Significance
Amyotrophic lateral sclerosis (ALS) is a devastating disorder inevitably resulting in paralysis and death. Mutations in genes encoding RNA binding proteins cause familial ALS, but wild-type (WT) versions of these proteins accumulate in sporadic disease. In this study, we identified and characterized human up-frameshift protein 1 (hUPF1) as a potent modifier of neurodegeneration in ALS models. hUPF1 dramatically reduced toxicity in primary mammalian neurons expressing WT and disease-associated mutant RNA binding proteins, demonstrating efficacy in familial and sporadic disease models, and verifying the existence of a conserved neurotoxic pathway targeted by hUPF1. The work has fundamental implications for ALS disease mechanisms and offers a novel perspective on effective therapies generalizable to both sporadic and familial disease.
Keywords: ALS, FTD, neurodegeneration, RNA binding proteins, RNA decay
Abstract
Over 30% of patients with amyotrophic lateral sclerosis (ALS) exhibit cognitive deficits indicative of frontotemporal dementia (FTD), suggesting a common pathogenesis for both diseases. Consistent with this hypothesis, neuronal and glial inclusions rich in TDP43, an essential RNA-binding protein, are found in the majority of those with ALS and FTD, and mutations in TDP43 and a related RNA-binding protein, FUS, cause familial ALS and FTD. TDP43 and FUS affect the splicing of thousands of transcripts, in some cases triggering nonsense-mediated mRNA decay (NMD), a highly conserved RNA degradation pathway. Here, we take advantage of a faithful primary neuronal model of ALS and FTD to investigate and characterize the role of human up-frameshift protein 1 (hUPF1), an RNA helicase and master regulator of NMD, in these disorders. We show that hUPF1 significantly protects mammalian neurons from both TDP43- and FUS-related toxicity. Expression of hUPF2, another essential component of NMD, also improves survival, whereas inhibiting NMD prevents rescue by hUPF1, suggesting that hUPF1 acts through NMD to enhance survival. These studies emphasize the importance of RNA metabolism in ALS and FTD, and identify a uniquely effective therapeutic strategy for these disorders.
Amyotrophic lateral sclerosis (ALS) is a progressive and lethal motor neuron disease that most often arises sporadically, but can be inherited in 10–15% of patients (1). Many of the genes implicated in familial ALS (fALS) encode RNA-binding proteins, including fused in sarcoma (FUS), transactive response element DNA/RNA-binding protein of 43 kDa (TDP43), and heteronuclear ribonuclear proteins (hnRNPs) (2), emphasizing RNA-based toxicity as a fundamental mechanism contributing to motor neuron degeneration in ALS.
More than 40 different mutations in the TDP43 gene (TARDBP) have now been associated with fALS (3). Disease-associated mutations in TARDBP affect the turnover (4), amount (5), and subcellular localization of TDP43 (6, 7), in many cases resulting in cytoplasmic TDP43 inclusions. Affected neurons in sporadic ALS (sALS) exhibit identical inclusions containing wild-type (WT) TDP43 (8), and WT TDP43 accumulation causes neurodegeneration in cellular and animal models (6, 9, 10), providing a pathogenic link between fALS and sALS. FUS mutations have also been linked to fALS (11, 12). Unlike TDP43, FUS-related pathology is limited to fALS due to FUS mutations (13). FUS and TDP43 bind largely nonoverlapping RNA targets (14), leading to the unexpected conclusion that, despite their homology and involvement in fALS, TDP43 and FUS have distinct roles in RNA metabolism.
TDP43 regulates its own expression through a negative feedback loop (15), but the mechanism by which it does so remains unclear. Conflicting data suggest that TDP43 autoregulation involves nonsense-mediated decay (NMD) (16) or exosome-mediated degradation (15). Excess TDP43 enhances splicing in the TARDBP 3′UTR, potentially targeting the transcript for NMD, whereas deficiencies in human up-frameshift protein 1 (hUPF1), an essential component of NMD (17), result in elevated TARDBP mRNA levels (16). As with TDP43, FUS also binds to its own pre-mRNA (14), reducing exon 7 inclusion and shifting the translational reading frame so that a premature termination codon appears in exon 8 (18). The resulting transcript is targeted for NMD, indicating that NMD might be a conserved mechanism for TDP43 and FUS regulation.
ECM2, a yeast homolog of hUPF1, and hUPF1 itself demonstrated cytoprotective properties in a yeast model of ALS involving FUS overexpression (19). We wondered if hUPF1 could prevent TDP43-related toxicity, and whether it was capable of doing so in mammalian neurons. Here, we show that hUPF1 can indeed rescue neurons from both FUS- and TDP43-associated toxicity through an NMD-dependent mechanism.
Results
hUPF1 Improves Survival in Neuronal Models of ALS.
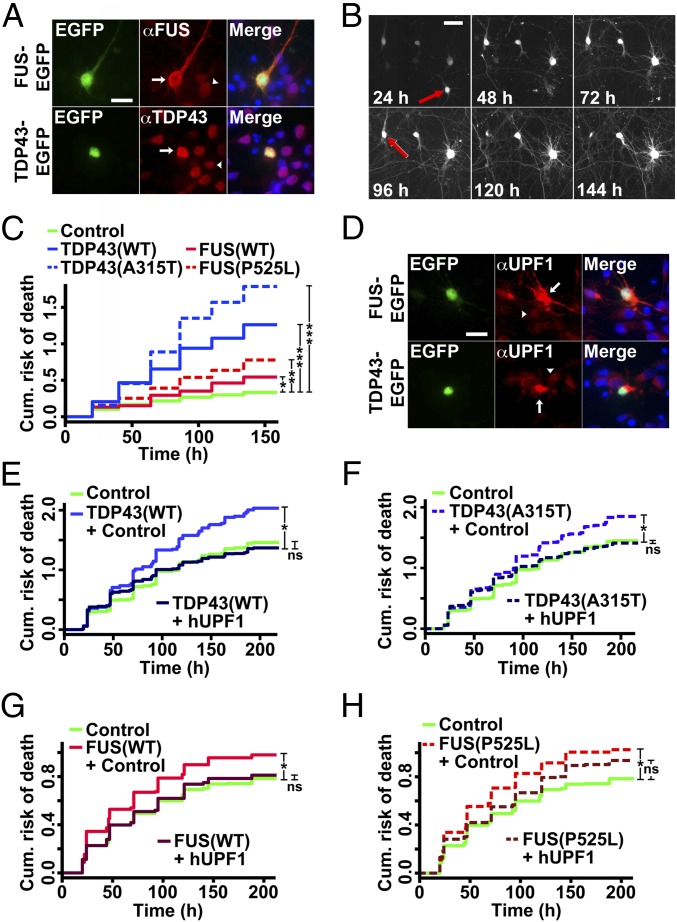
Primary rodent cortical neurons were transfected with WT TDP43 or FUS, or fALS-associated mutant versions of each protein. Each was fused at the carboxyl terminus to the red fluorescent protein, mApple, or enhanced green fluorescent protein (EGFP), facilitating in situ assessment of protein expression in living neurons at a single-cell level (Fig. 1A). Transfected neurons exhibited a 2-fold increase in anti-TDP43 or -FUS antibody reactivity by quantitative immunocytochemistry (ICC) (Fig. S1). We then imaged the neurons with a fully automated longitudinal fluorescence microscopy (LFM) system (6, 20) (Fig. 1B). Neurons were cotransfected with an untagged, diffusely localized fluorophore, enabling the determination of cell death by dissolution of the soma or loss of fluorescence (red arrows, Fig. 1B). In previous studies, these criteria proved to be at least as sensitive as annexin-V staining or methods based on the detection of intracellular enzymes (21). Kaplan–Meier survival plots (Fig. S1), hazard plots (Fig. 1C) depicting the cumulative risk of death, and hazard ratios (HRs) representing the relative risk of death were calculated for each population using Cox hazards analysis. We noted significant increases in the risk of death for neurons expressing WT and mutant TDP43 or FUS, in comparison with control neurons (Fig. 1C). Interestingly, WT and mutant TDP43 appeared to be substantially more toxic than WT and mutant FUS, consistent with recent studies demonstrating the stark dose-dependent neurotoxicity of TDP43 (4).
