Significance
Signaling pathways form complex networks of biochemical reactions, but inferring the topology of such networks and measuring how they are remodeled in disease is still challenging. Using MS, our study defined the circuitry and plasticity of a kinase signaling network de novo, with unprecedented depth and without prior assumptions of its topology. In addition, we observed a degree of stochasticity in how the network was remodeled upon chronic inhibition of phosphoinositide 3-kinase (PI3K) or mammalian target of rapamycin complexes 1/2 (mTORC1/2), suggesting that the initial condition of the system was not the only determinant of how cells become resistant to targeted therapies. These observations may have implications for our ability to predict the evolution of signaling networks during therapy to prevent the acquisition of resistance.
Keywords: signaling network, phosphoproteomics, systems biology, PI3K, MEK
Abstract
Our understanding of physiology and disease is hampered by the difficulty of measuring the circuitry and plasticity of signaling networks that regulate cell biology, and how these relate to phenotypes. Here, using mass spectrometry-based phosphoproteomics, we systematically characterized the topology of a network comprising the PI3K/Akt/mTOR and MEK/ERK signaling axes and confirmed its biological relevance by assessing its dynamics upon EGF and IGF1 stimulation. Measuring the activity of this network in models of acquired drug resistance revealed that cells chronically treated with PI3K or mTORC1/2 inhibitors differed in the way their networks were remodeled. Unexpectedly, we also observed a degree of heterogeneity in the network state between cells resistant to the same inhibitor, indicating that even identical and carefully controlled experimental conditions can give rise to the evolution of distinct kinase network statuses. These data suggest that the initial conditions of the system do not necessarily determine the mechanism by which cancer cells become resistant to PI3K/mTOR targeted therapies. The patterns of signaling network activity observed in the resistant cells mirrored the patterns of response to several drug combination treatments, suggesting that the activity of the defined signaling network truly reflected the evolved phenotypic diversity.
Cell signaling pathways form complex networks of biochemical reactions that integrate and decode extracellular signals into appropriate responses (1). The reconstruction of these networks, and systematic analyses of their properties, is important in the advancement of our molecular understanding of disease at the systems level (2). The topology and plasticity of cell signaling networks play major roles in fundamental (3–5) and disease physiology (6, 7). Attempts to characterize such molecular organization have relied on inference algorithms that obtain information on protein interactions and posttranslational modification (PTMs) from the literature (8–10). The accuracy of network reconstruction using such models is limited by the availability of data (10) and by the fact that signaling events are often cell-type specific. As a result, although they can provide insightful data, models that derive network topologies from studies that have used different cell types and organisms result in composite or averaged networks, which, critically, do not always reflect network structure in specific cell types, at specific stages of cell development, or under defined physiological conditions (10).
Reconstruction of signaling networks through the use of a single set of well-defined experimental data is appealing, because this approach does not commit to a preconception of how such networks may be wired in a given cell type under defined conditions (3). The maturation of phosphoproteomics techniques based on mass spectrometry (MS) is now allowing the simultaneous quantification of several thousands of phosphorylation sites per experiment, and approaches to derive kinase activity from these large-scale phosphoproteomics datasets have been reported (11–14). One such approach, named kinase substrate enrichment analysis (KSEA), is based on the premise that, because each phosphorylation site is the result of a kinase’s catalytic activity, phosphoproteomic profiling provides a means by which to capture and measure the activities of all kinases expressed in the system under investigation (14).
Here, we first used MS-based phosphoproteomics to define a kinase signaling network by systematically identifying phosphorylation sites downstream of kinases targeted by small-molecule kinase inhibitors of the PI3K/Akt/mTOR and MEK-ERK signaling axes. These two ubiquitous pathways form a network that regulates growth factor, antigen, and insulin signaling while also being deregulated in most cancers (15–17). We then measured the activity and plasticity of different routes within this experimentally defined kinase signaling network in cells chronically treated with small-molecule inhibitors of PI3K and mTORC1/2. We found that remodeling of kinase networks in resistant cells produced patterns of signaling activity linked to their evolved phenotypes.
Results
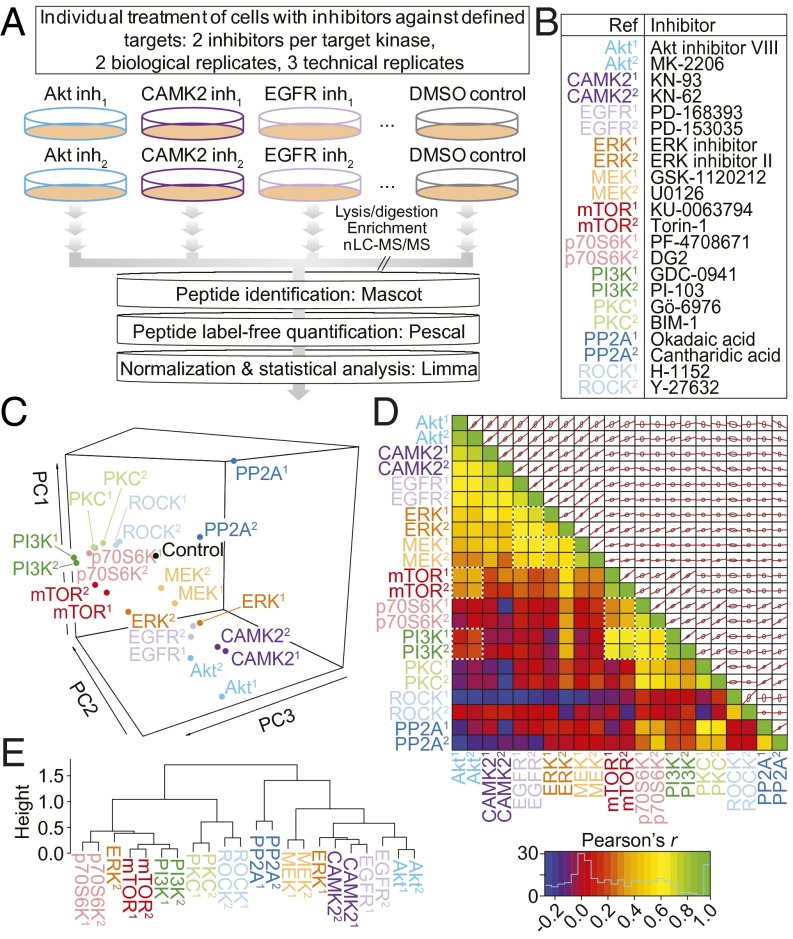
We set out to classify phosphorylation sites into groups defined by their patterns of modulation in response to inhibitors of cell signaling. We treated MCF7 cells with single small-molecule inhibitors against a panel of kinases and measured the resulting changes in phosphorylation through the use of MS-based phosphoproteomics (Fig. 1A). Twenty structurally distinct kinase inhibitors, two phosphatase inhibitors, or DMSO vehicle control (Fig. 1B) were the conditions used for the experiments. The kinases targeted in the experiments, alongside the inhibitors used, were chosen on the basis of their known involvement in growth factor and metabolic signaling, and their current therapeutic potential (SI Appendix, Table S1).
