Abstract
Background:
Pathology reports typically require manual review to abstract research data. We developed a natural language processing (NLP) system to automatically interpret free-text breast pathology reports with limited assistance from manual abstraction.
Methods:
We used an iterative approach of machine learning algorithms and constructed groups of related findings to identify breast-related procedures and results from free-text pathology reports. We evaluated the NLP system using an all-or-nothing approach to determine which reports could be processed entirely using NLP and which reports needed manual review beyond NLP. We divided 3234 reports for development (2910, 90%), and evaluation (324, 10%) purposes using manually reviewed pathology data as our gold standard.
Results:
NLP correctly coded 12.7% of the evaluation set, flagged 49.1% of reports for manual review, incorrectly coded 30.8%, and correctly omitted 7.4% from the evaluation set due to irrelevancy (i.e. not breast-related). Common procedures and results were identified correctly (e.g. invasive ductal with 95.5% precision and 94.0% sensitivity), but entire reports were flagged for manual review because of rare findings and substantial variation in pathology report text.
Conclusions:
The NLP system we developed did not perform sufficiently for abstracting entire breast pathology reports. The all-or-nothing approach resulted in too broad of a scope of work and limited our flexibility to identify breast pathology procedures and results. Our NLP system was also limited by the lack of the gold standard data on rare findings and wide variation in pathology text. Focusing on individual, common elements and improving pathology text report standardization may improve performance.
Keywords: Breast cancer, natural language processing, pathology, validation
BACKGROUND
Breast pathology reports are generated after tissue extraction to describe benign and malignant pathology findings. Pathology report data are important for many cancer screening, performance, and treatment studies to identify relevant procedures and outcomes in a standardized format.[1,2,3,4] Even with electronic medical records (EMRs), most pathology reports remain free-text and have varying information provided by different interpreting pathologists. To be used for research, a trained human abstractor must review individual pathology reports to code information to a normalized, structured form.[5]
Manual abstraction is time-consuming and costly in human hours potentially making it infeasible for very large research studies. To address these limitations, researchers have explored using natural language processing (NLP) for automated pathology data abstraction. NLP is a method currently developed for use within the clinical domain, such as EMRs, including pathology reports, with the goal of making the abstraction process more efficient.[6,7,8,9,10,11]
Natural language processing has had mixed results with pathology reports. Several studies have demonstrated its success in interpreting pathology reports, particularly when extracting a very limited number of findings or a single feature.[10,12,13] We are aware of only one study that tested NLP on breast pathology reports, and the authors concluded that the complexity, length, and variation in text of breast pathology reports limited the accuracy of NLP.[14] We hoped to further explore and improve upon the findings from this prior study.
We designed a NLP system that would address issues from prior efforts, such as error propagation, spelling mistakes, and variation in the language used to convey negation.[7,8,9,10,14,15] We sought to develop an all-or-nothing method to accurately extract a large number of breast pathology results and procedures using NLP alone. Our goal in using this all-or-nothing approach was to abstract breast pathology at a report level – meaning the entire report at once – thus, limiting the number of pathology reports that would require any additional human review.
METHODS
This study took place at Group Health Cooperative, a mixed-model delivery health system in Washington state that provides both healthcare and health insurance to approximately 600,000 members. Approximately 2500 breast pathology reports are abstracted annually at Group Health Research Institute,[16] one of the participating sites in the national Breast Cancer Surveillance Consortium (BCSC).[17] Manual abstraction is aided by the existence of an EMR from which programmers identify and download all breast pathology reports for review. Pathology reports are identified because the report contains a string of certain words, such as “breast,” lumpec,” “mastec,” or common misspellings or abbreviations of “breast” (e.g. “brest” or “brst”). The NLP system was only tested on free-text reports, which generally come from pathologists within the group health medical system. Ethics approval was obtained from the Group Health Human Subjects Review Committee along with a waiver of consent for the pathology report review.
Data Source
We used data from 3234 breast pathology reports from group health spanning November 2011 to December 2012 to develop and test the NLP system [Figure 1]. Only data from this time period could be used due to variations in the BCSC pathology codebook from 2003 to 2011. We randomly divided reports into two sets: A development set (n = 2910, 90%) and an evaluation set (n = 324, 10%). The development set was further randomly divided into a training set (n = 2637, 90.6%) and a test set (n = 273, 9.4%) for preliminary testing. This 90/10 split allowed the NLP system to fully develop using the bulk of the data before being tested on unseen data with the remaining 324 reports in the evaluation set.
Figure 1.
