Significance
MicroRNAs are key players in the regulation of gene expression. An understanding of human conditions caused by microRNA mutations provides insight into mechanisms of gene regulation and into the interplay between development and maintenance in tissue homeostasis. The eye represents a notable target tissue of genetic diseases. Inherited retinal degenerations and developmental eye disorders are two separate groups that represent leading causes of blindness worldwide. Identifying underlying genetic causes of such conditions is important for diagnosis, counseling, and potential therapy development. We identified a dominant mutation in microRNA-204 as the genetic cause of a unique phenotype of retinal degeneration and coloboma and thus highlight the importance of microRNA-204 as a master regulator of ocular development and normal maintenance.
Keywords: retinitis pigmentosa, coloboma, miR-204, microRNA, retinal degeneration
Abstract
Ocular developmental disorders, including the group classified as microphthalmia, anophthalmia, and coloboma (MAC) and inherited retinal dystrophies, collectively represent leading causes of hereditary blindness. Characterized by extreme genetic and clinical heterogeneity, the separate groups share many common genetic causes, in particular relating to pathways controlling retinal and retinal pigment epithelial maintenance. To understand these shared pathways and delineate the overlap between these groups, we investigated the genetic cause of an autosomal dominantly inherited condition of retinal dystrophy and bilateral coloboma, present in varying degrees in a large, five-generation family. By linkage analysis and exome sequencing, we identified a previously undescribed heterozygous mutation, n.37C > T, in the seed region of microRNA-204 (miR-204), which segregates with the disease in all affected individuals. We demonstrated that this mutation determines significant alterations of miR-204 targeting capabilities via in vitro assays, including transcriptome analysis. In vivo injection, in medaka fish (Oryzias latipes), of the mutated miR-204 caused a phenotype consistent with that observed in the family, including photoreceptor alterations with reduced numbers of both cones and rods as a result of increased apoptosis, thereby confirming the pathogenic effect of the n.37C > T mutation. Finally, knockdown assays in medaka fish demonstrated that miR-204 is necessary for normal photoreceptor function. Overall, these data highlight the importance of miR-204 in the regulation of ocular development and maintenance and provide the first evidence, to our knowledge, of its contribution to eye disease, likely through a gain-of-function mechanism.
The eye is a major target tissue for genetic disease. The main group of genetic disorders affecting the retina is represented by inherited retinal dystrophies that include, among others, retinitis pigmentosa, one of the leading causes of inherited blindness in the Western population, with a prevalence of 1:4,000 (1). The loss of vision in severe retinal dystrophies is most often a result of progressive loss or dysfunction of photoreceptor cells or retinal pigment epithelial (RPE) cells. In addition to degenerative disorders, the eye is also the target of a number of developmental genetic disorders, among which the group classified as microphthalmia, anophthalmia, and coloboma (MAC) features major structural eye malformations. Both inherited retinal dystrophies and MACs are characterized by extreme genetic heterogeneity. According to recent estimates, more than 150 retinal dystrophy genes (sph.uth.edu/Retnet/home.htm) and more than 25 MAC genes (www.omim.org) have been reported to date, including several that account for conditions in both groups. Many others remain to be identified for both conditions.
MicroRNAs (miRNAs) are emerging as key players in the control of fundamental biological processes in both physiological and pathological conditions. They are single-stranded, noncoding, short (20–25 nucleotides) RNAs that regulate gene expression through inhibiting translation of mRNAs or promoting their degradation. Their importance in regulating gene expression in retinal cells is highlighted by the high number of miRNAs that are preferentially expressed in the vertebrate retina (2–5). Evidence indicates that miRNAs are important for the development and maintenance of correct function within the eye, and in particular the retina (6–8). Mutations in miRNAs or their target genes may contribute to a range of ocular abnormalities. In this study we investigated the molecular basis for an unusual phenotype observed in a family with a history of varying degrees of retinal degeneration and MAC features (coloboma). We describe the identification of a dominant mutation in miR-204 and demonstrate the functional significance of this mutation to the retinal phenotype observed in this family.
Results
Clinical Information.
A diagnosis of bilateral coloboma and rod-cone dystrophy with or without cataract was made in nine individuals of a five-generation family of white British descent. Family history suggested an autosomal dominant pattern of inheritance, and full clinical examination demonstrated that the phenotype was not associated with extraocular manifestations.
Individual V-1 was diagnosed with retinal dystrophy and bilateral iris coloboma in early childhood. At age 18 y, he had reduced visual acuities (0.74 LogMar right, 0.84 LogMar left) and was registered as partially sighted. Mild hypermetropic astigmatism was observed (right eye, +1.75/+0.50 × 100; left eye, +2.25/+0.50 × 85). Color vision was significantly compromised bilaterally in the City University color vision test (third edition, 1998) and the Ishihara color vision screening test (38-plate edition, 1979). Bilateral iris colobomata with iridolenticular adhesions was present with no evidence of cataract (Fig. 1B). Dilated posterior segment examination revealed scattered RPE mottling with retinal atrophy and attenuation of the retinal vasculature. Optical coherence tomography imaging was consistent with severe photoreceptor loss (Fig. 1D). Electroretinography revealed extinguished right and left eye responses to all conditions except a minimal residual light-adapted 30-Hz flicker response (Fig. 1E); notably, 4 y before, electrodiagnostic testing had revealed less attenuated light-adapted responses. The proband’s mother, IV-I, aged 42 y, presented with a similar phenotype, having been diagnosed with bilateral iris colobomata and a progressive retinopathy in infancy. She underwent bilateral cataract surgery at age 30 y and subsequently had a, likely iatrogenic, retinal detachment in her left eye. She was registered as blind at age 30 y and at age 42 y has hand movement vision in her right eye and no perception of light in her left eye. Bilateral iris colobomata are noted (Fig. 1C). Individuals II-1, III-1, III-2, and III-3 were all registered blind. All had bilateral iris coloboma and a slowly progressive retinal dystrophy leading to marked loss of vision in late childhood or early adult life. Individual III-2 was also found to have congenital cataracts and underwent right cataract surgery at age 7 y.
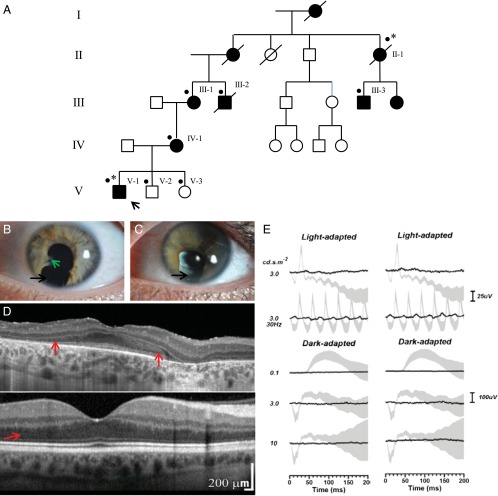
Fig. 1.
Ocular phenotype of family with inherited retinal dystrophy and iris coloboma. (A) Pedigree of the family. The proband is denoted by an arrow. *, patients affected with retinal dystrophy and coloboma who underwent exome sequencing; ●, all patients for whom DNA was available for testing. (B and C) Slit lamp biomicroscopy photographs of the left eye of affected family members are presented; a high degree of interocular symmetry was observed. The left eye of patient V-I (B) revealed inferior iris coloboma (black arrow) and iridolenticular adhesions (green arrow). The left eye (C) of patient IV-I demonstrated inferior iris coloboma. (D) OCT (Heidelberg Specralis) of the central macula of the right eye of patient V-1 is shown (Top); a high degree of interocular symmetry was observed, and an image from the right eye of an unaffected individual is also shown for comparison (Bottom). The OCT suggests extensive loss of the hyporeflective band corresponding to the photoreceptor nuclei (red arrow). (E) Electroretinography of left (Left) and right (Right) eye of patient V-1 is shown. The black lines correspond to the recorded traces and the gray areas represent the normal limits. Extinguished responses to all conditions are observed except for a minimal light-adapted 30-Hz flicker response corresponding to residual cone function.
Identification of a miR-204 Mutation by Linkage Analysis and Exome Sequencing.
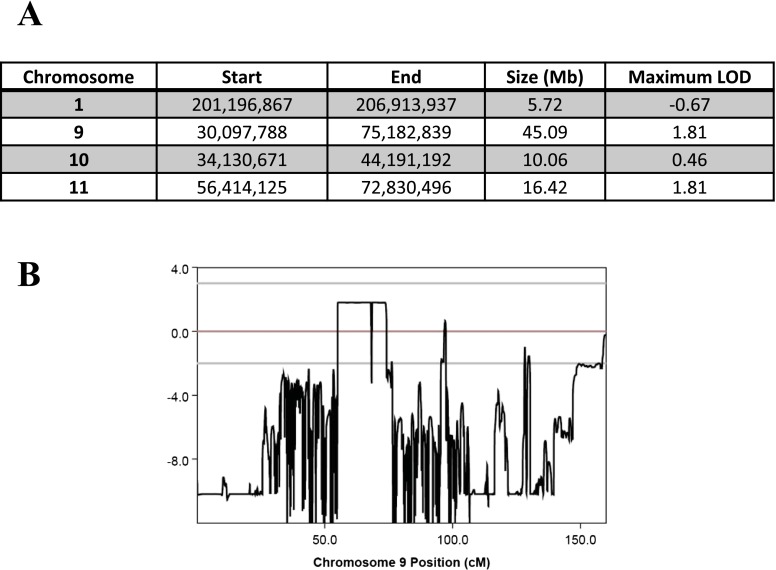
To identify possible chromosomal regions associated with the retinal dystrophy and iris coloboma phenotype in this family, parametric linkage analysis was performed for six individuals (patients II-1, III-1, III-2, V-1, V-2, and V-3), which provided eight informative meioses. These data achieved a maximal LOD (logarithm of odds) score of 1.81, which falls below the level of 3.0 required to confirm significance of linkage. The data were therefore analyzed on the basis of linkage exclusion for those regions with a LOD score less than −2. There were four regions remaining over 5 Mb, which had a LOD score of between −2 and 1.81 (Fig. S1). Subsequently, exome sequencing was performed on patients II-I and V-I to identify possible causal variants within the potential linked regions. This revealed 39 variant calls common to both patients. To prioritize the likely disease causing mutations, we assumed a dominant model of inheritance and excluded homozygous changes. All variants reported in the Exome Variant Server of the National Heart, Lung and Blood Institute Exome Sequencing Project (evs.gs.washington.edu/EVS/), dbSNP136, and the 1000 Genomes Project (www.1000genomes.org) were excluded. Variants that had been previously seen within an in-house database of 244 exomes and those variants confirmed by Sanger sequencing not to cosegregate with disease in the family were also excluded. This resulted in a single candidate variant. The n.37C > T mutation identified in the microRNA MIR204 (reference sequence NR_029621) was an excellent candidate for further study. MiR-204 is preferentially expressed in the eye, and several studies have determined its role in eye development (3, 9–11). Evidence of the critical function of miR-204 in eye development strongly suggests a pathogenic role for the mutation identified in this family.
Fig. S1.
Multipoint parametric linkage analysis for a dominant model. (A) Table shows details of potential linked regions. (B) LOD plot for analysis across chromosome 9 is shown.
