Significance
Many viruses, including those of global concern, are dependent on internalization for their entry. We found that ribonuclease kappa (RNASEK) is required for infection of every virus we tested that enters cells through an acid-dependent pathway, including dengue, West Nile, Sindbis, Rift Valley Fever, and influenza viruses. Mechanistically, we found that RNASEK has no effect on virus binding to cells but, rather, is required for their uptake. RNASEK was required for diverse viruses that are dependent on clathrin-mediated endocytosis for entry, but we found that RNASEK was dispensable for general endocytic uptake. Therefore, RNASEK appears to play a unique role in viral uptake and may be a therapeutically viable target to inhibit major human viral pathogens.
Keywords: arbovirus, entry, endocytosis, clathrin-mediated endocytosis, macropinocytosis
Abstract
Viruses must gain entry into cells to establish infection. In general, viruses enter either at the plasma membrane or from intracellular endosomal compartments. Viruses that use endosomal pathways are dependent on the cellular factors that control this process; however, these genes have proven to be essential for endogenous cargo uptake, and thus are of limited value for therapeutic intervention. The identification of genes that are selectively required for viral uptake would make appealing drug targets, as their inhibition would block an early step in the life cycle of diverse viruses. At this time, we lack pan-antiviral therapeutics, in part because of our lack of knowledge of such cellular factors. RNAi screening has begun to reveal previously unknown genes that play roles in viral infection. We identified dRNASEK in two genome-wide RNAi screens performed in Drosophila cells against West Nile and Rift Valley Fever viruses. Here we found that ribonuclease kappa (RNASEK) is essential for the infection of human cells by divergent and unrelated positive- and negative-strand-enveloped viruses from the Flaviviridae, Togaviridae, Bunyaviridae, and Orthomyxoviridae families that all enter cells from endosomal compartments. In contrast, RNASEK was dispensable for viruses, including parainfluenza virus 5 and Coxsackie B virus, that enter at the plasma membrane. RNASEK is dispensable for attachment but is required for uptake of these acid-dependent viruses. Furthermore, this requirement appears specific, as general endocytic uptake of transferrin is unaffected in RNASEK-depleted cells. Therefore, RNASEK is a potential host cell Achilles’ heel for viral infection.
Viral pathogens are quite diverse in their replication strategies; however, all viruses must enter cells to initiate their replication cycles. The first step involves binding of virus particles to the cell surface. Such interactions can involve attachment factors, which have low affinity but concentrate viruses on the surface of cells, and receptors that intricately interact with viral envelope glycoproteins, which, in addition to binding, promote other aspects of infection such as internalization. Although a plethora of receptors and pathways can be used, most viruses take advantage of the cellular endocytic machinery and penetrate from within the cytosol (reviewed in refs. 1–3). Clathrin-mediated endocytosis, macropinocytosis, and caveolin-mediated endocytosis are the best-studied forms of uptake used by viruses. Clathrin-mediated endocytosis is the most common mechanism used by small viruses, as clathrin-coated vesicles have a diameter of 60–200 nm and can be enlarged to fit even larger particles (4, 5). This pathway is constitutive on most cells, and some viruses use preexisting clathrin-coated pits for entry (e.g., dengue virus), whereas others induce the formation of these structures (e.g., influenza virus) (6, 7). Macropinocytosis is an actin-dependent endocytic process for the nonselective uptake of nutrients in response to receptor engagement. It is the predominant pathway for many larger viruses, including vaccinia virus, but is also used by others, including influenza virus under some conditions (8–10). It also remains unclear which pathways are used by some viruses, including Rift Valley Fever virus (RVFV) (11–13).
The molecular mechanisms involved in these uptake mechanisms are complex and rely on key molecules and organelles that are essential for cellular viability, as these uptake mechanisms bring nutrients and other metabolites into the cytosol for cellular growth and survival. Indeed, these processes and proteins involved are highly conserved from yeast to humans (14, 15). Depending on the virus entry requirements, some viruses fuse within early endosomal vesicles, whereas others traffic to more acidic compartments or macropinosomes for entry. Because many viruses are dependent on these endosomal trafficking pathways for entry, much effort has been made in identifying the specific cellular genes required for viral entry (16). Therapeutics targeting entry are appealing because it is the first step in the infection cycle, and many viruses use common pathways; thus, inhibition may be broadly antiviral, rather than active against only a specific virus. Furthermore, many viruses have high mutation rates and rapidly evolve resistance to therapeutics targeting virally encoded genes. Conversely, therapeutics against host encoded targets would likely be more difficult for the virus to evade.
Recent advances in functional genomic technologies have facilitated the use of unbiased genome-wide RNAi screens to identify cellular genes required for viral infection (17). Such approaches allow for the discovery of otherwise unknown genes that play essential roles in infection. We recently performed such screens in insect cells against two disparate insect-borne human pathogens: the flavivirus West Nile virus (WNV) and the bunyavirus RVFV (18, 19). These are both arthropod-borne human pathogens for which there are no vaccines or therapeutics. Furthermore, these viruses are quite divergent: WNV is a flavivirus that is a globally important cause of encephalitis (20), and RVFV is a bunyavirus that causes significant morbidity and mortality in livestock and humans in Africa (21). In our screens, there were only three genes that promoted infection by both viruses: dRAB5, dSTX7, and dRNASEK (CG40127). The functions of RAB5 and STX7 have been described, but little is known about ribonuclease kappa (RNASEK). Both RAB5 and STX7 are involved in endosomal transport and have roles in viral entry (22, 23). Both WNV and RVFV are enveloped RNA viruses that require an acidic compartment for entry (12, 13, 24). RNASEK is a single-copy, 137-aa protein conserved from insects to humans (25–27) with an unknown function. We set out to determine the role of RNASEK in viral infection and found that RNASEK is required for internalization of a diverse panel of viruses of medical concern.
Results
RNASEK Promotes Flaviviral Infection in Insect Cells.
