Significance
Nonphosphorus lipids produced by heterotrophic bacteria have been measured in marine ecosystems without an understanding of their origins or role. This work shows SAR11 chemoheterotrophic bacteria synthesize multiple nonphosphorus lipids in response to phosphate depletion. Because this process results in a reduced cellular P:C ratio, it impacts our understanding of ocean processes related to cellular elemental stoichiometry by showing how different environmental parameters alter P:C ratios in heterotrophs. Also, SAR11 grown with excess organophosphonate synthesized phosphorus-free lipids. This finding contrasts the contemporary view of organophosphorus utilization because organophosphate-derived phosphorus did not equally substitute for inorganic phosphate in lipids. Considering lipid phosphorus content was lower in cells using organophosphonate, phosphorus-based productivity estimates may vary as a function of phosphorus source.
Keywords: marine phosphorus cycle, lipids, glucuronic acid, cyanobacteria, methylphosphonate
Abstract
Phytoplankton inhabiting oligotrophic ocean gyres actively reduce their phosphorus demand by replacing polar membrane phospholipids with those lacking phosphorus. Although the synthesis of nonphosphorus lipids is well documented in some heterotrophic bacterial lineages, phosphorus-free lipid synthesis in oligotrophic marine chemoheterotrophs has not been directly demonstrated, implying they are disadvantaged in phosphate-deplete ecosystems, relative to phytoplankton. Here, we show the SAR11 clade chemoheterotroph Pelagibacter sp. str. HTCC7211 renovates membrane lipids when phosphate starved by replacing a portion of its phospholipids with monoglucosyl- and glucuronosyl-diacylglycerols and by synthesizing new ornithine lipids. Lipid profiles of cells grown with excess phosphate consisted entirely of phospholipids. Conversely, up to 40% of the total lipids were converted to nonphosphorus lipids when cells were starved for phosphate, or when growing on methylphosphonate. Cells sequentially limited by phosphate and methylphosphonate transformed >75% of their lipids to phosphorus-free analogs. During phosphate starvation, a four-gene cluster was significantly up-regulated that likely encodes the enzymes responsible for lipid renovation. These genes were found in Pelagibacterales strains isolated from a phosphate-deficient ocean gyre, but not in other strains from coastal environments, suggesting alternate lipid synthesis is a specific adaptation to phosphate scarcity. Similar gene clusters are found in the genomes of other marine α-proteobacteria, implying lipid renovation is a common strategy used by heterotrophic cells to reduce their requirement for phosphorus in oligotrophic habitats.
Microbes primarily assimilate phosphorus (P) in its +5 valence state (phosphate; Pi), which comprises ∼3% of total cellular mass as a structural constituent of nucleic acids and phospholipids, and is intimately involved in energy metabolism and some transport functions (via ATP hydrolysis) (1). In oligotrophic ocean gyres, Pi concentrations are extremely low (0.2–1.0 nM in the Sargasso Sea; ref. 2) and the availability of Pi can limit bacterial and primary production (2–5). Microbes inhabiting these low Pi environments have evolved numerous strategies to maintain growth and enhance their competitiveness for trace amounts of Pi. These mechanisms are commonly induced by Pi starvation and include one or more of the following: (i) expression of high affinity Pi transporters (6); (ii) reduction of cellular Pi quotas (7, 8); (iii) utilization of alternate phosphorus sources (9, 10); and (iv) polyphosphate storage and breakdown (11, 12). Such strategies facilitate survival in the face of Pi insufficiency.
Polar membrane lipids are a substantial cellular sink for phosphate in bacteria. Structural lipids consist of glycerol esterified to hydrophobic fatty acid chains (diacylglycerol) and a hydrophilic polar head group, which commonly contains Pi (phospholipids). When grown with sufficient Pi, phospholipids in Gram-negative marine bacteria frequently include phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and diphosphatidylglycerol (13). However, when Pi is low or limiting, many microbes replace phospholipids with those that lack phosphorus (14–16). Nonphosphorus polar head groups are structurally diverse and include sulfoquinovose (sulfolipids) (17), various monosaccharides and disaccharides (glycolipids; reviewed in ref. 18), ornithine, or other amino acids (reviewed in ref. 19). When Pi stressed, the marine cyanobacteria Prochlorococcus and Synechococcus reduce their Pi demand by 0.5–8.6 attomoles P per cell by substituting phosphorus-containing lipids with sulfolipids; depending on the strain, this reduction equates to 10–86% of the P bound in their nucleic acids (7). There is indirect evidence natural populations of marine chemoheterotrophic bacteria also use nonphosphorus lipids in response to Pi deprivation. Bacterioplankton collected in the Sargasso Sea had greater concentrations of nonphosphorus lipids than those from adjacent regions of the North Atlantic where Pi was relatively abundant (20).
