Significance
Mitochondria contain several hundreds of proteins. The mitochondrial content is regulated by the uptake and degradation of proteins. Stabilization of protein structure by disulfide bonds was proposed to drive protein accumulation in the intermembrane space of mitochondria. However, it remained unknown if structural alterations could lead to protein escape through the physiological barrier formed by the outer mitochondrial membrane. In this work, we present evidence for size-dependent retrograde movement of mitochondrial proteins to the cytosol. We identify the translocase of the outer mitochondrial membrane channel protein Tom40 as an exit route. Our results indicate that the retro-translocation serves as an important surveillance mechanism that regulates the abundance of intermembrane space proteins in response to changes in cellular physiology.
Keywords: mitochondria, protein biogenesis, protein transport, protein turnover, protein quality control
Abstract
The content of mitochondrial proteome is maintained through two highly dynamic processes, the influx of newly synthesized proteins from the cytosol and the protein degradation. Mitochondrial proteins are targeted to the intermembrane space by the mitochondrial intermembrane space assembly pathway that couples their import and oxidative folding. The folding trap was proposed to be a driving mechanism for the mitochondrial accumulation of these proteins. Whether the reverse movement of unfolded proteins to the cytosol occurs across the intact outer membrane is unknown. We found that reduced, conformationally destabilized proteins are released from mitochondria in a size-limited manner. We identified the general import pore protein Tom40 as an escape gate. We propose that the mitochondrial proteome is not only regulated by the import and degradation of proteins but also by their retro-translocation to the external cytosolic location. Thus, protein release is a mechanism that contributes to the mitochondrial proteome surveillance.
Mitochondrial biogenesis is essential for eukaryotic cells. Because most mitochondrial proteins originate in the cytosol, mitochondria had to develop a protein import system. Given the complex architecture of these organelles, with two membranes and two aqueous compartments, protein import and sorting require the cooperation of several pathways. The main entry gate for precursor proteins is the translocase of the outer mitochondrial membrane (TOM) complex. Upon entering mitochondria, proteins are routed to different sorting machineries (1–5).
Reaching the final location is one step in the maturation of mitochondrial proteins that must be accompanied by their proper folding. The mitochondrial intermembrane space assembly (MIA) pathway for intermembrane space (IMS) proteins illustrates the importance of coupling these processes because this pathway links protein import with oxidative folding (6–10). Upon protein synthesis in the cytosol, the cysteine residues of IMS proteins remain in a reduced state, owing to the reducing properties of the cytosolic environment (11, 12). After entering the TOM channel, precursor proteins are specifically recognized by Mia40 protein, and their cysteine residues are oxidized through the cooperative action of Mia40 and Erv1 proteins (7, 13–17). Mia40 is a receptor, folding catalyst, and disulfide carrier, and the Erv1 protein serves as a sulfhydryl oxidase. The oxidative folding is believed to provide a trapping mechanism that prevents the escape of proteins from the IMS back to the cytosol (10, 13, 18). Our initial result raised a possibility that the reverse process can also occur, as we observed the relocation of in vitro imported Tim8 from mitochondria to the incubation buffer (13). Thus, we sought to establish whether and how this process can proceed in the presence of the intact outer membrane (OM). Our study provides, to our knowledge, the first characterization of the mitochondrial protein retro-translocation. The protein retro-translocation serves as a regulatory and quality control mechanism for the mitochondrial IMS proteome.
Results and Discussion
Proteins That Are Unable to Fold Are Released From the IMS in Vivo.