Fig. 1.
hUPF1 rescues neuron loss in ALS models. (A) Primary neurons transfected with TDP43-EGFP and FUS-EGFP were probed using anti-TDP43 and -FUS antibodies, demonstrating selective overexpression in transfected (arrow) vs. untransfected (arrowhead) neurons. (B) Time of death, represented by the last time the cell was observed alive (red arrows), was used to create cumulative hazard plots (C) depicting risk of death over time. *P < 0.05, **P < 0.0001, ***P ≤ 1 × 10−10, by Cox hazards analysis. n = 98–139 neurons per genotype. (D) Immunocytochemistry using an anti-UPF1 antibody confirmed UPF1 overexpression in transfected (arrow) vs. untransfected (arrowhead) neurons. (E–H) The survival of neurons coexpressing hUPF1 and TDP43(WT) (E), TDP43(A315T) (F), FUS(WT) (G), or FUS(P525L) (H) was determined using LFM. In E and F, n = 1,150–1,205 neurons per genotype; *P < 1 × 10−9. In G and H, n = 312–595 neurons per genotype; *P < 0.01. ns, not significant, by Cox hazards analysis. Results pooled from three or more independent experiments. (Scale bars in A, B, and D, 50 µm.)
Fig. S1.
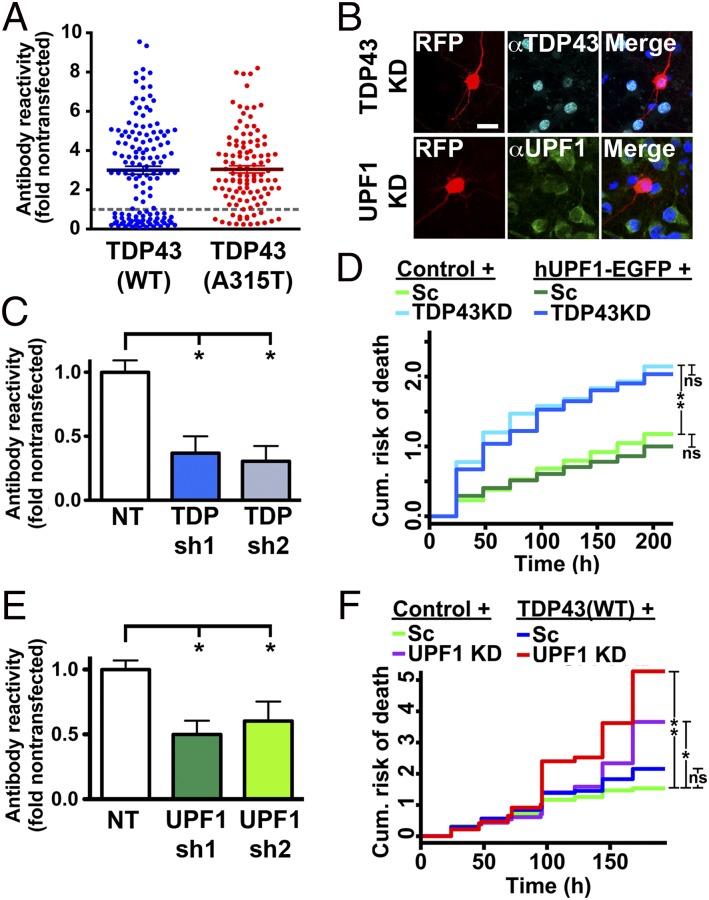
A primary neuron model of ALS. Primary rodent cortical neurons were dissected then transfected with plasmids encoding mApple or mApple-tagged TDP43 or FUS variants on day 4 in vitro. (A) Neurons transfected with TDP43 or FUS displayed an approximate 2-fold mean increase in antibody reactivity, as measured by quantitative fluorescence microscopy. n = 316–433 neurons per genotype. Error bars, ± SEM. (B) Survival of transfected neurons was followed by LFM as described in the text, and survival was plotted using Kaplan–Meier analysis. *P < 0.05, **P < 0.0001, ***P < 1 × 10−10, by Cox hazards analysis. n = 98–139 neurons per genotype, pooled from 8 wells each.
We next asked if hUPF1 could improve survival in primary neurons expressing TDP43 and FUS. We coexpressed hUPF1 in neurons transfected with WT and mutant TDP43 or FUS (Fig. 1D), and confirmed an approximate 3-fold increase in anti-UPF1 antibody reactivity in transfected cells (Fig. S2). We then tracked neuronal survival using LFM (Fig. 1 E–H). Coexpression of hUPF1 had a striking effect on the toxicity of WT and mutant TDP43, effectively reducing the risk of death by ∼50% in neurons transfected with TDP43(WT) and ∼40% in those expressing mutant (A315T) TDP43. In neurons expressing FUS(WT), hUPF1 transfection reduced the risk of death by ∼30%, whereas in cells expressing mutant (P525L) FUS, hUPF1 improved survival by ∼20%. Thus, hUPF1 potently prevents cell death due to WT and mutant TDP43 or FUS in mammalian neurons, extending prior results from yeast (19) and suggesting that hUPF1’s effects are conserved across highly divergent species. Moreover, the data imply that TDP43 and FUS act through at least one shared pathway and, importantly, modulation of this pathway by hUPF1 counteracts the toxicity of disease-associated TARDBP and FUS mutations.
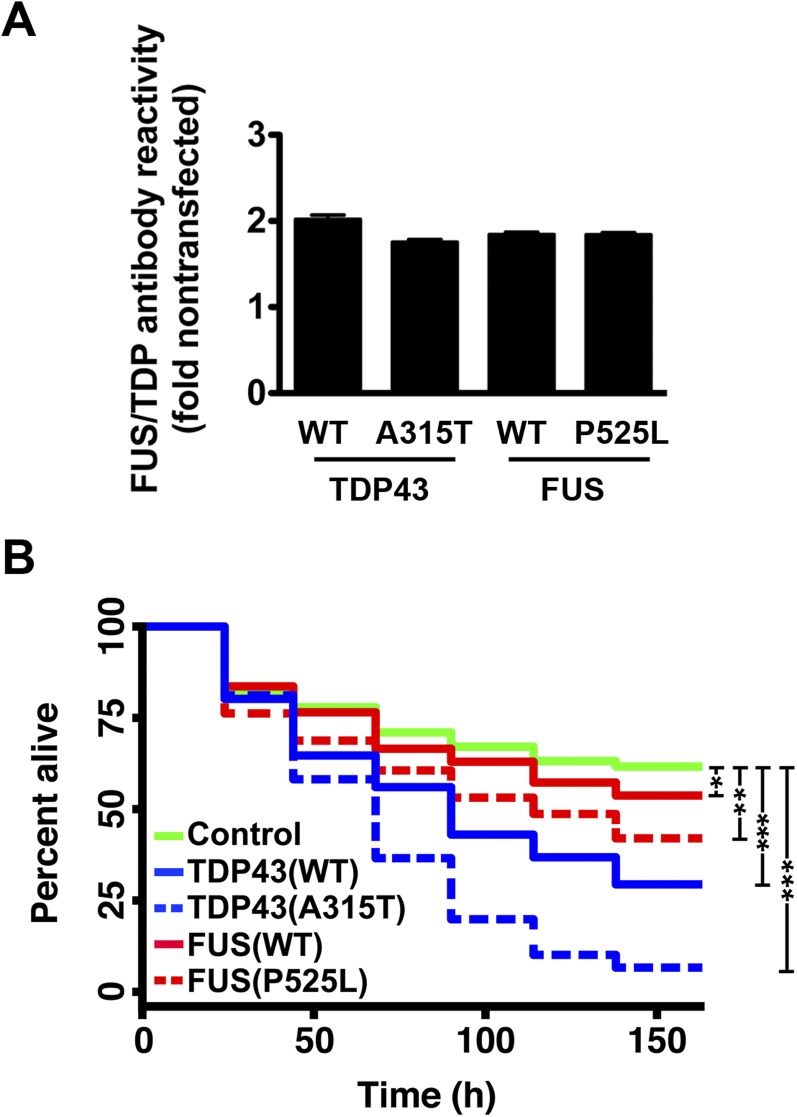
Fig. S2.
Quantitative immunocytochemistry of hUPF1-EGFP in transfected neurons. (A) Primary rodent cortical neurons were transfected with hUPF1-EGFP, fixed and probed using antibodies that recognize total UPF1. The amount of UPF1 in transfected neurons (arrow) was compared with endogenous UPF1 in untransfected neurons (arrowhead) to calculate the fold overexpression. (Scale bar, 50 µm.) (B) Primary neurons cotransfected with hUPF1-EGFP and WT and mutant versions of TDP43 or FUS demonstrated a 3-fold increase in UPF1 antibody reactivity by quantitative ICC. n = 316–433 neurons per genotype. (C) The relationship between hUPF1-EGFP intensity and total UPF1 levels was determined for individual neurons using linear regression analysis. Dotted lines, 95% confidence intervals. n = 76 neurons. Data were pooled from eight separate wells.
The Amount of hUPF1 Is Critical for Neuroprotection.