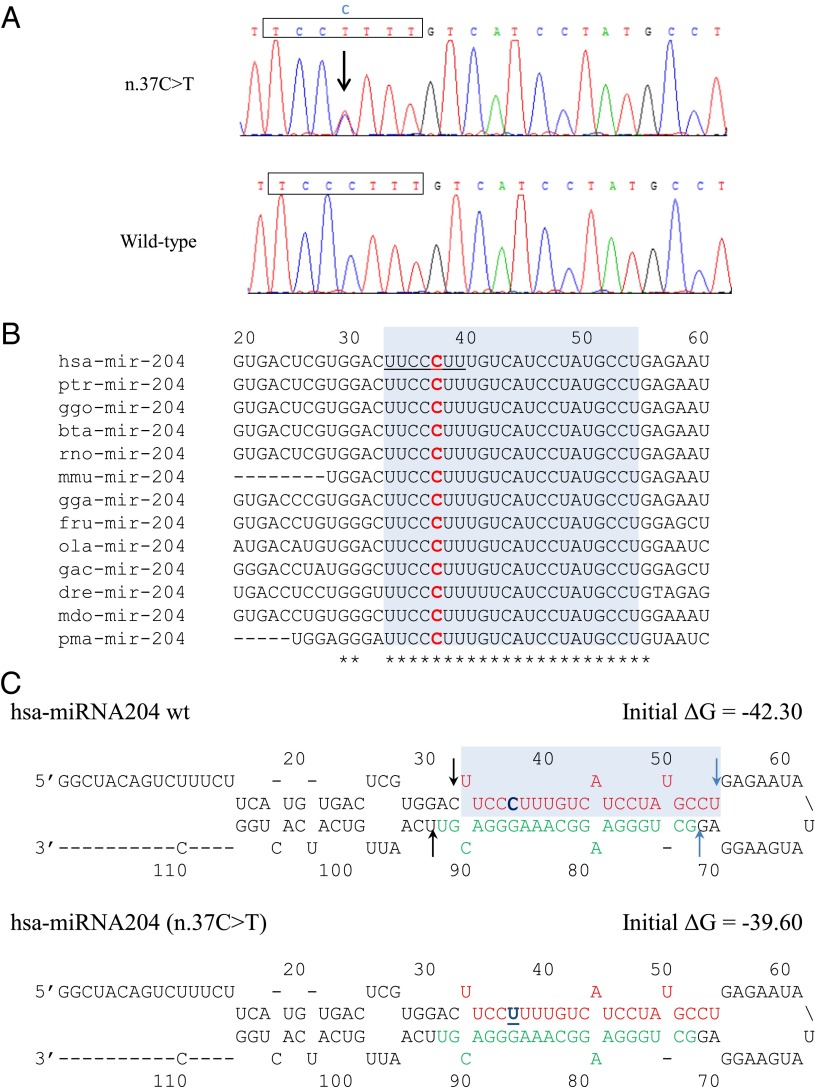
Sanger sequencing confirmed the MIR204 n.37C > T mutation segregates with the retinal dystrophy and coloboma phenotype in the family members who were tested (II-1, III-1, III-2, III-3, IV-1, and V-1) and were absent in the nonaffected individuals tested (V-2, V-3) (Fig. 2A). The mutation is positioned at the fourth nucleotide within the highly conserved seven-nucleotide seed region of the 5p arm of the precursor microRNA [annotated according to mirBase, mirbase.org (12)] (Fig. 2B). Coinheritance of retinal dystrophy and MAC is considerably rare. Mutation analysis of an additional 21 cases of known retinal dystrophy with MAC with no family history identified no variants in miR-204. Screening of an additional cohort of 457 patients with isolated MAC and 672 patients with autosomal dominant retinal degeneration phenotypes revealed no novel mutations in miR-204. Moreover, this variant was not present in the control databases of the Exome Variant Server or 1000 Genomes Project.
Fig. 2.
The novel miR-204 mutation (n.37C > T) identified in a family with autosomal dominant retinal dystrophy and coloboma. (A) Electropherograms showing the 22-nt mature sequence of mir-204. Patient sequence (Top) contains the heterozygous mutation (n.37C > T), which segregated among affected individuals and the wt sequence (Bottom) observed in unaffected individuals. The boxed sequence indicates the 7-bp seed region. (B) Multiple species alignment showing segment of the mir-204 sequence, including the 5p arm (shaded area), obtained using CLUSTALW software. The human mutation at nucleotide position +37 is indicated in red, and the underlined sequence denotes the seed region. bta, Bos taurus; dre, Danio rerio; fru, Fugu rubripes; gac, Gasterosteus aculeatus; gga, Gallus gallus; ggo, Gorilla gorilla; Hsa, Homo sapiens; mdo, Monodelphis domestica; mmu, Mus musculus; ola, Oryzias latipes; pma, Petromyzon marinus; ptr, Pan troglodytes; rno, Rattus norvegicus. (C) Predicted secondary structures of wt and mut (n.37C > T) miR-204 precursors obtained using the m-fold algorithm. The nucleotide mut in the family is indicated in blue and underlined. The 5p arm is shown in red, whereas the 3p arm is shown in green. Predicted sites for Drosha (black arrows) and Dicer (blue arrows) cleavage are indicated.
The Effect of the n.37C > T Mutation on miRNA Target Recognition.
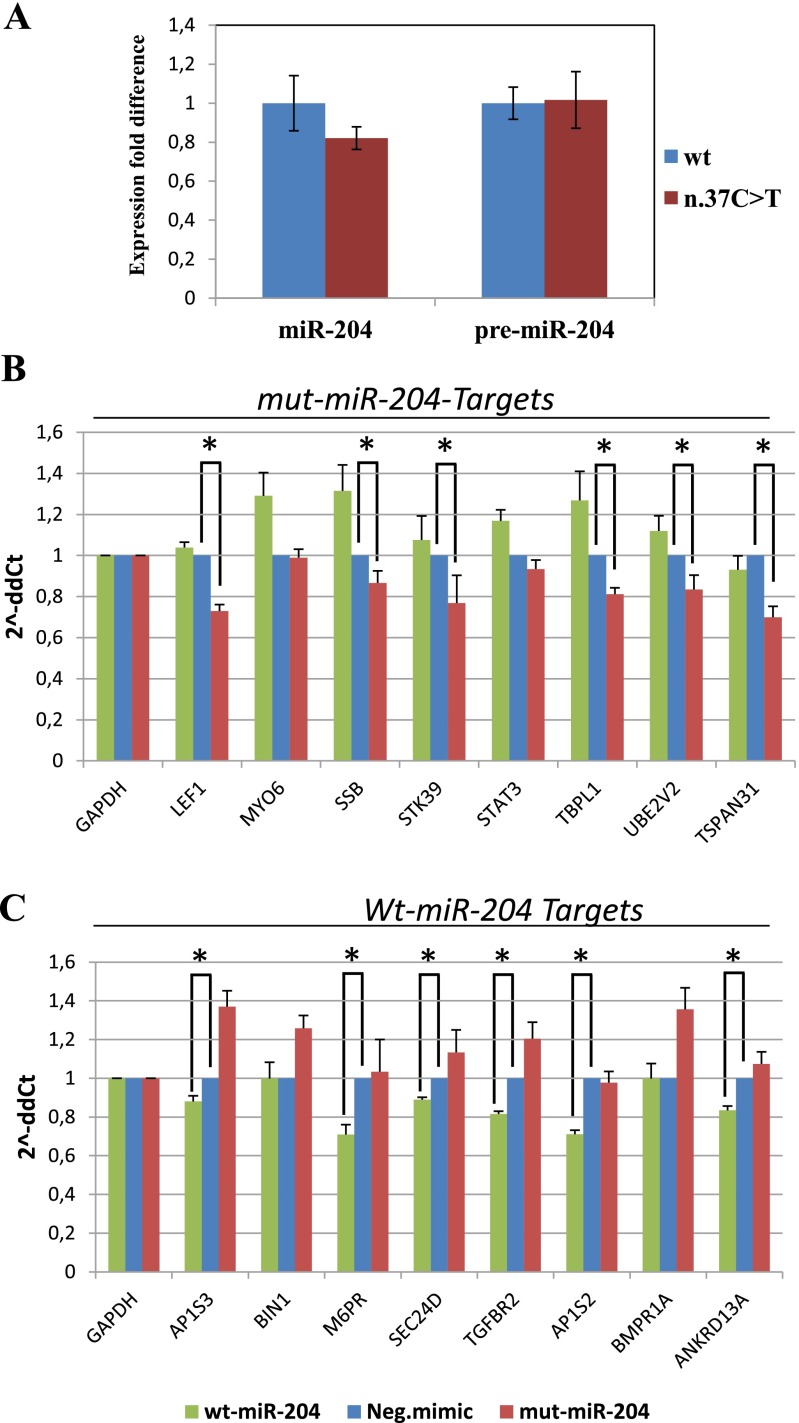
As the MIR204 mutation identified in our family lies within the seed region of the 5p arm, and therefore in the stem region of premiR-204, we used the m-fold algorithm (mfold.rit.albany.edu/) (13) to assess how the mutation identified might alter the predicted RNA secondary structure of this microRNA. Although the n.37C > T mutation introduced a base-pairing mismatch, the decrease in free energy value was small, no bulges were introduced into the RNA structure, and the regions critical for Drosha and Dicer processing of pri-miRNA were not affected (Fig. 2C; nucleotides 32 and 92 for Drosha and nucleotides 54 and 71 for Dicer) (14). Therefore, the n.37C > T mutation was not predicted to destabilize the secondary structure of premiR-204 (Fig. 2C). Because mutations in miRNA sequence have been previously suggested to alter premiRNA processing (15), we examined the effect of the variant on the biogenesis of miR-204. Expression vectors containing the wild-type (wt) or the mutated (mut) form of precursor sequences of hsa-miR-204 were transiently transfected into HeLa cells, and expression levels of mature processed miRNA (miR-204) and of the precursor species (premiR) for both the wt and mut forms were assessed by quantitative RT-PCR (qRT-PCR), using specific custom-designed assays. Notably, the expression levels of both the miR-204 and premiR-204 species appeared largely unaffected by the n.37C > T mutation (17% decrease and 1.6% increase, respectively) compared with wt (Fig. S2A). This would suggest that these mechanisms do not account for the pathogenesis of this dominantly inherited phenotype.
Fig. S2.
