Abstract
Amelogenins are proteins formed by alternative splicing of the amelogenin gene, and are essential for tooth enamel formation. However, the unique functions of various alternatively spliced amelogenins in enamel formation are not well understood. In this study, we determined the spatiotemporal location of amelogenins derived from transcripts containing exon4 (AMG+4) in the enamel matrix, and the relative binding of recombinant AMG+4 to hydroxyapatite (HAP). Immunohistochemistry and mass spectrometry analyses showed that AMG+4 proteins were secreted into the enamel matrix at the early maturation stage. A stage-specific increase in the synthesis of AMG+4 was further supported by our observation that in mice overexpressing leucine-rich amelogenin peptide (TgLRAP), in which ameloblasts differentiate earlier, AMG+4 transcripts were also upregulated earlier. In vitro binding studies, supported by in silico modeling of protein binding to calcium and phosphate, showed that more recombinant AMG+4 bound to hydroxyapatite (HAP) as compared with recombinant AMG-4. The temporal and spatial localization of amelogenins containing exon4 peptide, and their functional differences in HAP binding, suggests that the unique properties of amelogenins containing exon4 cause a specific enhancement of biomineralization related to stabilization of early-formed HAP at the maturation stage.
Keywords: mineralized tissue, protein expression, tooth development, matrix biology, craniofacial biology, cell-matrix interactions
Introduction
Amelogenins constitute the major protein component of the secretory dental enamel matrix, and many studies have been undertaken in an effort to understand their biochemical function during enamel formation and mineralization (Termine et al. 1980). Amelogenins are alternatively spliced (Gibson et al. 1992; Fincham et al. 1994; Lyngstadaas et al. 1995), though there is little information as to the function of the alternatively spliced amelogenins.
Fully translated human amelogenin is a protein that is the product of 7 exons, with exon1 being non-coding. Rodents can have 2 additional exons, 8 and 9, forming an alternate c terminus when exon7 is skipped (Li et al. 1998). Two primary amelogenin isoforms have been identified in the secretory-stage enamel matrix. One of these is the hydrophobic amelogenin (Eastoe 1964), encoded by exons2, 3, 5, 6, and 7, and known as M180 in mice and H175 in humans. The other is leucine-rich amelogenin peptide (LRAP) (Fincham et al. 1981), translated from exons2, 3, 5, 6D, and 7 (Gibson et al. 1991), and known as M59 in mice and H58 in humans. M180 is known to direct HAP formation, whereas LRAP has been shown to enhance proliferation and differentiation of mesenchymal cells (Boabaid et al. 2004; Ye et al. 2006; Warotayanont et al. 2009) and to direct ameloblast differentiation (Le et al. 2007; Stahl et al. 2013). Phosphorylated LRAP stabilizes mineral formation (Le Norcy et al. 2011), but overexpression of LRAP in an amelogenin knockout mouse model fails to rescue the enamel mineralization defect (Chen et al. 2003).
Amelogenin exon4 is present in the amelogenin gene of many species (Sire et al. 2012), and amelogenin proteins that contain exon4-encoded proteins have been reported in mouse (Simmer 1994) and porcine (Yamakoshi 2011) enamel matrix. Messenger RNAs (mRNAs) containing exon4 have been amplified from human (Salido et al. 1992), mouse (Hu et al. 1997), and rat (Li et al. 1995) enamel organs. The full-length human amelogenin mRNA (hX189) was reported to constitute 3.5% of the total tooth organ mRNAs (Salido et al. 1992).
The function of AMG+4 in amelogenesis is not clear. Iacob and Veis (2006) found that odontoblasts in P0 mouse molars (overlaid by predominantly presecretory-stage ameloblasts) express both m59 (LRAP) and m73 (LRAP plus exon4), whereas ameloblasts primarily express m180 (no exon4), while the stratum intermedium expresses the full-length isoform containing exon4, m194. Mice that overexpress m194 (Cho et al. 2014) form disorganized and hypoplastic enamel, not seen in the m180-overexpressing mice (Gibson et al. 2011), suggesting a specific role for AMG+4 in enamel biomineralization.
In this study, we examined the spatiotemporal synthesis and secretion of AMG+4 into the enamel matrix. To investigate the possible function of amelogenins containing exon4, we tested the relative binding of AMG+4 to hydroxyapatite, and found increased binding of recombinant rh189 as compared with recombinant amelogenin lacking exon4 (rh175). Our previous work (Stahl et al. 2013) showed that ameloblasts differentiate earlier in mice overexpressing LRAP, and we further used this model to investigate the relationship between the timing of AMG+4 expression and the progression of matrix mineralization.