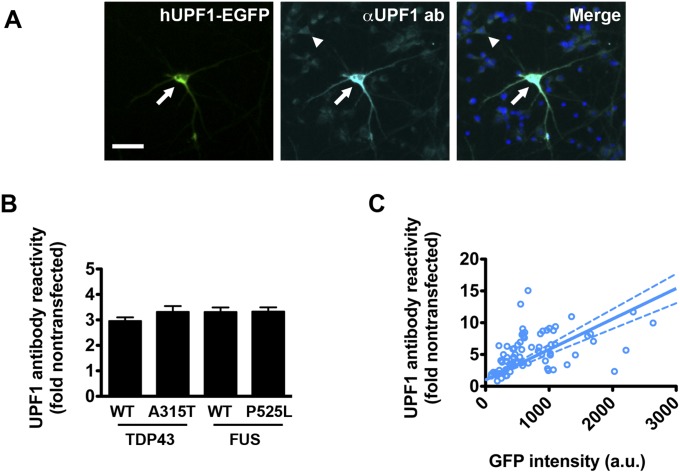
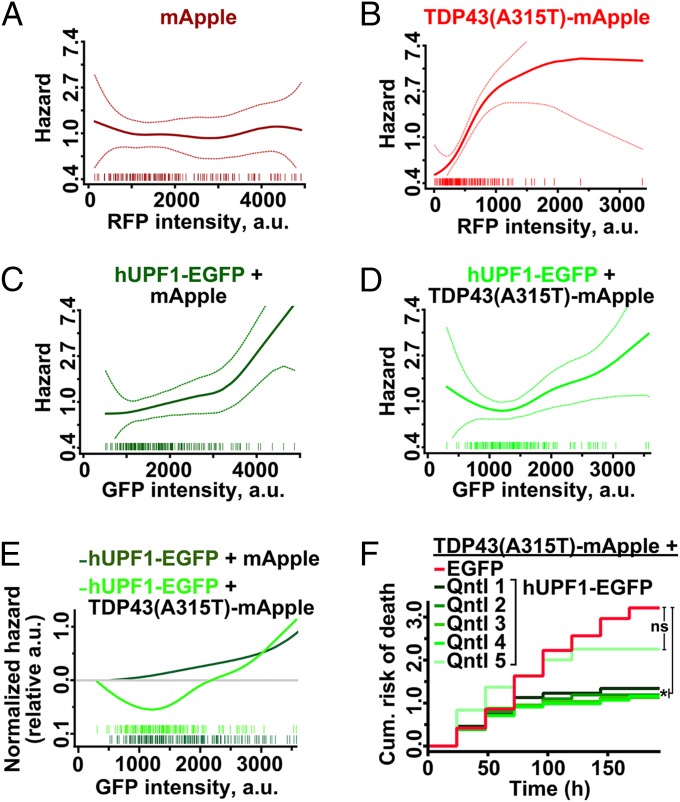
Elevated hUPF1 levels can repress translation (22) and trigger cell death, yet we observed improved survival in neurons overexpressing hUPF1, leading us to believe that the relationship between hUPF1 levels and survival is complex. Because protein amount is directly proportional to the intensity of the fluorophore to which it is fused (20), we labeled hUPF1 with EGFP and estimated TDP43-mApple and hUPF1-EGFP levels by measuring fluorescence intensity in the RFP and GFP channels, respectively. We then used Cox hazards analysis to relate hUPF1-EGFP levels to neuronal survival with penalized spline modeling. These models apply a penalty function to nonparametric regression analyses, thereby maximizing accuracy when fitting noisy observations to a curve (23). Notably, such methods have been successfully used to illustrate the dose-dependent toxicity of mutant huntingtin in neurons (24). In control neurons expressing mApple, hazard is unaffected by mApple levels (Fig. 2A). For TDP43(A315T)-mApple, a steady increase in hazard was observed with rising RFP intensities (Fig. 2B). These data are consistent with prior results showing that TDP43 levels strongly predict survival times in transfected neurons (4, 6).
Fig. 2.
The amount of hUPF1 is critical for neuroprotection. (A) mApple levels were unrelated to survival (n = 193, plinear = 0.94, pnonlinear = 0.53), but increasing amounts of TDP43(A315T)-mApple (B) magnified hazard (n = 164, plinear = 1 × 10−9, pnonlinear = 0.01). In control neurons (C), hUPF1-EGFP levels and hazard were linearly related (n = 193, plinear = 1 × 10−6, pnonlinear = 0.2), indicative of dose-dependent toxicity. In TDP43(A315T)-mApple transfected neurons, low levels of hUPF1-EGFP (D) reduced hazard, whereas high levels were toxic (n = 164, plinear = 0.005, pnonlinear = 0.3). (E) Splines for hUPF1-EGFP in neurons expressing mApple or TDP43(A315T)-mApple emphasize hUPF1’s neuroprotective properties at low levels. (F) Neurons expressing TDP43(A315T)-mApple and hUPF1-EGFP were separated into quintiles based upon GFP intensity, and survival was analyzed by LFM. *P < 0.04; ns, not significant, Cox hazards analysis. Dotted lines in A–D, 95% confidence interval.
In neurons expressing hUPF1-EGFP, we noted a muted increase in the risk of death with rising GFP intensities, indicative of mild dose-dependent toxicity (Fig. 2C). The shape of the spline was different in cells coexpressing TDP43(A315T)-mApple, however, with a prominent drop in hazard at low expression levels (Fig. 2D). To better illustrate this relationship, we combined splines for hUPF1-EGFP in neurons expressing either mApple or TDP43(A315T)-mApple (Fig. 2E); in this plot, the y axis represents the risk of death relative to the zero point (i.e., undetectable hUPF1-EGFP expression) for each population. Neuroprotection by hUPF1-EGFP in cells expressing TDP43(A315T)-mApple is evident at low expression levels, but toxicity ensues as GFP intensity rises above 2,000 a.u. At this point, neuroprotection might be masked by inherent toxicity of hUPF1-EGFP. We investigated this possibility further by splitting the TDP43(A315T)-mApple + hUPF1-EGFP cohort into five quintiles based upon single-cell GFP intensity (Fig. 2F). The risk of death was reduced in quintiles 1–4, representing neurons with GFP intensities <1,800 a.u., but not in quintile 5, confirming the dose-limiting toxicity of hUPF1-EGFP. Quintile 5 contained cells exhibiting GFP intensities >1,800 a.u., corresponding to an approximate 10-fold increase in anti-UPF1 antibody reactivity (Fig. S2). Thus, a “therapeutic window” for hUPF1 exists at less than 10-fold endogenous UPF1 levels—at higher levels, hUPF1 enhances the risk of death, instead of reducing it.
hUPF1 Expression Does Not Rescue Toxicity from Loss of TDP43.
Absence of TDP43 is lethal (25), and TDP43 knockdown recapitulates motor neuron loss in animal models (26). In addition, nuclear clearing of TDP43 is characteristic of ALS (27), and TDP43 overexpression triggers a reduction in endogenous (en)-TDP43 through autoregulation (15, 28), suggesting that TDP43 overexpression may cause toxicity through a paradoxical reduction in en-TDP43. We noted downregulation of TDP43 upon expression of WT and mutant TDP43-EGFP in ∼25% of transfected primary neurons, indicative of en-TDP43 autoregulation in these cells (Fig. 3A). If TDP43 overexpression induces toxicity by reducing en-TDP43, then hUPF1 should be able to rescue cell death caused by en-TDP43 knockdown. To address this hypothesis, we knocked down en-TDP43 in mouse neurons using shRNA (Fig. 3B) or artificial microRNAs (26) (data not shown), achieving 40–60% reduction of en-TDP43 in transfected neurons (Fig. 3C). We then coexpressed hUPF1-EGFP in these neurons and used LFM to determine neuronal survival (Fig. 3D). Knockdown of en-TDP43 was toxic (HR 1.82, P = 1 × 10−11), consistent with prior results (25). However, there was no protective effect of hUPF1-EGFP in en-TDP43 knockdown neurons, implying that hUPF1 cannot prevent cell death due to en-TDP43 deficiency. These results also show that TDP43-mediated toxicity in our model is unrelated to a loss of en-TDP43 function, because hUPF1 rescues the former but not the latter.