Effect of miR-204 mutation on miRNA biogenesis and targeting capabilities. (A) The effect of the n.37C > T mutation on premiR-204 processing was evaluated by qRT-PCR, using a standard curve method. Variations in the expression levels of the mature miRNA and premiR-204 in the mutant (n.37C > T) were compared with the wt levels (set as 1). Values are presented as the mean ± SD (represented as a percentage of variation) of three independent experiments, each performed in triplicate. Wt-miR-204 mimics (blue bars), mut-miR-204 mimics (red bars). (B and C) qRT-PCR validation of either mut-miR-204–specific (B) and wt-miR-204–specific (C) predicted target mRNAs on total RNA obtained from cells transfected with mut-miR-204 mimics (red bars), wt-miR-204 mimics (green bars), and negative (control) mimics (blue bars). The histograms show expression fold change vs. controls (2^ΔΔCt values, y axis). The GAPDH gene was used to normalize expression levels. Six of eight genes tested in B indeed behave as specific targets for the mut-miR-204, and six of eight genes tested in C behave as specific targets for the wt-miR-204. *Adjusted P < 0.05 in a one-tail t test. ANKRD13A, ankyrin repeat domain 13A; AP1S2, adaptor-related protein complex 1, sigma 2 subunit; AP1S3, adaptor-related protein complex 1, sigma 3 subunit; BIN1, bridging integrator 1; BMPR1A, bone morphogenetic protein receptor, type IA; LEF1, lymphoid enhancer-binding factor 1; MYO6, myosin VI; M6PR, mannose-6-phosphate receptor; SEC24D, SEC24 family member D; SSB, Sjögren syndrome antigen B; STAT3, signal transducer and activator of transcription 3; STK39, serine threonine kinase 39; TBPL1, TBP-like 1; TGFBR2, transforming growth factor, beta receptor II; TSPAN31, tetraspanin 31; UBE2V2, ubiquitin-conjugating enzyme E2 variant 2.
Because the mutation lies within the seed region of the mature miRNA, it was more likely that this change would affect recognition of target genes. This could occur through two different modalities; that is, either impaired recognition of bona fide miR-204 targets (loss-of-function mechanism) or creation of novel aberrant target sites (gain-of-function mechanism). To evaluate the relative contribution of either of these mechanisms, we carried out a microRNA target prediction with the TargetScan (www.targetscan.org) and Diana-microT (diana.cslab.ece.ntua.gr/microT/) tools, using as query the miR-204 sequence containing the n.37C > T variant. Intriguingly, we found that the mutated (mut-)miR-204 sequence was predicted to target a much higher number of mRNAs (mut-miR-204 targets; n = 1,129) compared with the wt sequence (wt-miR-204 targets; n = 557) (Dataset S1). Only 135 mRNA were predicted to be targeted by both the wt and the mut-miR-204 sequence, suggesting the n.37C > T sequence variant could indeed cause a notable alteration of miR-204 targeting properties.
To validate the above predictions and to obtain a more global assessment of the effect of the n.37C > T sequence variant in the miR-204 sequence, we carried out transcriptome analysis by RNAseq in a human RPE cell line (ARPE-19) (16) in which miR-204 is expressed at low levels (17). After transfecting ARPE-19 cells with microRNA mimics corresponding to either the wt- or mut-miR-204 sequence, we compared the results obtained from their RNAseq transcriptome profiles. We reasoned that if the target predictions were reliable, predicted wt-miR-204 targets should be enriched within the genes down-regulated in wt-transfected cells, whereas predicted mut-miR-204 targets should be enriched within the genes down-regulated in mut-transfected cells.
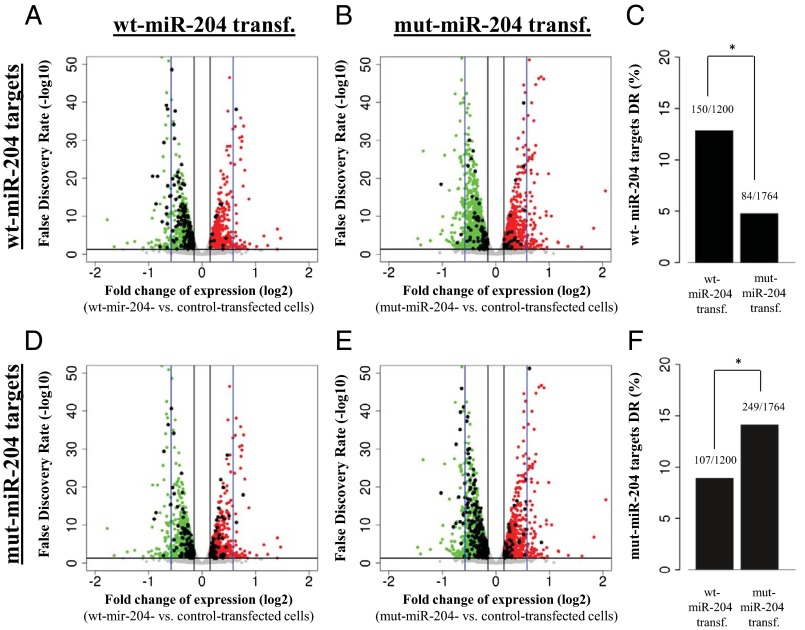
First, we found that the number of significantly down-regulated genes (fold change decrease of at least 10% at a false discovery rate < 0.05 with respect to a negative mimic control) was higher in mut-miR-204–transfected (n = 1,764) than in wt-miR-204–transfected (n = 1,200) ARPE-19 cells, supporting the notion that the mutation has a significant effect at the transcriptome level. We then looked at the representation of predicted targets within down-regulated genes and found that the predicted targets specific for wt-miR-204 were more significantly enriched in the genes down-regulated in cells transfected with wt mimics (150/1,200) (Fig. 3 A and C) than in genes down-regulated in cells transfected with mut mimics (84/1,764) (Fig. 3 B and C); P < 0.0001 (Fig. 3C). In contrast, predicted target genes specific for mut-miR-204 were more significantly enriched in the genes down-regulated in cells transfected with mut-mimics (249/1,764) (Fig. 3 E and F) than in the genes down-regulated in cells transfected with wt-mimics (107/1,200) (Fig. 3 D and F); P <0.0001 (Fig. 3F). The complete lists of the wt-miR-204 and mut-miR-204 predicted targets found to be down-regulated are available, respectively, as Datasets S2 and S3.
Fig. 3.
The n.37C > T sequence variation confers altered targeting capabilities to miR-204, as assessed by transcriptome analysis. (A–F) Schematic representations of the behavior of predicted targets for either wt-miR-204 (A–C) or mut-miR-204 (D–F) in RNA-Seq experiments carried out in ARPE-19 cells transfected with either wt-miR-204 (A and D) or mut-miR-204 mimics (B and E). (A, B, D, and E) X axis, expression fold changes (on a log2 scale) versus control ARPE-19 cells (i.e., negative mimic transfected). Y axis, statistical significance of expression changes represented as false discovery rate values on a log10 scale. The black horizontal line indicates a false discovery rate of 0.05, and the black vertical lines indicate the 10% expression fold change thresholds chosen to identify genes showing significant expression changes. The blue vertical lines mark the 50% expression fold change value. Red dots indicate the genes that show statistically significant up-regulation in each experiment. Green dots indicate the genes that show statistically significant down-regulation in each experiment. Gray dots represent genes that do not display significant expression changes. Black dots in A and B represent genes that are predicted to be targets of the wt-miR-204, whereas in D and E, they represent genes that are predicted to be targets of the mut-miR-204; that is, carrying the n.37C > T variation. Wt-miR-204–specific predicted targets are significantly more enriched in the genes down-regulated in cells transfected with wt mimics (A) than in genes down-regulated in cells transfected with mut mimics (B), as quantified in the graph shown in C (*P < 0.0001). Conversely, mut-miR-204–specific predicted targets are significantly more enriched in the genes down-regulated (DR) in cells transfected with mut-mimics (E) than in the genes down-regulated in cells transfected with wt-mimics (D), as quantified in the graph shown in F (*P <0.0001).
To provide independent validation of the presented RNAseq analysis, we carried out qRT-PCR assays on two small subsets, respectively, of “newly generated” predicted targets, that is, those specifically down-regulated in mut-miR-204–transfected cells, but not in wt-miR-204–transfected cells, and of “lost” predicted targets, that is, those specifically down-regulated in wt-miR-204–transfected but not in mut-miR-204 transfected cells. The qPCR assays indeed validated the RNAseq results for 6/8 newly generated predicted targets and for 6/8 of the predicted lost targets analyzed (Fig. S2 B and C). Together, these data strongly suggested that the n.37C > T mutation leads to a significant alteration of miR-204 function, both by generating a large number of novel and aberrant direct targets and by impairing the recognition of some wt targets.
Injection of the n.37C > T mut-miR-204 Causes Severe Ocular Malformations Associated with Retinal Dystrophy in Vivo.
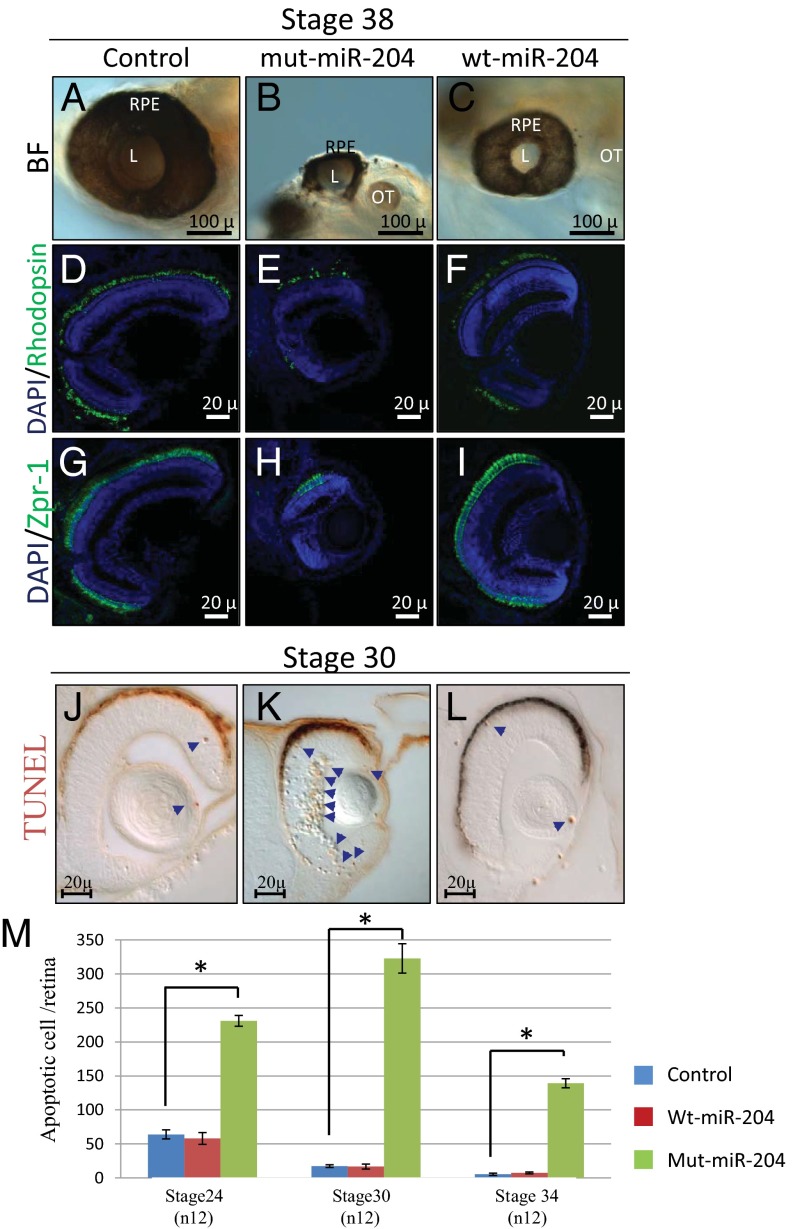
In the human genome, miR-204 is located within intron 8 of the transient receptor potential (TRP) channel gene, TRPM3 on chromosome 9q21.12. Deletions within 9q21, including those specifically encompassing the TRPM3 gene, have not been reported to cause ocular phenotypes; instead, features of mental retardation, epilepsy, speech delay, autistic behavior, and moderate facial dysmorphology have been reported (18–21). This suggests that the heterozygous n.37C > T mutation may not act via haploinsufficiency but, rather, through a gain-of-function mechanism. To further test this hypothesis, we overexpressed mut-miR-204 in medaka fish. Embryos injected with mut-miR-204 at the one-cell stage showed an aberrant eye phenotype (90 ± 5% of 1,500 injected embryos; Table S1) from St19, corresponding to optic vesicle formation, onward. In particular, growth of the eyecup was significantly impaired and culminated at St38 in evident microphthalmia and optic coloboma in comparison with control-injected embryos (Fig. 4 A and B). These phenotypic alterations were different and more severe than those observed after overexpression of the wt-miR-204 mimic (Fig. 4C).
