI Appendix, Fig. S6 A–D). To our knowledge, the role of SPI1 is not well studied in these cancers. Our result indicates SPI1 as a promising target for further study.
As another example of predictions from RABIT, we explored our results on breast cancer, which is an intensively studied cancer type in the past decade. We found that 80 out of 150 TFs analyzed have support from the National Cancer Institute (NCI) cancer gene index or Google search with gene name to be related to breast cancer. Among the rest, there are 21 TFs whose target genes show significant differential expression in TCGA and METABRIC cohorts (SI Appendix, Fig. S7), but little is known about their role in breast cancer from the literature. For example, the histone demethylase PHF8 target genes are up-regulated in more than 60% of breast tumors (SI Appendix, Fig. S7). There has been no study implicating PHF8 in breast cancer to the best of our knowledge, but PHF8 is a known oncogene and potential therapeutic target for esophageal squamous cell carcinoma and prostate cancer (24, 25). Our prediction suggests that PHF8 might also be an oncogene in breast cancer.
Quality Assessment of RABIT Result and Method Comparison.
Encouraged by the consistency of RABIT output with a few previous studies, we set out to systemically check the quality of our computational predictions. To measure the cancer relevance level of a TF, we computed the percentage of tumors with the TF target genes differentially regulated and averaged across all TCGA cancer types. We tested whether the cancer relevance levels defined above are consistent with cancer gene databases. The NCI cancer gene index project assigned a number for each gene as the count of sentences from MEDLINE abstracts in which the gene name and a cancer term cooccurred (26). We also included databases with annotations of cancer-related genes, including the Bushman Laboratory cancer driver gene list (27, 28), the COSMIC somatic mutation catalog (29), and the CCGD mouse cancer driver genes (30). We found TFs with higher percentage of tumors showing target differential expression are associated with the cancer gene annotations in all databases (Fig. 3A and SI Appendix, Fig. S8A).
Fig. 3.
Reliable performance of RABIT framework. (A) All TFs are classified into three categories by NCI cancer index. The category “Zero” includes all TFs with zero index value. We then ranked the rest of the TFs by their NCI cancer indices and assigned the top half to the “High” category and the lower half to the “Low” category. For each category, we plotted the percentage of tumors with target genes differentially regulated and averaged across all cancer types. The bottom and top of the boxes are the 25th and 75th percentiles (interquartile range). Whiskers on the top and bottom represent the maximum and minimum data points within the range represented by 1.5 times the interquartile range. The P value is computed by the Spearman’s rank correlation test. (B) As the gold standard of cancer-associated TFs, we took TFs annotated as cancer-related in at least two out of four cancer gene databases (NCI Cancer Index, Bushman, COSMIC, and CCGD). The performance of identifying cancer-related TFs is compared among several methods, and the areas under the ROC curve of each method are plotted. (C) For cell lines K562 and HL60, there are gene expression-profiling data profiled by ENCODE and genome-wide CRISPR-screening data available from previous studies. We applied RABIT to infer the TF regulatory impact in shaping the expression patterns in each cell line. The Spearman’s rank correlations between the TF regulatory activity scores and the CRISPR-screening scores are calculated, and the P values of the correlation test are attached after each correlation ratio. The result is shown for the K562 cell. (D) The CRISPR correlation result is shown for the HL60 cell.
Because our predicted TF cancer relevance is highly consistent with the annotations from cancer gene databases, we use the knowledge from these databases as the gold standard to compare RABIT performance with several other methods. The gold-standard positive set is defined as TFs annotated as cancer-associated in at least two out of four cancer gene databases described above, and the negative set is defined as the rest of the TFs. Using receiver-operating characteristic (ROC) curve and precision-recall (PR) curve, we compared the ability of predicting cancer-associated TFs among several methods (details provided in in SI Appendix, SI Methods). RABIT has the largest area under curve (AUC) among all methods (Fig. 3B and SI Appendix, Fig. S8 B and C). The second-best methods are LAR and LASSO, which are very popular regression-based feature selection algorithms (31, 32). Because the first two steps of RABIT framework (without step 3) also composed a general feature selection algorithm (Fig. 1B), we compared among RABIT, LAR, and LASSO on the performance of feature selection. Using each algorithm, we select the top 10 most significant TFs to predict the tumor gene expression patterns. RABIT achieved better cross-validation error and shorter running time than LAR and LASSO in all cases tested (SI Appendix, Fig. S9).