Oligotrophic bacteria belonging to the SAR11 clade (Pelagibacterales) of α-proteobacteria are numerically dominant chemoheterotrophs in marine euphotic zones worldwide (21). Pelagibacterales cells are small (volume of 0.01 μm3; ref. 22) and contain streamlined genomes (23, 24). The reduced cell and genome size likely stem from natural selection to reduce the overhead cost of replication in oligotrophic ocean gyres where P and N may periodically limit growth (25, 26). Despite their abundance in low Pi environments, it remains unknown whether Pelagibacterales synthesize phosphorus-free lipids to reduce their P quota. The genome of Pelagibacter ubique str. HTCC1062 lacks genes predicted to encode proteins used in sulfolipid, betaine, or ornithine lipid biosynthesis, suggesting this strain is unable to modulate lipid composition in response to Pi availability (7, 8). Previous laboratory experiments partially supported this prediction by showing that P. ubique lacks nonphosphorus lipids when grown under Pi replete conditions; however, lipids from Pi limited cells were not examined in that research (7). Relative to P. ubique, a Sargasso Sea isolate, Pelagibacter sp. str. HTCC7211 (str. HTCC7211) contains extended genetic inventory associated with Pi acquisition, storage, and metabolism (10). When Pi limited, str. HTCC7211 induces a suite of these genes, including both inorganic (pstSCAB) and organophosphate (phnCDEE2) ABC transporters and the C–P lyase complex (phnGHIJKLNM), required for phosphonate degradation (10). Laboratory experiments have distinctly linked the expression of these genes to the utilization of both phosphate esters and phosphonates (including methylphosphonate; MPn) (10). This finding indicates organophosphate utilization is one strategy str. HTCC7211 employs to evade Pi growth limitation.
While examining gene expression profiles of Pi starved str. HTCC7211 cultures, we observed the unexpected up-regulation of a four gene cluster proximal to the collection of P uptake genes on the str. HTCC7211 chromosome. Two of the genes were annotated as “putative hemolysins,” one as a “glycosyltransferase,” and a “metallophosphatase.” Comparative genomic examination of these genes led us to hypothesize that the four genes might be involved in the restructuring of lipid polar head groups and the synthesis of nonphosphorus lipids. Herein, we present the results of laboratory experiments designed to test the potential for synthesis of nonphosphorus lipids in response to Pi stress by Pelagibacter.
Results
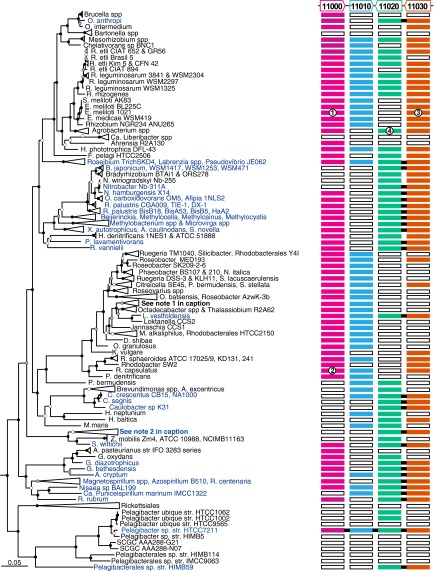
Previously we reported Pi starvation in str. HTCC7211 significantly induced the expression of 31 genes, including those encoding Pi and organophosphorus transport and catabolism proteins (10). Proximal to the genes encoding a high-affinity Pi transporter (pstSCAB), a four-gene cluster (HTCC7211_00011000–HTCC7211_00011030) was significantly up-regulated in Pi starved cultures, relative to Pi replete cultures (2.7- to 8.2-fold, reported in ref. 10). Two of these genes (HTCC7211_00011000 and HTCC7211_00011010) are oriented in a probable operon (+ strand; Fig. 1) and annotated as “putative hemolysins” belonging to COG3176. The other two genes (HTCC7211_00011020 and HTCC7211_00011030) are located downstream of HTCC7211_00011010 in a second probable operon (- strand; Fig. 1). HTCC7211_00011020 is annotated as a group 1 glycosyltransferase belonging to COG438 (RfaG). HTCC7211_00011030 is annotated as a metallophosphoesterase belonging to COG2908. Protein domain signatures in each predicted amino acid sequence were identified with InterProScan5 (27). HTCC7211_00011000 contains an acyl-CoA N-acyltransferase domain (IPR016181; residues 26–174). HTCC7211_00011010 contains a phospholipid/glycerol acyltransferase domain (IPR002123; residues 49–180). HTCC7211_00011020 contains a glycosyltransferase subfamily 4, N-terminal domain (IPR028098; residues 5–164), and a glycosyl transferase, family 1 domain (IPR001296; residues 177–285). HTCC7211_00011030 contains a calcineurin-like phosphoesterase domain, lpxH type domain (IPR024654; residues 10–249).
Fig. 1.
Distribution of nonphosphorus lipid synthesis genes in α-proteobacteria. Tree inferred from concatenated 16S/23S rRNA gene sequences. Most monophyletic groups where all members had the same patterns of gene distribution were collapsed. The outgroup taxa can be found in Dataset S1. Node labels represent Shimodaira–Hasegawa confidence test values: black filled, ≥0.9; gray filled, 0.7–0.9; white filled, ≤0.7. Pelagibacter sp. str. HTCC7211 gene orientation is depicted above colored bars; “HTCC7211_000” for each gene identifier has been omitted. The colored bars are a visual representation of Hal clustering results. Bars indicate ortholog presence (filled), absence (open), and chromosomal synteny (black line between bars), or lack of synteny (no black line between bars), for the four genes. Those taxa with adjacent glycosyltransferases and metallophosphoesterases are colored blue in the tree for ease of identification. Numbers inside of colored boxes indicate genes for which functions have been characterized, as follows: 1, SMc01116 (OlsB, E. meliloti); 2, RCAP_rcc02997 (OlsB, R. capsulatus); 3, SMc00171 (phospholipase C, E. meliloti); 4, atu2297 (GADG/MGDG glycosyltransferase, A. fabrum, in collapsed node). Note 1: Sulfitobacter spp., O. indolifex, R. denitrificans and litoralis, Rhodobacterales HTCC2083. Note 2: Erythrobacter HTCC2594/NAP1, and C. bathyomarinum JL354, N. aromaticivorans DSM 12444, Sphingobium spp, Sphingopyxis alaskensis RB2256, Sphingomonas SKA58.