Fig. 3.
hUPF1 rescues cell death arising from TDP43 overexpression but not knockdown. (A) Expression of TDP43(WT) (n = 136) and TDP43(A315T) (n = 111) down-regulated en-TDP43 in 15–36% of transfected neurons, as judged by anti-TDP43 antibody reactivity. (B and C) Knockdown of en-TDP43 or en-UPF1 in mouse primary neurons using shRNAs. (Scale bar, 25 µm.) (D) en-TDP43 knockdown increased the risk of death in primary neurons, as determined by LFM, but survival was unaffected by hUPF1. Reduction of 40–50% in en-UPF1 (E) exacerbated neurodegeneration due to TDP43 overexpression (F). In C and E, *P < 0.05, ANOVA with Dunnett’s test. In D, n = 305–513 neurons per genotype. In F, n = 249–332 neurons per genotype. For both D and F, ns, P > 0.05; *P < 0.006, **P < 2 × 10−7, Cox hazards analysis. Results were pooled from 8 to 24 wells per condition, in duplicate.
We were concerned that the observed neuroprotection by hUPF1 might represent an artifact of the model system, based upon hUPF1 overexpression. We therefore sought to determine if en-UPF1 is capable of protecting neurons from TDP43-induced cell death. Using shRNAs (Fig. 3 E and F), we knocked down en-UPF1 in primary mouse cortical neurons, then coexpressed TDP43(WT) in these cells and imaged them by LFM (Fig. 3G). For these experiments, we expressed TDP43(WT) at a low dose (4), to better discriminate modulation of TDP43-dependent toxicity by en-UPF1. Knockdown of en-UPF1 was itself toxic (HR 1.56, P = 7 × 10−5), and en-UPF1 deficiency exacerbated the toxicity of TDP43(WT) (HR 2.42, P = 1 × 10−15), indicating that both endogenous and exogenous UPF1 can protect neurons from TDP43-mediated toxicity.
hUPF1 Specifically Rescues Toxicity Related to RNA-Binding Proteins.
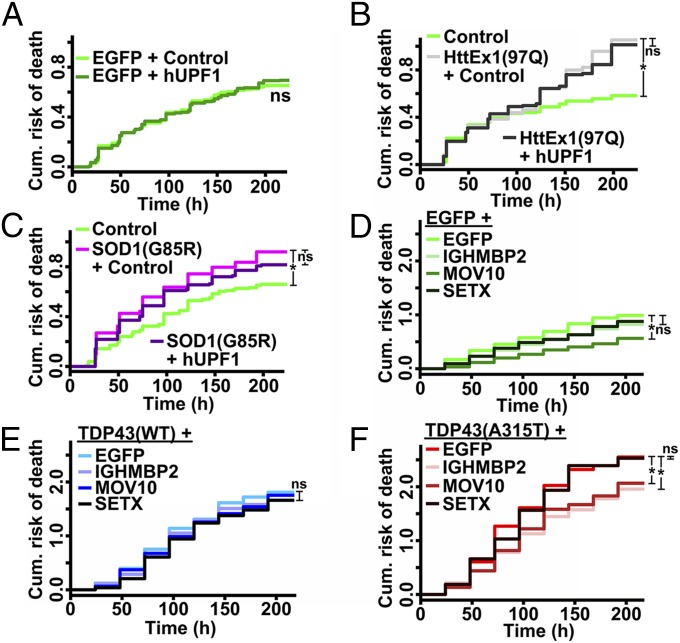
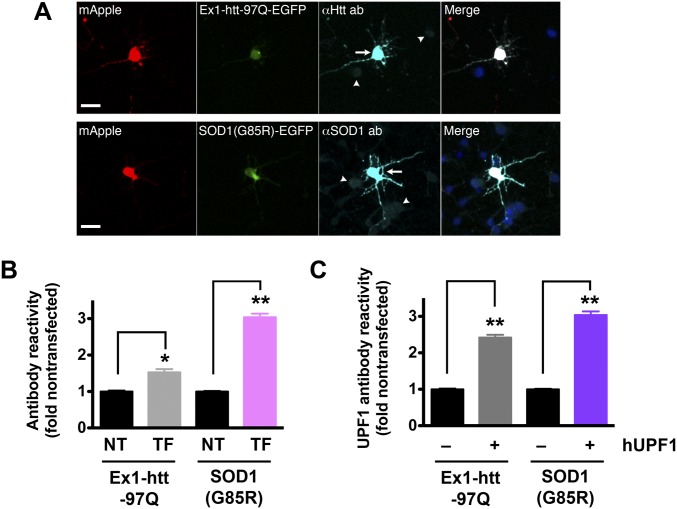
We next wondered if hUPF1 might be broadly therapeutic and nonspecific in its neuroprotective properties. Expression of hUPF1 failed to improve neuronal survival in control neurons (Fig. 4A), arguing against this hypothesis and indicating that hUPF1 expression fails to affect baseline viability. Could hUPF1 protect neurons more generally from aggregate-prone proteins such as TDP43 and FUS? To test this idea, we tested hUPF1 in a neuronal model of Huntington’s disease, a neurodegenerative condition that, like ALS, is characterized pathologically by neuronal inclusions (29). Neurons expressing an N-terminal fragment of mutant huntingtin (htt) carrying a 97-residue polyglutamine expansion (Ex1-Htt-97Q) (Fig. S3) displayed an elevated risk of death compared with control neurons (20) (Fig. 4B). Expression of hUPF1 failed to rescue toxicity from Ex1-Htt-97Q, suggesting that hUPF1’s actions may be specific to mechanisms of neurodegeneration in ALS.
Fig. 4.
SF1 helicases partially rescue TDP43-mediated neurodegeneration. (A) hUPF1 had no effect on the survival of control neurons expressing EGFP alone. n = 862–893 neurons per genotype. (B) Ex1-Htt-97Q increased neuronal risk of death, regardless of hUPF1 expression. n = 197–266 neurons per genotype. (C) hUPF1 expression also failed to improve survival in cells expressing SOD1(G85R). n = 253–1028 neurons per genotype. SF1 helicases were safe in control neurons (D), but none prevent cell death due to TDP43(WT) overexpression (E). In contrast, IGHMBP2 and MOV10 mitigated TDP43(A315T)-mediated toxicity (F). *P < 0.02, ns, not significant, by Cox hazards analysis. In D–F, n = 145–198 neurons per genotype. Results were pooled from eight wells per condition, performed in triplicate.
Fig. S3.
Quantitative immunocytochemistry of mutant htt and SOD1 in transfected neurons. (A) Primary cortical neurons were dissected, cultured, then transfected with mApple and either mutant htt (Ex1-htt-97Q) or SOD1(G85R) fused to EGFP. Transfected proteins were visualized by immunocytochemistry using antibodies that recognize both endogenous (arrowheads) and exogenous (arrows) htt or SOD1. (Scale bars, 25 µm.) (B) Fold overexpression of htt or SOD1 was calculated by comparing antibody reactivity in nontransfected cells (NT) to that in transfected neurons (TF). htt, 161 neurons each for NT and TF groups; SOD1, 146 neurons each for NT and TF groups. (C) Quantitative immunocytochemistry with antibodies against UPF1 was used to determine the UPF1 level in untransfected (−) cells and those transfected (+) with hUPF1. Ex1-htt-97Q, n = 124 neurons each for (−) and (+) groups; SOD1(G85R), n = 148 neurons each for (−) and (+) groups. In B and C, *P < 0.001, **P < 0.0001, vs. NT by two-tailed t test. Data were pooled from four experiments. Error bars represent ± SEM.
If this were the case, then hUPF1 might prevent cell death from genetic causes of ALS that are independent of the RNA-binding proteins TDP43 and FUS. To test this hypothesis, we examined the survival of neurons expressing mutant superoxide dismutase 1 (mSOD1) associated with fALS (2). Overexpression of mSOD1(G93A) (Fig. S3) significantly increased the risk of death in transfected neurons compared with the control (Fig. 4C), but we did not observe an improvement in survival with hUPF1 expression. These data have two important implications: first, hUPF1 effectively reduces toxicity caused by TDP43 or FUS, but not SOD1; and second, mSOD1 and the RNA-binding proteins TDP43 and FUS cause disease through at least partly distinct mechanisms.
Superfamily-1 Helicases Exhibit Limited Neuroprotective Properties.
hUPF1 is a member of the superfamily-1 (SF1) group of DNA/RNA helicases that also includes senataxin (SETX), Maloney leukemia virus 10 homolog (MOV10), and Ig helicase mu binding protein 2 (IGHMBP2) (30). We wondered if other SF1 helicases might act analogously to hUPF1 in protecting neurons from TDP43-mediated toxicity. To answer this question, we transfected primary neurons with WT or mutant TDP43, along with SETX, IGHMBP2, or MOV10, then imaged with LFM. Control neurons (Fig. 4D) transfected with MOV10 survived significantly better than controls (HR 0.53, P = 1 × 10−5), but survival was unaffected by other SF1 helicases. None reduced toxicity from TDP43(WT) overexpression (Fig. 4E), but two helicases (MOV10 and IGHMBP2) improved survival in TDP43(A315T)-transfected neurons (for MOV10, HR 0.75, P = 0.01; for IGHMBP2, HR 0.73, P = 0.007) (Fig. 4). These observations suggest that the neuroprotective properties of SF1 helicases are unique, with hUPF1 improving the survival of both WT and mutant TDP43, whereas MOV10 and IGHMBP2 protect against toxicity associated with mutant but not WT TDP43.
hUPF1 Acts Through Nonsense-Mediated mRNA Decay to Improve Neuron Survival.