Fig. 1.
Phosphoproteomics data elucidate the relationships between kinase inhibitors. (A) Experimental design. Inh, inhibitor. (B) Kinase inhibitors used in the study. (C) PCA of the 4,651 phosphorylation sites whose abundance was reduced significantly (adjusted P < 0.05) by at least one inhibitor treatment. PC, principal component. (D) Lower triangle, Pearson correlation coefficients between each of the inhibitor treatments. Known kinase–kinase relationships are highlighted with white, dashed boxes. Upper triangle, pair-wise alignments of the 4,651 phosphorylation site log2 fold ratios for each inhibitor combination. Red lines indicate the linear model formed between the two variables; gray ellipses represent one SD from the mean in both dimensions. (E) Unsupervised, hierarchical clustering (Pearson correlation distance metric) of the mean log2 ratios for peptides containing common phosphorylation motifs represented in the filtered 4,651 phosphorylation sites.
The MS experiments resulted in the identification of a total of 13,405 unique phosphopeptide ions across the six analytical replicates per condition (three technical and two biological). A quality control summary of these data is shown in SI Appendix, Fig. S1. Each phosphopeptide was quantified across all of the experimental conditions by using a previously described label-free methodology (18, 19), generating 1,930,320 data points (Dataset S1).
Phosphoproteomics Data Allow the Classification of Kinase Inhibitors Based on the Targets They Inhibit.
We observed 4,651 phosphorylation sites significantly reduced in abundance by at least one kinase inhibitor (adjusted P ≤ 0.05). To assess the global effects of inhibitors on these sites, we used principal component analysis (PCA). This multivariate statistical analysis method allows the separation of experimental conditions based on the overall structure of the underlying data. PCA of the inhibitor-treated phosphoproteomes demonstrated that inhibitors directed against the same kinase were closer to each other in principal component space than to the rest of the inhibitors (Fig. 1C), indicating that inhibitors against the same kinase produced similar effects on global phosphorylation. The only exception to this observation was the ERK inhibitors; these being close in the PC1 but not PC2 dimension, suggesting that these compounds had slightly different quantitative effects on the phosphoproteome. Whereas inhibitors against kinases related to the MAPK signaling cascade (EGFR, MEK, and ERK) separated from those related to the PI3K/mTOR signaling axis (PI3K, mTOR, p70S6K), inhibitors of Akt associated more closely with EGFR and CAMK2 inhibitors than to inhibitors of its well-known upstream activator, PI3K. As would be expected, inhibitors targeting PP2A (a protein phosphatase) separated well from the kinase inhibitors. Analysis using a correlation matrix reinforced the relationships observed among mTOR, Akt, and PI3K inhibitors and among EGRF, ERK, and MEK inhibitor pairs (Fig. 1D). Statistical significance of each correlation is shown in SI Appendix, Fig. S2. Motif analysis (14) further revealed that the inhibitor pairs exhibited strongly correlated impacts on specific phosphorylation motifs while mirroring the relationships seen in Fig. 1 C and D (Fig. 1E). Together, these data show that inhibitors against the same kinases produced more similar changes in the phosphoproteomes than to the rest of inhibitors and, with the exception of Akt, those against the same canonical pathways also affected a common set of phosphorylation sites.
Deriving Activity Markers of Inhibitor Targets from Phosphoproteomics Data.