We searched for orthologs to HTCC7211_00011000–HTCC7211_00011030 in α-proteobacterial genomes by using the Hal pipeline (28) and found all four genes differentially distributed across the clade (Fig. 1 and Dataset S1). HTCC7211_00011000 clustered with amino acid sequences of characterized enzymes from Ensifer (Sinorhizobium) meliloti 1021 (OlsB) and Rhodobacter capsulatus SB 1003 (OlsB) (Fig. 1). OlsB catalyzes the first step in ornithine lipid formation (29, 30) (Fig. S1). The second step of ornithine lipid biosynthesis is catalyzed by the protein product of olsA (Fig. S1), which is frequently found downstream of olsB (30, 31). Unexpectedly, the amino acid sequence of HTCC7211_00011010 did not cluster with OlsA sequences from characterized enzymes, despite all three having phospholipid/glycerol acyltransferase domain (IPR002123) signatures. HTCC7211_00011020 clustered with an Agrobacterium fabrum C58 gene (Fig. 1) recently demonstrated to encode a bifunctional glycosyltransferase that forms both monoglucosyl- and glucuronosyl-diacylglycerols (MGDG and GADG, respectively) by the addition of glucose or glucuronic acid moieties to diacylglycerols (32) (Fig. S2). HTCC7211_00011030 clustered with an E. meliloti gene shown to encode a phospholipase C (PlcP) (Fig. 1) that initiates lipid renovation during Pi starvation by cleaving phosphorus-containing polar head groups from diacylglycerols (8) (Fig. S2).
We hypothesized that HTCC7211_00011000–HTCC7211_00011030 encode proteins involved in the renovation of membrane lipids during Pi stress. Renovation is defined here as the combined result of: (i) the synthesis of new ornithine lipids; and (ii) the exchange of phospholipids for MGDG and/or GADG. We based this hypothesis on the increased expression of these genes under Pi limiting conditions (10), and the characterized functions of orthologous genes in other α-proteobacteria (Fig. 1). We tested this hypothesis by analyzing the lipid polar head group composition over time in Pi replete and Pi deplete growth conditions (Fig. 2). Total lipids extracted per sample ranged from 2.8 to 9.7 attomoles⋅cell−1 and varied as a function of growth state and P source (Dataset S2 and Fig. S3).
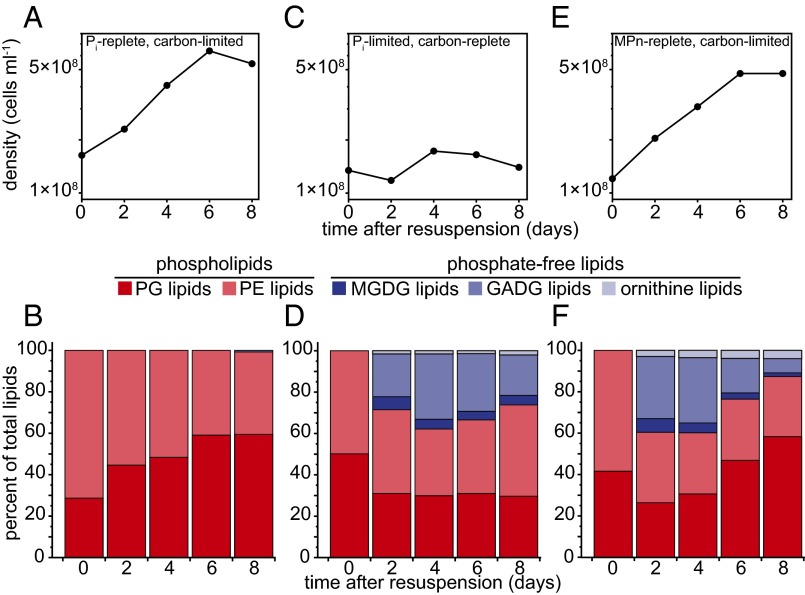
Fig. 2.
Pelagibacter sp. str. HTCC7211 renovates lipid composition in response to Pi limitation. Growth curves show cell densities in suspensions that were subsampled for lipid analyses during time courses: excess Pi (A); no Pi (C); or excess MPn (E). Time course of lipid composition of str. HTCC7211 cells in AMS1 containing: excess Pi (B); no Pi (D); or excess MPn (F).
Strain HTCC7211 cells growing exponentially with excess Pi (Fig. 2A) exclusively contained phospholipids (PG and PE) (Fig. 2B). In carbon-limited stationary phase, a low proportion of GADG lipids were detected (<1%; Fig. 2B). Cells incubated in growth medium without added Pi did not grow appreciably (Fig. 2C), but renovated 26–38% of total cellular phospholipids with multiple P-free lipids (GADG > MGDG > ornithine lipid; Fig. 2D). Although cells renovated membrane lipids in the absence of Pi, total cellular lipid content decreased, relative to Pi replete cells, suggesting cell size was slightly reduced (Fig. S3).