Table S1.
Summary of phenotypic effects of microRNA mimic injections in medaka embryos
| Injected mimic | Number of injected embryos | Concentration of injected mimic, μM | Frequency of phenotype observed, % ± SD | |||
| Microphthalmia | Coloboma | Anophthalmia | Lethality | |||
| Control | 1,250 | 4 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 2 ± 1 |
| Wt-miR-204 | 2,350 | 4 | 87 ± 3 | 0 ± 0 | 0 ± 0 | 3 ± 2 |
| Mut-miR-204 | 2,350 | 4 | 90 ± 2 | 56 ± 3 | 2 ± 3 | 3 ± 2 |
Fig. 4.
The n.37C > T miR-204 mutation has a deleterious effect in vivo. (A–C) Bright-field microscopy images of lateral views eyes from control-injected (A), mut-miR-204–injected (B), and wt-miR-204–injected (C) embryos. Representative frontal eye sections immunostained with an anti-Rhodopsin (D–F; green) and ZPR-1 (G–I; green) antibodies, from st38 control-injected (D and G), mut-miR-204–injected (E and H), and wt-miR-204–injected (F and I) medaka fish. Sections are counterstained with DAPI (blue). We observed a significant reduction in the staining of both photoreceptor rods and cones in mut-miR-204–injected medaka fish compared with control-injected and wt-miR-204–injected animals. (J–L) Frontal vibratome sections of TUNEL-stained control-injected (J), mut-miR-204–injected (K), and wt-miR-204–injected embryos. A significant increase of cell death in the eyes of mut-miR-204–injected embryos was observed compared with both wt-miR-204– and control-injected embryos. Some TUNEL-positive cells are marked by blue arrows. (M) Quantification of TUNEL-positive cells in the retina of control-injected (blue bars), wt-miR-204–injected (red bars), and mut-miR-204–injected (green bars) embryos at three different embryonic stages; *P < 0.0001 in likelihood ratio tests. L, lens; OT, otic vesicle; RPE, retinal pigment epithelium. (Scale bars, 100 and 20 μm.)
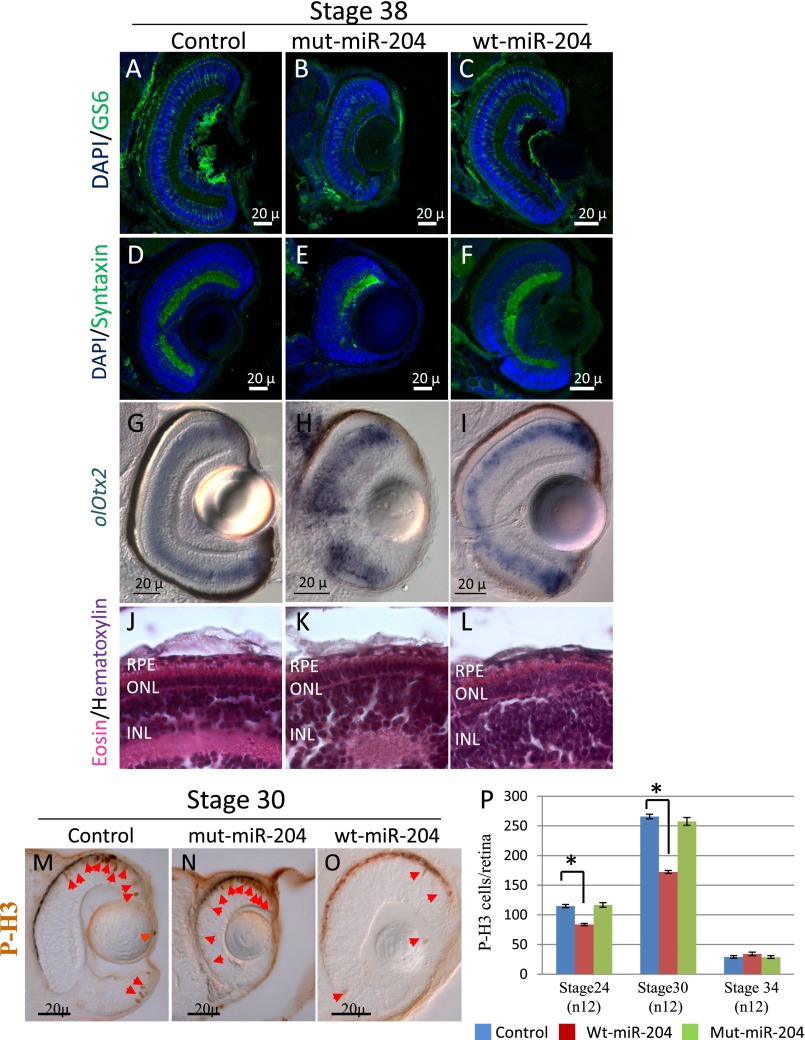
To assess whether the n.37C > T variation also had an effect on retinal development, we analyzed the retinas of mut-miR-204–injected embryos at St.38 (n = 20 eyes). Throughout the entire retina, we observed a notable reduction of both rod and cone photoreceptor cells in mut-miR-204 overexpression in comparison with both wt-miR-204 and control injected embryos, as assessed by immunofluorescence analysis with anti-Rhodopsin and anti-Zpr-1 antibodies (Fig. 4 D–I), respectively. We also observed by immunofluorescence a significant reduction of Muller glial, amacrine, and retinal ganglion cells (Fig. S3). Unlike photoreceptor cells, the latter reductions were only observed in the ventral part of the retina, suggesting the ventral retina alterations are mainly a result of the presence of the aberrant coloboma. We did not observe any apparent alteration of bipolar cells as assessed by RNA in situ hybridization (ISH) assays (Fig. S3). Moreover, we did not detect any apparent morphological defects in the retinal pigment epithelium of both mut-miR-204– and wt-miR-204–injected embryos with respect to controls, as determined by hematoxylin and eosin staining (Fig. S3 J–L). We next asked whether changes in apoptosis and/or cell proliferation were associated with the ocular phenotype observed in mut-miR-204 embryos. From the optic cup stage onward, the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay, a specific method to detect cell death, revealed a significant increase in the number of TUNEL-positive apoptotic cells in retinal tissue (but not in the RPE) in mut-miR-204–injected embryos in comparison with both wt-miR-204– and control-injected embryos (Fig. 4 J–L). Notably, no significant differences were observed in the number of TUNEL-positive cells between wt-miR-204–injected and control retinas (Fig. 4 J and L). In contrast, we did not observe any alteration in the number of proliferating retinal cells in mut-miR-204–injected in comparison with both wt-miR-204– and control-injected embryos, as determined by immunostaining for phosphorylated histone H3, a specific marker for cells in the M-phase (Fig. S3). In contrast, the onset of the morphologically visible phenotype of wt-miR-204 overexpression was associated with a significant decrease in the number of phosphorylated histone H3-positive cells (Fig. S3). These findings indicate that the ocular phenotype observed in mut-miR-204–injected embryos is likely a result of activation of cell apoptosis.
Fig. S3.
The n.37C > T miR-204 mutation has a deleterious effect in vivo. (A–F) Representative frontal eye sections immunostained with an anti-GS6 (A–C; green) and Syntaxin (D–F; green) antibodies, from st38 control-injected (A and D), mut-miR-204–injected (B and E), and wt-miR-204–injected (C and F) medaka fish. A significant signal reduction is visible in the sections stained with GS6 that marks Muller glial cells (A–C) and Syntaxin that marks both amacrine and ganglion cells (D–F). Sections are counterstained with DAPI (blue). (Scale bars, 20 μm.) (G–L) Frontal vibratome sections of st38 control-injected (G), mut-miR-204–injected (H), and wt-miR-204–injected (I) medaka fish processed for whole-mount RNA ISH with antisense probes for olOtx2. No apparent alterations in bipolar cells, marked by olOtx2, were observed. (J–L) Representative retinal frontal sections of control-injected (J), mut-miR-204–injected (K), and wt-miR-204–injected (L) medaka fish processed for hematoxylin/eosin staining. No apparent morphological differences are visible in the retinal pigment epithelium (RPE). ONL, outer nuclear layer; INL, inner nuclear layer. (M–O) Frontal vibratome sections of St30 control-injected (M), mut-miR-204–injected (N), and wt-miR-204–injected (O) medaka embryos immunostained with antiphospho-histone H3 (P-H3) antibodies. (P) Quantification of PH3-positive cells in the retina of control-injected, mut-miR-204–injected, and wt-miR-204–injected injected embryos at different embryonic stages, as indicated. No significant differences were detected between mut-miR-204–injected and control-injected embryos. In contrast, a significant decrease was detected in wt-miR-204 eyes compared with controls. Red arrows, P-H3-positive cells. *P < 0.05.
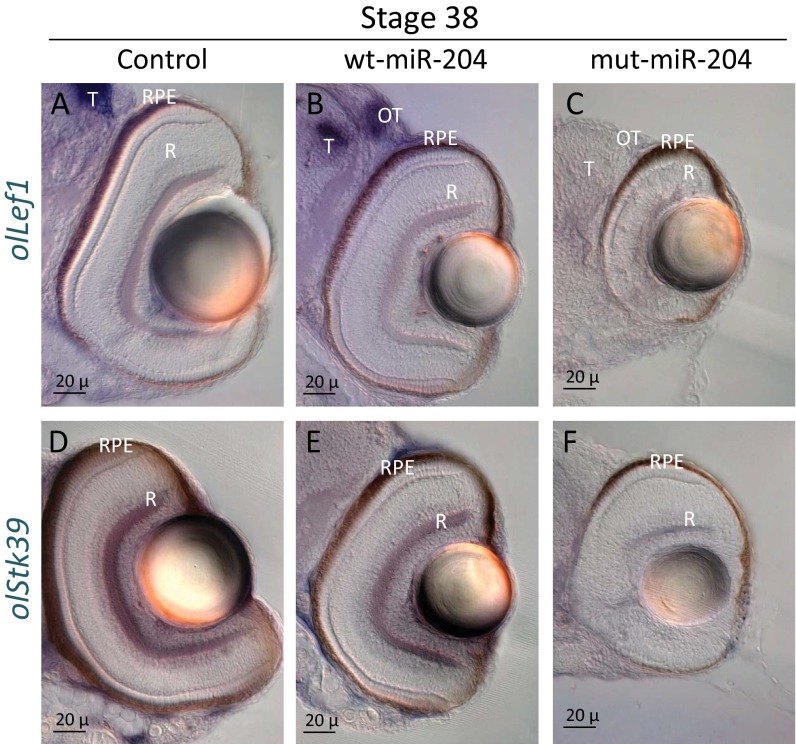
Finally, we decided to analyze in vivo the behavior of two of the mut-miR-204–specific predicted targets we had also validated by qRT-PCR assays in ARPE-19 cells; namely, Lef1 (lymphoid enhancer-binding factor-1) and Stk39 (serine threonine kinase 39) (Fig. S4). Lef1 encodes a transcription factor involved in the Wnt signaling pathway (22), whereas Stk39 encodes a serine/threonine kinase with a possible role in the cellular stress response pathway (23). Specifically, we analyzed by RNA ISH their expression in control, wt-miR-204–injected, and mut-miR-204–injected medaka embryos. Notably, we observed a reduction of their expression domains in mut-miR-204–injected embryos compared with both wt-miR-204 and control embryos (Fig. S4). Conversely, no apparent alterations of their expression domains were observed in wt-miR-204–injected compared with control embryos (Fig. S4). Of note, we did not observe in mut-miR-204–injected embryos any gross lens alteration, unlike that previously described in miR-204 knockdown in medaka (10). Overall, these data strongly support in vivo the pathogenic role of the n.37C > T variation in miR-204 in the complex phenotype observed in our family, which is likely exerted through a gain-of-function mechanism.