To check whether RABIT can accurately identify important TFs in a condition studied, we compared the RABIT results with the growth phenotypes after TF knockout in cell lines. There are two recent works of genome-wide CRISPR screening in K562 cell and HL60 cell (33, 34). In a screening experiment, each gene is assigned a score to represent whether the cell growth rate is affected after knocking out that gene. The ENCODE project also generated gene expression profiles for K562 and HL60, and we applied RABIT to identify TFs that shape the gene expression patterns in these cell lines. We found that for TFs selected by RABIT, the TF regulatory activity scores are significantly negatively correlated with the TF gene CRISPR screening scores (Fig. 3 C and D). This means if RABIT assigns a TF as highly active, knocking out that TF will significantly slow down the cell growth.
To check whether our results on TCGA datasets are consistent with other tumor-profiling cohorts, we applied RABIT on METABRIC breast cancer data (35), Rembrandt glioma data (36), Gravendeel glioma data (37), and Genotype-Tissue Expression (GTEx) normal tissue data (38) (SI Appendix, Fig. S10 A and B). As expected, the TCGA results show positive correlation with other tumor cohort results but negative correlation with GTEx normal tissue cohorts (SI Appendix, Fig. S10 C and D). For example, oncogenes in breast cancer (such as MYC, FOXM1, and RAD21) and oncogenes in GBM (such as SPI1) show the same direction of up- or down-regulation of their target genes in both TCGA and other tumor cohorts, but no regulatory activity is observed in GTEx normal tissue (SI Appendix, Fig. S10 A and B).
Besides ChIP-seq data, TF recognition motifs can also model TF-binding specificity. To check whether our findings based on ChIP-seq data are consistent with the TF regulatory motifs’ result, we collected recognition motifs of 505 TFs from several studies (9, 39, 40). We searched the matches of TF motifs near gene transcription start sites and applied RABIT to characterize TF activity in regulating tumor gene expression (SI Appendix, Fig. S11A). Between ChIP-seq results and regulatory motif results of a TF in each tumor, we computed the correlation of TF regulatory activity scores (t value of TF regulatory potential on target genes; example in Table 1). We found the correlations are positive for all tumors in each cancer type (SI Appendix, Fig. S11B), indicating consistency between ChIP-seq and recognition motif analysis. This result is not surprising because ChIP-seq–binding peaks and TF motif-binding sites significantly correlate with each other (SI Appendix, Fig. S11C). Thus, our result of TF activity correlation could be derived from the similarity of TF target genes.
Landscape of Posttranscriptional Regulation in Cancer.
Besides TFs, RBPs can also control gene expression through posttranscriptional regulation. Recent years saw increasing studies demonstrating RBPs as important players in tumorigenesis (41), although the function and targets of the vast majority of RBPs are still uncharacterized. To this end, we collected 172 recognition motifs for 133 RBPs (42) and predicted the putative targets of each by searching the recognition motifs over gene 3′UTR regions (43). We then analyzed the regulatory activity of RBPs in the same way as TFs, using RABIT.
As an example of RBP regulatory activity in cancer, there are four RBPs (RBFOX1, RBFOX2, RBFOX3, and EIF2S1) with almost the same binding preference of GCAUG sequence, and their motifs are clustered together (Fig. 4A). The gene targets of this motif cluster are strongly down-regulated in most cancer types, especially in GBM (Fig. 4B). We found that a higher level of motif targets down-regulation indicated worse patient survival in GBM (Fig. 4C and SI Appendix, Fig. S6 E and F). Because there are several RBP members in this cluster, we applied the forward selection algorithm and found RBFOX1, RBFOX2, and RBFOX3, but not EIF2S1, are the relevant factors driving regulatory activity (SI Appendix, Table S2). RBFOX1, RBFOX2, and RBFOX3 are known as evolutionarily conserved tissue-specific alternative splicing regulators in metazoans (44). A previous study showed that RBFOX1 suppresses malignancy in glioma by regulating the alternative splicing of TPM1 (45). Our analysis suggests that besides regulating alternative splicing, RBFOX1 and its homologs could bind the gene 3′UTR regions and increase mRNA stability (SI Appendix, Table S2). The loss of RBFOX1 target stabilization is universal among GBM tumors and serves as an indicator of poor patient survival (Fig. 4).