Lipid profiles of cells resuspended into growth medium with MPn as the sole P source were measured. Exponentially growing cells using MPn as the sole P source (Fig. 2E) replaced phospholipids with P-free lipids (Fig. 2F). In this growth condition, up to 40% of total cellular phospholipids were renovated to P-free analogs. Similar to Pi starved profiles, GADG was the most abundant P-free lipid (Fig. 2F). However, after an initial drop in abundance, the total percent of phospholipids increased to 87% as the cells entered carbon-limited stationary phase. Total cellular lipid contents in MPn-replete cells were comparable to those of Pi replete cultures, suggesting cell size was conserved during renovation (Fig. S3).
Cells harvested in midlogarithmic phase, grown with MPn as the sole P source for >20 generations, contained a standing stock of 11% P-free lipids (Fig. S4).
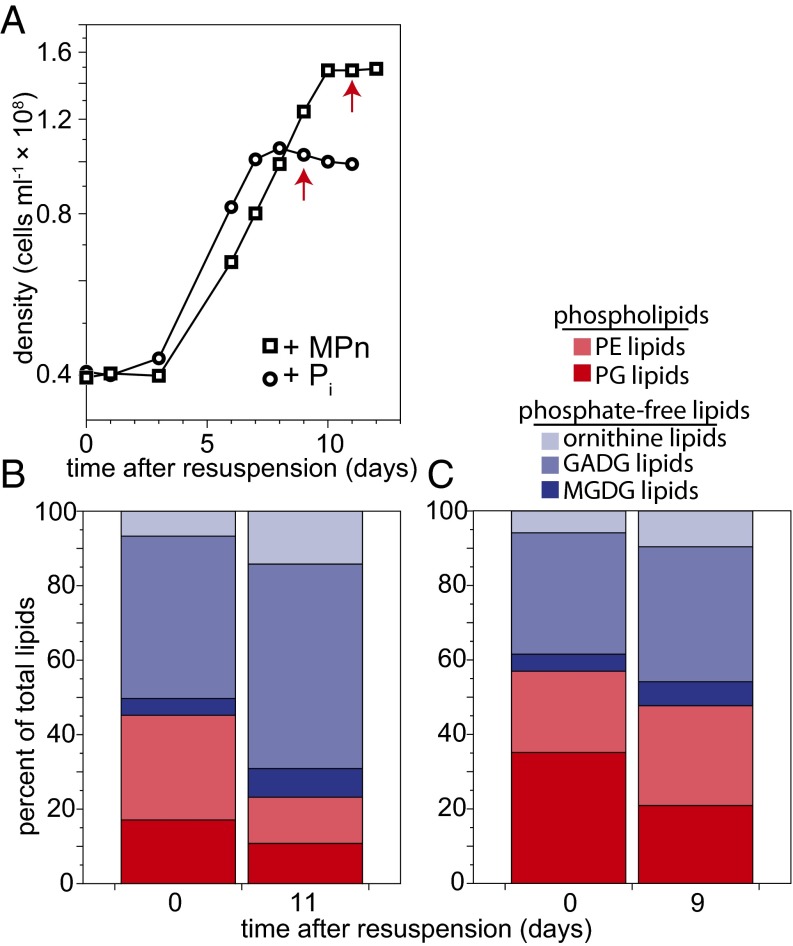
Pi starved cells exposed to MPn levels sufficient to sustain >1 generation, but insufficient for maximal growth (that is, cells were MPn-limited; Fig. 3A), contained 77% P-free lipids in MPn-limited stationary phase (Fig. 3B). A lesser degree of conversion was observed when Pi starved cells were exposed to Pi sufficient for >1 generation, but insufficient for maximal growth (Fig. 3 A and C).
Fig. 3.
Majority of lipids are phosphate-free analogs under conditions of methylphosphonate starvation. (A) Bacterial growth curves of previously Pi starved str. HTCC7211 cells in growth medium with growth-limiting amounts of Pi or MPn. Lipid profiles before and after reaching MPn (B) or Pi (C) limitation. Red arrows in A point to P limited sample points for B and C.
Previous measurements of the lipid composition of P. ubique str. HTCC1062 growing in Pi replete natural seawater medium showed that only PE and PG were present in lipid extractions (7). We conducted similar experiments with Pi replete and Pi starved str. HTCC1062 cells grown in synthetic medium. The lipids of str. HTCC1062 cells growing in Pi replete conditions were comprised exclusively of phospholipids (PG and PE). Nonphosphorus lipids were not detected under conditions of Pi starvation (Fig. S5).
Discussion
Similar to phytoplankton (7) and terrestrial α-proteobacteria (8), we show Pelagibacter sp. str. HTCC7211, but not P. ubique str. HTCC1062, modulates its phospholipid composition in response to P availability (Fig. 2). We identify the genes likely to confer the ability to renovate polar lipid head groups (Fig. 1) and link to studies of their regulation (10). Published metagenomic analysis show that three of these genes are overrepresented in the Sargasso Sea (HTCC7211_00011010–HTCC7211_00011030) relative to Pelagibacterales populations inhabiting the comparatively P-replete North Pacific subtropical gyre (11). In these studies, a metric called “multiplicity per cell” was used to infer that ∼28% of the Pelagibacterales population contained HTCC7211_00011010; ∼95% contained HTCC7211_00011020 and all cells contained HTCC7211_00011030 (∼117%, indicating one or more copies in some genomes). These data suggest that the capacity to convert phospholipids to glycolipids, as conferred by orthologs to HTCC7211_00011020 (MGDG/GADG glycosyltransferase) and HTCC7211_00011030 (phospholipase C), is widespread in Pelagibacterales lineages inhabiting the Sargasso Sea and is likely a specific adaptation to P scarcity. These findings likely explain reports of MGDG production by chemoheterotrophic bacteria from the Sargasso Sea (20).