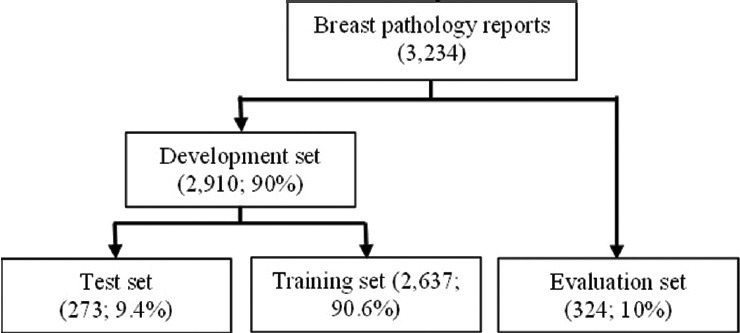
The number of reports used for natural language processing system training and testing, including validation and evaluation of the training set
A highly-trained abstractor uses a 55-page BCSC pathology codebook that defines three sets of BCSC standardized pathology findings on breast pathology: Biopsy and surgical procedures (21 findings), benign and malignant results (43 findings), and associated laterality (4 findings).[16] For each pathology report, the abstractor can code one procedure, one laterality, and up to five results, even if more than five results appear in that report. If a report contains more than one procedure, it is divided into multiple outputs to account for each finding; thus reports are manually abstracted on a procedure-level. Figure 2 is an example of an abstracted report. As the Figure shows, each pathology procedure finding is abstracted along with associated results and laterality findings. Abstraction quality is reviewed annually comparing 2% of reports to an expert pathologist review. In 2013, manual abstraction had an 85% accuracy rate when compared with expert pathology review.
Figure 2.
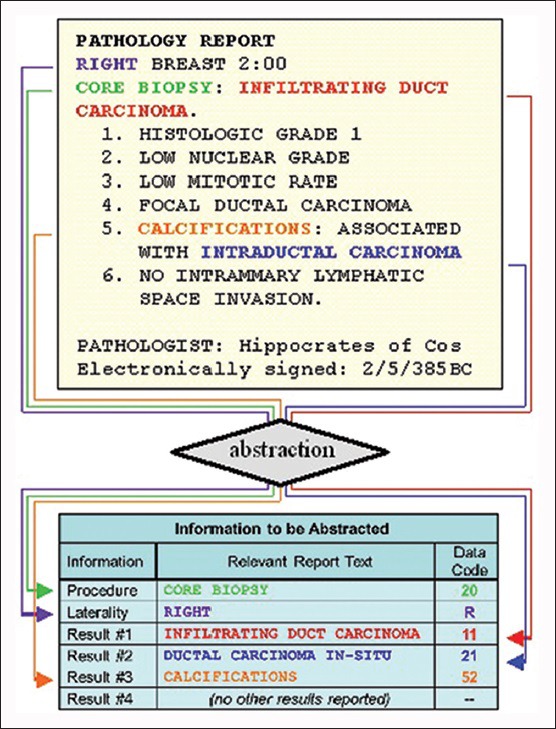
This is an example of an abstracted breast pathology report and associated natural language processing data codes. Relevant procedures and results are color-coded to show how they correspond between the report at the top and the data at the bottom
Natural Language Processing System Development: General Approach
We used an iterative process to develop, evaluate, and refine potential NLP system models to extract pathology findings from reports.[18] Because a single report can contain multiple procedures, the NLP system divided reports with more than one procedure into multiple outputs, similar to the manual abstraction process. A machine learning model was developed for each finding (procedure, result, and laterality) in the training data using the Python machine learning library scikit-learn.[19] For each finding, we used the scikit-learn implementations of the naïve Bayes and support vector machine classifiers with a variety of parameter settings to train candidate models using the training data. We selected this toolkit because it provided a vast range of optimized NLP tools.[19]
Each candidate model was evaluated against the test set. To increase candidate model accuracy, we reviewed the precision of each model, selecting the classifier with the highest positive predictive value (PPV) and the classifier with the highest negative predictive value (NPV) [Figure 3]. The classifiers that produced the most precise models on the training data were then run on the entire development set and evaluated against the held-out evaluation set. A particular procedure, result, or laterality finding was only assigned when both the high-PPV and high-NPV classifiers predicted the finding. When the two classifiers disagreed, that particular report is flagged for manual review. If a report contained both malignant and benign results, we prioritized malignant findings over benign findings because the malignant findings would typically be considered the worst outcome in research projects.
Figure 3.