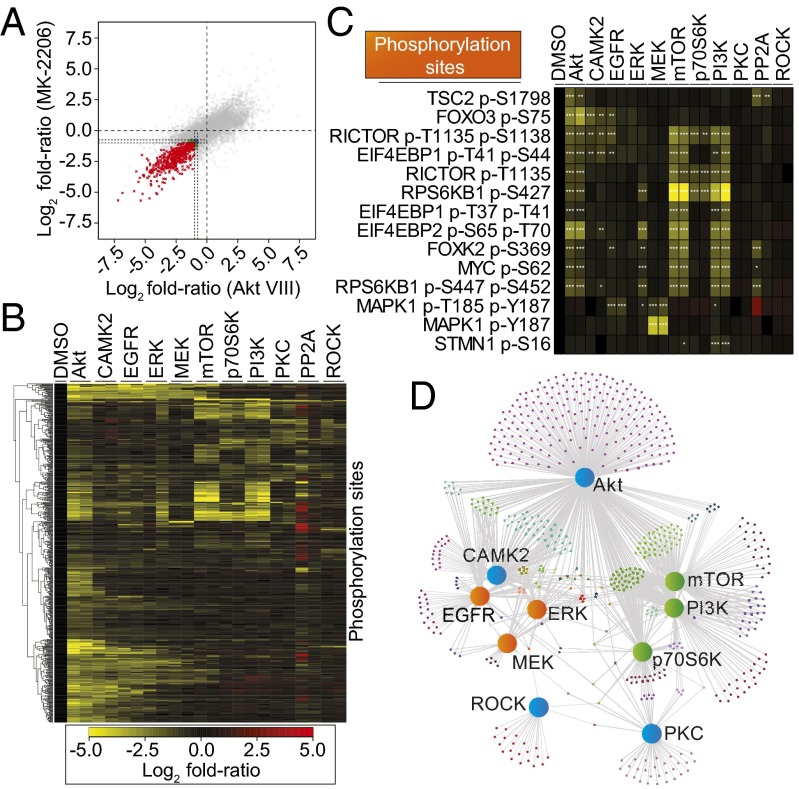
To provide an additional level of classification to the dataset, we further grouped the phosphorylation sites based on their behavior under treatment with inhibitors against the same kinase. The number of phosphorylation sites selected at this stage depended on the stringency of the thresholds used for selection (SI Appendix, Fig. S3). Most importantly however, we selected only those phosphorylation sites that reached the required statistical thresholds in both inhibitor treatments targeting the same kinase. The selection of phosphorylation sites inhibited by structurally distinct compounds targeting the same kinase should result in datasets enriched in phosphorylation sites specific to the intended kinase. This concept is illustrated in Fig. 2A for the phosphorylation sites modulated by the two different Akt inhibitors (MK-2206 and Akt Inhibitor VIII), which shows phosphorylation sites inhibited by both inhibitors (red data points in Fig. 2A) and sites specifically inhibited by one compound but not the other (blue and green data points in Fig. 2A). We hypothesized that sites inhibited by the Akt inhibitor MK-2206 but not by Akt inhibitor VIII, and vice versa, were off-target effects, whereas those inhibited by both compounds were more likely to be truly downstream of Akt. This analysis was performed for each of the 10 kinases targeted in the study (Fig. 1A) and revealed 610 phosphorylation sites reduced in abundance by at least one inhibitor pair (i.e., by both inhibitors against the same kinase). These sites, although not necessarily directly phosphorylated by the intended target kinase—as they could be phosphorylated by kinases acting downstream or by closely related kinases— are readouts of the actual kinases affected by the inhibitor/compound, and, thus, we referred to them as compound-target activity markers (CTAMs).
Fig. 2.
Inference of a kinase signaling network topology from phosphoproteomics data. (A) Phosphorylation site log2 fold ratios (versus DMSO control) for the two Akt inhibitors: MK-2206 and Akt inhibitor VIII. Dotted lines represent the thresholds for CTAM identification. The phosphorylation sites colored in the negative quadrant of the graph (bottom left) represent the CTAMs identified for Akt. Red, Log2 fold-ratio (FR) ≤ −1.0, adjusted (adj.) P ≤ 0.1 for both inhibitors; blue, FR ≤ −0.75, adj. P ≤ 0.05 for both inhibitors; green, mixed thresholds between inhibitors. (B) Six hundred ten phosphorylation sites identified as being markers of compound-target activity for at least one kinase. (C) Illustrative examples of phosphorylation sites arranged into CTAM groups. *FR ≤ −0.75, adj. P ≤ 0.05; **FR ≤ −1.0, adj. P ≤ 0.1; ***FR ≤ -1.0, adj. P ≤ 0.01. Colors as in B. (D) An undirected, bipartite network graph showing all of the identified activity markers. The layout of the network graph is based on a force-directed drawing algorithm. The large nodes represent the kinases targeted in the experiment (orange, MAPK-associated; green, PI3K/mTOR/p70S6K-associated; blue, mixed association). Smaller nodes represent individual phosphorylation sites. Gray edges denote whether the phosphorylation site is a CTAM of the kinase to which it is connected.
Inferring Signaling Network Topology from Phosphoproteomics Data.
Visualizing the 610 identified CTAM phosphorylation sites simultaneously revealed that a large number of them were identified as markers of more than one compound-target pair (Fig. 2B). Therefore, to investigate the relationships between inhibitor pairs further, and to allow inference of signaling network topology from the data, the 610 CTAM phosphorylation sites were further classified based on whether these were inhibited by one or more inhibitor pairs. A number of known patterns of kinase signaling topology emerged from this analysis (Fig. 2C). For example, we identified 41 phosphorylation sites that were inhibited by the inhibitor pairs against Akt, mTOR, p70S6K, and PI3K (Fig. 2C). Consistent with previous knowledge, these sites included those on BAD, Ser427 on KS6B1 (p70S6K1), and Thr1135 on RICTOR (Fig. 2C) (20–22). Alongside these known modifications, sites that have not yet been functionally annotated were also present in this group, and together these 41 sites were classified as members of the Akt-mTOR-PI3K-p70S6K CTAM group. Similarly, sites modulated by Akt, mTOR, and PI3K inhibitor pairs, but not by the p70S6K inhibitor pair, included GSK3β at Ser9, Myc at Ser62, and AKTS1 (also known as PRAS40) at Ser183 (SI Appendix, Fig. S4); a total of 55 phosphorylation sites were found to have this pattern of inhibition and defined an Akt-mTOR-PI3K group that was independent of p70S6K. In addition to these well-known kinase cascades, we also found evidence for the existence of as yet uncharacterized relationships between the kinases targeted by the inhibitors and/or the inhibitors themselves; examples included sites modulated by mTOR and PI3K inhibitor pairs without the involvement of Akt or p70S6K (37 substrates; Fig. 2 B and C). As Fig. 2 B and C illustrate, we also found evidence of sites inhibited by both Akt inhibitors but unaffected by PI3K and other inhibitors (284 substrates), and PI3K sites independent of Akt and mTOR (33 substrates). Overall, the 610 phosphorylation site activity markers found in this study (SI Appendix, Table S2) were grouped into 55 CTAM groups.