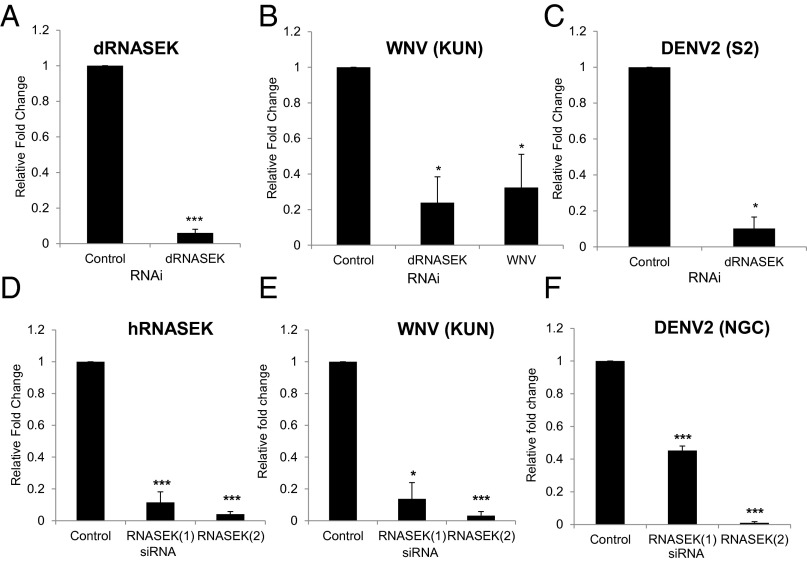
Because we had identified dRNASEK as promoting infection in our genome-wide RNAi screens, we set out to verify this outside of a screening format with independent RNAi reagents. First, we verified that we efficiently depleted dRNASEK, as measured by RT-quantitative PCR (qPCR), compared with control knockdown (Fig. 1A). We challenged these cells with WNV [Kunjin strain (KUN)], as it is closely related to WNV strains circulating globally (but can be used under biosafety level 2 containment), and monitored infection by RT-qPCR and automated microscopy. We found that depletion of dRNASEK significantly attenuated infection of WNV in Drosophila cells (Fig. 1B and Fig. S1 A and B). We extended this study to dengue virus because it is a pandemic threat (two-fifths of the world’s population is at risk, and it is the fastest-spreading tropical disease and arthropod-borne viral pathogen worldwide). We challenged control or RNASEK-depleted cells with dengue virus serotype 2 (DENV2) [S2 or New Guinea C (NGC)] and monitored infection by RT-qPCR or automated microscopy and observed that loss of dRNASEK significantly attenuates infection (Fig. 1C and Fig. S1 A and B). We observed no change in cell number or growth properties of dRNASEK-depleted cells (Fig. S1C).
Fig. 1.
RNASEK promotes flavivirus infection in both Drosophila and human cells. Drosophila DL1 cells were treated with the indicated dsRNAs for 3 d and subsequently infected with WNV (KUN) (MOI 1, 48 h) or DENV2 (S2) (MOI 0.5, 72 h), and RNA was subject to RT-qPCR for dRNASEK (A), WNV (B), or DENV2 (C). Mean ± SEM; n = 3; *P < 0.05, ***P < 0.001. Human U2OS cells were transfected with the indicated siRNAs for 3 d and subsequently infected with WNV (KUN) (MOI 0.5, 24 h) or DENV2 (NGC) (MOI 0.5, 40 h), and RNA was subject to RT-qPCR for hRNASEK (D), WNV (E), or DENV2 (F). Mean ± SEM; n = 3; *P < 0.05, ***P < 0.001.
Fig. S1.
RNASEK promotes flavivirus infection in Drosophila cells. (A) Representative images of Drosophila DL1 cells treated with dsRNA against dRNASEK and incubated for 3 d before infection by WNV (KUN) (MOI 10) or DENV2 (S2) (MOI 5) or DENV2 (NGC) (MOI 5) for 72 h. (B and C) Cells were processed for automated microscopy and automated image analysis with (B) fold change in percentage infection shown or (C) fold change in cell number shown as mean ± SEM; n = 3; *P < 0.05.
RNASEK Promotes Flavivirus Infection in Human Cells.
RNASEK is a highly conserved protein with 51% homology between Drosophila and humans (25). Therefore, we tested whether these flaviviruses require hRNASEK for infection in human cells. We obtained two independent siRNAs against hRNASEK and found that they efficiently deplete hRNASEK in human osteosarcoma cells (U2OS), as measured by RT-qPCR (Fig. 1D). We challenged these cells with WNV (KUN) or DENV2 (NGC) and monitored infection by RT-qPCR or automated microscopy. We found that depletion of hRNASEK significantly attenuates infection of these flaviviruses in human cells (Fig. 1 E and F and Fig. S2 A and B). Again, we observed no change in cell number or growth properties of hRNASEK-depleted cells (Fig. S2C). We also tested a different human cell line, HEK293T cells, and found that depletion of hRNASEK is efficient in these cells and attenuates WNV (KUN) infection (Fig. S3A). Our initial studies monitored infection of WNV at a late point postinfection, so we next tested whether depletion of hRNASEK affects infection at early points. We depleted hRNASEK in U2OS cells and monitored infection by WNV (KUN) 8 hours post infection (hpi), and observed significantly decreased infection at this early time (Fig. S3B).
Fig. S2.
RNASEK promotes flavivirus infection in human cells. (A) Representative images of U2OS cells transfected with either control siRNA or independent siRNAs targeting hRNASEK and, 3 d later, infected with WNV (KUN) (MOI 1) or DENV2 (NGC) (MOI 1) for 24 or 48 h, respectively. (B) Cells were processed for automated microscopy and image analysis with fold change in percentage infection or (C) cell number shown as mean ± SEM; n = 3; *P < 0.05, ***P < 0.001.
Fig. S3.
RNASEK is required early and in multiple cell lines. (A) HEK 293T cells were transfected with the indicated siRNAs. Three days later, cells were infected with WNV (KUN) (MOI 1) for 24 h and processed for RT-qPCR analysis of RNASEK or WNV RNA with mean ± SEM; n > 3; *P < 0.05, **P < 0.005, ***P < 0.001. (B) U2OS cells were transfected with siRNAs and, 3 d later, infected with WNV (KUN) (MOI 0.5) and processed at 8 hpi for RT-qPCR analysis. Results show relative levels of RNASEK mRNA or WNV RNA with mean ± SEM; n = 4; *P < 0.05, **P < 0.005, ***P < 0.001.
RNASEK Is Required for Infection of Diverse pH-Dependent Viruses.