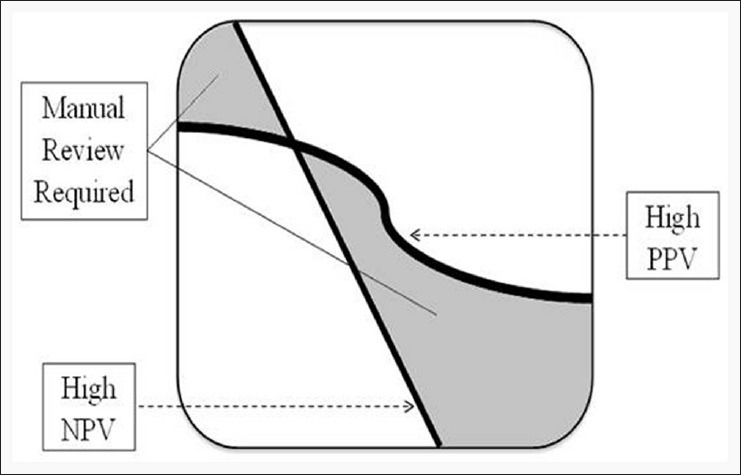
How we flagged reports for manual review. The squiggly line is the threshold for positive predictive value (PPV) (reports had to have a PPV above the line) and the diagonal line the threshold for negative predictive value (NPV) (reports had to have a NPV below the line). Reports that fell into the white areas were assigned a data finding. Reports that fell into the grey areas were flagged for manual review
Features Selection
We selected features to determine which findings to assign and to distinguish the pathology findings from one another. For instance, features such as “invasive ductal carcinoma” needed to be differentiated from “invasive lobular carcinoma,” “ductal hyperplasia,” “atypical ductal hyperplasia,” and “ductal carcinoma in situ”. We classified the free-text reports using two types of features for each document: Sequences of adjacent words (n-gram) and BCSC codebook-derived keywords.[5,16,20,21,22] The n-gram features consisted of sequences of one (unigram), two (bigram), and three (trigram) adjacent words, along with 1-skip-bigrams and 1-skip-trigrams, which skip an intervening word. For example, the phrase “carcinoma type infiltrating ductal” is represented by two 1-skip-bigrams (“carcinoma infiltrating,” “type ductal”) and two 1-skip-trigrams (“carcinoma type ductal,” “carcinoma infiltrating ductal”).
The BCSC pathology codebook includes keywords for each procedure and result to guide manual abstraction. For example, the codebook lists similar terms that belong to the finding “invasive ductal,” including: “adenocarcinoma infiltrating ductal,” “infiltrating ductal cancer,” “infiltrating ductal carcinoma,” and “intraductal papillary adenosarcoma with invasion". We identified the keywords in reports, allowing for up to two intervening words, ignoring nonessential words like “with,” and allowing for the words to appear in any order. The same feature (e.g. “invasive ductal”) was used for each finding regardless of which keyword (e.g. “adenocarcinoma infiltrating ductal”) was identified in the report. Thus, the feature for the finding “invasive ductal” was added if any of the keywords were found in the text.
The total number of unique features across all reports that we extracted was 4,211,995, and the average number of unique features per report was about 1302. To reduce this, we grouped related findings together into broader categories based on similar features. Figure 4 is an example of one of these groupings. The first algorithm run on every report determined whether the report was breast-related or whether it should be omitted from the review entirely. Those omitted from the review did not need to be reviewed for any other features. If the report was not omitted, we then used an NLP algorithm to determine if it belonged to a broad category of “invasive cancers”. The individual findings “invasive cancer, not otherwise specified (NOS),” “invasive ductal,” and “invasive lobular” all belonged to this larger group of “invasive cancers”. We determined whether a report belonged to the larger category before determining whether it represented a more specific finding. If it did not belong to “invasive cancers,” then we knew it could not be “invasive cancer, NOS,” “invasive ductal,” or “invasive lobular”. This method also provided a means of identifying rarer findings (e.g. “invasive cancer, NOS”), which had very limited (if any) training data available. If a result within the larger category of invasive cancers did not belong to either “invasive ductal” or “invasive lobular”), then we inferred that the result was “invasive cancer, NOS”.
Figure 4.