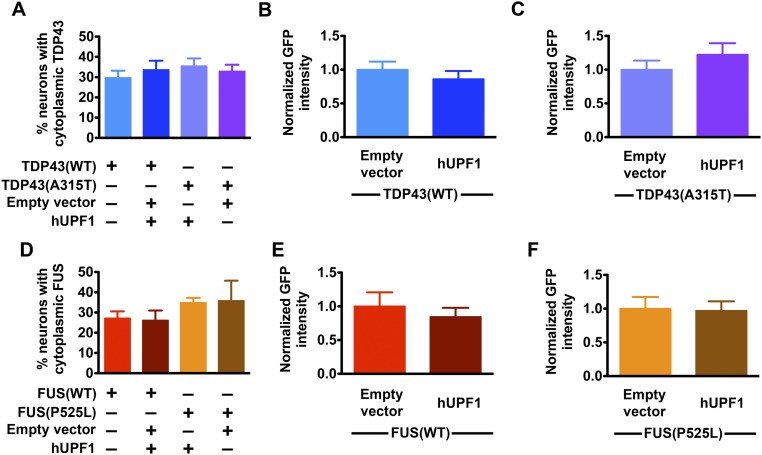
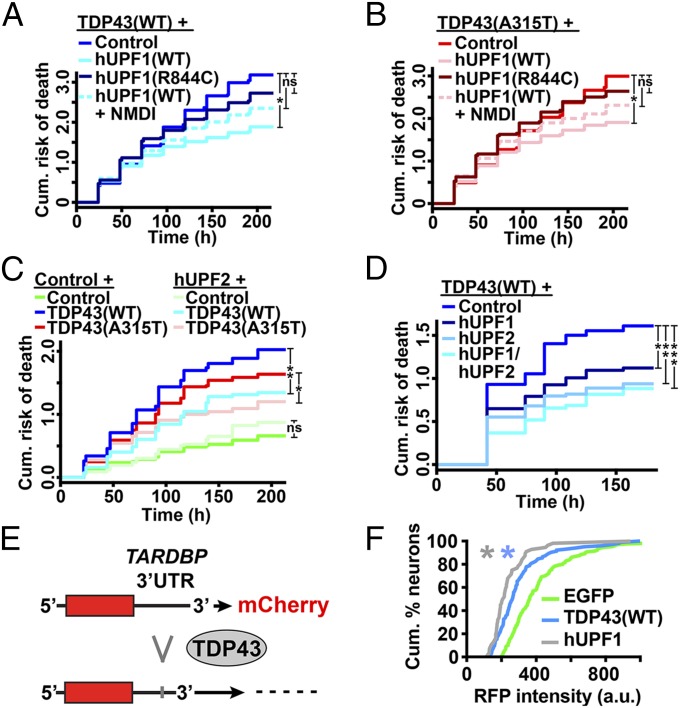
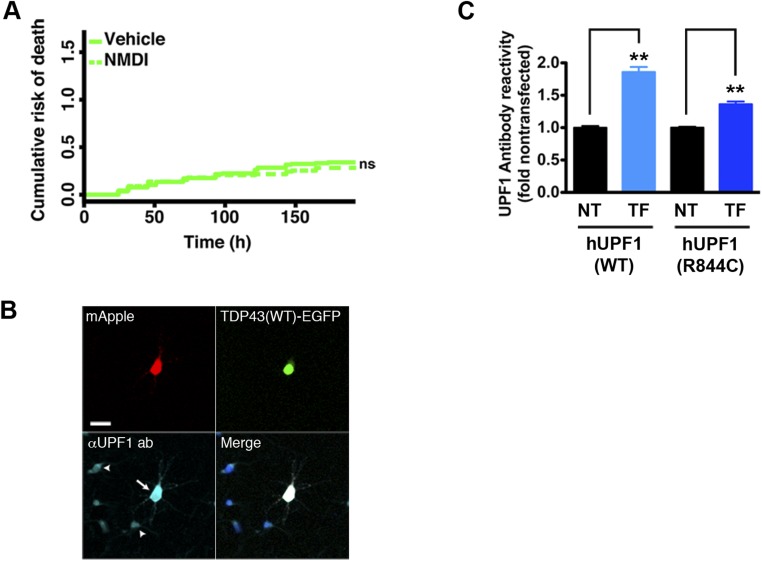
TDP43 levels and localization are significant predictors of death in ALS models (6), but neither was affected by hUPF1 (Fig. S4), arguing against promoter competition or effects on protein levels or localization as a means of rescue. hUPF1 functions in NMD, and data show that both TDP43 and FUS negatively regulate their own mRNAs via NMD (16, 18). Given this potential connection between TDP43, FUS, and hUPF1, we next asked whether hUPF1-mediated neuroprotection depends upon NMD. We expressed TDP43(WT)- or TDP43(A315T)-mApple in primary neurons together with hUPF1-EGFP and treated the cells with NMD inhibitor 1 (NMDI), a compound that blocks NMD by preventing dephosphorylation of UPF1 (31). We then followed the survival of transfected neurons using LFM (Fig. 5 A and B). NMDI by itself was not toxic (Fig. S5) but attenuated the rescue of WT and mutant TDP43-dependent toxicity by hUPF1, suggesting that the beneficial effect of hUPF1 depends at least in part on NMD.
Fig. S4.
Effect of hUPF1 on exogenous TDP43 and FUS. The percentage of cells exhibiting cytoplasmic TDP43-EGFP (A) or FUS-EGFP (D) was unchanged by expression of hUPF1. Likewise, the total levels of TDP43(WT)-EGFP (B), TDP43(A315T)-EGFP (C), FUS(WT)-EGFP (E), and FUS(P525L)-EGFP (F) were unaffected by hUPF1 expression. Data were pooled from eight wells per condition, in triplicate. For A–C, n = 319–386 neurons per genotype; for D–F, n = 98–112 neurons per genotype.
Fig. 5.
Neuroprotection by hUPF1 requires helicase activity and involves NMD. hUPF1 improved survival in cells expressing TDP43(WT) (A) and TDP43(A315T) (B), but helicase-null hUPF1(R844C) was ineffective, and rescue was attenuated with the addition of NMDI-1, an inhibitor of NMD. (C) hUPF2 also improved survival in neurons expressing TDP43(WT) and TDP43(A315T). (D) Expression of both hUPF1 and hUPF2 further reduced the risk of death in neurons transfected with TDP43(WT). In A and B, *P < 0.01; in C, *P = 0.02, **P ≤ 1 × 10−4; in D, *P = 0.02, **P < 0.001, ***P < 1 × 10−5; ns, not significant by Cox hazards analysis. In A and B, n = 360–686 neurons per genotype. In C and D, n = 87–258 neurons per genotype. Results were pooled from eight wells per condition, in three or more experiments. (E) Binding of TDP43 to the TARDBP 3′UTR triggers splicing and RNA decay of a fluorescent reporter. (F) TDP43(WT) (n = 104) and hUPF1 (n = 56) effectively reduce single-cell reporter intensity, compared with EGFP (n = 130). *P ≤ 0.05, Kolmogorov–Smirnov test. Results pooled from 8–16 wells per experiment, in duplicate.
Fig. S5.