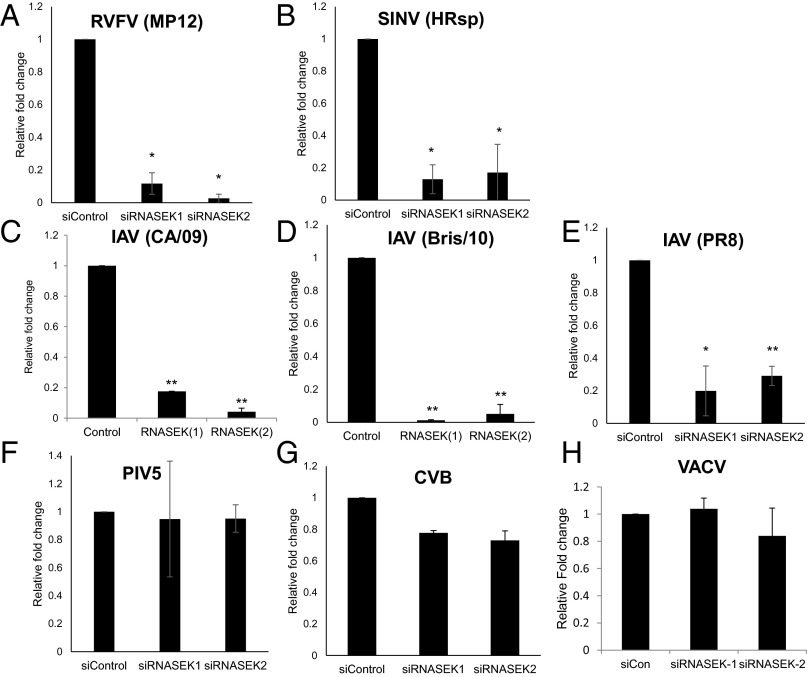
Our initial RNAi screens identified dRNASEK as required for both WNV (NY2000) and RVFV (MP12) infection. Therefore, we tested the role of hRNASEK in RVFV infection of human cells. Indeed, we found that depletion of hRNASEK significantly reduces infection by RVFV (MP12), as measured by RT-qPCR and immunoblot (Fig. 2A and Fig. S4A). Because humans are infected by three families of arthropod-borne viruses, the flaviviruses, the bunyaviruses, and the alphaviruses, and we found that both flaviviruses and bunyaviruses are sensitive to RNASEK, we tested the role of a hRNASEK in alphavirus infection. We used the prototypical alphavirus, Sindbis virus (SINV, HRsp strain), and found that depletion of hRNASEK attenuates infection, as measured by RT-qPCR and immunoblot (Fig. 2B and Fig. S4B). Aside from being arthropod-borne, these viruses all require endocytic uptake and acidification for entry (28–30). Therefore, we tested whether RNASEK was required for infection of influenza virus, an unrelated pH-dependent virus that is not transmitted by insects. Influenza viruses are diverse, causing 3–5 million cases of severe illness and about 250,000–500,000 deaths yearly, and even more during pandemics (31, 32). We tested two antigenically distinct influenza A viruses (IAV), H1N1 (A/Puerto Rico/8/1934 (PR8) and A/California/7/2009 (CA/09), and an H3N2 strain (A/Brisbane/10/2007 (Bris/10). Again, depletion of hRNASEK significantly attenuated infection, as measured by RT-qPCR and automated microscopy (Fig. 2 C–E and Fig. S4C).
Fig. 2.
RNASEK promotes infection of a broad spectrum of RNA viruses in human cells. Human U2OS cells were transfected with either the indicated siRNAs for 3 d and subsequently infected with (A) RVFV (MP12) (MOI 1, 24 h), (B) SINV (HRsp) (MOI 0.5, 24 h), (C) IAV (CA/09) (MOI 0.05, 24 h), (D) IAV (Bris/10) (MOI 0.05, 24 h), (E) IAV (PR8) (MOI 0.05, 2 4 h), (F) PIV5 (MOI 0.1, 24 h), or (G) CVB (MOI 3, 8 h) and processed for RT-qPCR analysis of viral RNA. Mean ± SEM; n = 3; *P < 0.05, **P < 0.005. (H) Human U2OS cells were transfected with either the indicated siRNAs for 3 d and subsequently infected with VACV (MOI 0.5, 8 h) and processed for automated microscopy with the percentage of infected cells quantified. Mean ± SEM; n = 3.
Fig. S4.
RNASEK promotes diverse viral infections in human cells. U2OS cells were transfected with the indicated siRNAs. Three days later, cells were infected with (A) RVFV (MP12) (MOI 0.5) or (B) SINV (HRsp) (MOI 0.5) for 24 h and probed for immunoblot with quantification of three independent experiments shown at right. (C) Three days posttransfection with the indicated siRNAs, U2OS cells were infected with the indicated strains of IAV (MOI 0.5) for 24 h and processed for automated microscopy and image analysis. The percentage of infected cells was quantified and is shown normalized to control. Mean ± SEM; n = 3; *P < 0.05, **P < 0.05.
All these viruses are enveloped and use endocytic mechanisms to access low-pH compartments for entry (22). To determine whether the commonality between these viruses was their dependence on intracellular modes of entry, we tested two viruses that penetrate at the plasma membrane: the paramyxovirus parainfluenza virus 5 (PIV5) and the picornavirus Coxsackie B virus (CVB) (33, 34). We found that depletion of hRNASEK had no effect on the infection of either PIV5 or CVB measured by RT-qPCR (Fig. 2 F and G). We also tested vaccinia virus (VACV), as it is dependent on macropinocytosis for entry (35, 36). We found that RNASEK was dispensable for infection by VACV (Fig. 2H). Therefore, our data suggest that hRNASEK is required for virus infection by viruses that undergo endocytic entry and fuse within internal compartments.
Requirements for RNASEK Can Be Bypassed by Fusion at the Membrane.