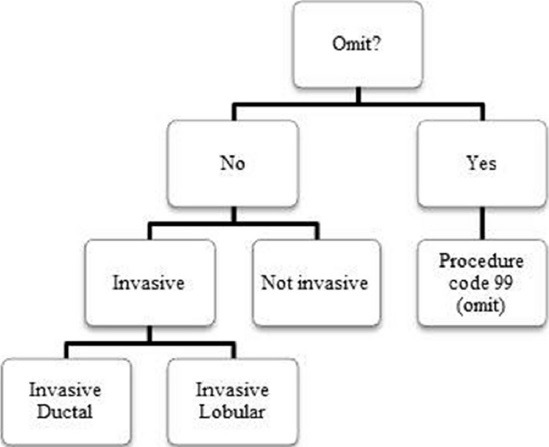
An example of a grouping of results used to improve natural language processing (NLP) system performance. “Omit” was at the top, which allowed the NLP system to exclude any nonbreast-related or irrelevant reports. If the reports were not omitted, we next determined if the results belonged to a large category of invasive or not invasive, and then individual categories of ductal or lobular among invasive reports
In clinical text, many of these keywords do not actually represent the presence of a finding because they are qualified by negation, uncertainty, or reference to the past. To help identify these cues (Appendi × 1 for a specific list of negation cues), we relied on a modification of the NegEx algorithm, which locates words before and after the keywords to determine whether or not the keywords are positively asserted.[23] We used 287 sets of terms to identify words before and after the keywords to determine whether or not the keywords were positively asserted. When the negation algorithm determined that a keyword was not positively asserted (it was qualified by negation, uncertainty, or reference to the past), the keyword was added as a separate feature than when the keyword was positively asserted.
Once all the features were identified in the training set, we used the Chi-squared test to order the features according to how predictive they were of each finding. We retained the top 1% of features and used them in our machine learning algorithm to extract findings from the evaluation set.
Evaluation
Our primary evaluation used an all-or-nothing approach to determine whether our NLP system could abstract a pathology report in its entirety. This meant that we compared the NLP system results with the gold standard at the report level instead of looking at individual procedure outputs. We compared NLP findings against the gold standard at the report level by identifying how many reports processed by NLP contained the exact same findings annotated by the human abstractor. The evaluation data were used to confirm the NLP system's accuracy as a final test on unseen data.
We also reviewed the NLP system's accuracy for identifying individual procedures, results, and laterality using precision and sensitivity measures. Specificity, accuracy, and f-score performance values were also recorded based on the amounts of true positives (hits), false positives (false hits), true negatives (TNs) (correct rejections), and false negatives (misses). These performance measures are a set of equations defined as follows: Precision (reproducibility, PPV); sensitivity (recall or hit rate); specificity (TN rate); accuracy (closeness of measured value to gold standard); and f-score (harmonic mean of precision and sensitivity).
RESULTS
Half (49.1%) of the evaluation set was flagged for manual review (n = 159) [Figure 5]. The remaining 50.9% of the set was either omitted (7.7%) or fully processed by the NLP system (43.2%), but only 41 reports from this fully processed subset (12.7%) were completely correctly abstracted at the report level. The remaining 99 reports (30.6%) were not flagged for manual review, but were incorrectly coded, meaning at least one procedure, laterality, or result finding within a report did not match the gold standard.
Figure 5.
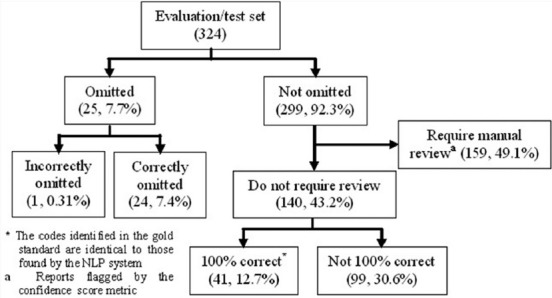
The results of our evaluation, or test, set. Among reports that were not omitted, 49.1% were flagged for manual review, 30.6% were assigned incorrect codes, and 12.7% were completely coded correctly following our all-or-nothing approach
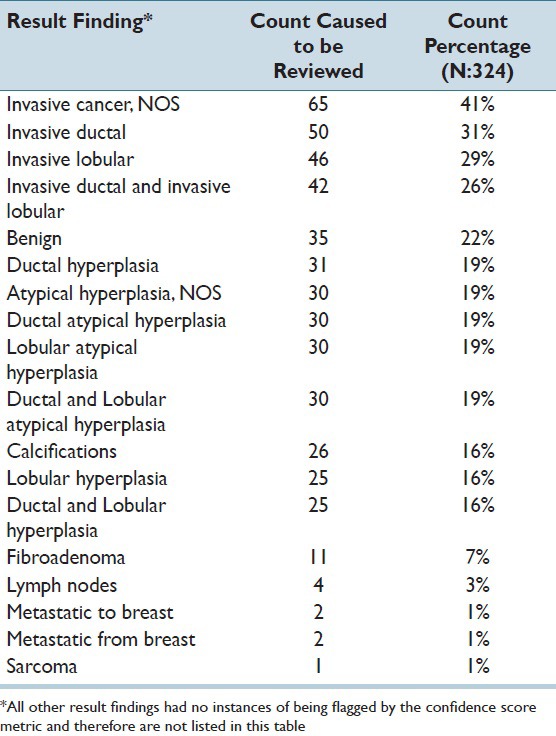
Pathology findings that caused reports to be flagged for manual review, along with the number of reports in which these particular findings were flagged, are shown in Table 1. Sometimes, multiple findings were flagged in the same report requiring manual review. The counts were higher for malignant findings than benign findings because benign results were not considered if a malignant finding within a report was already flagged for manual review.