Materials and Methods
All animal procedures were carried out with approval by the University of California San Francisco and University of Pennsylvania Institutional Animal Care and Use Committees. The experiments reported herein were conducted in compliance with the Animal Welfare Act and in accordance with the principles set forth in the “Guide for the Care and Use of Laboratory Animals” (Institute of Laboratory Animal Resources, National Resource Council, National Academy Press, 1996).
Mass Spectrometry (MS) Analysis of Amelogenins in the Developing Rat Enamel Matrix
Nine-week-old female Wistar rats were sacrificed by CO2 inhalation followed by cervical dislocation. Mandibular incisors were removed from the alveolar bone, and enamel matrix was dissected from the underlying dentin surface at 2 locations. Zone 1, which is primarily secretory-stage enamel, included enamel beginning at the apical end of the incisor, to enamel underlying the distal surface of the root of the first molar. Zone 2, which is primarily early-maturation-stage enamel, included enamel matrix from the distal root of the first molar continuing through early maturation until the enamel was too hard to dissect (Den Besten 1986; Smith and Nanci 1989) (Fig. 1A).
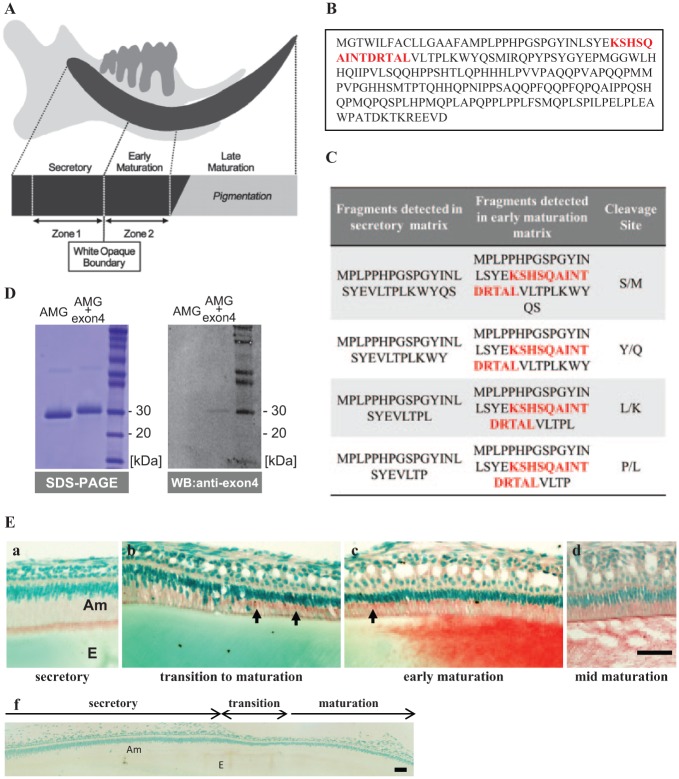
Figure 1.
Identification of amelogenin fragments containing exon4 in enamel matrix. (A) Diagram of a 9-week-old rat incisor showing the location of zone 1, which is primarily a secretory-stage enamel incisor, and zone 2, which is primarily early-maturation-stage enamel. (B) The protein sequence for the full-length amelogenin isoform containing 194 amino acids, with the sequence derived from exon4 shown in red. (C) Protein fragments detected by MS show that exon4 peptide (red font) is only found in the early maturation enamel matrix. (D) Purified RH189 (AMG+4) and RH175 (AMG-4) were separated by SDS PAGE, and then analyzed by Western blot to confirm that the anti-exon4 antibody detected only amelogenin containing exon4 (RH189). (E) Immunodetection of exon4 in incisor ameloblasts showed (a) secretory ameloblasts with positive staining at the apical border, but no obvious staining of the enamel matrix. Beginning in transition and early-maturation-stage ameloblasts (b, c), intense exon4 peptide immunoreaction was seen in the Golgi region (arrows) and in the enamel matrix, consistent with the MS analysis. Pre-immune staining of the incisor shows no nonspecific staining of matrix cells (f). Am, ameloblasts; E, enamel. Bars: 50 μm.