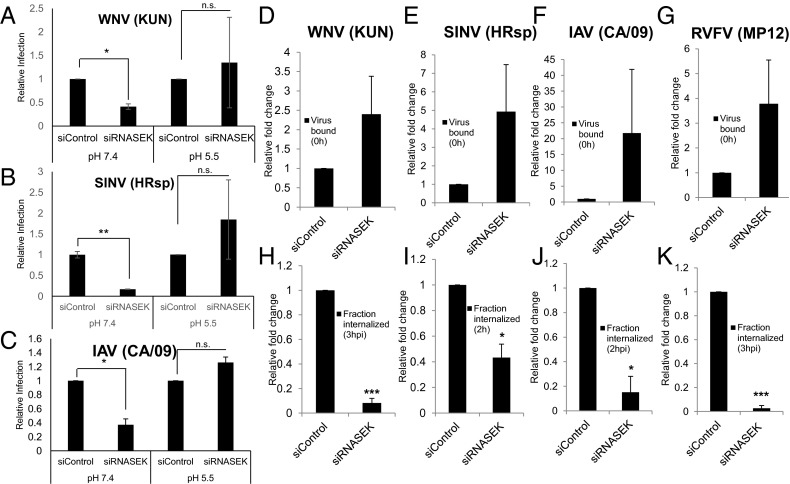
If the requirement for hRNASEK was during intracellular entry, then we reasoned that we may be able to bypass the requirement for RNASEK by inducing virus fusion at the plasma membrane. Many viruses, including WNV, SINV, and IAV, can enter cells at the membrane if the pH in the environment is acidified, allowing envelope triggering and cell fusion (37–40). This has been termed an acid bypass assay, allowing us to test whether RNASEK is required for steps in the entry pathway downstream of cellular attachment. For these studies, we bound virus [WNV (KUN), SINV (HRsp), or IAV (CA/09)] in the cold to siRNA-transfected cells for 1 h. Next, we incubated the cells in either neutral or acidic PBS for 10 min to allow plasma membrane fusion at the low pH. Cells were washed, ammonium chloride was added to block secondary infections, and infection was monitored 24 hpi by automated microscopy. We found that depletion of hRNASEK at neutral pH leads to decreased infection of all three viruses (Fig. 3 A–C, Left). In contrast, under acidic conditions that bypass endocytic requirements, depletion of hRNASEK has no effect on infection (Fig. 3 A–C, Right). These data suggest that hRNASEK is required downstream of cellular attachment during entry. These data also demonstrate that hRNASEK is not required at a step downstream of entry, as infection was normal once the viruses fused at the plasma membrane.
Fig. 3.
RNASEK is required for virus internalization. (A–C) U2OS cells were transfected with indicated siRNA for 72 h and infected with (A) WNV (KUN) (MOI 5), (B) SINV (HRsp) (MOI 50), or (C) IAV (CA/09) (MOI 5) for 1 h at 4 °C. Cells were then treated for 10 min with PBS (pH 5.5 or 7.4), washed, and incubated at 37 °C for 24 h in the presence of ammonium chloride to block spread (added 2 hpi) and processed for automated microscopy. The normalized percentage of infected cells per well for four wells per experiment for three independent experiments is shown. Mean ± SEM; *P < 0.05, **P < 0.01. (D–K) U2OS cells were transfected with the indicated siRNAs for 3 d and infected with (D and H) WNV (KUN) (MOI 5), (E and I) SINV (HRsp) (MOI 5), or (F and J) IAV (CA/09) (MOI 5), or RVFV (MP12) (MOI 5) at 4 °C. Samples were untreated or trypsinized and processed for RT-qPCR at 0 h. The amount of bound virus (total minus signal remaining posttrypsinization) normalized to control is shown (D–G). Alternatively, the cells were shifted to 37 °C for 2–3 h as indicated, trypsinized to remove external virus, and processed for RT-qPCR, and the amount internalized normalized to control is shown (H–K). Mean ± SEM; n = 3; *P < 0.05, **P < 0.01, ***P < 0.001.
Viral Uptake Is Dependent on RNASEK.
Our acid bypass assay suggests that RNASEK is required either for viral uptake or for the acidification of endosomal compartments. Therefore, we set out to determine whether virus internalization was dependent on RNASEK. For these studies, we used a RT-qPCR-based assay to monitor virus binding and internalization of WNV (KUN), SINV (HRsp), IAV (CA/09), and RVFV (MP12). Viruses were bound at 4 °C and then extensively washed to remove unbound virus. Next, we either left the cells untreated or trypsinized the cells to remove bound virus and verified that we successfully removed bound virus (Fig. S5). We quantified the cell-bound virus by subtracting the background signal from the total binding quantified by RT-qPCR and found that RNASEK-depleted cells had no statistically significant effect on binding, (Fig. 3 D–G), although there was a trend toward increased binding. Next, we monitored the amount of virus uptake by binding the viruses at 4 °C and washing unbound virus as earlier, but in this case, we subsequently shifted the cells to 37 °C for 2–3 h to allow internalization of bound virions. After internalization, cells were treated with trypsin to remove viral particles bound to the outside of cells. Here we found that cells lacking hRNASEK are significantly attenuated in their ability to internalize all four viruses (Fig. 3 H–K).
Fig. S5.
Trypsin treatment effectively clears viral particles from the cell surface. U2OS cells were transfected with the control siRNAs for 3 d and infected with WNV (KUN) (MOI 5), SINV (HRsp) (MOI 5), IAV (CA/09) (MOI 5), or RVFV (MP12) (MOI 5) at 4 °C. Samples were then mock treated or treated with trypsin for 3 min, washed twice, and processed for RT-qPCR. Mean ± SEM; n = 3; *P < 0.05, **P < 0.05, ***P < 0.001.