Visualizing the data as a bipartite, undirected network graph (Fig. 2D) further revealed the way in which the investigated kinase inhibitors related to each other in the signaling network. As expected and in concordance with published data and their canonical associations, Akt, PI3K, p70S6K, and mTOR inhibitor pairs affected a large number of common phosphorylation sites and, hence, were grouped together. Similarly, inhibitor pairs targeting the MAPK pathway (EGFR, MEK, and ERK) also grouped together and with CAMK2 (Fig. 2D). Randomization of the network’s topology revealed that these associations were not likely to have occurred by chance (SI Appendix, Fig. S5). These data therefore show that, although canonical associations between kinases were well represented in our dataset (e.g., EGFR-MEK-ERK and PI3K-Akt-mTOR-p70S6K), the existence of unexpected signaling routes also emerged from these data, including the existence of PI3K-mTOR signaling independent of Akt. Moreover, this analysis demonstrated the high degree of connectivity between these kinases.
Characterization of the Identified CTAM Groups’ Behavior and Probing of Network Plasticity.
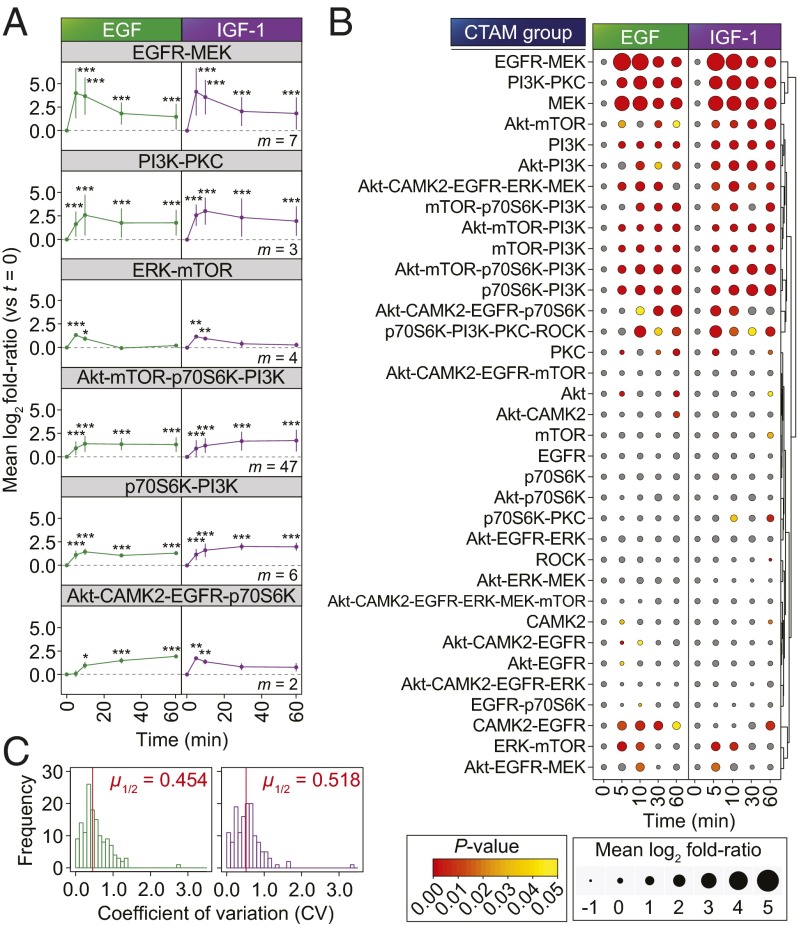
We next sought to confirm whether the CTAM groups could be used to measure the biochemical activation of pathways within the network and, thus, provide a snapshot of the network’s activation status at any given time, under any given condition. We hypothesized that, should these groups provide reliable readouts of network branch activity, each CTAM group should show the expected behavioral changes when the network is perturbed or stimulated under well-characterized experimental conditions. In addition, we reasoned that individual members within each group should demonstrate similar quantitative behavior to one another. We thus monitored the dynamics of phosphorylation of the CTAM groups across cells treated with either EGF or IGF1 at five independent time-points (Dataset S2). A quality control summary for this dataset is shown in SI Appendix, Fig. S6.
We observed that the temporal phosphorylation dynamics of CTAM groups commonly associated with EGFR and IGF-1R signaling (relative to the 0 min control in each case) were in-line with the previously reported effects of EGF and IGF1 on kinase signaling (Fig. 3A) (23, 24). For example, consistent with the known temporal dynamics of MAPK pathway activation, the EGFR-MEK group underwent significant, acute up-regulation upon stimulation with both growth factors for 5 min, before beginning to decline to a lower level at 60 min (Fig. 3 A and B). These data were in agreement with individual MS and Western blot data for the canonical EGFR-responsive MAPK (Thr202/Tyr204) and Akt sites (Ser473) (SI Appendix, Fig. S7). The median relative SDs (i.e., coefficient of variations) of individual phosphorylation sites within CTAM groups were 0.454 and 0.518 for EGF and IGF1 respectively (Fig. 3C), thus reflecting that these behaved similarly upon cell stimulation with the two growth factors. Taken together, the data shown in Fig. 3 provide evidence to support the notion that CTAM groups were readouts of the functional activation of branches within the network.