The complete four-gene cluster conferring glycolipid and ornithine lipid biosynthesis in str. HTCC7211 is not found in the same syntenic arrangement in other α-proteobacteria (Fig. 1). However, tandem genes encoding phospholipase C and the MGDG/GADG glycosyltransferase were identified in a number of α-proteobacteria, including the marine strains Pelagibacterales sp. str. HIMB59, “Candidatus Puniceispirillum marinum” str. IMCC1322 (SAR116), Erythrobacter litoralis HTCC2594, and Loktanella vestfoldensis SKA53 (Fig. 1). This finding expands our knowledge of the distribution of glycolipid synthesis genes to cosmopolitan marine bacteria and implies lipid renovation in response to Pi scarcity may be a relatively common feature of marine chemoheterotrophic bacteria.
HTCC7211_00011010 likely catalyzes the final step of ornithine lipid biosynthesis in str. HTCC7211. Ornithine lipids are synthesized by the N-acylation of ornithine with a hydroxy-fatty acyl group (by OlsB) (29), followed by an O-acylation of lyso-ornithine (by OlsA) to form ornithine lipid (Fig. S1). In most ornithine lipid synthesizing bacteria, olsA and olsB genes form an operon. We identified a probable olsB gene in the str. HTCC7211 genome (Fig. 1; HTCC7211_00011000) but were unable to identify an ortholog to known olsA genes with all vs. all BLASTP and Markov clustering (MCL). To further examine this question, we searched the str. HTCC7211 genome with a hidden Markov model (HMM) trained on amino acid sequences of characterized olsAs from E. melilotii and R. capsulatus plus the best BLAST hits to each (SI Methods). The analysis returned low E-value hits to HTCC7211_0001770 and did not provide additional evidence that HTCC7211_00011010 is an olsA ortholog. Thus, the primary amino acid sequence of HTCC7211_00011010 does not likely encode an ortholog to OlsA. However, OlsA has homology to glycerol acyltransferases, the protein domain we identified in HTCC7211_00011010. These similarities, plus the syntenic arrangement akin to other olsBA pairs, suggest HTCC7211_00011010 has an activity similar to that of characterized OlsAs. Regarding the str. HTCC7211 ornithine lipid synthesis gene cluster, we did not find other α-proteobacteria with the same chromosomal arrangement, implying this arrangement may be unique to a subset of organisms in the Pelagibacterales.
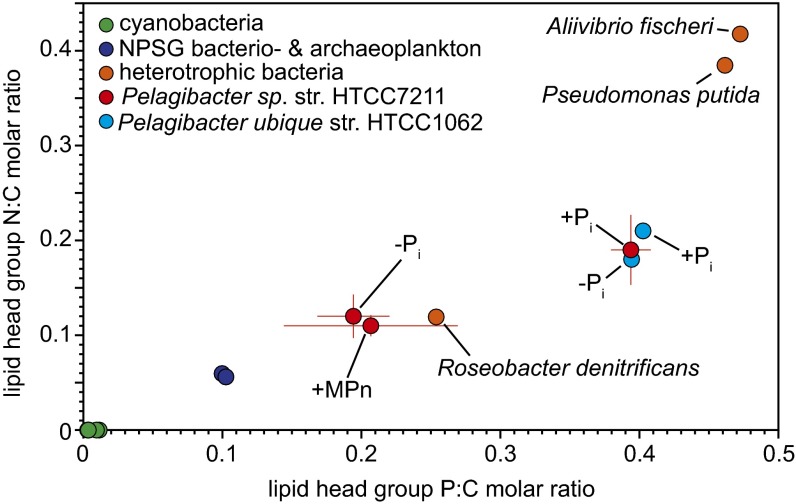
The use of lipids that lack P, together with the utilization of assorted organophosphorus sources (10), may enable certain Pelagibacterales members to cope with patchiness in the dissolved organic phosphorus pool and maintain growth in low Pi waters. In Pi replete conditions, the lipid polar head groups of str. HTCC7211 and str. HTCC1062 had a molar P:C ratio of ∼0.4 (Fig. 4). Under conditions of Pi deprivation or growth on MPn, the average molar P:C ratio decreased to ∼0.2 in str. HTCC7211 (Fig. 4). From the change in composition of polar head groups during Pi starvation, we calculated that lipid renovation reduces the P content of str. HTCC7211 by 2.7–3.9 amoles P⋅cell−1 (Dataset S2). The genome of str. HTCC7211 is 1,456,888 bp long and contains 4.8 attomoles P. Thus, the savings in lipid P is ∼56–81% of the P contained in the genome. A similar savings was also observed when cells were grown with MPn (Fig. 4), indicating that growth with organophosphorus sources may have unexpected effects on cellular stoichiometry. Although HTCC7211 appeared to use MPn-derived P for phospholipid synthesis in pure culture studies (Fig. 2F), we measured a standing stock of ∼11% nonphosphorus lipids in cultures grown exclusively on MPn (Fig. S4). Moreover, when MPn-adapted cells were starved for MPn (Fig. 3A), the majority of lipids were converted to non-P lipids (Fig. 3B). Previous studies showed that str. HTCC7211 used a broad spectrum of organophosphorus compounds. Although we only show the lipid composition with MPn, our data suggests deposition of P into, and withdrawal from, structural lipids may act as a P reserve for cells, effectively enabling survival in low Pi ecosystems where the bioavailability of Pi or organophosphorus is variable or patchy.