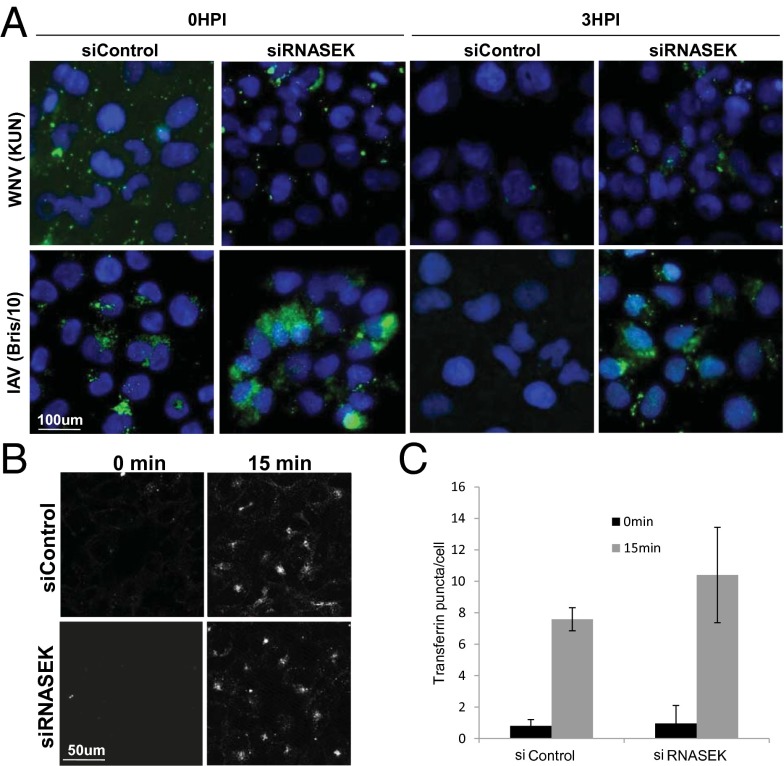
These data suggest that RNASEK is required for the initial stages of internalization from the plasma membrane for a diverse set of viruses that use different receptors for entry. To further verify this hypothesis, we performed a microscopy-based virus uptake assay with WNV (KUN) and IAV (Bris/10) because we have antibodies that efficiently recognize the viral glycoproteins for these viruses. We incubated control or RNASEK-depleted cells with virions at 4 °C, washed extensively, and fixed the cells (0 hpi) or shifted the cells to 37 °C for 3 h to allow internalization (3 hpi) and fixed the cells. Cells were then processed for microscopy in the absence of permeabilization to exclusively visualize extracellular virions, using antibodies against the viral glycoproteins. We found that there were similar levels of virus binding at 0 hpi comparing control with RNASEK-depleted cells for either WNV or IAV (Fig. 4A). At 3 hpi, we observed the loss of external virus staining in control cells, whereas extracellular virions were retained in RNASEK-depleted cells for both WNV and IAV infections (Fig. 4A). This was quantified in Fig. S6. Furthermore, to verify that the loss in signal observed in the control siRNA-treated cells at 3 hpi was a result of internalization, we compared unpermeabilized with permeabilized cells and found that we readily detected viral antigens inside the cells on permeabilization (Fig. S6). Therefore, in this microscopy-based assay, we observed that RNASEK has no effect on viral attachment, but is required for virus uptake.
Fig. 4.
Viral uptake, but not transferrin uptake, is disrupted in RNASEK-depleted cells. (A) siRNA-transfected U2OS cells were incubated with WNV (KUN) (MOI 15) or IAV (Bris/10) (MOI 15) for 1 h at 4 °C, washed, and either immediately processed or incubated for 3 h at 37 °C for immunofluorescence under nonpermeabilized conditions. (B) siRNA-transfected U2OS cells were incubated with 50 μg/mL fluorescently labeled transferrin in the cold for 1 h and shifted to 37 °C for 15 min before an acid wash to quench extracellular transferrin and subsequent processing for confocal microscopy. (C) Automated image analysis quantified the number of puncate per cell and was normalized to control at 0 min. Mean ± SEM; n = 3.
Fig. S6.
RNASEK is required for viral internalization. (A and B) Quantification of microscopy virus uptake assay in Fig. 4A for (A) WNV (KUN) or (B) IAV (Bris/10). Mean ± SEM; n = 3; ***P < 0.001. (C and D) U2OS cells were transfected with the control or RNASEK siRNAs for 3 d and inoculated with either (C) WNV (KUN) or (D) IAV (Bris/10) at MOI 15 for 3 h. Samples were either unpermeabilized or permeabilized with Triton X-100 during processing for immunofluorescence confocal microscopy.
RNASEK Is Dispensable for General Uptake.
Because the uptake of WNV, SINV, and IAV at the plasma membrane is clathrin-dependent (2), we assayed whether clathrin-mediated endocytosis is generally disrupted by RNASEK depletion. If RNASEK was required for clathrin-mediated endocytosis, it would not be an ideal drug target because clathrin-mediated endocytosis is required for essential cellular processes. A classical assay for clathrin-dependent endocytosis entails monitoring of transferrin uptake (41). Transferrin binds to the transferrin receptor, which requires clathrin-dependent endocytosis for internalization. Therefore, we assayed fluorescent transferrin uptake in control and RNASEK-depleted cells. We treated cells with Alexa594-Transferrin for 15 min and quenched extracellular transferrin, using an acid wash (42). We observed no difference in uptake between control and RNASEK-depleted cells (Fig. 4 B and C). As a positive control, we treated cells with the dynamin inhibitor dynasore, which completely blocks transferrin uptake (Fig. S7). Therefore, all clathrin-dependent endocytosis is not dependent on RNASEK. Macropinocytosis is another major mechanism for viral uptake. VACV entry is dependent on this process, and in some situations, RVFV and IAV can enter cells via macropinocytosis (8). Because VACV infection is RNASEK-independent, we expected that RNASEK is not required for macropinocytosis, but nevertheless, we tested whether the uptake of dextran was affected by the loss of RNASEK. We treated control or RNASEK-depleted cells with fluorescently labeled dextran for 30 min and observed no defect in the uptake in RNASEK-deficient cells, whereas treatment with the macropinocytosis inhibitor 5-(N-ethyl-N-isopropyl) amiloride (EIPA) blocked uptake (Fig. S8). These data suggest that RNASEK is not required for all clathrin-dependent endocytosis or macropinocytosis.
Fig. S7.

Dynasore blocks transferrin uptake. (A) U2OS cells were treated with Dynasore (50 μM) for 1 h before incubation with 50 μg/mL Alexa594-transferrin in the cold for 1 h (0 min) or shifted to 37 °C for 30 min and processed for confocal microscopy. (B) Automated image analysis was used to quantify the number of punctae per cell and normalized to 0 min for 10 independent fields. Mean ± SEM; n = 3.
Fig. S8.
RNASEK is not required for dextran uptake. (A) U2OS cells were siRNA-transfected for 3 d and incubated with vehicle or EIPA (0.1 μM) for 1 h before incubation with 15 μg/mL Texas Red-dextran (70 kD) in the cold for 1h (0 min) and shifted to 37 °C for 30 min and processed for confocal microscopy. (B) Automated image analysis was used to quantify the number of punctae per cell normalized to 0 min for 10 independent fields. Mean ± SEM; n = 3. (C) U2OS cells were siRNA transfected for 3 d and incubated with 250 μg/mL FITC-dextran (70 kD) for 0 or 30 min. Next, 12.5 μg/mL Alexa647-wheat germ agglutinin (WGA) was added before fixation and processing for confocal microscopy. Images are representative of two independent experiments.