Table 1.
Result findings that were flagged for manual review by the confidence score metric, including the number of reports that was flagged for review. Note that some reports were flagged by multiple codes.
Precision levels obtained by individual procedure and result findings are shown in Table 2. Common findings, such as “breast closed percutaneous biopsy/core biopsy, NOS,” and “invasive ductal” had high frequencies (54.0% and 28.1%, respectively) and performed well on all measures, with precision scores of 0.90 and 0.95, and sensitivities of 1.00 and 0.94, respectively. However, in a few instances, common findings performed poorly, such as the code “benign” (frequency 50.0%) with a precision score of 0.81 and sensitivity of 0.38.
Table 2.
Performance of several procedure, laterality, and result findings from the evaluation set (N:324) and processed by the final version of the NLP systema
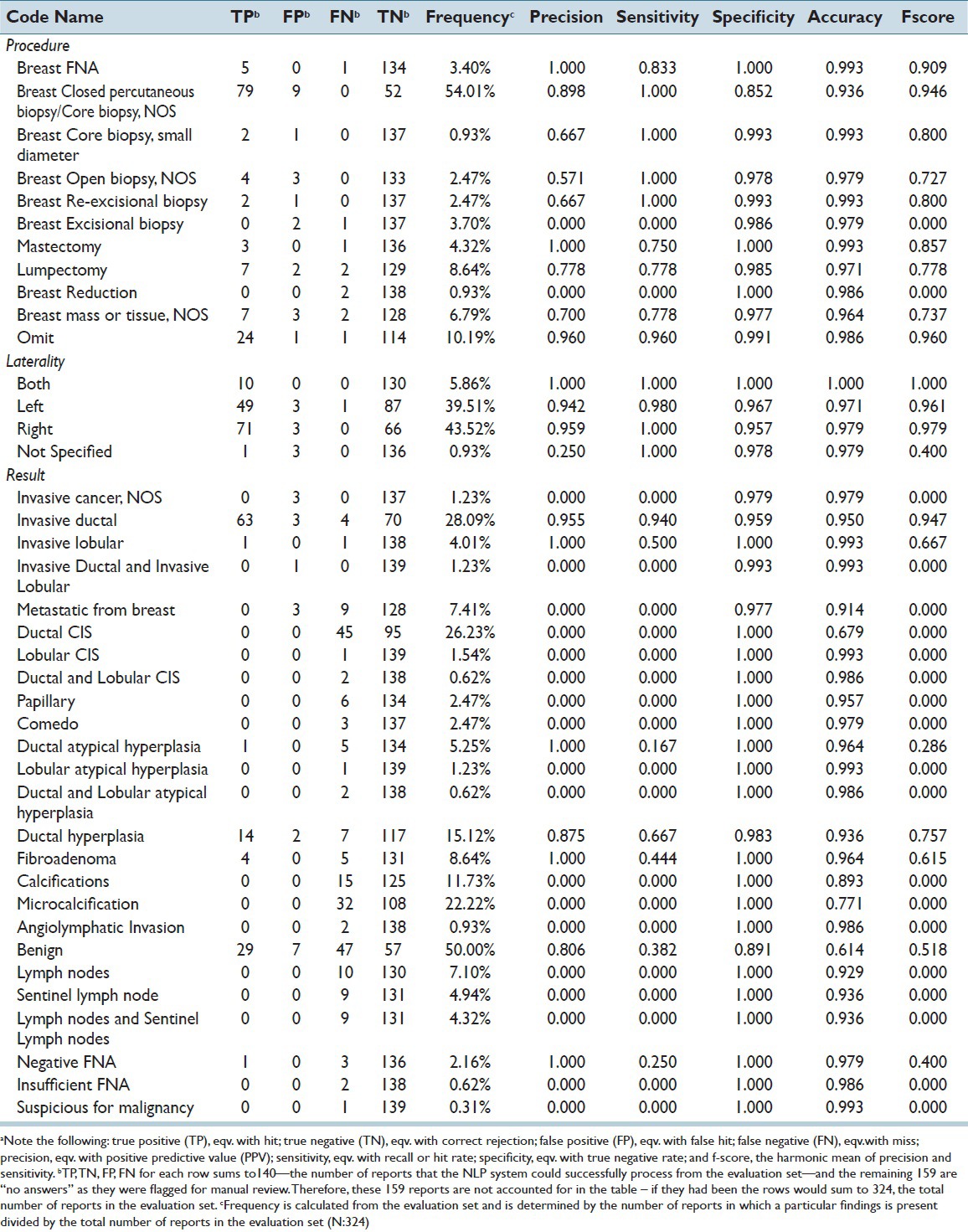
The “omit” category fared better in terms of number of reports accurately assigned the “omit” code in the evaluation set (n = 24, 7.4%) [Figure 5] and its high level of precision (0.96) and accuracy (0.99) as reported in Table 2. Overall, the NLP system was able to identify 13.6% (n = 440) of the full set of pathology reports as not requiring abstraction because they did not contain relevant breast pathology information.