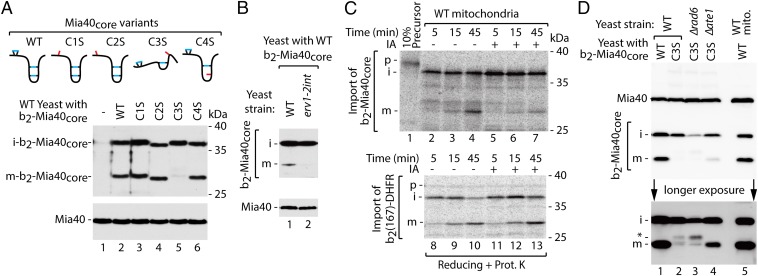
The folding trap was proposed as a mechanism for the IMS import of apocytochrome c (Cyc1) by the attachment of heme through the interaction with cytochrome c heme lyase (19). The oxidative folding trap also provides the commonly accepted rationale by which the MIA pathway acts to accumulate proteins in the IMS. This model indicates that protein accumulation in the IMS might be lost because of the unfolding caused by disulfide bond reduction. This, however, was difficult to experimentally prove because the in vivo accumulation of proteins and in organello import both demonstrate net results of the opposing reactions (i.e., oxidative folding by MIA and hypothetical backward movement of inefficiently oxidized precursors to the cytosol). We uncoupled inward from backward transport by fusing a MIA-dependent substrate protein with a presequence. The C-terminal domain of yeast Mia40, termed Mia40core, was fused with the bipartite presequence of cytochrome b2, resulting in b2-Mia40core (20). Native yeast Mia40 uses the translocase of the inner mitochondrial membrane 23 (TIM23) pathway for import and sorting into the inner membrane (IM), and mature Mia40 remains membrane-bound. In contrast, Mia40core uses the MIA import pathway and is soluble in the IMS (20). Fusion of the cytochrome b2 presequence to Mia40core allowed its MIA-independent import. Moreover, maturation of the fusion protein could be monitored based on size changes of the processed forms: the IM-bound intermediate form (i-form) cleaved by the mitochondrial processing peptidase MPP and the soluble mature form (m-form) additionally cleaved by the IM peptidase IMP (20). In such an experimental setup, inward transport no longer depends on MIA-driven oxidative folding, and backward movement from mitochondria could be dissected. The processing of b2-Mia40core and its four single cysteine-to-serine variants was monitored in cellular protein extracts (Fig. 1A). Two forms of the fusion protein, i-form and m-form, were detected. The mutant of b2-Mia40core, in which the third cysteine residue was replaced by serine (C3S), was an exception because this protein is likely unable to fold properly (21). No accumulation of the m-form was observed, whereas the i-form of b2-Mia40core-C3S accumulated at levels similar to other b2-Mia40core variants (Fig. 1A, lane 5). This indicates specific disappearance of m-b2-Mia40core-C3S. When wild-type b2-Mia40core was expressed in the strain with impaired oxidative folding (erv1-2int), the m-form was not accumulated, mimicking the behavior of b2-Mia40core-C3S (Fig. 1B). Similarly, when the radiolabeled b2-Mia40core precursor was imported into isolated mitochondria in the presence of iodoacetamide (IA), which inhibits oxidative folding by blocking thiol groups, the decrease in the m-form but not i-form was observed (Fig. 1C, Top). As no effect of the IA treatment on the import and processing of MIA-unrelated b2 fusion proteins was observed (Fig. 1C, Bottom, and Fig. S1A), we concluded that inhibition of oxidative folding by removing or blocking thiol groups resulted in the inefficient maintenance of proteins in the IMS.
Fig. 1.
Proteins that are defective in oxidative folding can be released from the IMS to the cytosol. (A) Schematic representation and cellular levels of b2-Mia40core variants. (B) Cellular levels of b2-Mia40core expressed in WT and erv1-2int strains. (C) Import of [35S]b2-Mia40core and [35S]b2(167)-DHFR into WT mitochondria with or without IA (50 mM). (D) Cellular levels of b2-Mia40core-C3S in WT, Δrad6, and Δate1 strains. Proteins were analyzed by SDS/PAGE and immunodetection (A, B, and D) or autoradiography (C). i, intermediate; m, mature; p, precursor; WT, wild type; *, altered migration of b2-Mia40core-C3S m-form.
The depletion of m-b2-Mia40core-C3S could be caused either by IMS proteases (22) or backward movement to the cytosol for proteasomal degradation (23). The m-b2-Mia40core-C3S form was not rescued by the deletion of any of the known yeast IMS proteases (i.e., Yme1, Atp23, and Prd1; Fig. S1B). Because the cleavage by IMP exposes glutamic acid residue at the N terminus of m-b2-Mia40core-C3S, we tested strains that are defective in the cytosolic Arg/N-end rule pathway involved in the proteasomal degradation of proteins with the N-terminal glutamic acid residue (Fig. 1D) (24). To enter the Arg/N-end rule pathway, the N-terminal glutamic acid residue requires arginine conjugation by the arginyl transferase Ate1. In Δate1 cells, the increased accumulation of the b2-Mia40core-C3S m-form was observed (Fig. 1D, lane 4). With deletion of the E2 ubiquitin-conjugating enzyme Rad6, the accumulation of a band of slightly slower migration was visible (Fig. 1D, lane 3), which indicated possible arginylation by Ate1 preceding ubiquitination by the Ubr1/Rad6 heterodimer and degradation (24, 25). The m-b2-Mia40core-C3S rescue in the Δrad6 and Δate1 cells indicated exposure of the m-form to the cytosolic quality control machinery supporting the release hypothesis. This process was incomplete owing to a possible contribution of other degradation mechanisms.
Disulfide Bond Reduction Causes Protein Escape from the IMS.