Discussion
By mining our RNAi screening data (18, 19), we found that RNASEK was potentially required for infection of WNV and RVFV in insect cells. Given that RNASEK is conserved but poorly characterized, we set out to dissect the role of RNASEK in viral infection. We found that RNASEK promotes viral uptake of a diverse panel of viruses including flaviviruses, alphaviruses, bunyaviruses, and orthomyxoviruses. These viruses all enter cells using endocytic routes and are dependent on acidification for entry. We tested whether viruses that fuse at the membrane, including a paramyxovirus and a picornavirus, were dependent on RNASEK, and found that they were not. We also found that VACV, which enters by macropinocytosis, was not dependent on RNASEK. By performing a series of assays to narrow down the requirement, we found that RNASEK promotes the earliest steps of virus uptake but has no effect on virus binding to cells. RNASEK was dispensable once viruses delivered their payload to the cytoplasm, suggesting it has no additional roles in viral infection downstream of entry.
The panel of viruses sensitive to RNASEK uses disparate receptors and endocytic pathways for entry. WNV, DENV, SINV, and IAV are thought to be largely dependent on clathrin-mediated endocytosis (2). RVFV and IAV are thought to use macropinocytosis at least under some conditions; however, VACV, which is dependent on macropinocytosis, was RNASEK-independent (7, 8, 12). Therefore, our data suggest that RNASEK selectively affects clathrin-mediated endocytosis of viruses. Thus, we tested whether RNASEK is required for the uptake of other cargo dependent on clathrin-mediated endocytosis or macropinocytosis. As expected, we found that RNASEK was dispensable for the macropinocytic uptake of dextran. However, surprisingly, we found that RNASEK was dispensable for the uptake of the classic cargo transferrin, which is dependent on clathrin-mediated endocytosis. Therefore, RNASEK is seemingly uniquely required for the internalization of viruses, and not canonical cargo. Such characteristics differentiate RNASEK from other recently identified factors, such as FUZ and TER94, which have been shown to be required for the uptake of both alphaviruses and cargo (38, 43).
RNASEK is a ubiquitous protein that arose in metazoans. Most canonical endocytosis genes are conserved in yeast, further suggesting that RNASEK has more specialized functions. RNASEK is thought to encode a protein with two putative transmembrane domains that may anchor the protein at the plasma membrane for its function. How RNASEK promotes the internalization of virions, but not other cargo, such as transferrin, is unclear, but there are a number of possibilities. First, it may be required for uptake of large cargo. The typical biomolecule internalized by clathrin-mediated endocytosis, including transferrin, is small, whereas viruses are much larger. Second, it may be required for signaling events that promote internalization. After attachment and receptor engagement, many viruses induce signaling cascades that promote entry (3, 44). This is most clearly established for viruses that engage receptors that induce macropinocytosis for uptake, including IAV (3, 8, 9, 44–46). Informatic analysis suggests that RNASEK has an SH2 domain that may be interacting with signaling events engaged by virus binding at the membrane. However, many endogenous cargoes also induce signaling events including transferrin binding to its receptor (47, 48).
Importantly, we found that RNASEK is not required for clathrin-mediated endocytosis or macropinocytosis of transferrin or dextran, respectively. Therefore, targeted inhibition of RNASEK is not expected to interfere with these normal cellular functions. Thus, RNASEK may present a previously unknown target for pan-antiviral therapeutic interventions because major viral pathogens such as influenza and dengue virus are dependent on RNASEK for infection.
Materials and Methods
Cell Lines, Viruses, Antibodies, and Reagents.
Human cells and Drosophila DL1 cells were grown and maintained as previously described (49). The WNV-KUNV isolate 550 (CH16532) was a gift of R. Tesh and propagated as previously described (50). DENV2-S2 was provided by M. Garcia-Blanco and propagated as described (51). GFP-expressing Sindbis virus, a gift from R. Hardy, University of Indiana, was propagated as detailed previously. Coxsackie B virus was provided by J. Bergelson, Children's Hospital of Pennsylvania (CHOP), and influenza stocks [H1N1 (California/7/2009), H1N1 (PR8), and H3N2 (Brisbane/10/2007)] were grown at the Wistar Institute. DENV2 (New Guinea C) and PIV5 stocks were purchased (ATCC) and propagated in BHK (DENV2) or Vero (PIV5) cells. VACV-YFP was a gift of S. Isaacs, University of Pennsylvania. Monoclonal antibodies against flavivirus NS1 and E proteins (4G2 and 9NS1, respectively) were provided by M. Diamond (Washington University). RVFV GN monoclonal antibody (4D4) was provided by R. Doms (CHOP). Influenza A antibody to NP was provided by BEI Resources (NR-4282). Other antibodies were obtained as follows: GFP (Santa Cruz Biotechnology [SCBT]), beta-actin (SCBT), and tubulin (Sigma). Fluorescent secondary antibodies and transferrin were obtained from Invitrogen, and Horse radish peroxidase-conjugated secondary antibodies were from Amersham. Other reagents were from Sigma.
RNAi.
Drosophila cells were treated with dsRNA for 3 d, as described (52). U2OS cells or 293T cells were transfected with 30 nM either negative control siRNA (Ambion) or one of two independent siRNAs targeting RNASEK (Ambion s54102, s45103), both of which target the major isoform of RNASEK, but only s54102 targets the poorly conserved isoform 2, using HiPerfect according to the manufacturers protocol (Qiagen) and infected at 72 hours post transfection (hpt), as indicated.
RT-qPCR Assays.
Total RNA was extracted using TRIzol reagent (Invitrogen). cDNA was generated using random hexamers to prime reverse transcription reactions using M-MLV reverse transcriptase. qPCR was performed with the cDNA, using Power SYBR Green PCR Master Mix and a StepOne Plus RT-PCR system (Applied Biosystems). Reactions were initially run at initial 95 °C for 5 min and then 40 cycles of 95 °C for 20 s, 52 °C for 30 s, and 72 °C for 30 s Analysis by ΔΔCT method. Relative copy numbers were generated by normalizing to cells to rp49 for Drosophila cells or 28S RNA for human cells and compared with control siRNA. Primers are described in Table S1.
Table S1.