Fig. 4.
Phosphorus, nitrogen, and carbon molar ratios in lipid polar head groups from str. HTCC7211 and str. HTCC1062 under different phosphorus regimes. Strain HTCC7211 data points are the mean elemental ratios ± SD from days 2–8 in Fig. 2 (n = 4) (Dataset S2). Cyanobacteria, non-Pelagibacterales heterotrophs and North Pacific Subtropical Gyre (NPSG) values were calculated from published data (44).
We have demonstrated that some Pelagibacterales strains can modify their lipids in response to P stress, replacing phospholipids with GADG, MGDG, and ornithine lipids, thereby reducing cellular demands for P. The genes associated with this adaptive response are found in hypervariable genomic regions and appear to be most abundant in Pelagibacterales strains that inhabit Pi limited ocean regions, like the Sargasso Sea. This discovery is significant because it shows that, like phytoplankton, chemoheterotrophic bacterioplankton have mechanisms that cause them to deviate from expected elemental ratios, thereby changing estimates of the impacts of P limitation on productivity and biomass accumulation. In this case, the mechanism identified is associated with specific genes that previously had poorly assigned functions. Because many genes in microbial genomes have incompletely assigned functions, studies like this improve the accuracy of annotations and are important to improving the long-term impact of genome data on the prediction geochemical processes. In future work, it will be important to understand why cells prefer phospholipids and to identify changes in fitness associated with the substitution of glycolipids.
Methods
Organism Source and Growth Conditions.
P. ubique str. HTCC1062 and Pelagibacter sp. str. HTCC7211 were revived from frozen stocks and cultivated on AMS1 synthetic growth medium without added phosphate as described (10, 33, 34). Cell growth was monitored by flow cytometry as described (35). Phosphorus was added as NaH2PO4 or methylphosphonate (MPn), as indicated in the text.
Cell Harvesting for Lipid Profiles.
Strain HTCC7211 cells, grown in AMS1 with excess Pi (100 μM), were harvested in late-logarithmic growth-phase (approximately 1.0 × 108 cells⋅ml−1) by centrifugation (17,664 × g for 1.0 h at 20 °C). Pellets were washed twice with growth medium and resuspended in one of the following conditions: (i) Pi replete (100 μM); (ii) MPn-replete medium (100 μM); or (iii) Pi deplete growth medium (no Pi added). Suspensions were monitored for growth (reported in Fig. 2 A, C, and E) and subsampled (40 mL) by centrifugation (48,298 × for 1.0 h at 4 °C) at t = 0, 2, 4, 6, and 8 d. The supernatant was removed and cell pellets were frozen at −80 °C until lipid extraction.
To study the effect of sequential Pi and MPn limitation on lipid composition, str. HTCC7211 cells were grown with excess Pi to late-logarithmic growth phase and harvested by centrifugation. Pellets were washed with growth medium, resuspended in growth medium without added P (to approximately 4 × 107 cells⋅mL−1), and incubated for 4 d to induce the Pi starvation response. After starvation, P-starved cells were used to inoculate growth media containing a growth-limiting amount of Pi (1.2 μM) or MPn (1.5 μM). The carbon and nitrogen constituents of this medium support cell yields in excess of 5.0 × 108 cells⋅mL−1. Therefore, Pi or MPn were growth-limiting, as calculated from the cellular P quotas of 11 amol P⋅cell−1 (Pi-grown) or 10 amol P⋅cell−1 (MPn-grown) (10). Cell suspensions were subsampled by centrifugation at t = 0 d and after P-limited stationary phase had been reached, as determined from direct cell counts: 9 d for Pi-grown cells; 12 d for MPn-grown cells.
Lipid Analysis.
Polar lipids were extracted from the cell pellets as described (36). Published methods were also used as a basis for polar lipid separation by normal-phase high performance liquid chromatography (20) and concomitant analysis by positive electrospray ionization ion-trap mass spectrometry (37). Additional structural elucidation of GADG and ornithine lipids was conducted by using the same HPLC method in conjunction with positive electrospray ionization high mass-resolution mass spectrometry (Figs. S6 and S7 and SI Methods).
Lipid Polar Head Group Elemental Stoichiometry.
The relative elemental ratios of polar head groups under different P regimes were calculated from the molar proportion of each membrane lipid type in each treatment and the molecular formulas of each polar head group: C2H8NO4P for phosphoethanolamine; C3H7O7P for phosphoglycerol; C6H9O7 for glucuronic acid; C6H11O6 for glucose; and C5H11N2O2 for ornithine (Dataset S2).
Gene Identifiers.
All gene identifiers in this study are presented as Integrated Microbial Genomes (IMG) Gene ID numbers. In previous works (10, 11, 38), the genes HTCC7211_00011000–HTCC7211_00011030 were listed with different identifiers [listed as: previous identifier (IMG Gene ID)]: PB7211_1302 (HTCC7211_00011000); PB7211_635 (HTCC7211_00011010); PB7211_960 (HTCC7211_00011020); PB7211_983 (HTCC7211_00011030).
Identification of Putative Lipid Renovation Gene Orthologs.