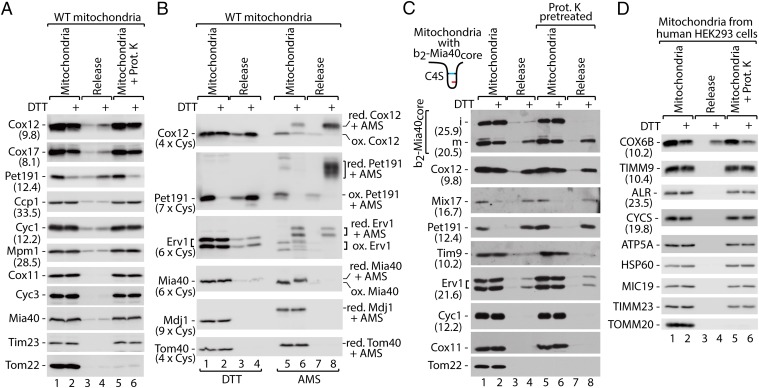
We established an in organello assay for the release of IMS proteins. The isolated mitochondria were treated with the reducing agent DTT to allow the reduction of disulfide bonds (Fig. 2A). The MIA substrate proteins Cox12, Cox17, and Pet191 were found to be decreased in the mitochondria and increased in the release fraction (Fig. 2A, lanes 1 and 2 vs. 3 and 4). The OM integrity was unaffected by DTT because the control IMS-soluble and MIA-independent proteins (Ccp1, Cyc1, and Mpm1) remained unaffected (Fig. 2A), whereas they were efficiently released upon hypoosmotic swelling (Fig. S2A, lanes 1–4). In addition, the OM integrity was demonstrated by treatment with the protease. Proteins, which were sensitive to protease treatment upon swelling of mitochondria (Cox11, Cyc3, Mia40, Mpm1, and Tim23; Fig. S2 A and B), remained protease-protected (Fig. 2A, lanes 5 and 6 vs. 1 and 2). The OM protein Tom22 was digested as expected (Fig. 2A). Thus, the release of proteins was not due to a compromised integrity of mitochondrial membranes.
Fig. 2.
Monitoring the release of MIA substrate proteins in organello. (A) Protein levels in WT mitochondria and corresponding supernatants (release) upon treatment with DTT. (B) Protein levels and redox state in WT mitochondria and supernatants (release) upon treatment with DTT followed by denaturation in the presence of thiol-modifying agent AMS. (C) Levels of b2-Mia40core-C4S in mitochondria and supernatants (release) upon reduction with DTT with or without protease pretreatment. (D) Protein levels in human HEK293 mitochondria and supernatants (release) upon reduction with DTT. (A–D) Proteins were analyzed by SDS/PAGE and immunodetection. Protein molecular weight (A, C, and D; kDa) or number of cysteine residues (B) is given in parentheses. i, intermediate; m, mature; ox, oxidized; red, reduced; WT, wild type.
The mitochondrial and release fractions were treated with an agent that reacts with free thiol groups causing the migration shift (Fig. S3A). The released Cox12, Pet191, and also Erv1 proteins were found exclusively in the reduced state (i.e., modified by AMS) in contrast to their more oxidized state observed in the mitochondria (Fig. 2B, lane 8 vs. 5). In the case of Pet191, the relocation was almost complete, whereas Cox12 and Erv1 were partially released (Fig. 2B, lane 4 vs. 2 and 8 vs. 6). The unreleased remainder of Cox12 and Erv1 in the mitochondrial fraction was partially reduced (Fig. 2B, lane 6). As expected, the DTT treatment also affected the redox state of the membrane-anchored Mia40 protein, whereas Mdj1 and Tom40 remained unaffected (Fig. 2B). A similar approach was used to test release susceptibility of small Tim proteins, a distinct class of MIA substrates. Tim8, Tim10, and Tim13 were found largely unaffected by the DTT treatment (Fig. S3B). This is in agreement with the observation that mature small Tim proteins, which are assembled in heterohexameric complexes, are resistant to reducing agents (13, 26).
Next, we used an in organello release assay to confirm the release of the b2-Mia40core m-form (Fig. 2C). The b2-Mia40core-C4S variant was used because the accumulation of its m-form was similar to wild-type protein (Fig. 1A), but it was likely more prone to reduction. The m-b2-Mia40core-C4S form was efficiently released similarly to Cox12, Mix17, and Pet191 (Fig. 2C). Mitochondria were pretreated with protease to exclude release of proteins, which due to incomplete import might have accumulated in transit—that is, at the OM. The comparison of samples with and without protease pretreatment excluded such a scenario (Fig. 2C, lanes 1–4 vs. 5–8). The in organello assay was also applied to mitochondria isolated from a human cell line (HEK293). The human MIA substrate protein COX6B was released from mitochondria and recovered in the release buffer (Fig. 2D; for analysis of control proteins, see Fig. S4 A and B). Thus, protein release is also a feature of human mitochondria.