Fig. 4.
The landscape of posttranscriptional regulation in cancer. (A) As an example of RNA-binding protein (RBP) motif clusters, there are five motifs with similar binding preference of GCAUG. We grouped them together as cluster 9. (B) The percentage of tumors with RBP motif target genes differentially regulated is shown for each cancer type in the same way as Fig. 2. Each RBP motif cluster is labeled with the consensus sequence of centroid motif averaged among all members, followed with RBP name or cluster index if there are multiple members. Besides the TCGA data result, we also included METABRIC breast tumor data and Rembrandt and Gravendeel glioma data results for comparison. (C) The GBM patients are ordered by the levels of target down-regulation of motif cluster 9. The top half of patients are classified as “High,” and the bottom half are classified as “Low.” The overall survival days are plotted by a Kaplan–Meier curve, and the P value is estimated by the Weibull model, with age and sex as background factors.
Besides the RBFOX1 regulatory motif cluster, there are many other RBP motifs showing significant regulatory impact in cancer. For example, cluster 1, which is composed of nine RBPs, has its target genes significantly up-regulated in most cancer types (Fig. 4B). Among cluster 1 members, HuR (ELAV1) is a well-known oncogenic RBP that promotes tumor proliferation and malignancy in many human cancers (46). Besides these examples of cancer-associated RBPs, the RABIT framework provides a regulatory map between 133 RBPs and 18 cancer types, which will facilitate further exploration of posttranscriptional regulation in cancers.
Discussion
This study comprehensively integrated TF ChIP-seq and binding motifs with TCGA tumor-profiling data for systematic identification of cancer-associated TFs. RABIT has shown superior performance compared with other state-of-the-art methods. We also applied RABIT to identify a set of RBPs that might play important roles in shaping gene expression in tumors, demonstrating RABIT as a versatile framework for finding cancer-associated gene expression regulators. Notably, there are abundant previous works on integrating ChIP-seq and gene expression data to understand gene regulatory mechanisms (15). For example, ChIP-seq profiles of 12 TFs and RNA-seq expression profiles in mouse embryonic stem cells have been analyzed together, using the regression method (16–18). However, these previous studies were conducted when ChIP-seq and expression-profiling data were generated in the same condition, without further requirement of removing any background confounding effect. Thus, compared to previous works, RABIT is specially designed for large-scale regulatory analysis across diverse cancer types.
As a limitation of RABIT, the linear model used assumes a TF either up-regulates or down-regulates its target genes, which can be represented by one regression coefficient with a positive or negative sign. However, certain TFs may up-regulate and down-regulate target genes depending on different contexts or cofactors. Thus, more versatile models considering the binding context of TFs will be necessary as future works.
With the development of high-throughput sequencing technologies, high-quality transcriptome and mutation profiles of tumor samples have been rapidly generated for diverse cancer types. However, a big gap still exists between getting the tumor profiles and understanding the molecular mechanism of tumorigenesis. Public resources such as ChIP-seq and recognition motifs provide a rich base for bridging this gap and understanding how cancer genes are regulated. Our study provides a cost-effective and systematic framework for integrating regulatory genomics resources with tumor-profiling data to better understand gene regulation in cancers.
Methods
Background Factors of Tumor Gene Expression.