The Hal software package (28) was used to generate orthologous protein clusters from 272 α-proteobacteria from the IMG database (39). All vs. all BLASTP was followed by MCL at 13 inflation parameters. Clusters generated with the inflation parameter of 1.5 were used to identify orthologs of the genes predicted to be involved in lipid remodeling in str. HTCC7211 (HTCC7211_00011000–HTCC7211_00011030). The complete distribution for all four genes in all α-proteobacteria can be found in Dataset S1.
α-Proteobacterial Phylogeny.
Concatenated 16S-23S rRNA genes from almost all of the above α-proteobacteria and six outgroups (including members of the β-, γ-, and δ-proteobacteria) were used to manually construct a maximum likelihood tree similar to that in ref. 40. Some rRNA gene sequences from organisms included in the Hal analysis were excluded from the 16–23S tree because of poor quality or truncated rRNA genes (Dataset S1). All identifiers for taxa represented in the tree are provided in Dataset S1. The 16S and 23S rRNA genes were aligned separately with MUSCLE (41) by using default settings, and curated with Gblocks (42) using the following settings: -b1 = (n/2)+1; -b2 = (n/2) + 1; -b3 = (n/2); -b4 = 2; -b5 = h, where n = number of taxa. Alignments were normalized and concatenated with normalize_alignments.py and catPhylip.pl, respectively, included in the Hal package. The final alignment contained 261 taxa and 4,096 characters. The tree was inferred by using FastTree2 (43) with default settings.
Supplementary Material
Acknowledgments
This work was supported by Marine Microbiology Initiative of the Gordon and Betty Moore Foundation Grant GBMF607.01 (to S.J.G.), National Science Foundation [Division of Ocean Sciences Grant 0962362 (to A.W.)], and an Alfred P. Sloan Foundation Research Fellowship (to A.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505034112/-/DCSupplemental.
References
- 1.Lengeler JW, Drews G. In: Biology of the Prokaryotes. Schlegel HG, editor. Blackwell Science Ltd; Oxford: 1998. [Google Scholar]
- 2.Wu J, Sunda W, Boyle EA, Karl DM. Phosphate depletion in the western North Atlantic Ocean. Science. 2000;289(5480):759–762. doi: 10.1126/science.289.5480.759. [DOI] [PubMed] [Google Scholar]
- 3.Tyrrell T. The relative influences of nitrogen and phosphorus on oceanic primary production. Nature. 1999;400:525–531. [Google Scholar]
- 4.Sañudo-Wilhelmy SA, et al. Phosphorus limitation of nitrogen fixation by Trichodesmium in the central Atlantic Ocean. Nature. 2001;411(6833):66–69. doi: 10.1038/35075041. [DOI] [PubMed] [Google Scholar]
- 5.Thingstad TF, et al. Nature of phosphorus limitation in the ultraoligotrophic eastern Mediterranean. Science. 2005;309(5737):1068–1071. doi: 10.1126/science.1112632. [DOI] [PubMed] [Google Scholar]
- 6.Krumhardt KM, et al. Effects of phosphorus starvation versus limitation on the marine cyanobacterium Prochlorococcus MED4 I: Uptake physiology. Environ Microbiol. 2013;15(7):2114–2128. doi: 10.1111/1462-2920.12079. [DOI] [PubMed] [Google Scholar]
- 7.Van Mooy BAS, et al. Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature. 2009;458(7234):69–72. doi: 10.1038/nature07659. [DOI] [PubMed] [Google Scholar]
- 8.Zavaleta-Pastor M, et al. Sinorhizobium meliloti phospholipase C required for lipid remodeling during phosphorus limitation. Proc Natl Acad Sci USA. 2010;107(1):302–307. doi: 10.1073/pnas.0912930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyhrman ST, et al. Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature. 2006;439(7072):68–71. doi: 10.1038/nature04203. [DOI] [PubMed] [Google Scholar]
- 10.Carini P, White AE, Campbell EO, Giovannoni SJ. Methane production by phosphate-starved SAR11 chemoheterotrophic marine bacteria. Nat Commun. 2014;5:4346. doi: 10.1038/ncomms5346. [DOI] [PubMed] [Google Scholar]
- 11.Coleman ML, Chisholm SW. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA. 2010;107(43):18634–18639. doi: 10.1073/pnas.1009480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin P, Dyhrman ST, Lomas MW, Poulton NJ, Van Mooy BAS. Accumulation and enhanced cycling of polyphosphate by Sargasso Sea plankton in response to low phosphorus. Proc Natl Acad Sci USA. 2014;111(22):8089–8094. doi: 10.1073/pnas.1321719111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver JD, Colwell RR. Extractable lipids of gram-negative marine bacteria: Phospholipid composition. J Bacteriol. 1973;114(3):897–908. doi: 10.1128/jb.114.3.897-908.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minnikin DE, Abdolrahimzadeh H, Baddiley J. Replacement of acidic phosphates by acidic glycolipids in Pseudomonas diminuta. Nature. 1974;249(454):268–269. doi: 10.1038/249268a0. [DOI] [PubMed] [Google Scholar]
- 15.Minnikin DE, Abdolrahimzadeh H. The replacement of phosphatidylethanolamine and acidic phospholipids by an ornithine-amide lipid and a minor phosphorus-free lipid in Pseudomonas fluorescens NCMB 129. FEBS Lett. 1974;43(3):257–260. doi: 10.1016/0014-5793(74)80655-1. [DOI] [PubMed] [Google Scholar]
- 16.Geiger O, Röhrs V, Weissenmayer B, Finan TM, Thomas-Oates JE. The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti. Mol Microbiol. 1999;32(1):63–73. doi: 10.1046/j.1365-2958.1999.01325.x. [DOI] [PubMed] [Google Scholar]