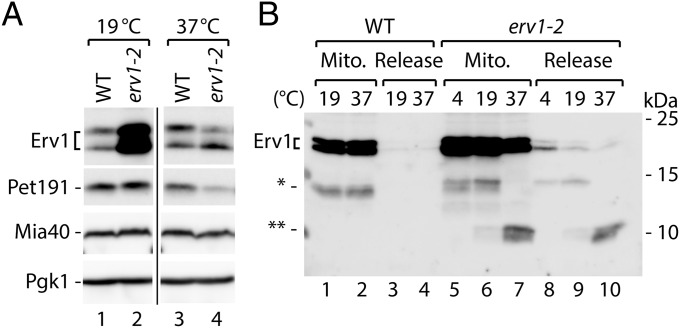
The temperature-sensitive variant of Erv1 protein Erv1-2 (27) was accumulated in the cells to higher levels than wild-type Erv1 under permissive conditions (Fig. 3A). Upon shifting to the restrictive temperature, the Erv1-2 levels dropped significantly (Fig. 3A). To test whether this effect was connected with release, mitochondria were subjected to incubation at various temperatures (Fig. 3B). During incubation at the restrictive temperature (37 °C), Erv1-2 was found to be processed and its degradation product was partially released from mitochondria (Fig. 3B, lanes 7 and 10). In this case, the increase in temperature, without reducing treatment, was sufficient to cause destabilization of the mutant protein, accompanied by the release of its small degradation derivative.
Fig. 3.
Erv1-2 is destabilized and released from mitochondria upon heat shock. (A) Cellular protein levels of WT and erv1-2 strains grown at 19 °C and 37 °C. (B) Levels of Erv1 and Erv1-2 proteins in mitochondria and supernatants (release) upon incubation at the indicated temperatures. (A and B) Proteins were analyzed by SDS/PAGE and immunodetection. WT, wild type; *, cleaved forms of Erv1.
Small Proteins Escape Mitochondria More Efficiently via the Tom40 Channel.
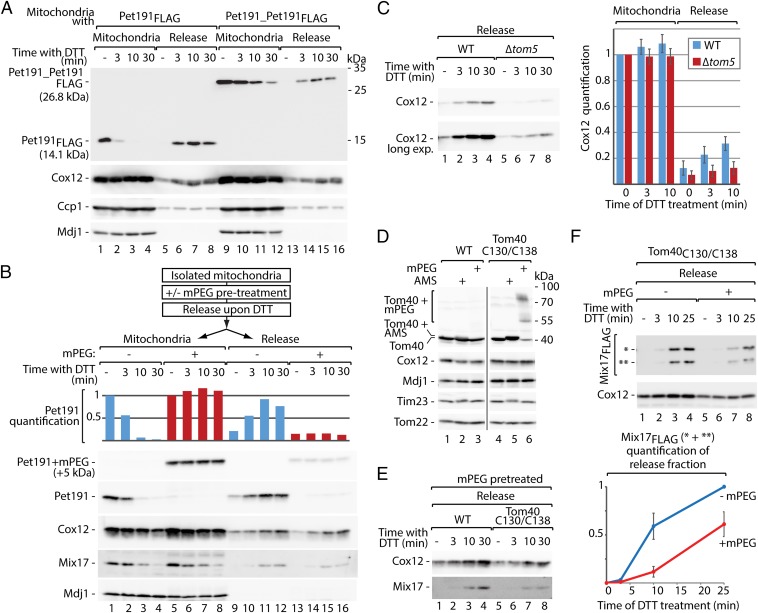
To analyze the impact of protein size on its ability to escape mitochondria, a tandem head-to-tail fusion of two Pet191 molecules and FLAG tag was created (Pet191_Pet191FLAG), the molecular mass of which was doubled. The tandem protein was released from mitochondria less efficiently than the corresponding monomer Pet191FLAG protein (Fig. 4A). Pet191 contains an odd number of cysteine residues, at least one of which is likely reduced and thus should be available for thiol-modifying agents. This property of Pet191 allowed modification with methoxypolyethylene glycol maleimide (mPEG) (5,000 Da), adding both molecular mass and structural complexity through the formation of a side chain. The release of the modified protein was almost completely abolished, in contrast to Cox12 and Mix17 proteins, which did not undergo modifications under these conditions (Fig. 4B). These results demonstrate that both linear protein length and 3D complexity are among the limiting factors for protein retro-translocation.