CONCLUSIONS
We attempted to develop an all-or-nothing NLP system using machine learning to classify all breast pathology procedures, laterality, and results within an entire report the same way a manual abstractor classified them with little success. We tried to maximize our NLP system's performance via four major computational methods: (1) Using both coding manual keywords and n-gram to identify features;[21,22] (2) constructing categories of related findings; (3) removing rare findings that were considered less important (meaning less useful for further research and analysis); and (4) creating a method to flag reports for manual review. Even with these improvements, only 12.7% of the evaluation set of reports could be processed entirely and accurately by our NLP abstraction system. Below we discuss the reasons our NLP system did not perform well in our setting, which include problems related to the scope of the work, our NLP system design, and the gold standard data.
Scope of Work
While our NLP system performed well on some common individual procedure, result, and laterality codes, the all-or-nothing approach reduced overall performance. The all-or-nothing approach is desirable in research settings that want to eliminate reviewing reports that do not need human eyes. The cost and time required for reading a pathology report to abstract a number of results are not proportionally larger than the cost and time required to abstract just a single result. If chart abstractors need to review pathology for even a few items, this requires a programmer to identify the report, an abstractor to open the report and read it in its entirety, and various overhead costs associated with these tasks. While this kind of approach may be appropriate for some NLP systems, it was not for ours. Our all-or-nothing system required NLP to code nearly 70 procedures and results correctly in each pathology report. The scope of this project may have been too broad. Our NLP system might have performed better if we had coded a smaller set of results; however, this would have been insufficient for our research purposes. Balancing the need to reduce manual abstraction with NLP accuracy is an important trade-off. A partial abstraction approach may fare better than all-or-nothing in demonstrating the potential benefits of developing and implementing an NLP system for breast pathology reports, but may not be sufficient for research study implementation.
Natural Language Processing System Design
Many individual classifiers for codes were strong on their own, but when they were analyzed simultaneously, the NLP system performed poorly due to propagation of errors. Error propagation is a cascading effect in which residual error is generated after a first error has been made. For instance, if a pathology finding was incorrectly not omitted and subsequently coded as “invasive ductal,” this resulted in two classification errors – one for not omitting and another for assigning the wrong code. While common findings performed well, our all-or-nothing approach meant that their high performance was essentially hidden behind the poor performance of other findings. However, this approach most aligned with our aim of determining the extent to which NLP could accurately process entire reports.
Splitting a single report into multiple separate outputs led to several complications. Due to our all-or-nothing evaluation approach, we attempted to re-combine the procedures that had been separated during preprocessing into complete reports so that they could be compared with the gold standard at a report level. In those instances when re-combination was necessary, it was difficult to compare the gold standard and NLP system results against one another, as the gold standard did not involve any explicit procedure-separation rules, unlike our NLP outputs. For example, the NLP system separated a report into four different procedure outputs while the gold standard had only been separated into three. Then, when re-combined into a single report, some codes did not match up with one another in an identical manner, making the overall report appear incorrect. This issue often affected the comparison of our results with the gold standard by lowering the performance values of our results.
Gold Standard Data
There are several aspects of our gold standard data that lowered the success of the NLP process. Due to the small training dataset, there was a scarcity of certain pathology findings within the data. The rarity of certain findings in training data made it difficult to develop high-performing models. The inclusion of broad-brush stroke categories, such as the “benign” finding, which incorporated too many divergent concepts at once, further reduced accuracy. Further, our gold standard had 85% accuracy, which established an upper limit to what we would expect for NLP system performance. The magnitude of these problems and their exact influence on the results are unknown; however, these issues likely lowered the performance of our NLP system for two reasons: They taught the model to look for the wrong things and in the evaluation stage, penalized the model for choosing the right answer when the gold standard had the wrong answer.