Fig. 3.
Kinetics of CTAM group behavior upon growth factor stimulation. (A) Temporal profiles of each of selected CTAM groups in cells treated with growth factors (EGF, green; IGF1, purple; m = number of phosphorylation sites quantified in the named CTAM group). Data points represent mean ± SD. ***P < 0.001; **P < 0.01; *P < 0.05. (B) Temporal profiles for each of the CTAM groups represented in the dataset (with m ≥ 2). Data point sizes are proportional to the mean log2 fold ratio (versus t = 0 min) and colored according to the statistical significance of enrichment. Unsupervised hierarchical clustering was based on the Euclidean distance metric. (C) Distribution of coefficient of variations for each of the quantified CTAM groups at each of the time-points. Red lines indicate the median.
Analysis of Network Plasticity in Models of Acquired Resistance to Kinase Inhibitors.
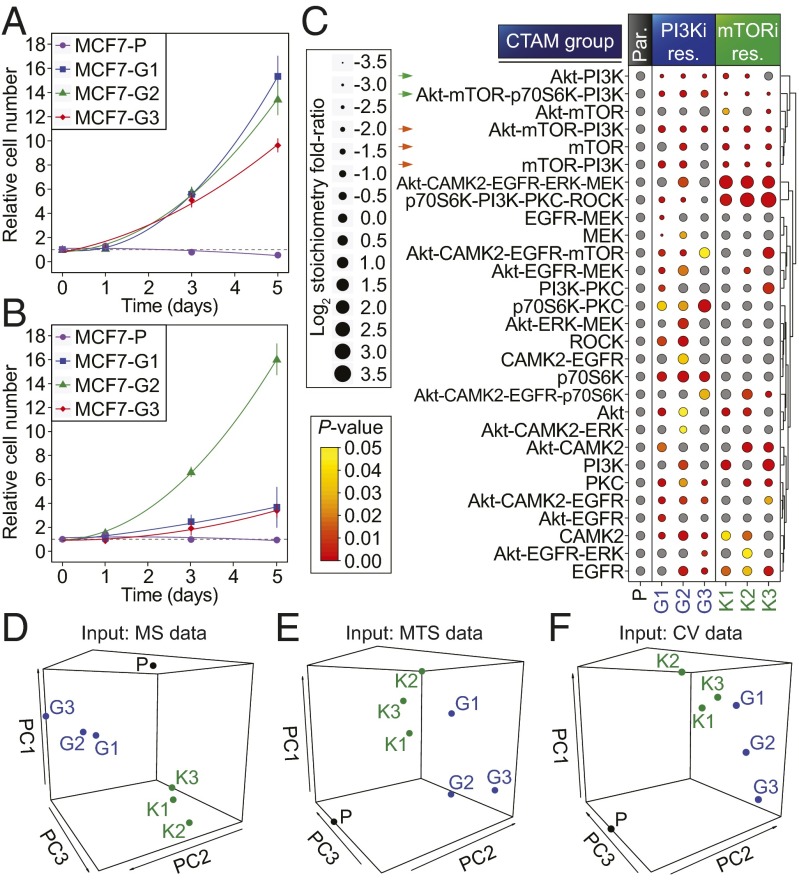
To further investigate kinase signaling plasticity in our CTAM-defined signaling network, we measured the phosphorylation sites that define the network in cancer cell-line models of acquired resistance to two kinase inhibitors in clinical development; namely, GDC-0941 (a pan class I PI3K inhibitor) and KU-0063794 (an mTORC1/2 inhibitor) (25, 26). We obtained six independent cell cultures resistant to each of the inhibitors compared with the parental cells from which they were derived (three per drug: MCF7-G and MCF7-K resistant to GDC-041 and KU-0063794, respectively). To achieve this aim, we chronically exposed the cells to an increasing concentration of the relevant inhibitor up to a maximum of 1 µM. The cells were initially challenged with a low concentration of each drug (100 nM) so as not to bias the resistance selection for intrinsically resistant cells. The resultant cell lines were able to proliferate in the presence of 1 µM of inhibitor, whereas parental cells were unable to do so under the same conditions (Fig. 4 A and B). We quantified the phosphoproteomes of these cells and normalized these measurements to total protein by simultaneously analyzing the total proteome (Dataset S3). A summary of the quantitative and qualitative data are shown in SI Appendix, Fig. S8.
Fig. 4.
Evolution of signaling network activity in cells resistant to mTORC1/2 and PI3K inhibitors. Growth of parental and drug-resistant MCF7 cell cultures in the presence of GDC-0941 (A) or KU-0063794 (B). Data points represent the mean ± SD. (C) CTAM group (with m ≥ 2) profiles for each of the resistant (res.) cell-lines compared with the parental (par.) cell-line. Dot sizes represent the mean log2 fold ratio of each CTAM group relative to parental cell line, normalized to the unmodified protein abundance. Colors represent the significance of enrichment. Hierarchical clustering of the CTAM groups was based on the Euclidean distance metric. (D) PCA of the phosphoproteomic data shown in C. (E) PCA of cell viability data (measured by MTS) as a function of treatment with a panel of kinase inhibitors (shown in SI Appendix, Fig. S10A). (F) As in E, however, cell viability was measured by crystal violet staining (shown in SI Appendix, Fig. S10B).