A large portion of tumor gene expression variation is derived from the gene CNA and promoter DNA methylation, which are not direct effects of TF regulation (14). We also found promoter degree (total number of ChIP-seq peaks near the gene TSS) is positively correlated with tumor expression patterns in most cancer types (SI Appendix, Fig. S1A). The promoter CpG content, defined as (CpG dinucleotide frequency)/(C frequency × G frequency) 1kb around gene TSS, also has a strong positive correlation with gene expression in cancer types such as lung squamous cell carcinoma (LUSC) (SI Appendix, Fig. S1B). When testing the TF regulatory impact on gene expression, RABIT controls the effect from these background factors (gene CNA, promoter DNA methylation, promoter degree, and CpG content).
Search TFs Driving Tumor Gene Expression Patterns.
RABIT uses three steps to identify TFs that drive tumor-specific gene expression patterns at both the individual tumor level and the whole cancer-type level. In step 1, RABIT screens for TFs that significantly affect the gene expression patterns in each tumor and selects the most relevant ChIP-seq profile, if multiple profiles exists for the same TF. In step 2, RABIT further selects a subset of TFs among those screened in step 1 to achieve an optimized model error. In step 3, RABIT investigates how well the public ChIP-seq profiles can capture the active TF targets in each cancer type and cleans up insignificant TFs.
In step 1, RABIT runs a TF screening by testing the regulatory impact for each individual TF with a linear regression (Fig. 1B). The regression units are human genes, and the response variable is gene expression difference between tumor and normal sample (SI Appendix, Fig. S2A). The regression covariates include the regulatory potential scores of an individual TF over gene promoters and four background factors defined in the section above (five covariates in total). The regression coefficients and their SEs are estimated by the least squares method. We defined the t value (coefficient/SE) as the regulatory activity score for each TF and assessed its significance by the t test (Table 1). The P values are converted to false discovery rates (FDR) by the Benjamini–Hochberg procedure. We screen a set of significant TFs with an FDR threshold of 0.05. If several ChIP-seq profiles exist for the same TF, we select the profile with the highest absolute value of TF regulatory activity score (Fig. 1B and SI Appendix, Fig. S3).
In step 2, to achieve an optimized error in predicting tumor gene expression patterns, we apply stepwise forward selection to find a subset of TFs among those screened in step 1 (47) (Fig. 1B). We use Mallow’s Cp as a model selection criterion to decide which covariates (screened TFs) should be included in the model (47). For each tumor, we start with four covariates of gene expression background factors (promoter degree, CpG content, gene CNA, promoter methylation), and search through the TFs screened in step 1. At each round, one TF is selected from the candidate set to best minimize the Mallow’s Cp. The process is repeated until no TFs can be added to further reduce the Mallow’s Cp.
In step 3, RABIT cleans up cases where the target genes decided from public ChIP-seq profiles cannot represent active TF target genes in a cancer type because most public ChIP-seq data were generated under experimental conditions distinct from those in cancers. For each TF, RABIT computed its regulatory activity score in each tumor in step 1, which measures the level of TF target genes’ differential expression (Table 1). We regressed the TF regulatory activity score against the two covariates, TF gene expression value and TF somatic mutation count, across all tumor samples in the same cancer type (Table 2 and SI Appendix, Fig. S2B). The significance level of each covariate is assessed by the t test, and the P values of all covariates are grouped together and converted to FDRs by the Benjamini–Hochberg procedure. Those TFs with insignificant coefficients on both covariates (gene expression and somatic mutation) are excluded from results (FDR threshold, 0.05).