List of primers used
| Target | Forward primer | Reverse primer |
| dRP49 | AAGAAGCGCACCAAACACTTCATC | TCTGTTGTCGATACCCTTCGGCTT |
| dRNASEK | CCCTCATTGAGGACTTACCT | GTAGGCATTCTGATTGTATGCT |
| hRNASEK | TCATGTTGATAATGCTCGGA | AATCTTTCTCCGTGAAGGG |
| h28s rRNA | GGGTGGTAAACTCCATCTAAGG | GCCCTCTTGAACTCTCTCTTC |
| RVFV N | CAAGCAGTGGACCGCAATGAGA | GGGCTTGTTGCCACGAGTTAGA |
| DENV2 | TGAGGACTACATGGGCTCTG | AAACCTCCCTGGATTTCCTT |
| WNV | CCTGTGTGAGCTGACAAACTTAGT | GCGTTTTAGCATATTGACAGCC |
| IAV M | TTAGGATTTGTGTTCACGCTCACCG | CCAGCCATTTGCTCCATAGCCTTG |
| SINV NSP1 | GCTGAAACACCATCGCTCTGCTTT | TGGTGTCGAAGCCAATCCAGTACA |
Acid Bypass Assay.
U2OS cells were transfected with siRNAs and infected with SINV (HRsp) (multiplicity of infection [MOI] 5), WNV (KUN) (MOI 5), or IAV (CA/09) (MOI 5) in the cold for 1 h to allow binding. Cells were then washed and incubated for 10 min in either PBS at pH 5.5 or PBS at pH 7.2. Culture media was then replaced and cells were incubated at 37 °C with 20 mM NH4Cl and processed for automated microscopy at 24 hpi. Cells were fixed with PBS/4% (vol/vol) formaldehyde for 15 min and washed 3× with PBS/1% Triton X-100. Cells were processed and imaged, using an automated microscope (ImageXpress Micro) and automated analysis, as previously described (53). Four wells per condition with four sites per well were collected and quantified (MetaXpress).
Viral Entry Assays.
siRNA-transfected cells were incubated with WNV (KUN, MOI 5 for qPCR, MOI 15 for microscopy), SIN (HRsp, MOI 5), IAV (H1N1, MOI 5 for qPCR assay, MOI 15 for microscopy), or RVFV (MP12, MOI 5) in the cold (t = 0) or incubated at 37 °C for 2–3 h (t = 2 or 3 h). WNV (KUN) and RVFV (MP12) were spinoculated onto the cells for the qPCR assay. For the qPCR assay, cells were also left untreated or treated with trypsin for 3 min, as indicated. For the microscopy assay, cells were washed twice with PBS at the end of each point, fixed in PBS/4% (vol/vol) formaldehyde (15 min, room temperature), and blocked in PBS/2% (wt/vol) BSA and processed for confocal microscopy using anti-E for WNV or anti-HA for IAV (53).
Transferrin Uptake Assay.
siRNA-transfected U2OS cells were treated with 50 µg/mL Alexa594-labeled transferrin for 1h at 4 °C and then incubated at 37 °C for 15 min. After washing, cells were placed in acid strip buffer, as described, to monitor intracellular transferrin (42). Automated image analysis was used to quantify the number of punctae per cell of 10 fields per condition in three independent experiments.
Supplementary Material
Acknowledgments
We thank members of the S.C. laboratory for helpful discussions and technical advice throughout. We thank BEI. This work was supported by National Institutes of Health Grants R01AI074951 (to S.C.), U54AI057168 (to S.C.), R21AI103441 (to S.C.), R01AI095500 (to S.C.), R01AI113047 (to S.E.H.), and R01AI108686 (to S.E.H.). S.C. is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424098112/-/DCSupplemental.
References
- 1.Mercer J, Helenius A. Virus entry by macropinocytosis. Nat Cell Biol. 2009;11(5):510–520. doi: 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- 2.Mercer J, Schelhaas M, Helenius A. Virus entry by endocytosis. Annu Rev Biochem. 2010;79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 3.Yamauchi Y, Helenius A. Virus entry at a glance. J Cell Sci. 2013;126(Pt 6):1289–1295. doi: 10.1242/jcs.119685. [DOI] [PubMed] [Google Scholar]
- 4.Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009;5(4):e1000394. doi: 10.1371/journal.ppat.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- 6.Rust MJ, Lakadamyali M, Zhang F, Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat Struct Mol Biol. 2004;11(6):567–573. doi: 10.1038/nsmb769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Schaar HM, et al. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 2008;4(12):e1000244. doi: 10.1371/journal.ppat.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercer J, Helenius A. Gulping rather than sipping: Macropinocytosis as a way of virus entry. Curr Opin Microbiol. 2012;15(4):490–499. doi: 10.1016/j.mib.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 9.de Vries E, et al. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 2011;7(3):e1001329. doi: 10.1371/journal.ppat.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edinger TO, Pohl MO, Stertz S. Entry of influenza A virus: Host factors and antiviral targets. J Gen Virol. 2014;95(Pt 2):263–277. doi: 10.1099/vir.0.059477-0. [DOI] [PubMed] [Google Scholar]
- 11.de Boer SM, et al. Acid-activated structural reorganization of the Rift Valley fever virus Gc fusion protein. J Virol. 2012;86(24):13642–13652. doi: 10.1128/JVI.01973-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filone CM, et al. Rift valley fever virus infection of human cells and insect hosts is promoted by protein kinase C epsilon. PLoS ONE. 2010;5(11):e15483. doi: 10.1371/journal.pone.0015483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harmon B, et al. Rift Valley fever virus strain MP-12 enters mammalian host cells via caveola-mediated endocytosis. J Virol. 2012;86(23):12954–12970. doi: 10.1128/JVI.02242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrifield CJ, Kaksonen M. 2014. Endocytic accessory factors and regulation of clathrin-mediated endocytosis. Cold Spring Harb Perspect Biol 3;6(11):a016733. [DOI] [PMC free article] [PubMed]
- 15.Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- 16.Kilcher S, Mercer J. Next generation approaches to study virus entry and infection. Curr Opin Virol. 2014;4:8–14. doi: 10.1016/j.coviro.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Panda D, Cherry S. Cell-based genomic screening: Elucidating virus-host interactions. Curr Opin Virol. 2012;2(6):784–792. doi: 10.1016/j.coviro.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasunaga A, et al. Genome-wide RNAi screen identifies broadly-acting host factors that inhibit arbovirus infection. PLoS Pathog. 2014;10(2):e1003914. doi: 10.1371/journal.ppat.1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopkins KC, et al. A genome-wide RNAi screen reveals that mRNA decapping restricts bunyaviral replication by limiting the pools of Dcp2-accessible targets for cap-snatching. Genes Dev. 2013;27(13):1511–1525. doi: 10.1101/gad.215384.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suthar MS, Diamond MS, Gale M., Jr West Nile virus infection and immunity. Nat Rev Microbiol. 