Analysis of the kinase network in the presence of inhibitor revealed that the large majority of the CTAM groups containing mTOR were down-regulated in all of the mTOR inhibitor resistant (MCF7-K) cell lines (green and orange arrows in Fig. 4C). Similarly, CTAM groups containing PI3K were down-regulated in all of the PI3K inhibitor-resistant (MCF7-G) cell lines (green arrows in Fig. 4C). We confirmed these data by measuring well-known markers of pathway activities, which showed that our results were consistent with the levels of key regulatory phosphorylation sites governing these pathways on Akt (Ser473) and p70S6K (Thr389), as determined by Western blot (SI Appendix, Fig. S9). These data suggested that the pathways targeted by the inhibitors remained inhibited in resistant cells in the presence of the drug. We therefore reasoned that resistance was not the result of differences in how resistant cells metabolized the inhibitors and that instead, consistent with other studies (27), resistance more likely arose as a consequence of a rewiring of kinase signaling. Of interest, this rewiring was markedly dissimilar between the MCF7-K and MCF7-G cell lines (Fig. 4C), suggesting that the resistance mechanisms that had evolved against the mTOR inhibitor were distinct to those evolved in response to the PI3K inhibitor. This hypothesis was reinforced through the use of an unbiased multivariate analysis of the normalized phosphoproteomics data, which highlighted the differences between the parental and resistant cells, and the differences between the rewiring of MCF7-K and MCF7-G cells, because these separated clearly in principal component space (Fig. 4D). Unexpectedly, however, cells resistant to the same inhibitor also separated in PC space, suggesting that the activation state of the signaling network was heterogeneous between individual resistant lines (Fig. 4 C and D) despite these being derived from the same parental culture, at the same passage number, and being exposed to identical experimental conditions for the same amount of time.
Because the resistant cell lines seemed to differ in the way in which they had rewired their signaling network compared with parental cells and to each other (Fig. 4 C and D), we hypothesized that each cell line should respond differently to a panel of small-molecule kinase inhibitors alone and in combination, because their response would be a function of their signaling network’s activity. To test this prediction, we treated each of the cell lines with a panel of small-molecule inhibitors (targeting PI3K, mTOR, CAMK2, Akt, MEK, and EGFR) and measured their relative cell viability through the use of the MTS and crystal violet assays (SI Appendix, Fig. S10 A and B, respectively). These inhibitors were chosen because the CTAMs of their associated kinases were increased in abundance in some of the resistant cells relative to parental (Fig. 4C), thus suggesting that these kinases may be involved in the resistance phenotype. An unbiased, multivariate analysis of the resulting data revealed that the resistant and parental cells responded differently to the inhibitors, as they separated in PC space. Moreover, this analysis separated the MCF7-G and MCF7-K cells and the individual resistant cell lines from one another in a manner reflecting that observed in the PCA of the phosphoproteomics data (Fig. 4 E and F). Taken together, these data indicate that the heterogeneous rewiring of the signaling network in resistant cells observed by CTAM analysis (Fig. 4C) resulted in functional differences in how cells responded to perturbations to the network (Fig. 4 E and F).
Discussion
In this study, we first performed a thorough analysis of the connections that exist between the nodes of the PI3K-MEK kinase network (Figs. 1 and 2). This initial study revealed both expected and unexpected links among kinases, signaling pathways and the pharmacological agents targeting them. For example, the known PI3K-Akt-mTOR-p70S6K, PI3K-Akt-mTOR, and MEK-ERK relationships were well represented in our data; however, we also found evidence for as yet uncharacterized connections between kinase inhibitor targets, such as those defined by mTOR-PI3K associations without the involvement of Akt and Akt inhibitor-dependent but PI3K inhibitor-independent sites. Overall, our data exemplify the complex relationship between kinases in signaling networks and illustrate that our knowledge of this complexity is still limited.
An advantage of defining signaling routes using a set of experimental data derived from a defined system—in contrast to approaches that compile information from the literature (28, 29)—is that cell signaling events are often cell-type and cell-context dependent (30). Therefore, “averaged” signaling networks, derived from disparate cell types and organisms, as shown in canonical signaling pathway schematics, are not always representative of how signaling networks are in fact wired in specific cellular systems. Although efforts have been made to overlay empirical transcriptional data onto these averaged networks (31), a key aspect of our study is that we not only provide evidence of as yet uncharacterized signaling routes but also identified phosphorylation sites markers of such routes’ activities specific to our cell-line model, which could then be used to measure the dynamics and circuitry of the kinase network in a systematic manner. The CTAM approach to define signaling network branches, which can then be measured in subsequent experiments, has conceptual similarities to approaches that derive cell biological information from gene expression patterns by examining how such patterns correlate with compendia of profiles obtained from systematic gene inactivation experiments (32). The observation that CTAM groups were modulated by growth factors with the expected kinetics (refs. 23 and 24; Fig. 3) and that these changes were similar for members of such groups (Fig. 3) provided evidence to suggest that these CTAM groups are biochemical readouts of signaling activity.