Supplementary Material
Acknowledgments
We thank Myles Brown, David Louis, Mario Suva, Jing Mi, Di Wu, Bo Li, and Eric Severson for helpful discussions. The work was supported by NIH Grants NHGRI U41 HG7000, NCI U01 CA180980, R01 GM113242-01, and NSF DMS1208771.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424272112/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Ell B, Kang Y. Transcriptional control of cancer metastasis. Trends Cell Biol. 2013;23(12):603–611. doi: 10.1016/j.tcb.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152(6):1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev Cancer. 2009;9(11):785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halasi M, Gartel AL. Targeting FOXM1 in cancer. Biochem Pharmacol. 2013;85(5):644–652. doi: 10.1016/j.bcp.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 7.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerstein M. Genomics: ENCODE leads the way on big data. Nature. 2012;489(7415):208. doi: 10.1038/489208b. [DOI] [PubMed] [Google Scholar]
- 9.Jolma A, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152(1–2):327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Weinstein JN, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoadley KA, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma X, Xiao L, Wong WH. Learning regulatory programs by threshold SVD regression. Proc Natl Acad Sci USA. 2014;111(44):15675–15680. doi: 10.1073/pnas.1417808111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Liang M, Zhang Z. Regression analysis of combined gene expression regulation in acute myeloid leukemia. PLOS Comput Biol. 2014;10(10):e1003908. doi: 10.1371/journal.pcbi.1003908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152(3):633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angelini C, Costa V. Understanding gene regulatory mechanisms by integrating ChIP-seq and RNA-seq data: Statistical solutions to biological problems. Frontiers Cell Dev Biol. 2014;2:51. doi: 10.3389/fcell.2014.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouyang Z, Zhou Q, Wong WH. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc Natl Acad Sci USA. 2009;106(51):21521–21526. doi: 10.1073/pnas.0904863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLeay RC, Lesluyes T, Cuellar Partida G, Bailey TL. Genome-wide in silico prediction of gene expression. Bioinformatics. 2012;28(21):2789–2796. doi: 10.1093/bioinformatics/bts529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budden DM, et al. Predicting expression: The complementary power of histone modification and transcription factor binding data. Epigenet Chromatin. 2014;7(1):36. doi: 10.1186/1756-8935-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013;8(12):2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frisch R, Waugh FV. Partial time regressions as compared with individual trends. Econometrica. 1933;1(4):387–401. [Google Scholar]
- 21.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes JM, McEwan M, Horsfield JA. Gene regulation by cohesin in cancer: Is the ring an unexpected party to proliferation? Mol Cancer Res. 2011;9(12):1587–1607. doi: 10.1158/1541-7786.MCR-11-0382. [DOI] [PubMed] [Google Scholar]
- 23.Xu H, et al. Enhanced RAD21 cohesin expression confers poor prognosis and resistance to chemotherapy in high grade luminal, basal and HER2 breast cancers. Breast Cancer Res. 2011;13(1):R9. doi: 10.1186/bcr2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun X, et al. Oncogenic features of PHF8 histone demethylase in esophageal squamous cell carcinoma. PLoS ONE. 2013;8(10):e77353. doi: 10.1371/journal.pone.0077353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Björkman M, et al. Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion. Oncogene. 2012;31(29):3444–3456. doi: 10.1038/onc.2011.512. [DOI] [PubMed] [Google Scholar]
- 26.National Cancer Institute 2014. Cancer Gene Index End User Documentation. National Cancer Institute Wiki. Available at https://wiki.nci.nih.gov/x/hC5yAQ. Accessed August 22, 2014.
- 27.Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2012;12(1):51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 28.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbott KL, et al. The Candidate Cancer Gene Database: A database of cancer driver genes from forward genetic screens in mice. Nucleic Acids Res. 2015;43(Database issue):D844–D848. doi: 10.1093/nar/gku770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Ann Stat. 2004;32(2):407–499. [Google Scholar]
- 32.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 33.Gilbert LA, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159(3):647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madhavan S, et al. Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gravendeel LA, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 38.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathelier A, et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42(Database issue):D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matys V, et al. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31(1):374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wurth L. Versatility of RNA-binding proteins in cancer. Comp Funct Genomics. 2012;2012:178525. doi: 10.1155/2012/178525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang P, Singh M, Coller HA. Computational assessment of the cooperativity between RNA binding proteins and MicroRNAs in transcript decay. PLOS Comput Biol. 2013;9(5):e1003075. doi: 10.1371/journal.pcbi.1003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66(24):3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu J, et al. From the cover: Neutralization of terminal differentiation in gliomagenesis. Proc Natl Acad Sci USA. 2013;110(36):14520–14527. doi: 10.1073/pnas.1308610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, et al. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci. 2013;14(5):10015–10041. doi: 10.3390/ijms140510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.James G, Witten D, Hastie T, Tibshirani R. 2013. An Introduction to Statistical Learning: With Applications in R (Springer, New York), 1st Ed, pp xvi, 426 pp.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.