Evaluation of NMD inhibitor 1 (NMDI) and mutant hUPF1(R844C) in primary neurons. (A) Primary cortical neurons were treated with NMDI or vehicle (DMSO) at 5 µM, transfected with EGFP, and followed by LFM. Vehicle, n = 170 cells; NMDI, n = 132 cells. ns, not significant (P > 0.05), Cox proportional hazards analysis. Results were combined from eight wells per condition in duplicate. (B) Expression of hUPF1(R844C) in transfected neurons (arrow) was confirmed by immunocytochemistry using antibodies that recognize endogenous (arrowheads) as well as exogenous UPF1. (Scale bar, 25 µm.) (C) Fold overexpression of hUPF1(WT) or hUPF1(R844C) was determined by comparison of antibody reactivity in transfected (TF) vs. nontransfected (NT) neurons. hUPF1(WT), n = 197 neurons each for NT and TF groups; hUPF1(R844C), n = 144 neurons each for NT and TF groups. Error bars, ± SEM. **P < 0.0001 vs NT by 2-tailed t test. Data were pooled from four experiments.
hUPF1 possesses RNA helicase activity that is necessary for disassembly of RNA particles, displacement of RNA-binding proteins, and efficient NMD (32, 33). To determine if helicase ctivity is required for neuroprotection by hUPF1, we transfected primary rodent cortical neurons with WT or mutant TDP43-mApple and a helicase-null variant of hUPF1, hUPF1(R844C) (34), then imaged the cells using LFM (Fig. 5 A and B). Unlike WT hUPF1, expression of hUPF1(R844C) failed to improve neuronal survival. The level of mutant hUPF1(R844C) in transfected neurons was slightly lower than that of hUPF1(WT) (Fig. S5), but given the steep relationship between hUPF1 levels and neuroprotection seen in Fig. 2, we would have expected a more pronounced reduction in toxicity if hUPF1(R844C) acted similarly to hUPF1(WT). The absence of neuroprotection by hUPF1(R844C) implies that RNA helicase activity is required for rescue by hUPF1.
If hUPF1 acts through NMD to reduce toxicity, then additional NMD effectors might be equally protective. We tested this hypothesis by expressing hUPF2, a protein that activates NMD by derepressing hUPF1 (32), in neurons that were transfected with WT and mutant TDP43 (Fig. 5C). hUPF2 expression improved survival in neurons transfected with TDP43(WT) (HR 0.68, P = 0.003) and TDP43(A315T) (HR 0.63, P = 0.001), suggesting that NMD modulation is indeed an effective strategy for preventing TDP43-related toxicity. Because NMD requires both hUPF1 and hUPF2 (32), we wondered if coexpression of both factors would be even more beneficial. Indeed, coexpressing hUPF1 and hUPF2 in neurons transfected with TDP43(WT) (Fig. 5D) produced an even greater reduction in the risk of death than either hUPF1 or hUPF2 alone (hUPF1, HR 0.70, P = 0.01; hUPF2, HR 0.59, P = 0.001; hUPF1 + hUPF2, HR 0.51, P = 1 × 10−5). Taken together, these data indicate that hUPF1 mitigates the toxicity of TDP43 through its role in NMD.
TDP43 negatively regulates itself by binding to its pre-mRNA transcript and triggering RNA decay (15, 16). We surmised that hUPF1 might be accelerating this process. To answer this question, we constructed a fluorescent reporter of TARDBP mRNA decay (Fig. 5 E and F) by amplifying and inserting 3 kb of the human TARDBP 3′UTR, including several TDP43 binding sites (15), downstream of the mCherry ORF. We tested the reporter in primary neurons by measuring red fluorescence intensity on a single-cell basis in response to cotransfection with TDP43 or hUPF1 (Fig. 5 E and F). As expected, expression of WT TDP43 significantly reduced the intensity of the mCherry-TARDBP reporter. hUPF1 also effectively reduced reporter intensity, indicating that hUPF1 is indeed capable of enhancing the decay of TARDBP mRNA.
Discussion
In vitro and in vivo models of ALS provide compelling evidence that accumulation of either TDP43 or FUS causes neurodegeneration and motor neuron disease (6, 35–37). In healthy cells, TDP43 (16) and FUS (18) are tightly regulated by a negative feedback loop involving NMD, preventing otherwise toxic protein deficiency or accumulation. Here, we show that hUPF1, a requisite factor for NMD, improves survival in neuronal models of ALS. Neuroprotection by hUPF1 is specific for neurodegenerative diseases caused by the accumulation of the RNA-binding proteins TDP43 and FUS, requires hUPF1 helicase activity, and is blocked by inhibition of NMD. Our results therefore implicate NMD as a previously unidentified therapeutic target in ALS.
Using nonparametric models (23, 24) we observed optimal survival at moderate to low amounts of hUPF1. In contrast, hUPF1 expression in excess of ∼10-fold endogenous levels was associated with enhanced toxicity. High-level hUPF1 overexpression and phosphorylation enhance the interaction between hUPF1 and eIF3, preventing the 60S ribosomal subunit from joining with the 40S ribosomal subunit at initiation codons and down-regulating translation on a global scale (22). Consistent with these results, neuroprotection waned and hazard increased in neurons expressing the highest amounts of hUPF1.
hUPF1 expression had little effect upon control neurons, and it likewise failed to reduce toxicity in a model of Huntington’s disease. Furthermore, neuroprotection by hUPF1 was limited to ALS models involving the RNA-binding proteins TDP43 and FUS, and did not extend to mSOD1-mediated neurodegeneration. The specificity of hUPF1’s rescue has several important implications for potential disease mechanisms in ALS and therapy development. First, hUPF1 reduces toxicity associated with both WT and mutant RNA-binding proteins. Because the accumulation of WT TDP43 is a hallmark of sALS (27), hUPF1 may be beneficial for both sALS and fALS due to TARBDP or FUS mutations. The large majority of ALS occurs sporadically (1), making it essential that therapeutic strategies show efficacy in models of sALS as well as fALS. Second, hUPF1 affects a conserved pathway in TDP43- and FUS-related disease, a particularly salient observation given the disparate roles played by these proteins in RNA metabolism (14). Lastly, neuron loss in mSOD1-associated ALS may occur through mechanisms that are fundamentally dissimilar to those involving TDP43 and FUS. Considering that >90% of ALS exhibits TDP43-based pathology (27), these observations may partially explain the poor efficacy of therapies originally developed and tested in transgenic mSOD1 mice.
TDP43 and FUS bind to and trigger alternative splicing of their own pre-mRNA transcripts. In each case, the alternative splicing event results in transcripts that are substrates for NMD (14, 16, 18). Because both TDP43 and FUS are regulated by similar mechanisms, hUPF1 is intricately poised to affect their expression through NMD and therefore represents a potential therapeutic target for ALS associated with either TDP43 or FUS deposition.
Nuclear exclusion of TDP43 and FUS in ALS occurs concomitantly with cytoplasmic accumulation of the proteins (27), raising the question of whether neurodegeneration is primarily due to loss of nuclear function, gain of cytoplasmic function, or a combination of both. Overexpression of TDP43 or FUS is sufficient to induce neurodegeneration in cellular and animal models of ALS (6, 37). Moreover, induced pluripotent stem cell (iPSC)-derived neurons and neuroprogenitor cells from patients with fALS harboring TARDBP mutations exhibit increased levels of total and cytoplasmic TDP43 relative to control neurons (5, 7), consistent with gain of function toxicity. Still, exogenous TDP43 effectively down-regulates en-TDP43 (15, 28), raising the possibility that even in overexpression models, toxicity arises from a loss of en-TDP43 function. Whereas hUPF1 effectively rescued neurons from TDP43 overexpression, it failed to affect survival in TDP43 knockdown neurons. These results provide strong evidence that loss of en-TDP43 is not responsible for toxicity associated with TDP43 overexpression and imply that distinct mechanisms cause cell death from absence or overabundance of TDP43.
Null mutations in UPF1 are embryonic lethal (38), and knocking down UPF1 in mature neurons is likewise toxic, testifying to the essential nature of the protein. hUPF1 is a member of the SF1 helicases containing homologous ATPase and RNA helicase domains (30). Mutations affecting the helicase domain of two SF1 members, IGHMBP2 and SETX, cause motor neuron disease (39, 40), indicating a selective vulnerability of motor neurons to functional deficiencies in certain RNA helicases. IGHMBP2 participates in mRNA decay, transcription, and translation (41, 42), whereas SETX functions in transcription and the DNA damage response (43). MOV10 localizes to P-bodies, cytoplasmic sites of RNA processing (44), and directly promotes NMD by unwinding pre-mRNA transcripts and removing bound proteins (45). Of the SF1 helicases, all but SETX participate in RNA decay, and are capable of reducing mutant TDP43-mediated toxicity. Our results therefore emphasize RNA clearance as a necessary property of neuroprotective SF1 helicases in this model.
ALS is a progressive and ultimately lethal neurodegenerative disease. More effective therapies are urgently needed, but the results of human clinical trials involving candidate therapies have so far been disappointing (46). Here, we showed that hUPF1 dramatically improves neuronal survival in ALS models through a mechanism involving NMD. In related work (47), hUPF1 demonstrated therapeutic effects in a rat sALS model, providing valuable in vivo confirmation of the neuroprotective activity of hUPF1. Because accumulations of TDP43 or FUS account for the large majority of disease burden in ALS, it is our hope that strategies based on hUPF1 might prove successful in halting the inevitable progression of ALS.