Fig. 4.
Protein release is size dependent and involves the TOM complex. (A) Release of Pet191FLAG and Pet191_Pet191FLAG from mitochondria upon treatment with DTT. (B) Release of Pet191 upon modification with mPEG (5,000 Da) and quantification. Signals from mitochondrial fractions not treated with DTT were set to 1. (C) Protein release from WT and Δtom5 mitochondria upon treatment with DTT and quantification. Signals from the mitochondrial fractions of WT and Δtom5 not treated with DTT were set to 1. Mitochondria fractions are shown in Fig. S5A. Mean ± SEM (n = 3). (D) Susceptibility of Tom40 and Tom40C130/C138 to thiol-modifying agents (AMS or mPEG) under native conditions. (E) Protein release from WT and Tom40C130/C138 mitochondria pretreated with mPEG upon treatment with DTT. Mitochondria fractions are shown in Fig. S5E. (F) Mix17FLAG release from Tom40C130/C138 mitochondria, with or without pretreatment with mPEG, upon treatment with DTT and quantification. The signal from the sample not treated with mPEG and incubated with DTT for 25 min was set to 1. Signals from DTT untreated samples were subtracted. Mean ± SEM (n = 3). Mitochondria fractions are shown in Fig. S5G. (A–F) Proteins were analyzed by SDS/PAGE and immunodetection. WT, wild type; *, full-size Mix17FLAG; **, Mix17FLAG degradation product.
The release of Cox12 upon DTT was decreased in the mitochondria isolated from Δtom5 compared with the wild-type cells (Fig. 4C; see Fig. S5A for mitochondrial samples). This raised the possibility that the TOM complex is responsible for the backward transfer of proteins. First, we excluded a direct effect of DTT on the cysteine residues of Tom40 by comparing the release from mitochondria with wild-type and cysteine-free variants of Tom40 (Fig. S5B). No difference in release efficiency was observed between Tom40CFREE and wild-type mitochondria (Fig. S5B). Next, to test the involvement of Tom40 in the release process, the Tom40C130/C138 variant was used, in which cysteine residues were placed in positions 130 and 138 (28) (Fig. S5C). The steady-state levels of mitochondrial proteins were not altered (Fig. S5D). The thiol groups of the engineered cysteine residues faced the lumen of Tom40 (Fig. S5C). This allowed for modification of Tom40C130/C138 with thiol-reacting agents in the intact mitochondria (Fig. 4D). Upon modification with mPEG (5,000 Da), spatial blockade was generated in the lumen of the Tom40C130/C138 channel that was recently demonstrated by us to inhibit protein import into mitochondria (29). Thus, we pretreated mitochondria with mPEG before the in organello release assay. The release of Cox12 and Mix17 from mitochondria with Tom40C130/C138 modified by mPEG was decreased compared with wild type (Fig. 4E). The efficient modification of Tom40C130/C138 in this experiment was confirmed by analysis of mitochondrial fractions (Fig. S5E). The Mix17FLAG fusion protein derivatives, which are present in mitochondria (Fig. S5F), were also released less efficiently upon channel blockade (Fig. 4F; see Fig. S5G for mitochondrial samples). These results show the involvement of the Tom40 channel protein in the mitochondrial protein release.
Protein Release Contributes to Mitochondrial Protein Quality Control.