Fig. S4.
Expression of olLef1 and olStk39 is altered in mut-miR-204–injected medaka embryos (A–F) Frontal vibratome sections of st38 control-injected (A and D), mut-miR-204–injected (B and E), and wt-miR-204–injected (C and F) medaka fish processed for whole-mount RNA ISH with antisense probes for olLef1 (A–C) and olStk39 (D–F). The expression domains of both olLef1 and olStk39 are not altered in wt-miR-204 (C and F) but are reduced in mut-miR-204–injected embryos (B and E) compared with control-injected embryos (A and D). OT, optic tectum; R, retina; RPE, retinal pigment epithelium; T, telencephalon.
MiR-204 Has a Physiological Role in Photoreceptor Function.
MiR-204 has previously been shown to be expressed in the developing eye of medaka fish and mouse (3, 10), and we previously reported that knockdown of miR-204 in medaka fish was linked to microphthalmia, abnormal lens development, eye coloboma, and defects in axon projections of retinal ganglion cells (9, 10, 24). To confirm a physiological role of miR-204 also in the human eye, we performed RNA ISH experiments on adult human retina. This confirmed that miR-204 is expressed in the adult human retina and revealed expression in the retinal ganglion cell layer, inner nuclear layer, and retinal pigment epithelium. Importantly, this also demonstrated expression in the human photoreceptor cell layer (Fig. S5).
Fig. S5.
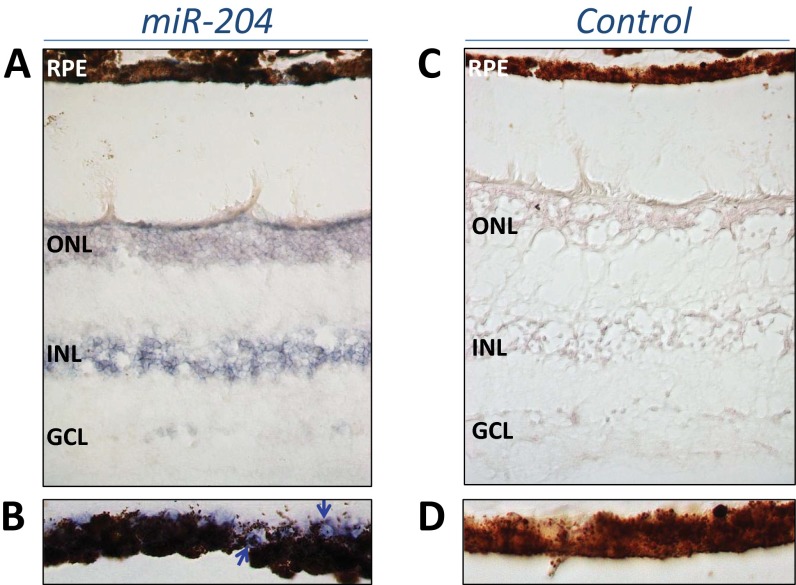
Expression of miR-204 in the human retina. (A) RNA ISH with a miR-204 probe on sections from human adult retina. Expression signal is detected in the ganglion cell layer (GCL), in the inner nuclear layer (INL), and in the outer nuclear layer (ONL). (B) Details, at higher magnification, of a retinal pigment epithelium (RPE) area. MiR-204 signal can be detected also in RPE cells (blue arrows). Please note that the pigmentation interferes with a proper RNA ISH signal detection in the RPE. (C) RNA ISH with a scrambled miRNA probe (negative control). (D) Higher magnification of an RPE area from the control experiment.
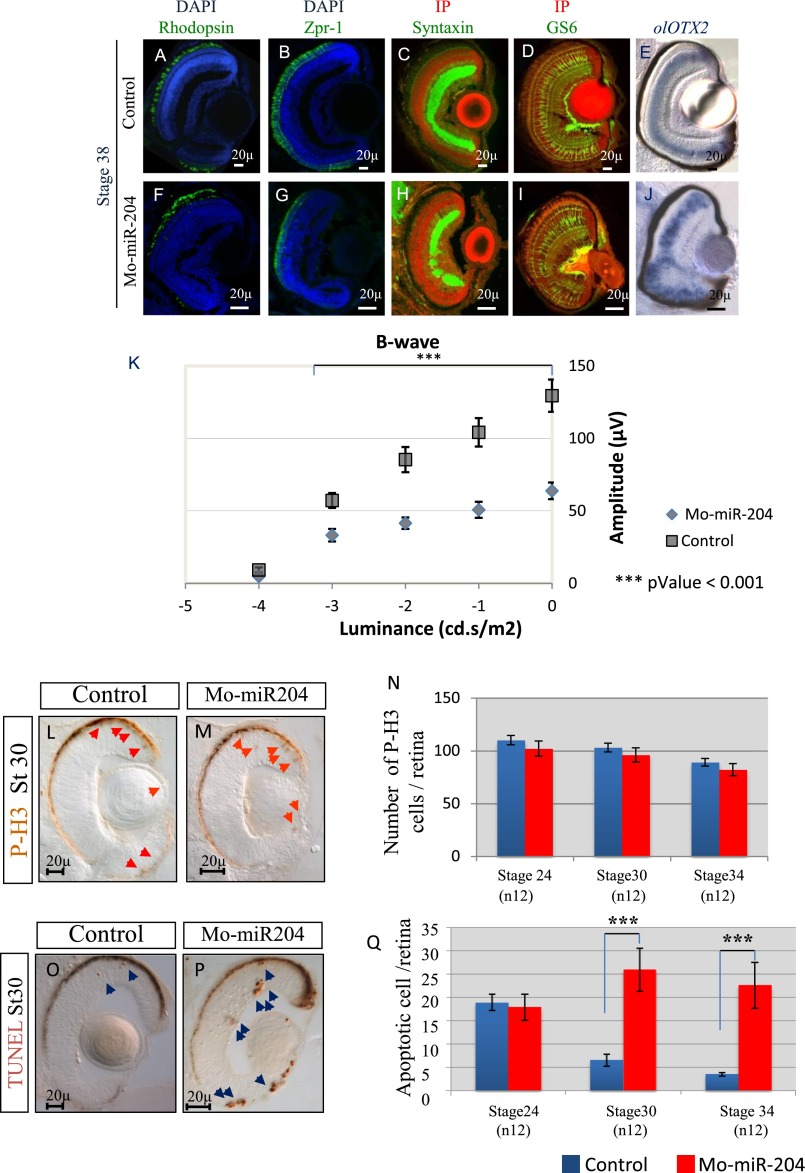
To test the possible contribution of miR-204 in photoreceptor homeostasis, we therefore interfered with miR-204 processing and activity in medaka fish, using a multiblocking morpholino (Mo), as previously described (9, 25), and assessed the consequences in the differentiated retina. Comparison of the morphology of the differentiated retina of control and Mo-miR-204–injected embryos revealed that morphant retinas displayed significant alteration in photoreceptor marker staining, which was more evident in ventral regions (Fig. S6). In particular, we observed by immunofluorescence a moderate reduction of rhodopsin-positive mature rod photoreceptors and of Zpr-1-positive mature cone photoreceptors (Fig. S6). In contrast, we did not observe in morphant fish any apparent alterations in the number of other retinal neuronal cells, as determined by expression analysis of Syntaxin, GS6, and olOtx2 (Fig. S6). We also observed an increased apoptosis, as determined by TUNEL assays, and no alterations in cell proliferation by phosphorylated histone H3 staining (Fig. S6). Finally, we recorded flash electroretinogram (ERG) under scotopic conditions from both control and miR-204 morphant larvae. We observed a significant reduction of the amplitude of the b-wave in miR-204 morphant compared with control medaka larvae (n = 30) (Fig. S6). These data confirm that miR-204 is necessary for normal photoreceptor differentiation and function in vertebrates.
Fig. S6.
Loss of function of miR-204 has a deleterious effect on photoreceptor maintenance and function in vivo. Representative frontal eye sections of St38 control-injected (A and B) and Mo-miR-204–injected (F and G) medaka fish embryos immunostained with anti-Rhodopsin and Zpr-1 antibodies (green). Sections are counterstained with DAPI (blue). We observed a significant reduction in the intensity of both rhodopsin and Zpr-1 staining in Mo-miR-204–injected medaka fish compared with controls. Representative frontal eye sections of St38 control-injected (C and D) and Mo-miR-204–injected (H and I) medaka fish embryos immunostained with anti-Syntaxin and GS6 antibodies (green). No significant differences in GS6 staining were detected between morphants and control embryos. In Mo-miR-204–injected embryos, amacrine and ganglion cells do not form a proper inner plexiform layer, as described in ref. 22) (green signal). Sections are counterstained with propidium iodide (IP; red signal). (E and J) Frontal vibratome sections of wt and Mo-miR-204–injected medaka embryos processed for whole-mount RNA ISH with antisense probe for olOtx2. No apparent alterations in bipolar cells were observed, as determined by expression analysis of olOtx2 (E and J). (K) Intensity response curves of mean b-wave amplitude (±SEM) of control-injected and Mo-miR-204 injected larvae (n = 30). The b-wave amplitudes of morphant larvae are significantly smaller with respect to controls over the 5 units of luminance intensity tested (shown on the x axis in a logarithmic scale). (L and M) Frontal vibratome sections of St30 control-injected and Mo-miR-204–injected embryos immunostained with antiphospho-histone H3 (P-H3) antibodies. (N) Quantification of PH3-positive cells in the retina of the control-injected or Mo-miR-204–injected embryos at different embryonic stages as indicated. No significant changes were detected between morphants and controls. Red arrows, P-H3-positive cells. (O and P) Frontal vibratome sections of St30 TUNEL-stained control and miR-204 morphant embryos. (O and P) A significant increase of cell death in miR-204 morphant eyes was observed compared with control. (Q) Quantification of TUNEL-positive cells in the retina of control-injected or Mo-miR-204–injected embryos at different embryonic stages; blue arrows, TUNEL-positive cells. ***P < 0.0001 in in likelihood ratio tests. (Scale bars, 20 μm.)