Signaling networks are not static, but rather highly dynamic structures that are extremely plastic in response to external stimuli. A comparison of the network between parental and cells resistant to either a PI3K or mTORC1/2 inhibitor revealed widespread differences in CTAM group abundances in three separate resistant cell cultures per inhibitor (Fig. 4C). Our data are therefore consistent with published studies showing that kinase signaling is remodeled in response to chronic kinase inhibition (27, 33). Because of the depth of our analyses, however, our data emphasize the extent to which signaling networks are modulated as a whole to overcome chronic inhibition of single nodes. Concurrent with these observations, comparison of the data on network status (phosphoproteomics) and phenotypes (responses to inhibitors) for each cell line using multivariate analyses indicated that the activity of the signaling network was a reflection of the cells’ newly acquired phenotypes. This phenomenon was evidenced in the PCAs of all these disparate sets of data, which separated the resistant from parental cells, and the MCF7-K from the MCF7-G cells in a similar manner (Fig. 4 D–F). Our data therefore accentuate the complex relationship between PI3K and mTORC1/2, as the cells treated with the two inhibitors changed their signaling differently, thus suggesting different mechanisms of resistance in response to the two inhibitors (Fig. 4 D–F). This result was unexpected because PI3K and mTOR are often placed in the same canonical signaling pathway. However, these data are consistent with the observation that phosphorylation sites modulated by mTORC1/2 or PI3K inhibitors only partially overlapped (Fig. 2E), and with a recent study highlighting mTOR’s independence from PI3K in some systems (34). The simple maintenance of cells in culture could have contributed to the evolution of cell networks shown in Fig. 4; however, the observation that chronic exposure to mTORC1/2 or PI3K inhibitors produced marked differences in network remodeling argues that drug exposure had a greater effect on how signaling evolved than that which would be observed as a result of long-term culture without the application of such a selective pressure.
Recent publications indicate that, rather than there being a single mechanism of acquired resistance for a given therapy, there is a plethora of ways in which signaling networks can be rewired in cancer cells as these become resistant to targeted therapies (35, 36). It is not known, however, whether the way in which cancer cells develop resistance is predetermined by the molecular imprinting of cancer cells at presentation (i.e., before the resistance phenotype ensues). This understanding has potential implications for cancer therapeutics because, if the evolutionary pathways that lead to the acquisition of resistance were reliant on genomic or other molecular factors present in tumor cells at presentation, then analyzing the initial conditions of cancer cells could, at least in principle, be used to predict the resistance mechanism more likely to occur in such a tumor.
Contrary to this idea, our data suggest that it may not be possible to predict the particular mechanism of resistance of a given tumor by analyzing it at the molecular level at presentation. Indeed, in our study, the same cell line was split into six identical populations, which were then maintained in the same concentrations of inhibitors by the same operator for the same amount of time and passage numbers. Unexpectedly, the CTAM network profiles were highly heterogeneous across cells resistant to the same compound (Fig. 4 C and D). This heterogeneity in network status had a functional consequence in that cells resistant to the same compound responded differently to inhibitors of signaling at the level of cell proliferation (Fig. 4 E and F). Intratumoral heterogeneity and evolutionary divergence of initially clonal systems has been documented at the genetic level in both mammalian and bacterial systems (37, 38). Although we cannot entirely exclude the impact of long-term cell culture, our study suggests that chronic treatment with targeted kinase inhibitors profoundly influences the divergence of kinase network signaling. Thus, because identical experimental conditions can result in the evolution of distinct signaling networks (Fig. 4 C and D) and drug-resistance phenotypes (Fig. 4 E and F), perhaps due to stochastic effects, the initial conditions of the system may not be accurate predictors of the evolutionary route that may lead to resistance. Therefore, if the acquisition of resistance is truly indeterministic and cannot be predicted at disease onset, identification of the resistance mechanisms relevant to each individual patient will most likely require the ability to measure the signaling network in individual tumors after resistance has ensued with depth and without a preconception of how signaling may have been rewired as a result of therapy.
Methods
Cell Lines.
The parental MCF7 cell line and MCF7-G1, MCF7-G2, MCF7-G3, MCF7-K1, MCF7-K2, and MCF7-K3 resistant cells were cultured in DMEM (supplemented with 10% (vol/vol) fetal-bovine serum and 100 U⋅mL−1 penicillin/streptomycin) at 37 °C in a humidified atmosphere at 5% CO2. After treatment, as indicated in the text, cells were lysed in urea lysis buffer and proteins digested with trypsin.
Mass Spectrometry-Based Phosphoproteomics.
Phosphorylated peptides were enriched through the use of TiO2 beads (GL Sciences) in a similar manner to that previously described (14, 19) with some modifications, and analyzed by LC-MS/MS using a LTQ-Orbitrap mass spectrometer. Peptides were identified by means of Mascot searches against the SwissProt human protein database. Peptide quantification was achieved by using Pescal as previously described (14).
Statistical Analysis.
Following quantile normalization of the data (39), the magnitude and statistical significance of differences between conditions were computed by means of empirical Bayes shrinkage of SDs (40) using the limma package within the R computing environment (41, 42). The abundance of CTAMs was monitored systematically by using KSEA (14, 18, 19, 30).
More detailed description of these methods is provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of both past and present research groups; P. Faull and A. Montoya for their technical assistance; F. Iorio for help with the network randomization; and J. Fitzgibbon, A. Cameron, R. Grose, and members of the Integrative Cell Signaling and Proteomics group for helpful discussion. This work was supported by Barts and the London Charity Grant 297/997 and a Cancer Research UK Barts Cancer Institute Centre Grant C236/A11795.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423344112/-/DCSupplemental.