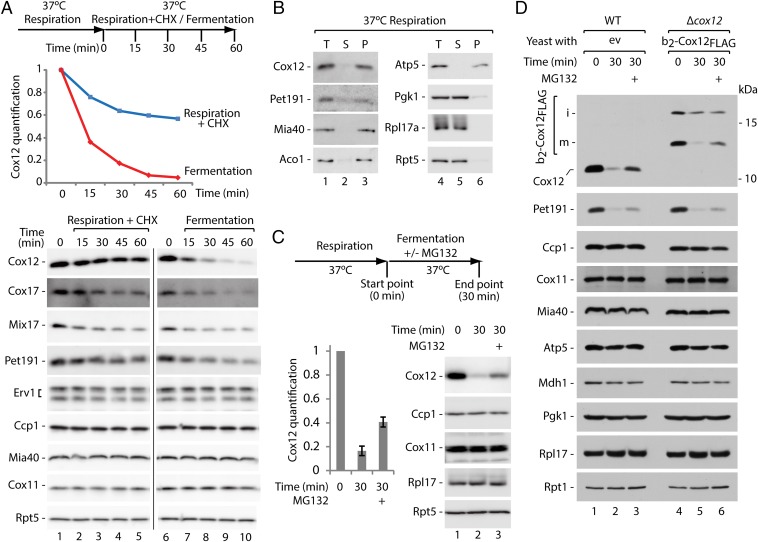
We sought to determine whether protein release is also important for cell physiology. The cells that were grown in respiratory medium were transferred to fermentative media. Under these conditions, mitochondrial biogenesis slows because oxidative phosphorylation is not needed. The redox environment in the cytosol has been reported to be more reducing during fermentation than during respiration (30), likely affecting the environment of the IMS (31). The in vivo analysis of protein steady-state levels revealed a decrease in MIA substrate proteins (i.e., Cox12, Cox17, Mix17, and Pet191) upon both inhibition of protein synthesis with cycloheximide (CHX) under respiratory conditions or transfer to glucose (Fig. 5A). Additionally, the decrease in protein levels (Cox12, Cox17, and Pet191) was faster upon transfer to glucose, and the protein levels were significantly reduced (aprox. 50% or less) only after 15 min (Fig. 5A, lanes 6–10). Cox12 and Pet191 were properly localized in mitochondria, similar to other mitochondrial proteins Mia40, Aco1, and Atp5 but in contrast to the cytosolic proteins Pgk1, Rpl17a, and Rpt5 (Fig. 5B). These observations indicated an active mechanism engaged in IMS protein removal during the switch from respiration to fermentation. We assessed whether Cox12 was accessible to the proteasome upon a shift to fermentation. Treatment with MG132 partially rescued the decrease in Cox12 (Fig. 5C). Similar stabilization was achieved in the presence of another proteasome inhibitor, PS341 (Fig. S6A). Thus, Cox12 was accessible to the ubiquitin-proteasome system in the cytosol. Next, we transformed wild-type cells with plasmids encoding FLAG-tagged MIA substrate proteins (Cox12FLAG or Pet191FLAG) under the control of a galactose-inducible promotor (23) to exclude a possibility that only newly synthesized proteins are substrates for the proteasome. The expression induced by galactose under respiratory conditions followed by the expression shutdown due to galactose removal was performed to minimize an effect of the new synthesis of ectopically expressed proteins (Fig. S6B, T1 vs. T0). Subsequently, cells were shifted to fermentation with or without the proteasome inhibitor. The levels of accumulated fusion proteins and their native counterparts were decreased upon a shift to fermentation and were rescued by proteasomal inhibition (Fig. S6B). These observations support the involvement of proteasomal degradation in the clearance of mature proteins that were released from mitochondria. Finally, the fusion protein of Cox12FLAG with the cytochrome b2 presequence was expressed in the strain lacking COX12. The b2-Cox12FLAG was functional because it complemented the respiratory growth defect of ∆cox12 (Fig. S6C). The fusion protein was present in the cells in two forms, i and m, similar to b2-Mia40core (Fig. 5D, lanes 4–6). When cells expressing b2-Cox12FLAG were shifted from respiratory to fermentative media, the IM-anchored i-form was decreased less efficiently and no rescue by the proteasome inhibition was observed. However, the m-form was strongly decreased upon fermentation and rescued upon proteasomal inhibition (Fig. 5D), indicating that the mitochondrially processed and IMS-soluble m-form of protein was released from mitochondria and cleared by the proteasome. These experiments show that the retro-translocation of mitochondrial proteins and their subsequent degradation in the cytosol is a physiological process that serves to deplete IMS proteins.
Fig. 5.
Respiration-to-fermentation shift induces protein escape to the cytosol and proteasomal clearance. (A) Cellular protein levels upon inhibition of protein synthesis or shift from respiration to fermentation. Experimental scheme and quantification of Cox12 are provided. Signals from time point 0 were set to 1. (B) Subcellular localization of MIA substrate proteins in the cells grown under respiratory conditions. (C) Protein levels upon shift from respiration to fermentation with or without MG132 treatment. Experimental scheme and quantification of Cox12 are provided. Signals from time point 0 were set to 1. Mean ± SEM (n = 7). (D) Protein levels in WT and ∆cox12 cells with plasmid expressing b2-Cox12FLAG upon shift from respiration to fermentation with or without MG132 treatment. Experiment was done based on the scheme as in C. (A–D) Proteins were analyzed by SDS/PAGE and immunodetection. CHX, cycloheximide; ev, empty vector; i, intermediate; m, mature; P, mitochondrial pellet; S, postmitochondrial supernatant, T, total; WT, wild type.