- 17.Benson AA, Daniel H, Wiser R. A sulfolipid in plants. Proc Natl Acad Sci USA. 1959;45(11):1582–1587. doi: 10.1073/pnas.45.11.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hölzl G, Dörmann P. Structure and function of glycoglycerolipids in plants and bacteria. Prog Lipid Res. 2007;46(5):225–243. doi: 10.1016/j.plipres.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Geiger O, González-Silva N, López-Lara IM, Sohlenkamp C. Amino acid-containing membrane lipids in bacteria. Prog Lipid Res. 2010;49(1):46–60. doi: 10.1016/j.plipres.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Popendorf KJ, Lomas MW, Van Mooy BAS. Microbial sources of intact polar diacylglycerolipids in the Western North Atlantic Ocean. Org Geochem. 2011;42:803–811. [Google Scholar]
- 21.Morris RM, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420(6917):806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 22.Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418(6898):630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- 23.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309(5738):1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 24.Grote J, et al. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. MBio. 2012;3(5):e00252–e12. doi: 10.1128/mBio.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giovannoni SJ, Cameron Thrash J, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014;8(8):1553–1565. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grzymski JJ, Dussaq AM. The significance of nitrogen cost minimization in proteomes of marine microorganisms. ISME J. 2012;6(1):71–80. doi: 10.1038/ismej.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones P, et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robbertse B, Yoder RJ, Boyd A, Reeves J, Spatafora JW. Hal: An automated pipeline for phylogenetic analyses of genomic data. PLoS Curr. 2011;3:RRN1213. doi: 10.1371/currents.RRN1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J-L, et al. Identification of a gene required for the formation of lyso-ornithine lipid, an intermediate in the biosynthesis of ornithine-containing lipids. Mol Microbiol. 2004;53(6):1757–1770. doi: 10.1111/j.1365-2958.2004.04240.x. [DOI] [PubMed] [Google Scholar]
- 30.Aygun-Sunar S, et al. Ornithine lipid is required for optimal steady-state amounts of c-type cytochromes in Rhodobacter capsulatus. Mol Microbiol. 2006;61(2):418–435. doi: 10.1111/j.1365-2958.2006.05253.x. [DOI] [PubMed] [Google Scholar]
- 31.Weissenmayer B, Gao J-L, López-Lara IM, Geiger O. Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol Microbiol. 2002;45(3):721–733. doi: 10.1046/j.1365-2958.2002.03043.x. [DOI] [PubMed] [Google Scholar]
- 32.Semeniuk A, Sohlenkamp C, Duda K, Hölzl G. A bifunctional glycosyltransferase from Agrobacterium tumefaciens synthesizes monoglucosyl and glucuronosyl diacylglycerol under phosphate deprivation. J Biol Chem. 2014;289(14):10104–10114. doi: 10.1074/jbc.M113.519298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carini P, et al. Discovery of a SAR11 growth requirement for thiamin’s pyrimidine precursor and its distribution in the Sargasso Sea. ISME J. 2014;8(8):1727–1738. doi: 10.1038/ismej.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carini P, Steindler L, Beszteri S, Giovannoni SJ. Nutrient requirements for growth of the extreme oligotroph ‘Candidatus Pelagibacter ubique’ HTCC1062 on a defined medium. ISME J. 2013;7(3):592–602. doi: 10.1038/ismej.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tripp HJ. 2008. Counting marine microbes with Guava Easy-Cyte 96 well plate reading flow cytometer. Nat Protoc Exchange. Available at www.nature.com/protocolexchange/protocols/422.
- 36.Popendorf KJ, Fredricks HF, Van Mooy BAS. Molecular ion-independent quantification of polar glycerolipid classes in marine plankton using triple quadrupole MS. Lipids. 2013;48(2):185–195. doi: 10.1007/s11745-012-3748-0. [DOI] [PubMed] [Google Scholar]
- 37.Van Mooy BAS, Fredricks HF. Bacterial and eukaryotic intact polar lipids in the eastern subtropical South Pacific: Water-column distribution, planktonic sources, and fatty acid composition. Geochim Cosmochim Acta. 2010;74:6499–6516. [Google Scholar]
- 38.Sowell SM, et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2009;3(1):93–105. doi: 10.1038/ismej.2008.83. [DOI] [PubMed] [Google Scholar]
- 39.Markowitz VM, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42(Database issue):D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith DP, et al. Proteomic and transcriptomic analyses of “Candidatus Pelagibacter ubique” describe the first PII-independent response to nitrogen limitation in a free-living Alphaproteobacterium. MBio. 2013;4(6):e00133–e12. doi: 10.1128/mBio.00133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 43.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Mooy BAS, Rocap G, Fredricks HF, Evans CT, Devol AH. Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc Natl Acad Sci USA. 2006;103(23):8607–8612. doi: 10.1073/pnas.0600540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Ferguson-Miller SM, Reid GE. Characterization of ornithine and glutamine lipids extracted from cell membranes of Rhodobacter sphaeroides. J Am Soc Mass Spectrom. 2009;20(2):198–212. doi: 10.1016/j.jasms.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eddy SR. Accelerated profile HMM searches. PLOS Comput Biol. 2011;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.