References
- 1.Du W, Elemento O. Cancer systems biology: Embracing complexity to develop better anticancer therapeutic strategies. Oncogene. September 15, 2014 doi: 10.1038/onc.2014.291. [DOI] [PubMed] [Google Scholar]
- 2.Papin JA, Hunter T, Palsson BO, Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol. 2005;6(2):99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- 3.Bodenmiller B, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 2010;3(153):rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jørgensen C, Linding R. Simplistic pathways or complex networks? Curr Opin Genet Dev. 2010;20(1):15–22. doi: 10.1016/j.gde.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332(6035):1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goltsov A, et al. Features of the reversible sensitivity-resistance transition in PI3K/PTEN/AKT signalling network after HER2 inhibition. Cell Signal. 2012;24(2):493–504. doi: 10.1016/j.cellsig.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 7.Goltsov A, et al. Compensatory effects in the PI3K/PTEN/AKT signaling network following receptor tyrosine kinase inhibition. Cell Signal. 2011;23(2):407–416. doi: 10.1016/j.cellsig.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 8.Kholodenko BN, et al. Untangling the wires: A strategy to trace functional interactions in signaling and gene networks. Proc Natl Acad Sci USA. 2002;99(20):12841–12846. doi: 10.1073/pnas.192442699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee S, Speed TP. Network inference using informative priors. Proc Natl Acad Sci USA. 2008;105(38):14313–14318. doi: 10.1073/pnas.0802272105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prill RJ, Saez-Rodriguez J, Alexopoulos LG, Sorger PK, Stolovitzky G. Crowdsourcing network inference: The DREAM predictive signaling network challenge. Sci Signal. 2011;4(189):mr7. doi: 10.1126/scisignal.2002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linding R, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129(7):1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson SM, et al. Large-scale discovery of ERK2 substrates identifies ERK-mediated transcriptional regulation by ETV3. Sci Signal. 2011;4(196):rs11. doi: 10.1126/scisignal.2002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bensimon A, Heck AJ, Aebersold R. Mass spectrometry-based proteomics and network biology. Annu Rev Biochem. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- 14.Casado P, et al. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci Signal. 2013;6(268):rs6. doi: 10.1126/scisignal.2003573. [DOI] [PubMed] [Google Scholar]
- 15.Posch C, et al. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc Natl Acad Sci USA. 2013;110(10):4015–4020. doi: 10.1073/pnas.1216013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renshaw J, et al. Dual blockade of the PI3K/AKT/mTOR (AZD8055) and RAS/MEK/ERK (AZD6244) pathways synergistically inhibits rhabdomyosarcoma cell growth in vitro and in vivo. Clin Cancer Res. 2013;19(21):5940–5951. doi: 10.1158/1078-0432.CCR-13-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts PJ, et al. Combined PI3K/mTOR and MEK inhibition provides broad antitumor activity in faithful murine cancer models. Clin Cancer Res. 2012;18(19):5290–5303. doi: 10.1158/1078-0432.CCR-12-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casado P, et al. Phosphoproteomics data classify hematological cancer cell lines according to tumor type and sensitivity to kinase inhibitors. Genome Biol. 2013;14(4):R37. doi: 10.1186/gb-2013-14-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montoya A, Beltran L, Casado P, Rodríguez-Prados JC, Cutillas PR. Characterization of a TiO2 enrichment method for label-free quantitative phosphoproteomics. Methods. 2011;54(4):370–378. doi: 10.1016/j.ymeth.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 21.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 22.Kumar JK, Ping RYS, Teong HF, Goh S, Clément M-V. Activation of a non-genomic Pim-1/Bad-Pser75 module is required for an efficient pro-survival effect of Bcl-xL induced by androgen in LNCaP cells. Int J Biochem Cell Biol. 2011;43(4):594–603. doi: 10.1016/j.biocel.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 23.Vincent AM, Feldman EL. Control of cell survival by IGF signaling pathways. Growth Horm IGF Res. 2002;12(4):193–197. doi: 10.1016/s1096-6374(02)00017-5. [DOI] [PubMed] [Google Scholar]
- 24.Ciaccio MF, Wagner JP, Chuu C-P, Lauffenburger DA, Jones RB. Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat Methods. 2010;7(2):148–155. doi: 10.1038/nmeth.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García-Martínez JM, et al. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421(1):29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munugalavadla V, et al. The PI3K inhibitor GDC-0941 combines with existing clinical regimens for superior activity in multiple myeloma. Oncogene. 2014;33(3):316–325. doi: 10.1038/onc.2012.594. [DOI] [PubMed] [Google Scholar]
- 27.Liu P, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17(9):1116–1120. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Sys Biol. 2005;1:2005.0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alcolea MP, Casado P, Rodríguez-Prados JC, Vanhaesebroeck B, Cutillas PR. Phosphoproteomic analysis of leukemia cells under basal and drug-treated conditions identifies markers of kinase pathway activation and mechanisms of resistance. Mol Cell Proteomics. 2012;11(8):453–466. doi: 10.1074/mcp.M112.017483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutta B, et al. A network-based, integrative study to identify core biological pathways that drive breast cancer clinical subtypes. Br J Cancer. 2012;106(6):1107–1116. doi: 10.1038/bjc.2011.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102(1):109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 33.Muranen T, et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21(2):227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elkabets M, et al. mTORC1 inhibition is required for sensitivity to PI3K p110 alpha inhibitors in PIK3CA-mutant breast cancer. Sci Transl Med. 2013;5(196):196ra99. doi: 10.1126/scitranslmed.3005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klempner SJ, Myers AP, Cantley LC. What a tangled web we weave: Emerging resistance mechanisms to inhibition of the phosphoinositide 3-kinase pathway. Cancer Discov. 2013;3(12):1345–1354. doi: 10.1158/2159-8290.CD-13-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013;19(11):1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 37.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Gac M, Plucain J, Hindré T, Lenski RE, Schneider D. Ecological and evolutionary dynamics of coexisting lineages during a long-term experiment with Escherichia coli. Proc Natl Acad Sci USA. 2012;109(24):9487–9492. doi: 10.1073/pnas.1207091109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 40.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Molec Biol. 2004;3(1):3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 41.Smyth GK. Limma: Linear Models for Microarray Data. Springer; New York: 2005. [Google Scholar]
- 42.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria; 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.