Discussion
In this study we describe a five-generation family with an overlapping phenotype of inherited retinal dystrophy and optic fissure closure defect, iris coloboma. To elucidate the underlying molecular basis of this phenotype, we undertook a combined linkage and exome sequencing approach identifying four potentially linked regions, one of which, on chromosome 9q21, contained a previously undescribed heterozygous mutation in MIR204 (n.37C > T) that segregated with the phenotype observed in this family. Given the well-documented role of miR-204 in eye development, this mutation was a strong candidate for further investigation.
MiR-204 has discrete and dynamic expression domains in the eye throughout development, being expressed in the RPE, neural retina, lens, and ciliary body in fish, mouse, and human (3, 4, 10, 11, 26–28). This miRNA has a closely related paralog in mammals; namely, miR-211, with which it shares the same seed sequence and very likely a similar set of targets and tissue expression pattern, although, in the latter respect, precise information is still lacking. A large body of evidence indicates that miR-204 plays a crucial role in the differentiation and function of all of the ocular structures in which it is expressed. In vitro assays have shown that miR-204 may be involved in the correct differentiation and function of RPE cells (27, 29, 30). MiR-204 is also required for proper lens development (9, 10). We previously reported that this action is exerted through a complex cross-talk with the transcription factor Pax6, a “master regulator” of eye development. We found that although Pax6 controls the expression of miR-204 (11), miR-204 can itself repress Pax6 through the targeting of the transcription factor Meis2 (10). Additional target genes that play a role in miR-204 action in lens differentiation are Ankrd13a (9), Sox11 (11), and Hmx1 (31). Finally, miR-204 controls the establishment of the proper dorsal-ventral axis, and its inactivation was linked to coloboma in medaka fish (10).
Despite its pervasive role in many aspects of ocular differentiation, miR-204 has not previously been reported to be significantly expressed in photoreceptor cells (3, 11, 25, 26, 28) or to have an effect on photoreceptor function. Here, we provide for the first time to our knowledge RNA ISH data for miR-204 in the human retina, which also reveals its expression in photoreceptors (Fig. S5). We demonstrate that knockdown of miR-204 function in medaka fish leads to progressive alteration and death of photoreceptor cells (Fig. S6). These data strongly indicate that miR-204 is also required for photoreceptor development and function. The significant expression of this miRNA in the photoreceptor cell layer in human (Fig. S5) supports the hypothesis that miR-204 could exert its role on photoreceptor function through a cell autonomous mechanism. However, a contribution to a noncell autonomous action mediated by the strong expression of miR-204 in the RPE is also likely (3, 11, 28). There are many examples of RPE-specific genes that play essential roles in photoreceptor function and whose mutations have a pathogenic role in inherited retinal dystrophies (32, 33). The dissection of the molecular mechanisms through which miR-204 regulates photoreceptor function requires further study.
This is the first report to our knowledge of a pathogenic mutation within miR-204 identified as the cause of an ocular disease. It is supported by two main considerations: First, the n.37C > T mutation falls within the mature miR-204 seed region, which is considered to be the most essential sequence domain for target recognition and down-regulation (34). We demonstrate in vitro that this variation has an effect on target mRNA recognition (Fig. 3 and Fig. S2), which would be anticipated to lead to significant dysregulation of gene expression in eye tissues both via natural or aberrantly generated miR-204 targets. Second, we demonstrate that the n.37C > T variation has a deleterious effect in vivo. In particular, we found that the overexpression of a miR-204 mimic carrying the n.37C > T mutation in medaka fish generates a dramatic eye phenotype characterized by gross malformations (including microphthalmia and coloboma) and photoreceptor abnormalities (Fig. 4). These results not only clearly support the deleterious role played by the n.37C > T mutation but also support its pathogenic role in the complex form of retinal dystrophy observed in the family we report in this study.
We have identified a miR-204 mutation in a single large, family. We did not identify this mutation, or additional miR-204 mutations, in patients with either isolated inherited retinal dystrophies or MAC; that is, phenotypes partly overlapping that observed in our family. However, the combination of retinal dystrophy and ocular coloboma is very rarely observed in clinical practice, in particular when inherited as an autosomal dominant trait. It is therefore conceivable that this particular miR-204 mutation underlies the pathogenesis of an extremely rare ocular condition. This is in line with the limited number of reports of mutations within microRNAs associated with Mendelian disorders to date. The first report of mutations within a miRNA contributing to a disease phenotype was by Mencía et al., who identified two point mutations within the seed region of miR-96, which segregated with the affected patients from two separate Spanish families with autosomal dominant progressive hearing loss (35). The resulting mutations directly affected the miR-96 biogenesis and resulted in a significant reduction in the silencing of target genes. Soldà et al. reported a mutation in the 3p arm of MIR96 in a family with nonsyndromic inherited hearing loss (15). This mutation was found to impair correct maturation of miR-96, leading to a significant decrease in the expression level of the mature 3p and 5p microRNA arms. The only other example of a microRNA with point mutations being associated with a Mendelian disorder is represented by MIR184. Hughes et al. identified a mutation in the central nucleotide of the seed region of MIR184 as the cause of keratoconus and early-onset anterior polar cataracts in a large Irish family, although the underlying pathogenic mechanism was not extensively delineated (36). Iliff et al. reported the same mutation as the cause of EDICT syndrome, an autosomal dominant anterior segment dysgenesis syndrome characterized by endothelial dystrophy, iris hypoplasia, congenital cataract and stromal thinning (37). A further two novel heterozygous substitution variants, neither of which were located within the seed region, were later identified in MIR184 in two patients with isolated keratoconus (38).
There are several reasons to explain the paucity of genetic diseases caused by miRNA mutations. First, given their shortness, it is expected that the number of potential mutations affecting miRNA mature sequences will be significantly lower compared with the number of mutations falling within functional sequence elements of protein coding genes. Second, given the fact that each miRNA can target hundreds of different genes, mutations affecting critical nucleotides of mature sequences may give rise to phenotypes that depend on the target specificity affected by each mutation. Mutations altering miRNA function may not only be localized within their mature sequences but also in their binding sites within the 3′-UTR of target genes, as described in the cases of spastic paraplegia (39) and X-linked chondrodysplasia (40). Such mutations in the 3′-UTR would be expected to selectively affect specific aspects of miRNA actions, and consequently to give rise to disease phenotypes that will differ from those observed when mutations lie in the miRNA sequence itself. The assessment of the true effect of 3′-UTR mutations on genetic disorders will require analysis of gene regions thus far neglected in mutation screens.
The n.37C > T mutation results in an autosomal dominant phenotype and thus could, in principle, act either through loss of function (i.e., by impairing recognition of wt-miR-204 targets) or gain of function (by creating novel aberrant recognition sites in genes that should not normally be targeted by miR-204) mechanisms. We hypothesize that the gain-of-function mechanism exerts a relatively greater role for the following three reasons: First, both bioinformatic predictions and transcriptome studies indicate that the n.37C > T mutation is able to generate a much larger dataset of predicted targets compared with the wt-miR-204 mature sequence (Fig. 3 and Dataset S1). Some of these were also validated both in vitro and in vivo (Figs. S2 and S4), including among the latter, Lef1. Lef1 has previously been shown to play a role in retina (41–43) and RPE differentiation (44). Second, the injection of the mut-miR-204 in medaka fish leads to a striking eye phenotype that is much more severe than that obtained with the wt-miR-204. In this respect, it is important to underline that the notable difference in phenotypic severity between the mut-miR-204–injected medaka and the patients carrying the n.37C > T mutation will be contributed to by the fact that although in medaka there is ectopic overexpression of the mut miRNA throughout the developing embryo, in patients the mutation exerts its deleterious effect only in the tissues and in the stages in which miR-204 is normally expressed.
Finally, there are previous clinical data that seem to further exclude haploinsufficiency. MiR-204 is located within intron 8 of the TRP channel gene, TRPM3, on chromosome 9q21.12. Deletions within 9q21 and those specifically encompassing the TRPM3 gene have not been reported to cause ocular phenotypes, instead causing features of mental retardation, epilepsy, speech delay, autistic behavior, and moderate facial dysmorphology (18–21). Deletion of this region has been described in association with an autism phenotype in a family in which a paternal deletion of exons 1–9 of TRPM3, which included MIR204, was shared by two affected sons and an unaffected daughter who were not described to have an ocular phenotype or visual symptoms. Furthermore, at least 30 patients with deletions in this region are listed on the Decipher database, a web-based resource and database of genomic copy number variation data from analysis of patient DNA; none is described with coloboma or retinal disease, although one had optic nerve hypoplasia. Altogether, the above data indicate that the n.37C > T mutation mostly acts with a gain-of-function mechanism, although we cannot exclude that the loss of some miR-204 wt targets may still contribute to the phenotype observed in our family.
This study represents the first example to our knowledge of a microRNA-caused genetic disease most likely occurring via a gain-of-function mechanism; further studies will need to be undertaken to examine the possibility that this mechanism applies to other micro-RNA-caused genetic disease. This is also the first example to our knowledge of a microRNA mutation with a causative role in inherited retinal dystrophies in patients, as to date, microRNAs have been linked to such diseases only in mouse (45).
In summary, we have identified a new genetic cause for the pathogenesis of complex forms of eye diseases particularly characterized by the combination of retinal dystrophy and developmental abnormalities. Our findings contribute to highlighting miR-204 as a new putative “master regulator” of eye development and, in particular, have been instrumental in uncovering its newly identified role in photoreceptor survival and function. Finally, our results shed further light on the recognition of the role of miRNAs as primary pathogenic agents in human genetic diseases.
Materials and Methods
Subjects.
A single five-generation family with a highly unusual and visually disabling phenotype consisting of retinal dystrophy and bilateral iris coloboma presented at the retinal and genetic clinic of the Manchester Royal Eye Hospital in the United Kingdom. A total of nine family members exhibited a similar phenotype (six female and three male) (Fig. 1A). To elucidate the molecular basis of this phenotype, we enrolled eight family members for clinical and molecular genetic analysis, including six affected individuals. Ethical approval and informed consent were obtained from all study participants. The research adhered to the tenets of the Declaration of Helsinki.
Disease status was determined by a full medical, ophthalmic, and family history combined with clinical examination and electrophysiological evidence in proband V-1 and his mother, patient IV-1. Medical records for other family members were reviewed with the patients’ written consent. Ophthalmic examination included anterior segment examination by slit lamp biomicroscopy with evaluation of anterior chamber depth using the Von Herrick method, intraocular pressure measurements, and dilated fundus examination and photography. Optical coherence tomography (OCT) for retinal thickness assessment using the Heidelberg spectral-domain OCT (Heidelberg engineering) was performed with autofluorescence and color imaging. Full-field flash ERGs were also carried out in individual V-1. Individual IV-1 underwent ERG testing in the 1980s. ERGs were recorded following the standards of the International Society for Clinical Electrophysiology of Vision (46). Both scotopic rod-driven responses and cone-driven photopic single flash and 30-Hz flicker stimuli were recorded sequentially.