Additional pathology reporting standardization could lead to improvements in NLP system abstraction.[7] Standardized pathology reporting exists, but pathologists are not required to use standardized language and reporting guidelines are not standardized across professional pathology organizations.[24,25,26,27,28] Widespread adoption of these guidelines could potentially improve the accuracy of an NLP system to code pathology reports. The data set also came from a single institution with fewer than 10 pathologists, which did not allow for the full linguistic variation of free text reports or different forms of standardization for free text breast pathology reports that may appear in other healthcare settings.[7]
Project Takeaways and Future Implementation
This project highlighted several lessons regarding the development and use of NLP to abstract breast pathology reports. First, it showed that within the scope of projects similar to this one, NLP cannot be expected to perform well when applied to an existing, complex manual process. Attempting to code 43 results, 21 procedures, and 4 laterality findings within the same report may have been overly ambitious, but was necessarily for our research purposes. Second, this project revealed the importance of high quality training data that contains examples of both common and rare pathology findings with as few inconsistencies as possible. We found features with sufficient high-quality data performed well while underperforming features suffered from insufficient training data. Third, this project helped demonstrate the circumstances under which NLP automated abstraction may or may not be useful. For instance, NLP could be useful when looking for clearly defined, common categories like “invasive ductal carcinoma”. However, manual review may still be required in instances when the NLP system is uncertain about a particular finding.
Future NLP research targeting a large number of findings may need to prioritize either precision or sensitivity, as it does not appear possible, based on this study, to have both high quality precision and sensitivity at the same time. Using NLP in this context may be better suited for research studies where one has access to resources for manual review for reports flagged by the NLP system and high performance is unnecessary for every pathology report. Future research on this subject could include using a different approach than the all-or-nothing approach taken by our team to evaluate the success of the NLP system. Instead of looking at success in replicating the abstraction process, perhaps the NLP system might function better when looking for only a few well-performing findings rather than 70 different findings within a single report.
In conclusion, we developed and evaluated an NLP system with the aim of creating a better abstraction system in which NLP could replace manual review. Our system faced many issues due to the complexity of pathology reports, such as language and reporting style variation, and a limited amount of high-quality training data to enable us to accurately classify report findings. Overall, the results of our all-or-nothing NLP system were not deemed satisfactory to develop a clear plan for implementation for research purposes.
ACKNOWLEDGMENTS
This research was supported by the grant P01CA154292 from the National Cancer Institute. The National Cancer Institute had no involvement in the study design; collection, analysis and interpretation of data; the writing of the report; or the decision to submit the article for publication.
Appendix
Appendix 1.

Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2015/6/1/38/159215
REFERENCES
- 1.Tice JA, O’Meara ES, Weaver DL, Vachon C, Ballard-Barbash R, Kerlikowske K. Benign breast disease, mammographic breast density, and the risk of breast cancer. J Natl Cancer Inst. 2013;105:1043–9. doi: 10.1093/jnci/djt124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison KH, Reisch LM, Carney PA, Weaver DL, Schnitt SJ, O’Malley FP, et al. Understanding diagnostic variability in breast pathology: Lessons learned from an expert consensus review panel. Histopathology. 2014;65:240–51. doi: 10.1111/his.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg RD, Yankaskas BC, Abraham LA, Sickles EA, Lehman CD, Geller BM, et al. Performance benchmarks for screening mammography. Radiology. 2006;241:55–66. doi: 10.1148/radiol.2411051504. [DOI] [PubMed] [Google Scholar]
- 4.Weaver DL, Rosenberg RD, Barlow WE, Ichikawa L, Carney PA, Kerlikowske K, et al. Pathologic findings from the breast cancer surveillance consortium: Population-based outcomes in women undergoing biopsy after screening mammography. Cancer. 2006;106:732–42. doi: 10.1002/cncr.21652. [DOI] [PubMed] [Google Scholar]
- 5.Zhu F, Patumcharoenpol P, Zhang C, Yang Y, Chan J, Meechai A, et al. Biomedical text mining and its applications in cancer research. J Biomed Inform. 2013;46:200–11. doi: 10.1016/j.jbi.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Friedman C. A broad-coverage natural language processing system. Proc AMIA Symp. 2000:270–4. [PMC free article] [PubMed] [Google Scholar]