The proteome of the IMS is well defined within boundaries of mitochondrial membranes—OM and IM. These boundaries allow for the efficient retention of soluble proteins unless the membrane is permeabilized chemically or by programmed cell death-related events (32). We found that IMS proteins escape from mitochondria, broadening our understanding of the means by which the mitochondrial proteome is maintained. Our recent study identified the role of the ubiquitin proteasome system in controlling the levels of precursor proteins destined to the IMS (23). The present study indicates that proteins can also relocate from the IMS back to the cytosol and thus become reexposed to the cytosolic quality control machinery. This mechanism can prevent the IMS accumulation of proteins with folding defects and also allow flexible adjustment of the proteome in response to changing needs. Thus, we propose a concept that, in addition to protein import and degradation, the mitochondrial IMS proteome is also maintained by protein retro-translocation to the external cytosolic milieu.
Materials and Methods
Yeast Strains, Growth Conditions, and Biochemical Procedures.
The Saccharomyces cerevisiae strains used in this study are listed in Table S1. The growth conditions are described in SI Materials and Methods. The nomenclature for the proteins is according to the Saccharomyces Genome Database. Theoretical mass of proteins is presented in Figs. 1 A–D and 3A (for Ccp1 and b2-Mia40core, processed forms). The cell fractionation, protein and mitochondrial isolations, generation of mitoplasts, protease sensitivity assays, and import of radiolabeled precursor proteins into isolated mitochondria were performed according to the standard procedures and are described in SI Materials and Methods.
In Organello Protein Release Assay.
Isolated mitochondria were resuspended in the release buffer (250 mM sucrose, 5 mM MgCl2, 80 mM KCl, 10 mM Mops/KOH, 5 mM methionine, and 10 mM KH2PO4/K2HPO4 pH 7.2 for yeast mitochondria; 250 mM sucrose, 5 mM MgOAc, 80 mM KOAc, 10 mM sodium succinate, 5 mM methionine, and 20 mM Hepes/KOH pH 7.4 for human mitochondria) and incubated at 30 °C or 37 °C for yeast and human samples, respectively, in the presence of the protease inhibitors (P8215, Sigma) for 10 min followed by treatment with or without 50 mM DTT for 30 min or as indicated. Mitochondria were then reisolated. Supernatant fractions were precipitated with 10% (wt/vol) TCA, and mitochondrial pellets were denatured in 2× Laemmli buffer with 100 mM DTT at 65 °C for 15 min. For proteinase K treatment (25 μg/mL), the samples were incubated at 4 °C for 15 min and protease inhibitors were omitted.
Cysteine Residue Modification in Intact Mitochondria.
Isolated mitochondria were resuspended in the release buffer supplemented with 1.6–2 mM mPEG (5,000 Da) or 2 mM 4'-acetamido-4'-maleimidylstilbene-2,2'-disulfonic acid (AMS) for 30 min at 30 °C. The corresponding volume of the solvent was used as a mock control. The samples that were used in the subsequent protein release assay were further diluted with the release buffer to ≤1 mM mPEG concentration. Alternatively, the mitochondria were reisolated by centrifugation, denatured in reducing Laemmli buffer, and analyzed directly.
Supplementary Material
Acknowledgments
We thank Bernard Guiard, Judith Müller, and Christophe Wirth for materials and discussion. Research in the A.C. laboratory is supported by the Foundation for Polish Science-Welcome Programme, cofinanced by the European Union within the European Regional Development Fund and Polish National Science Centre (NCN) Grant 2011/02/B/NZ2/01402. P.B. and A.G. were supported by the Foundation for Polish Science-Welcome Programme stipends. P.B., M.W., and P.S. were supported by NCN Grants DEC-2013/11/D/NZ1/02294, 2012/05/B/NZ3/00781, and 2011/03/N/NZ3/01614, respectively. Research in the N.W. laboratory is supported by the Excellence Initiative of the German Federal & State Governments (EXC 294 BIOSS).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504615112/-/DCSupplemental.