Linkage Analysis.
Genomewide SNP analysis was carried out using the Affymetrix Genome-Wide SNP6.0 microarray. Genotypes were generated using the genotyping console software provided by Affymetrix, using the Birdseed V2 algorithm and a confidence threshold of 0.005. A subset of SNPs was selected for linkage analysis based on having a minor allele frequency of 0.2 to reduce the problems associated with linkage disequilibrium when using dense marker sets. Parametric multipoint linkage analysis was performed using Merlin (csg.sph.umich.edu/abecasis/Merlin/index.html), using a dominant model with full penetrance.
Exome Sequencing and Variant Prioritization.
Whole-exome target enrichment and sequencing were performed on 3 μg of DNA extracted from peripheral blood of two affected family members, proband V-I and patient II-I (Fig. 1A). For patient V-I, enrichment was performed using the SureSelect Human All Exon v.1 Enrichment kit (Agilent Technologies) for the Illumina HiSeq system, according to the manufacturer’s protocol. Sequencing was carried out on a HiSEq. 2000 sequencer (Illumina Inc.), following the manufacturer’s protocols (Hong Kong Co.). Sequence data were mapped to the hg19 reference human genome, using BWA software. Variants were called using GATK v2.4.7 software and then filtered for those SNPs with ≤5× coverage. Approximately 4 GB of sequence mapped uniquely to the genome reference hg19, with 69.4% of the exome covered at 20-fold or higher. SNPs and insertions/deletions were initially annotated to genes using Ensembl v68, and the functional consequences were then defined against an in-house list of Refseq transcripts (www.ncbi.nlm.nih.gov/refseq). Using Ensembl’s defined consequence hierarchically system, the highest affecting consequence for a variant in a gene was retained. To find likely pathogenic changes, an in-house hierarchy system of functional consequence was used to prioritize variants, as detailed in ref. 47. Briefly, variants were filtered out if they were present in dbSNP136 (unless seen in the Human Gene Mutation Database, www.hgmd.org), seen more than once in our in-house variant database, or present in the 1000 Genomes Project or the Exome Variant Server. Novel changes were analyzed further using the in-silico tools SIFT (sorting intolerant from tolerant) (48), PolyPhen-2 (49), and splicing prediction tools via the software Alamut (Interactive Biosoftware, LLC) to assess their pathogenicity.
For patient II-1, enrichment was performed using TargetSeq exome enrichment (Life Technologies), following the manufacturer’s protocols. Emulsion PCR was conducted on the resultant enriched sample library, and the sample was run indexed with unrelated samples on a SOLiD 5500xl sequencer following the manufacturer’s protocols. Sequence data were mapped to the hg19 reference human genome, using Lifescope software. Approximately 4.5 GB of sequence mapped uniquely to the genome reference hg19, with 68.2% of the exome covered at 20-fold or higher. Variants were called using a combination of the Lifescope software suite and SAMtools (freeware) (50) and then filtered for those SNPs with ≤5× coverage. SNPs were annotated and filtered as for patient V-I.
PCR and Sanger Sequencing to Confirm Novel Variants.
Mutations identified in candidate genes by exome sequencing were confirmed by direct Sanger sequencing. PCR amplification of MIR204 (MI0000284) was performed using the following oligonucleotide primers: F-5′-GGACTTCCTGATCGCGTA-3′ and R-5′-TTTCACTCCTTCCTAATTCCAGA-3′. Amplicons were sequenced using the BigDye Terminator v3.1 system (Life Technologies) and analyzed on an ABI 3730 DNA Analyzer (Life Technologies).
Cell Transfections, RNA Extraction, and Analysis.
ARPE-19 cells (16) were cultured at 37 °C in a humidified chamber supplemented with 5% (vol/vol) CO2. Cells were seeded at 135,000 cells/mL in 24-multiwell plates and transfected with DNA constructs using the PolyFect transfection reagent (Qiagen) according to the manufacturer’s instructions. Total RNA was extracted using TRIZOL (Invitrogen) according to the manufacturer’s instructions and used for subsequent RNA-Seq-based profiling, as previously described (24). RNA-Seq libraries were generated following the standard Illumina RNASeq protocol and sequenced using an Illumina HiSEq 1000 at the Next Generation Sequencing Facility of the Telethon Institute of Genetics and Medicine. Differential expression analysis of read counts was performed using the Generalized Linear Model approach for multiple groups implemented in the Bioconductor package “edgeR” (51, 52). The expression cutoff used was 1 count per million in at least three samples. qRT-PCR was performed to validate subsets of RNA-seq results, as previously described (10). Oligonucleotide primers used are listed in Table S2.
Table S2.
Oligonucleotide primer sequences used for qRT-PCR-validation of RNA-seq data
| Gene | Primer sequence |
| GAPDH | Forward 5′ ACT TCA ACA GCG ACA CCC ACT3′ |
| Reverse 5′ GCC AAA TTC GTT GTC ATA CCA G 3′ | |
| LEF1 | Forward 5′ CGA GAT CAG TCA TCC CGA AG 3′ |
| Reverse 5′ AGG TTT GTG CTT GTC TGG CC 3′ | |
| MYO6 | Forward 5′ TGG GAC AAG ACC ACC TCA TG 3′ |
| Reverse 5′ TTT GCC GGC TCC TGA TTC TC 3′ | |
| SSB | Forward 5′ AAA ACT GGA TGA AGG CTG GG 3′ |
| Reverse 5′ GAT TTG CTC AAT GCT TCC AC 3′ | |
| STK39 | Forward 5′ GTG CCG TGA ACC TCG TTT TG 3′ |
| Reverse 5′ AGC AGA GAA GAG CTC CTG AG 3′ | |
| STAT3 | Forward 5′ CTC TAT CAG CAC AAT CTA CG 3′ |
| Reverse 5′ TGC AGT CTG TAG AAG GCG TG 3′ | |
| TBPL1 | Forward 5′ CAT CCT GCT GTG TGC TAT CG 3′ |
| Reverse 5′ GTA AAT CTG TTC CAC AGC AG 3′ | |
| UBE2V2 | Forward 5′ CAC TTA CAA GGT GGA CAG GC 3′ |
| Reverse 5′ AAT CTA ACT GAC GGA GGA GC 3′ | |
| TSPAN31 | Forward 5′ TTT GCG CTC TCA ACG TGG TC 3′ |
| Reverse 5′ AAG ACT CCC ACA GCA ATG AC 3′ | |
| AP1S3 | Forward 5′ TCA CAG GAC AAG CAG TTT TG 3′ |
| Reverse 5′ GCG TCA AGA GCT CAT TGT CC 3′ | |
| BIN1 | Forward 5′ ACG ACT ACA CGG CCA CTG AC 3′ |
| Reverse 5′ GGT TCC AGT CGC TCT CCT TC 3′ | |
| M6PR | Forward 5′ CTC TTT GAG ATG GAT AGC AG 3′ |
| Reverse 5′ TTT GGC TCC CAC TAC CAG TC 3′ | |
| SEC24D | Forward 5′ TCC AAA TGC CCA GTA CCA GC 3′ |
| Reverse 5′ TGG GTA AGA CAG TTG TGC GC 3′ | |
| TGFBR2 | Forward 5′ GCA TGA GCA ACT GCA GCA TC 3′ |
| Reverse 5′ ATA AAG TCA TGG TAG GGG AG 3′ | |
| AP1S2 | Forward 5′ ATG TGC AGC TTC CTT GAG TG 3′ |
| Reverse 5′ CCA GGG TAA TTA GTT CAT TG 3′ | |
| BMPR1 | Forward 5′ CTC ATC GAG ACC TAA AGA GC 3′ |
| Reverse 5′ TTC AAG GGC ACA TCA ACT TC 3′ | |
| ANKRD13A | Forward 5′ ACT ACC CCC TGC ACC TCC TA 3′ |
| Reverse 5′ GTC CCA AGG AAA CAG CAA GA 3′ |
qRT-PCR to Assess Biogenesis of miR-204.
psiUx plasmid constructs containing the wt or the mut form of precursor sequences of hsa-miR-204, generated as previously described (9, 10), were transfected into HeLa cells using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s instructions. RNA was extracted after 48 h, using the miRNeasy isolation kit (Qiagen). Reverse transcription was performed using the Taqman MicroRNA reverse transcription kit (Life Technologies) with specific custom-designed stem loop RT primers for the mature mir-204 (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGCAT-3′) and precursor mir-204 (5′-GGTCGTATGCAAAGCAGGGTCCGAGGTATCCATCGCACGCAT CGCACTGCATACGACCACGTCCCT-3′) products. Real-time PCR was performed using FastStart Universal SYBR Green Master mix (Roche Applied Sciences) and miRNA-specific forward and reverse primers (mature miR-204: F-5′- CGGCGTTTGTCATCCTATG-3′ and R-5′- GTGCAGGGTCCGAGGT-3′; PremiR-204: F-5′- CGGCGTTTGTCATCCTATG-3′ and R-5′- GAGCAGGGTCCGAGGT-3′). Expression of U6 snRNA, used to normalize RNA input, was assessed using a TaqMan Small RNA assay (Life Technologies), according to the manufacturer’s instructions. Expression values were generated using a standard curve method. In all experiments, EGFP was cotransfected as an internal control to normalize for transfection efficiency, and the expression was assessed using a Taqman gene expression assay (Life Technologies) according to the manufacturer’s instructions. Each assay was performed in triplicate, and data are presented as the mean ± SD of three individual experiments.
Medaka Stocks and miR-204 Mimic Mo Injections.
The Cab-strain of wt medaka fish (Oryzias latipes) was maintained as previously described (9). Embryos were staged according to Iwamatsu et al. (53). Mos (Gene Tools, LLC) and both wt and mut miRIDIAN Dharmacon miR-204 mimics were designed and injected into one-cell fertilized embryos, as previously described (10). The miR-204 Mo used in this study targets both identical copies of the mature miR-204 present in the medaka genome (10). All fish studies were conducted in strict accordance with the institutional guidelines for animal research and approved by the Italian Ministry of Health.
RNA in Situ Hybridization and Immunofluorescence.
Human retinal sections were obtained from cornea donors collected by the Fondazione Banca degli Occhi del Veneto) in compliance with the tenets of the Declaration of Helsinki. For miR-204 detection, we used the miRCURY detection miR-204 probe (Exiqon). In situ hybridization of 30 nM of probe was performed using the miRCURY LNA microRNA ISH Optimization kit at 53 °C, according to the manufacturer’s protocol (Exiqon), with minor modifications (3). As a negative control, a double-DIG-labeled LNA scrambled microRNA probe, included in the above kit, was hybridized in parallel at the same concentration and optimal hybridization temperature. Whole-mount RNA ISHs were performed, photographed, and sectioned (25-μm vibratome sections), as described by Conte et al. (25). Digoxigenin-labeled anti-sense and sense riboprobes for olCrx, olPax6, olOtx2, and olRhodopsin were used (25). Medaka embryos were cryostat-sectioned (12-μm), and immuno-labeling was performed as previously described (25), using mouse monoclonal antibodies to Rhodopsin (1:2,000; Sigma) and Zpr-1 (1:2,000; ZIRC) and the rabbit polyclonal antibody to Prox1 (1:200; Chemicon). Alexa-488-conjugated goat α-mouse and goat α-rabbit (1:1,000; Invitrogen) secondary antibodies were used. For the cell proliferation analysis, rabbit α-phospho-histone H3 (1:100; Cell Signaling Technology) and peroxidase-conjugated anti-rabbit antibody (1:200; Vector Laboratories) were used followed by 3,3′-diaminobenzidine staining, as previously described (54). Confocal images were acquired using the Zeiss microscope, LSM 710.