- 7.Hazlehurst BL, Lawrence JM, Donahoo WT, Sherwood NE, Kurtz SE, Xu S, et al. Automating assessment of lifestyle counseling in electronic health records. Am J Prev Med. 2014;46:457–64. doi: 10.1016/j.amepre.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu ST, Sohn S, Ravikumar KE, Wagholikar K, Jonnalagadda SR, Liu H, et al. Automated chart review for asthma cohort identification using natural language processing: An exploratory study. Ann Allergy Asthma Immunol. 2013;111:364–9. doi: 10.1016/j.anai.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehrotra A, Dellon ES, Schoen RE, Saul M, Bishehsari F, Farmer C, et al. Applying a natural language processing tool to electronic health records to assess performance on colonoscopy quality measures. Gastrointest Endosc. 2012;75:1233–9.e14. doi: 10.1016/j.gie.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imler TD, Morea J, Kahi C, Imperiale TF. Natural language processing accurately categorizes findings from colonoscopy and pathology reports. Clin Gastroenterol Hepatol. 2013;11:689–94. doi: 10.1016/j.cgh.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrell DS, Halgrim S, Tran DT, Buist DS, Chubak J, Chapman WW, et al. Using natural language processing to improve efficiency of manual chart abstraction in research: The case of breast cancer recurrence. Am J Epidemiol. 2014;179:749–58. doi: 10.1093/aje/kwt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim BJ, Merchant M, Zheng C, Thomas AA, Contreras R, Jacobsen SJ, et al. A natural language processing program effectively extracts key pathologic findings from radical prostatectomy reports. J Endourol. 2014;28:1474–8. doi: 10.1089/end.2014.0221. [DOI] [PubMed] [Google Scholar]
- 13.Thomas AA, Zheng C, Jung H, Chang A, Kim B, Gelfond J, et al. Extracting data from electronic medical records: Validation of a natural language processing program to assess prostate biopsy results. World J Urol. 2014;32:99–103. doi: 10.1007/s00345-013-1040-4. [DOI] [PubMed] [Google Scholar]
- 14.Buckley JM, Coopey SB, Sharko J, Polubriaginof F, Drohan B, Belli AK, et al. The feasibility of using natural language processing to extract clinical information from breast pathology reports. J Pathol Inform. 2012;3:23. doi: 10.4103/2153-3539.97788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harkema H, Chapman WW, Saul M, Dellon ES, Schoen RE, Mehrotra A. Developing a natural language processing application for measuring the quality of colonoscopy procedures. J Am Med Inform Assoc. 2011;18(Suppl 1):i150–6. doi: 10.1136/amiajnl-2011-000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehman C, Holt S, Peacock S, White E, Urban N. Use of the American College of Radiology BI-RADS guidelines by community radiologists: Concordance of assessments and recommendations assigned to screening mammograms. AJR Am J Roentgenol. 2002;179:15–20. doi: 10.2214/ajr.179.1.1790015. [DOI] [PubMed] [Google Scholar]
- 17.Ballard-Barbash R, Taplin SH, Yankaskas BC, Ernster VL, Rosenberg RD, Carney PA, et al. Breast Cancer Surveillance Consortium: A national mammography screening and outcomes database. AJR Am J Roentgenol. 1997;169:1001–8. doi: 10.2214/ajr.169.4.9308451. [DOI] [PubMed] [Google Scholar]
- 18.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine learning in python. J Mach Learn Res. 2011;12:2825–30. [Google Scholar]
- 20.Huang Y, Lowe HJ. A novel hybrid approach to automated negation detection in clinical radiology reports. J Am Med Inform Assoc. 2007;14:304–11. doi: 10.1197/jamia.M2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.FitzHenry F, Murff HJ, Matheny ME, Gentry N, Fielstein EM, Brown SH, et al. Exploring the frontier of electronic health record surveillance: The case of postoperative complications. Med Care. 2013;51:509–16. doi: 10.1097/MLR.0b013e31828d1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sidorov G, Velasquez F, Stamatatos E, Gelbukh AF, Chanona-Hernandez L. Syntactic N-grams as machine learning features for natural language processing. Expert Syst Appl. 2014;41:853–60. [Google Scholar]
- 23.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 24.No. 58 ed. NHS Cancer Screening Programmes and The Royal College of Pathologists; 2005. Pathology Report of Breast Disease: A Joint Document Incorporating the Third Edition of the NHS Breast Screening Programme's Guidelines for Pathology Reporting in Breast Cancer Screening and the Second Edition of The Royal College of Pathologists’ Minimum Dataset for Breast Cancer Histopathology. [Google Scholar]
- 25.Wilkinson NW, Shahryarinejad A, Winston JS, Watroba N, Edge SB. Concordance with breast cancer pathology reporting practice guidelines. J Am Coll Surg. 2003;196:38–43. doi: 10.1016/s1072-7515(02)01627-7. [DOI] [PubMed] [Google Scholar]
- 26.Amin MB. Letter to Frederick L. Green of American College of Surgeons, Commission on Cancer. CAP Today [eLetter] 2009 [Google Scholar]
- 27.Washington K. Letter to Stephen Edge of American College of Surgeons, Commission on Cancer. CAP Today [eLetter] 2011 [Google Scholar]
- 28.Paxton A. Cancer Protocols: Leaner, Later, More Lenient. CAP Today [eLetter] 2004 [Google Scholar]