Materials and Methods
SI Materials and Methods contains detailed descriptions of (i) all plasmids used for these experiments and their construction; (ii) an ethics statement on the treatment of animals; (iii) rodent primary neuron isolation, culturing and transfection techniques; (iv) quantitative immunocytochemistry and the antibodies used for these studies; (v) components and organization of the longitudinal fluorescence microscopy platform; and (vi) all software used for image and statistical analyses. All animal experiments were performed in accordance with the University of California, San Francisco, Assurance of Compliance with Public Health Service Policy on Humane Care and Use of Laboratory Animals by Awardee Institutions number A3400-01; the University of Michigan Policy and Procedures Concerning the Use of Vertebrate Animals as Subjects in Research, Testing, or Instruction; and the American Veterinary Medical Association.
SI Materials and Methods
Plasmids.
TDP43(WT)-mApple and TDP43(A315T)-mApple were created by inserting a HindIII site downstream of the ORF encoding TDP43 in pGW1-TDP43(WT)-EGFP and pGW1-TDP43(A315T)-EGFP (4) using site-directed mutagenesis and the following PCR primers: sense, 5′-ATA TAT AAG CTT CCA TGT CTG AAT ATA TTC GGG TAA CCG A-3′; antisense, 5′-ATA TAT AAG CTT GGA GAT CTC TTG TAC AGC TCG TCC ATG C-3′. The TDP43 ORF was then excised using HindIII and inserted into the HindIII site just upstream of mApple in pGW1-mApple (4). For FUS(WT)-mApple and FUS(P525L)-mApple, the ORFs encoding FUS(WT) and FUS(P525L) were PCR amplified from pGW1-FUS(WT)-EGFP (37) and pGW1-FUS(P525L)-EGFP (37) using the following primers: sense, 5′-TAT CTT AAG GCC ACC ATG GCC TCA AAC GAT TAT ACC CAA CAA GCA A-3′; antisense for FUS(WT), 5′-TAT AAA GCT TGT ATA CGG CCT CTC CCT GCG ATC CTG TCT GT-3′; antisense for FUS(P525L), 5′-TAT AAA GCT TGT ATA CAG CCT CTC CCT GCG ATC CTG TCT G-3′. The PCR products were digested using HindIII and AflII and inserted into pGW1-mApple cut with these enzymes.
To generate hUPF1 in FUGW, pCMV-Myc-hUPF1(WT) and pCMV-Myc-hUPF1(R844C) (34) were digested with ScaI and XbaI, and the 3.8-kb fragments separated by agarose gel electrophoresis. Separately, FUGW was cut with XbaI, releasing the EGFP insert. The 3.8-kb band was ligated into FUGW lacking EGFP, creating hUPF1-FUGW. To create hUPF1-EGFP in FUGW, FUGW was cut with XhoI and BstXI, releasing a 2.2-kb band. This insert was then ligated into hUPF1-FUGW that had been digested with the same enzymes.
For mouse UPF1 knockdown, we synthesized the following seed sequences as oligonucleotides: UPF1 Sh1, sense 5′-GAT CCC GGC AAG AAG TGG TTC TGC AAT TCA AGA GAT TGC AGA ACC ACT TCT TGC CTT TTT GG-3′ and antisense 5′-AGC TTT TCC AAA AAG GCA AGA AGT GGT TCT GCA ATC TCT TGA ATT GCA GAA CCA CTT CTT GCC GG-3′; UPF1 Sh2, sense 5′-GAT CCC GGA CTA TGA CAA GAA GTT GAT TCA AGA GAT CAA CTT CTT GTC ATA GTC CTT TTT GG-3′ and antisense 5′-AGC TTT TCC AAA AAG GAC TAT GAC AAG AAG TTG ATC TCT TGA ATC AAC TTC TTG TCA TAG TCC GG-3′. For mouse TDP43 knockdown, we synthesized the following seed sequences as oligonucleotides: TDP43 Sh1, sense 5′-GAT CCC GCA TTG AAA TTG TTA ATG AAT TCA AGA GAT TCA TTA ACA ATT TCA ATG CTT TTT GG-3′ and antisense 5′-AGC TTT TCC AAA AAG CAT TGA AAT TGT TAA TGA ATC TCT TGA ATT CAT TAA CAA TTT CAA TGC GG-3′; and TDP43 Sh2, sense 5′-GAT CCC GGT TTC TCT GTA ATA TTC TAT TCA AGA GAT AGA ATA TTA CAG AGA AAC CTT TTT GG-3′ and antisense 5′-AGC TTT TCC AAA AAG GTT TCT CTG TAA TAT TCT ATC TCT TGA ATA GAA TAT TAC AGA GAA ACC GG-3′. The oligonucleotides were phosphorylated with T4 polynucleotide kinase (New England Biolabs) according to the manufacturer’s protocol, and the enzyme was heat inactivated by incubation at 65 °C for 20 min. The oligonucleotides were then resuspended in annealing buffer (10 mM Tris, pH 7.5–8.0, 50 mM NaCl, 1 mM EDTA) at 100 µM each, and incubated at 95 °C for 10 min, then cooled back to room temperature over 45–60 min. The annealed oligonucleotides were digested with BamHI and HindIII, then inserted into pSilencer2.0 (Life Technologies) that had been cut with the same enzymes. Scrambled oligonucleotide was used as a control. Artificial microRNAs acting on TDP43 were made as described previously (26). pGW1-Ex1-Htt-97Q, pDEST54-SETX, and pCMV-Tag4b-IGHMBP2 were created as described (20, 43, 48), and pMyc-MOV10 was obtained from Addgene.
Ethics Statement.
Rat (Rattus norvegicus) colonies were maintained in accordance with the University of California, San Francisco, “Assurance of Compliance with PHS Policy on Humane Care and Use of Laboratory Animals by Awardee Institutions” number A3400-01, and the University of Michigan Policy and Procedures Concerning the Use of Vertebrate Animals as Subjects in Research, Testing or Instruction. The animal care facilities at The J. David Gladstone Institutes and the University of Michigan are fully approved and under the supervision of full-time veterinarians. There were no manipulations that might have produced discomfort, distress, pain, or injury. The animals were euthanized using CO2, resulting in rapid and painless death, consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association.
Cell Culture and Transfection.
Cortical neurons were dissected from embryonic day 20–21 rat pups and cultured at 0.6 × 106 cells per milliliter for 4 d in vitro before transfection, as described previously (4). Transfection of primary neurons was accomplished using Lipofectamine 2000 (Invitrogen). All transfections involved 0.2 µg DNA (total) and 0.5 µL Lipofectamine 2000 per well in 96-well plates, unless otherwise noted. Cells were incubated with Lipofectamine/DNA complexes for no more than 20 min at 37 °C before rinsing. The remainder of the transfection protocol was per the manufacturer’s suggestions, resulting in an overall transfection efficiency of <5%. For experiments involving NMD inhibitor (NMDI) (31), drug was added to a final concentration of 5 µM immediately after transfection.
Quantitative Immunocytochemistry.
Cortical neurons from rats were dissected and cultured as described (4) on 12-mm glass coverslips or 96-well plates coated with laminin (BD Biosciences) and poly-d-lysine (Millipore), then transfected at 4 d in vitro with TDP43, FUS, and/or hUPF1, or shRNAs directed against endogenous rat TDP43 or hUPF1. Immunocytochemistry was performed as described (4) using rabbit anti-TDP43 polyclonal antibodies (ref. 4; 1:5,000), rabbit anti-FUS polyclonal antibodies (Sigma, HPA008784; 1:100), mouse monoclonal anti-htt antibodies (Millipore, EM48 MAB5374; 1:100), rabbit polyclonal anti-SOD1 antibodies (Enzo Life Sciences, SOD-100; 1:100), or rabbit anti-UPF1/RENT1 monoclonal antibodies (Abcam, ab133564; 1:100). Anti-rabbit Cy5-labeled secondary antibodies (Jackson Immunochemical; 1:250) were used for detection of primary antibody/antigen complexes. Nuclei were stained by a brief incubation in PBS supplemented with Hoechst dye (10 µM final concentration). Immunocytochemical samples were imaged using LFM, as described below. Fold overexpression was determined by comparing the intensity of immunolabeling for each protein in transfected cells to that in nontransfected cells within the same field. Results were plotted using GraphPad Prism software.
Longitudinal Fluorescence Microscopy.
Neuronal survival analyses were accomplished using an automated microscopy platform described previously (4, 6, 20, 24). Briefly, images were obtained with an inverted microscope (Nikon TE2000) equipped with the PerfectFocus system, a high-numerical aperture 20× objective lens, and a 16-bit Andor iXon digital camera with an electron multiplied ultracooled charge-coupled device. Illumination was provided by a Lambda XL Xenon lamp (Sutter) with a 5-mm liquid light guide. An ASI 2000 stage, controlled by rotary encoders in the x and y axes, was used to perform stage movements. All components were encased in a custom-designed and climate-controlled environmental chamber (In Vivo Scientific). The illumination, filter wheels, focusing, stage movements, and image acquisitions were fully automated and coordinated with publicly available (ImageJ, µManager) software.