2013;11(2):115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- 21.Ikegami T, Makino S. The pathogenesis of Rift Valley fever. Viruses. 2011;3(5):493–519. doi: 10.3390/v3050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh M, Helenius A. Virus entry: Open sesame. Cell. 2006;124(4):729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirooz SD, et al. UVRAG is required for virus entry through combinatorial interaction with the class C-Vps complex and SNAREs. Proc Natl Acad Sci USA. 2014;111(7):2716–2721. doi: 10.1073/pnas.1320629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brinton MA. Replication cycle and molecular biology of the West Nile virus. Viruses. 2014;6(1):13–53. doi: 10.3390/v6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Economopoulou MA, Fragoulis EG, Sideris DC. Molecular cloning and characterization of the human RNase kappa, an ortholog of Cc RNase. Nucleic Acids Res. 2007;35(19):6389–6398. doi: 10.1093/nar/gkm718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiritsi MN, Fragoulis EG, Sideris DC. Essential cysteine residues for human RNase κ catalytic activity. FEBS J. 2012;279(7):1318–1326. doi: 10.1111/j.1742-4658.2012.08526.x. [DOI] [PubMed] [Google Scholar]
- 27.Rampias TN, Fragoulis EG, Sideris DC. Genomic structure and expression analysis of the RNase kappa family ortholog gene in the insect Ceratitis capitata. FEBS J. 2008;275(24):6217–6227. doi: 10.1111/j.1742-4658.2008.06746.x. [DOI] [PubMed] [Google Scholar]
- 28.Smit JM, Moesker B, Rodenhuis-Zybert I, Wilschut J. Flavivirus cell entry and membrane fusion. Viruses. 2011;3(2):160–171. doi: 10.3390/v3020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sánchez-San Martín C, Liu CY, Kielian M. Dealing with low pH: Entry and exit of alphaviruses and flaviviruses. Trends Microbiol. 2009;17(11):514–521. doi: 10.1016/j.tim.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guu TS, Zheng W, Tao YJ. Bunyavirus: Structure and replication. Adv Exp Med Biol. 2012;726:245–266. doi: 10.1007/978-1-4614-0980-9_11. [DOI] [PubMed] [Google Scholar]
- 31.Anonymous; Centers for Disease Control and Prevention (CDC) Estimates of deaths associated with seasonal influenza --- United States, 1976-2007. MMWR Morb Mortal Wkly Rep. 2010;59(33):1057–1062. [PubMed] [Google Scholar]
- 32.World Health Organization 2003 Influenza. Available at www.who.int/mediacentre/factsheets/2003/fs211/en/. Accessed November 3, 2014.
- 33.Bergelson JM, Coyne CB. Picornavirus entry. Adv Exp Med Biol. 2013;790:24–41. doi: 10.1007/978-1-4614-7651-1_2. [DOI] [PubMed] [Google Scholar]
- 34.Chang A, Dutch RE. Paramyxovirus fusion and entry: Multiple paths to a common end. Viruses. 2012;4(4):613–636. doi: 10.3390/v4040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercer J, et al. Vaccinia virus strains use distinct forms of macropinocytosis for host-cell entry. Proc Natl Acad Sci USA. 2010;107(20):9346–9351. doi: 10.1073/pnas.1004618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandgren KJ, et al. A differential role for macropinocytosis in mediating entry of the two forms of vaccinia virus into dendritic cells. PLoS Pathog. 2010;6(4):e1000866. doi: 10.1371/journal.ppat.1000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao M, Kielian M. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus membrane fusion. J Cell Biol. 2005;171(1):111–120. doi: 10.1083/jcb.200507075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panda D, et al. Genome-wide RNAi screen identifies SEC61A and VCP as conserved regulators of Sindbis virus entry. Cell Reports. 2013;5(6):1737–1748. doi: 10.1016/j.celrep.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stauffer S, et al. Stepwise priming by acidic pH and a high K+ concentration is required for efficient uncoating of influenza A virus cores after penetration. J Virol. 2014;88(22):13029–13046. doi: 10.1128/JVI.01430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson BS, et al. A therapeutic antibody against west nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog. 2009;5(5):e1000453. doi: 10.1371/journal.ppat.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayle KM, Le AM, Kamei DT. The intracellular trafficking pathway of transferrin. Biochim Biophys Acta. 2012;1820(3):264–281. doi: 10.1016/j.bbagen.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu JJ, Ng ML. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J Virol. 2004;78(19):10543–10555. doi: 10.1128/JVI.78.19.10543-10555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ooi YS, Stiles KM, Liu CY, Taylor GM, Kielian M. Genome-wide RNAi screen identifies novel host proteins required for alphavirus entry. PLoS Pathog. 2013;9(12):e1003835. doi: 10.1371/journal.ppat.1003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greber UF. Signalling in viral entry. Cell Mol Life Sci. 2002;59(4):608–626. doi: 10.1007/s00018-002-8453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eierhoff T, Hrincius ER, Rescher U, Ludwig S, Ehrhardt C. The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 2010;6(9):e1001099. doi: 10.1371/journal.ppat.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meertens L, et al. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe. 2012;12(4):544–557. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao H, Chen J, Krueger EW, McNiven MA. SRC-mediated phosphorylation of dynamin and cortactin regulates the “constitutive” endocytosis of transferrin. Mol Cell Biol. 2010;30(3):781–792. doi: 10.1128/MCB.00330-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelkmans L, et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005;436(7047):78–86. doi: 10.1038/nature03571. [DOI] [PubMed] [Google Scholar]
- 49.Cherry S, Perrimon N. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat Immunol. 2004;5(1):81–87. doi: 10.1038/ni1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanna SL, et al. N-linked glycosylation of west nile virus envelope proteins influences particle assembly and infectivity. J Virol. 2005;79(21):13262–13274. doi: 10.1128/JVI.79.21.13262-13274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sessions OM, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458(7241):1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cherry S. RNAi screening for host factors involved in viral infection using Drosophila cells. Methods Mol Biol. 2011;721:375–382. doi: 10.1007/978-1-61779-037-9_23. [DOI] [PubMed] [Google Scholar]
- 53.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30(4):588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]