Detection of Apoptotic Cell Death.
The extent and distribution of apoptotic cell death was determined by TUNEL, using the In Situ Cell Death Detection Kit, POD (Roche), following the manufacturer’s protocol.
Fish ERG.
ERGs were recorded as described previously (55). Briefly, medaka larvae were dark adapted for 30 min. For recording, a reference electrode was placed in a 1% agarose in ddH2O. The larva was placed dorsal-up on a moist paper covering the reference electrode. The recording electrode with a tip diameter of ∼20 μm was filled with buffer E3 (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, and 0.33 mM MgSO4) and placed on the cornea of the larva. Light stimuli of 100 ms with interstimulus intervals of 7 s were applied. The light stimulus intensity was 665 l×.
Supplementary Material
Acknowledgments
We thank Francesco Salierno and Jinjing Zang for technical assistance; Frans Cremers, Carmen Ayuso, Susanne Kohl, Bernd Wissinger, Carlo Rivolta, Chris Inglehearn, Carmel Toomes, Dror Sharon, and Robert K. Koenekoop and the whole European Retinal Dystrophy Consortium for sharing with us genetic data and for support in mutation analysis; the other members of the UK Inherited Retinal Disease Consortium, including Chris Inglehearn, Alison Hardcastle, and Andrea Nemeth; David Fitzpatrick and Daniel Schorderet for the study of ocular developmental disorders; Diego Ponzin, Stefano Ferrari, and the Fondazione Banca degli occhi del Veneto for providing eye tissue samples; Dr. Neil Parry, who performed and interpreted the patient ERGs; and the Telethon Institute of Genetics and Medicine Bioinformatics and Next Generation Sequencing cores for technical assistance in the generation and analysis of the transcriptome data. This work was supported by funding from the National Institute for Health Research’s Manchester Biomedical Research Centre; a program grant from Fight for Sight (Grant 1801); funding support from Retinitis Pigmentosa Fighting Blindness; from the Italian Telethon Foundation (Grant TGM11SB2); from the European Union (FP7-People-2012-ITN, Programme EyeTN, agreement 317472); and from Regione Campania (L.R. 5/2002, year 2007). S. Barbato was the recipient of a European Molecular Biology Organization short-term fellowship and of a fellowship from the Company of Biologists Ltd.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401464112/-/DCSupplemental.
References
- 1.Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. Photoreceptor degeneration: Genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010;11(4):273–284. doi: 10.1038/nrg2717. [DOI] [PubMed] [Google Scholar]
- 2.Arora A, et al. Prediction of microRNAs affecting mRNA expression during retinal development. BMC Dev Biol. 2010;10:1. doi: 10.1186/1471-213X-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karali M, Peluso I, Marigo V, Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest Ophthalmol Vis Sci. 2007;48(2):509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- 4.Karali M, et al. miRNeye: A microRNA expression atlas of the mouse eye. BMC Genomics. 2010;11:715. doi: 10.1186/1471-2164-11-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 6.Damiani D, et al. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci. 2008;28(19):4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.La Torre A, Georgi S, Reh TA. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc Natl Acad Sci USA. 2013;110(26):E2362–E2370. doi: 10.1073/pnas.1301837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conte I, Banfi S, Bovolenta P. Non-coding RNAs in the development of sensory organs and related diseases. Cell Mol Life Sci. 2013;70(21):4141–4155. doi: 10.1007/s00018-013-1335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avellino R, et al. miR-204 targeting of Ankrd13A controls both mesenchymal neural crest and lens cell migration. PLoS ONE. 2013;8(4):e61099. doi: 10.1371/journal.pone.0061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conte I, et al. miR-204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci USA. 2010;107(35):15491–15496. doi: 10.1073/pnas.0914785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaham O, et al. Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet. 2013;9(3):e1003357. doi: 10.1371/journal.pgen.1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 15.Soldà G, et al. A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing. Hum Mol Genet. 2012;21(3):577–585. doi: 10.1093/hmg/ddr493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62(2):155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- 17.Kutty RK, et al. MicroRNA expression in human retinal pigment epithelial (ARPE-19) cells: Increased expression of microRNA-9 by N-(4-hydroxyphenyl)retinamide. Mol Vis. 2010;16:1475–1486. [PMC free article] [PubMed] [Google Scholar]
- 18.Boudry-Labis E, et al. A novel microdeletion syndrome at 9q21.13 characterised by mental retardation, speech delay, epilepsy and characteristic facial features. Eur J Med Genet. 2013;56(3):163–170. doi: 10.1016/j.ejmg.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Bartnik M, et al. Application of array comparative genomic hybridization in 102 patients with epilepsy and additional neurodevelopmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(7):760–771. doi: 10.1002/ajmg.b.32081. [DOI] [PubMed] [Google Scholar]
- 20.Pua HH, et al. Novel interstitial 2.6 Mb deletion on 9q21 associated with multiple congenital anomalies. Am J Med Genet A. 2014;164A(1):237–242. doi: 10.1002/ajmg.a.36230. [DOI] [PubMed] [Google Scholar]
- 21.Baglietto MG, et al. RORB gene and 9q21.13 microdeletion: Report on a patient with epilepsy and mild intellectual disability. Eur J Med Genet. 2014;57(1):44–46. doi: 10.1016/j.ejmg.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Huber O, et al. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59(1):3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 23.Johnston AM, et al. SPAK, a STE20/SPS1-related kinase that activates the p38 pathway. Oncogene. 2000;19(37):4290–4297. doi: 10.1038/sj.onc.1203784. [DOI] [PubMed] [Google Scholar]
- 24.Conte I, et al. The combination of transcriptomics and informatics identifies pathways targeted by miR-204 during neurogenesis and axon guidance. Nucleic Acids Res. 2014;42(12):7793–7806. doi: 10.1093/nar/gku498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conte I, et al. Proper differentiation of photoreceptors and amacrine cells depends on a regulatory loop between NeuroD and Six6. Development. 2010;137(14):2307–2317. doi: 10.1242/dev.045294. [DOI] [PubMed] [Google Scholar]
- 26.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282(34):25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 27.Wang FE, et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24(5):1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deo M, Yu J-Y, Chung K-H, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235(9):2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- 29.Li W-B, et al. Development of retinal pigment epithelium from human parthenogenetic embryonic stem cells and microRNA signature. Invest Ophthalmol Vis Sci. 2012;53(9):5334–5343. doi: 10.1167/iovs.12-8303. [DOI] [PubMed] [Google Scholar]
- 30.Adijanto J, et al. Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J Biol Chem. 2012;287(24):20491–20503. doi: 10.1074/jbc.M112.354761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X-Y, Zhang K, Jiang Z-Y, Cai L-H. MiR-204/miR-211 downregulation contributes to candidemia-induced kidney injuries via derepression of Hmx1 expression. Life Sci. 2014;102(2):139–144. doi: 10.1016/j.lfs.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Davidson AE, et al. Missense mutations in a retinal pigment epithelium protein, bestrophin-1, cause retinitis pigmentosa. Am J Hum Genet. 2009;85(5):581–592. doi: 10.1016/j.ajhg.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson DA, et al. Mutations in the gene encoding lecithin retinol acyltransferase are associated with early-onset severe retinal dystrophy. Nat Genet. 2001;28(2):123–124. doi: 10.1038/88828. [DOI] [PubMed] [Google Scholar]
- 34.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 35.Mencía A, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41(5):609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 36.Hughes AE, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. 2011;89(5):628–633. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iliff BW, Riazuddin SA, Gottsch JD. A single-base substitution in the seed region of miR-184 causes EDICT syndrome. Invest Ophthalmol Vis Sci. 2012;53(1):348–353. doi: 10.1167/iovs.11-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lechner J, et al. Mutational analysis of MIR184 in sporadic keratoconus and myopia. Invest Ophthalmol Vis Sci. 2013;54(8):5266–5272. doi: 10.1167/iovs.13-12035. [DOI] [PubMed] [Google Scholar]
- 39.Züchner S, et al. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am J Hum Genet. 2006;79(2):365–369. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon D, et al. A mutation in the 3′-UTR of the HDAC6 gene abolishing the post-transcriptional regulation mediated by hsa-miR-433 is linked to a new form of dominant X-linked chondrodysplasia. Hum Mol Genet. 2010;19(10):2015–2027. doi: 10.1093/hmg/ddq083. [DOI] [PubMed] [Google Scholar]
- 41.Kubo F, Takeichi M, Nakagawa S. Wnt2b controls retinal cell differentiation at the ciliary marginal zone. Development. 2003;130(3):587–598. doi: 10.1242/dev.00244. [DOI] [PubMed] [Google Scholar]
- 42.Cho S-H, Cepko CL. Wnt2b/beta-catenin-mediated canonical Wnt signaling determines the peripheral fates of the chick eye. Development. 2006;133(16):3167–3177. doi: 10.1242/dev.02474. [DOI] [PubMed] [Google Scholar]
- 43.Fotaki V, Smith R, Pratt T, Price DJ. Foxg1 is required to limit the formation of ciliary margin tissue and Wnt/β-catenin signalling in the developing nasal retina of the mouse. Dev Biol. 2013;380(2):299–313. doi: 10.1016/j.ydbio.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westenskow P, Piccolo S, Fuhrmann S. Beta-catenin controls differentiation of the retinal pigment epithelium in the mouse optic cup by regulating Mitf and Otx2 expression. Development. 2009;136(15):2505–2510. doi: 10.1242/dev.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lumayag S, et al. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc Natl Acad Sci USA. 2013;110(6):E507–E516. doi: 10.1073/pnas.1212655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marmor MF, et al. International Society for Clinical Electrophysiology of Vision ISCEV Standard for full-field clinical electroretinography (2008 update) Doc Ophthalmol. 2009;118(1):69–77. doi: 10.1007/s10633-008-9155-4. [DOI] [PubMed] [Google Scholar]
- 47.Gillespie RL, et al. Personalized diagnosis and management of congenital cataract by next-generation sequencing. Ophthalmology. 2014;121(11):2124, e1–e2. doi: 10.1016/j.ophtha.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 49.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwamatsu T. Stages of normal development in the medaka Oryzias latipes. Mech Dev. 2004;121(7-8):605–618. doi: 10.1016/j.mod.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 54.Beccari L, Conte I, Cisneros E, Bovolenta P. Sox2-mediated differential activation of Six3.2 contributes to forebrain patterning. Development. 2012;139(1):151–164. doi: 10.1242/dev.067660. [DOI] [PubMed] [Google Scholar]
- 55.Makhankov YV, Rinner O, Neuhauss SCF. An inexpensive device for non-invasive electroretinography in small aquatic vertebrates. J Neurosci Methods. 2004;135(1-2):205–210. doi: 10.1016/j.jneumeth.2003.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.