Image Analyses.
Relevant data were extracted from the raw, digital images in a sequential process using an original script written for ImageJ. Between 9 and 25 images of each well were first stitched together to represent a larger total area for each well. Background fluorescence was calculated and subtracted from all stitched images. The stitched and corrected images were sequenced and aligned automatically, and neuron survival was determined by a software algorithm developed in Pipeline Pilot (Accelrys) or ImageJ, or counted by hand using a program written in MatLab. The time of death for each neuron was recorded as the last time a neuron was confirmed to be alive (left censoring). Intensities of transfected proteins were determined automatically in Figs. 2, 3, and 5, and Figs. S1–S5 by segmentation of neuronal cell bodies in ImageJ and measurement of mean pixel intensity within each region of interest. Statistical analyses, the generation of cumulative hazard plots, and penalized spline modeling were accomplished using custom-written scripts and the survival package within R, whereas scatter plots and bar graphs were created using GraphPad Prism.
Supplementary Material
Acknowledgments
We thank G. Yu and J. Herz (University of Texas, Southwestern) for antibodies; Z. Xu (University of Massachusetts Medical School) for microRNAs; S. West and K. Radhakrishnan (Genetic Recombination Lab, London Research Institute) for SETX constructs; D. Bedwell and S. Velu (University of Alabama) for NMDI; U. Fischer (Biocenter of the University of Wuerzburg) for IGHMBP2 constructs; K. Nelson and H. Oishi for administrative assistance; and M. Moore (University of Massachusetts Medical School), L. Maquat, B. Lucas, and MW-L. Popp (University of Rochester Medical Center) for their comments, suggestions, and insight. This work was supported by National Institutes of Health (NIH) Grants 1K08NS072233-01A1 (to S.J.B.), 3R01 NS039074, 2R01 NS045091, and NS083390 (to S.F.) from the National Institutes of Neurological Disorders and Stroke and by the Robert Packard Center for ALS Research, Target ALS, the Roddenberry Stem Cell Program, the Koret/Taube Center for Neurodegenerative Disease, the Hellman Foundation Alzheimer’s Disease Research Program (to S.F.), the Protein Folding Diseases Initiative at the University of Michigan, and the Department of Neurology at the University of Michigan (to S.J.B.). The Gladstone Institutes animal care facility was partly supported by an NIH Extramural Research Facilities Improvement Program Project (C06 RR018928).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509744112/-/DCSupplemental.
References
- 1.Huisman MHB, et al. Population based epidemiology of amyotrophic lateral sclerosis using capture-recapture methodology. J Neurol Neurosurg Psychiatry. 2011;82(10):1165–1170. doi: 10.1136/jnnp.2011.244939. [DOI] [PubMed] [Google Scholar]
- 2.Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barmada SJ, Finkbeiner S. Pathogenic TARDBP mutations in amyotrophic lateral sclerosis and frontotemporal dementia: Disease-associated pathways. Rev Neurosci. 2010;21(4):251–272. doi: 10.1515/revneuro.2010.21.4.251. [DOI] [PubMed] [Google Scholar]
- 4.Barmada SJ, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10(8):677–685. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilican B, et al. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc Natl Acad Sci USA. 2012;109(15):5803–5808. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barmada SJ, et al. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30(2):639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishimura AL, et al. Allele-specific knockdown of ALS-associated mutant TDP-43 in neural stem cells derived from induced pluripotent stem cells. PLoS ONE. 2014;9(3):e91269. doi: 10.1371/journal.pone.0091269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 9.Wils H, et al. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA. 2010;107(8):3858–3863. doi: 10.1073/pnas.0912417107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang DB, et al. Expansive gene transfer in the rat CNS rapidly produces amyotrophic lateral sclerosis relevant sequelae when TDP-43 is overexpressed. Mol Ther. 2010;18(12):2064–2074. doi: 10.1038/mt.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 13.Neumann M, et al. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132(Pt 11):2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagier-Tourenne C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15(11):1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayala YM, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30(2):277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14(4):459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popp MW-L, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Liu S, Liu G, Oztürk A, Hicks GG. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet. 2013;9(10):e1003895. doi: 10.1371/journal.pgen.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ju S, et al. A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol. 2011;9(4):e1001052. doi: 10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 21.Arrasate M, Finkbeiner S. Automated microscope system for determining factors that predict neuronal fate. Proc Natl Acad Sci USA. 2005;102(10):3840–3845. doi: 10.1073/pnas.0409777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isken O, et al. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133(2):314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skibinski G, Finkbeiner S. Longitudinal measures of proteostasis in live neurons: Features that determine fate in models of neurodegenerative disease. FEBS Lett. 2013;587(8):1139–1146. doi: 10.1016/j.febslet.2013.02.043. [DOI] [PubMed] [Google Scholar]
- 24.Miller J, et al. Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into huntington’s disease molecular pathogenesis. J Neurosci. 2010;30(31):10541–10550. doi: 10.1523/JNEUROSCI.0146-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sephton CF, et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285(9):6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C, et al. Partial loss of TDP-43 function causes phenotypes of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2014;111(12):E1121–E1129. doi: 10.1073/pnas.1322641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann M. Molecular neuropathology of TDP-43 proteinopathies. Int J Mol Sci. 2009;10(1):232–246. doi: 10.3390/ijms10010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Igaz LM, et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest. 2011;121(2):726–738. doi: 10.1172/JCI44867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finkbeiner S. Huntington’s Disease. Cold Spring Harb Perspect Biol. 2011;3(6):pii: a007476. doi: 10.1101/cshperspect.a007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26(1):253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durand S, et al. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol. 2007;178(7):1145–1160. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakrabarti S, et al. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell. 2011;41(6):693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 33.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell. 2010;143(6):938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun X, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci USA. 1998;95(17):10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang C, et al. FUS transgenic rats develop the phenotypes of amyotrophic lateral sclerosis and frontotemporal lobar degeneration. PLoS Genet. 2011;7(3):e1002011. doi: 10.1371/journal.pgen.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janssens J, et al. Overexpression of ALS-associated p.M337V human TDP-43 in mice worsens disease features compared to wild-type human TDP-43 mice. Mol Neurobiol. 2013;48(1):22–35. doi: 10.1007/s12035-013-8427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu H, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124(3):981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medghalchi SM, et al. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10(2):99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y-Z, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74(6):1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim SC, Bowler MW, Lai TF, Song H. The Ighmbp2 helicase structure reveals the molecular basis for disease-causing mutations in DMSA1. Nucleic Acids Res. 2012;40(21):11009–11022. doi: 10.1093/nar/gks792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Planell-Saguer M, Schroeder DG, Rodicio MC, Cox GA, Mourelatos Z. Biochemical and genetic evidence for a role of IGHMBP2 in the translational machinery. Hum Mol Genet. 2009;18(12):2115–2126. doi: 10.1093/hmg/ddp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molnar GM, et al. Association of the mammalian helicase MAH with the pre-mRNA splicing complex. Proc Natl Acad Sci USA. 1997;94(15):7831–7836. doi: 10.1073/pnas.94.15.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yüce Ö, West SC. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol Cell Biol. 2013;33(2):406–417. doi: 10.1128/MCB.01195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Izumi T, et al. Mov10 and APOBEC3G localization to processing bodies is not required for virion incorporation and antiviral activity. J Virol. 2013;87(20):11047–11062. doi: 10.1128/JVI.02070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregersen LH, et al. MOV10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell. 2014;54(4):573–585. doi: 10.1016/j.molcel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 46.Turner BJ, Talbot K. Transgenics, toxicity and therapeutics in rodent models of mutant SOD1-mediated familial ALS. Prog Neurobiol. 2008;85(1):94–134. doi: 10.1016/j.pneurobio.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Jackson KL, et al. Preservation of forelimb function by UPF1 gene therapy in a rat model of TDP-43-induced motor paralysis. Gene Ther. 2015;22(1):20–28. doi: 10.1038/gt.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grohmann K, et al. Characterization of Ighmbp2 in motor neurons and implications for the pathomechanism in a mouse model of human spinal muscular atrophy with respiratory distress type 1 (SMARD1) Hum Mol Genet. 2004;13(18):2031–2042. doi: 10.1093/hmg/ddh222. [DOI] [PubMed] [Google Scholar]