References
- 1.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 2.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: Machineries and mechanisms. Cell. 2009;138(4):628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: From proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11(9):655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 4.Endo T, Yamano K, Kawano S. Structural insight into the mitochondrial protein import system. Biochim Biophys Acta. 2011;1808(3):955–970. doi: 10.1016/j.bbamem.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 5.Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: Common principles and physiological networks. Biochim Biophys Acta. 2013;1833(2):274–285. doi: 10.1016/j.bbamcr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 6.Chacinska A, et al. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23(19):3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mesecke N, et al. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121(7):1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Sideris DP, Tokatlidis K. Oxidative protein folding in the mitochondrial intermembrane space. Antioxid Redox Signal. 2010;13(8):1189–1204. doi: 10.1089/ars.2010.3157. [DOI] [PubMed] [Google Scholar]
- 9.Riemer J, Fischer M, Herrmann JM. Oxidation-driven protein import into mitochondria: Insights and blind spots. Biochim Biophys Acta. 2011;1808(3):981–989. doi: 10.1016/j.bbamem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Stojanovski D, Bragoszewski P, Chacinska A. The MIA pathway: A tight bond between protein transport and oxidative folding in mitochondria. Biochim Biophys Acta. 2012;1823(7):1142–1150. doi: 10.1016/j.bbamcr.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Durigon R, Wang Q, Ceh Pavia E, Grant CM, Lu H. Cytosolic thioredoxin system facilitates the import of mitochondrial small Tim proteins. EMBO Rep. 2012;13(10):916–922. doi: 10.1038/embor.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banci L, Barbieri L, Luchinat E, Secci E. Visualization of redox-controlled protein fold in living cells. Chem Biol. 2013;20(6):747–752. doi: 10.1016/j.chembiol.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Müller JM, Milenkovic D, Guiard B, Pfanner N, Chacinska A. Precursor oxidation by Mia40 and Erv1 promotes vectorial transport of proteins into the mitochondrial intermembrane space. Mol Biol Cell. 2008;19(1):226–236. doi: 10.1091/mbc.E07-08-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milenkovic D, et al. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol Biol Cell. 2009;20(10):2530–2539. doi: 10.1091/mbc.E08-11-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banci L, et al. Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR. Proc Natl Acad Sci USA. 2011;108(12):4811–4816. doi: 10.1073/pnas.1014542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von der Malsburg K, et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21(4):694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Böttinger L, et al. In vivo evidence for cooperation of Mia40 and Erv1 in the oxidation of mitochondrial proteins. Mol Biol Cell. 2012;23(20):3957–3969. doi: 10.1091/mbc.E12-05-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann JM, Riemer J. Mitochondrial disulfide relay: Redox-regulated protein import into the intermembrane space. J Biol Chem. 2012;287(7):4426–4433. doi: 10.1074/jbc.R111.270678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer A, Neupert W, Lill R. Translocation of apocytochrome c across the outer membrane of mitochondria. J Biol Chem. 1995;270(21):12390–12397. doi: 10.1074/jbc.270.21.12390. [DOI] [PubMed] [Google Scholar]
- 20.Chacinska A, et al. Mitochondrial biogenesis, switching the sorting pathway of the intermembrane space receptor Mia40. J Biol Chem. 2008;283(44):29723–29729. doi: 10.1074/jbc.M805356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch JR, Schmid FX. Mia40 targets cysteines in a hydrophobic environment to direct oxidative protein folding in the mitochondria. Nat Commun. 2014;5:3041. doi: 10.1038/ncomms4041. [DOI] [PubMed] [Google Scholar]
- 22.Baker MJ, et al. Impaired folding of the mitochondrial small TIM chaperones induces clearance by the i-AAA protease. J Mol Biol. 2012;424(5):227–239. doi: 10.1016/j.jmb.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Bragoszewski P, Gornicka A, Sztolsztener ME, Chacinska A. The ubiquitin-proteasome system regulates mitochondrial intermembrane space proteins. Mol Cell Biol. 2013;33(11):2136–2148. doi: 10.1128/MCB.01579-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20(8):1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang CS, Shemorry A, Auerbach D, Varshavsky A. The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat Cell Biol. 2010;12(12):1177–1185. doi: 10.1038/ncb2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koehler CM. The small Tim proteins and the twin Cx3C motif. Trends Biochem Sci. 2004;29(1):1–4. doi: 10.1016/j.tibs.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Rissler M, et al. The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins. J Mol Biol. 2005;353(3):485–492. doi: 10.1016/j.jmb.2005.08.051. [DOI] [PubMed] [Google Scholar]
- 28.Qiu J, et al. Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell. 2013;154(3):596–608. doi: 10.1016/j.cell.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 29.Gornicka A, et al. A discrete pathway for the transfer of intermembrane space proteins across the outer membrane of mitochondria. Mol Biol Cell. 2014;25(25):3999–4009. doi: 10.1091/mbc.E14-06-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayer A, et al. Distinct redox regulation in sub-cellular compartments in response to various stress conditions in Saccharomyces cerevisiae. PLoS ONE. 2013;8(6):e65240. doi: 10.1371/journal.pone.0065240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojer K, et al. Glutathione redox potential in the mitochondrial intermembrane space is linked to the cytosol and impacts the Mia40 redox state. EMBO J. 2012;31(14):3169–3182. doi: 10.1038/emboj.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vögtle FN, et al. Intermembrane space proteome of yeast mitochondria. Mol Cell Proteomics. 2012;11(12):1840–1852. doi: 10.1074/mcp.M112.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.