The enamel matrix collected from zones 1 and 2 of each incisor was homogenized in 200 μL of 0.5 M acetic acid with protease inhibitor cocktail (Roche, Indianapolis, IN, USA), to solubilize amelogenins and other matrix proteins. The extract was then centrifuged at 10,000 g for 10 min to remove the undissolved particles. For each zone, 3 enamel extracts from 3 different incisors were diluted to 0.2 mg/mL and combined at equal volume before mass spectrometry (MS) analysis. Each group was desalted on a C18 ZipTip (Millipore, Billerica, MA, USA), and mixed at a 1:1 vol ratio with matrix-assisted laser desorption ionization (MALDI). Matrix α-Cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.1% trifluoroacetic acid (40 fmol/μL of [Glu1]-fibrinopeptide B [m/z 1570.677 Da]) was spiked into the MALDI matrix for internal mass calibration. Mass spectrometry analyses were carried out on a 4800 MALDI TOF/TOF (Applied Biosystems [AB] SCIEX, Foster City, CA, USA). MS and tandem MS (MS/MS) data were acquired with 4000 Series Explorer software (AB SCIEX). The sequences of amelogenin fragments were determined by manual interpretation of the MS/MS data.
Mouse Models
The mice overexpressing LRAP (TgLRAP) were provided by Dr. Carolyn Gibson, University of Pennsylvania. TgLRAP mice were generated by the incorporation of a plasmid containing the amelogenin promoter with cDNA encoding for LRAP (Chen et al. 2003). Wild-type (WT) mice were generated as littermates of TgLRAP mice and sacrificed at the same time-point as controls.
Immunohistochemistry
Maxillae and mandibles were dissected from P21 postnatal mice sacrificed by CO2 inhalation followed by decapitation, and immersed in 4% paraformaldehyde (PFA) for 1 day at 4°C. Following decalcification in 8% ethylene diamine tetraacetic acid (EDTA) (pH 7.3) at 4°C for 10 days, the samples were dehydrated through a graded series of ethanol followed by routine paraffin embedding and sectioning. After deparaffinization, sections were immunostained as previously described (Stahl et al. 2013) with a rabbit anti-exon4 antibody. The rabbit anti-exon4 polyclonal antibody (IgG) was raised against a synthesized murine amelogenin exon4 peptide with the amino acid sequence KSHSQAINTDRTAL (Genscript Inc., Piscataway, NJ, USA) and purified by Protein A affinity chromatography. Pre-immune IgG purified in the same manner was used as a negative control. To verify the specificity of the antibody, we performed a Western blot analysis of recombinant human amelogenin proteins with or without the exon4 region, using an Odyssey Western blotting kit and imaging system (LI-COR Biosciences, Lincoln, NE, USA) (Fig. 1D).
Von Kossa Stain and Gomori’s Trichrome Stain
Maxillae from P5 (secretory stage) WT and TgLRAP mice were fixed as described above, and undecalcified sections were obtained as previously described (Nakano et al. 2004). Mineralization of the tissue was visualized by 2.5% silver nitrate staining (Von Kossa stain) (Bleicher et al. 1999). Maturation of enamel matrix mineralization was assessed by Gomori’s Trichrome stain (Gomori 1950; Duailibi et al. 2004).
Polymerase Chain-reaction Amplification of Amelogenin mRNA with or without Exon4
Maxillary first molars were dissected from postnatal day 0 (P0) and day 5 (P5) WT and TgLRAP mice. Total RNA was isolated by means of an RNeasy Mini kit (Qiagen, Germantown, MD, USA). An aliquot containing 1 µg of total RNA was reverse-transcribed to cDNA with SuperScript® III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). Primer sequences for exon4 were: forward, 5′AAGTCA CATTCTCAGGCTATCAATACT3′; and reverse, 5′-ACTTCTTCCCGCT TGGTCTTGTCT-3′. Polymerase chain-reaction (PCR) amplification for reverse transcriptase (RT)-PCR was performed with the Hot Start Taq kit (Qiagen) by initial incubation of the reaction mixture at 95°C for 5 min, followed by 94°C, 57°C, and 72°C for 1 min each for 30 cycles and then 72°C for 10 min. The products were visualized on a 2% agarose gel with ethidium bromide staining.
Binding of Recombinant Amelogenins to Apatite
Hydroxyapatite was synthesized as previously described and characterized by x-ray diffraction (Tanimoto et al. 2008). Apatite powders were sequentially passed through 30- and 60-μm meshes, and only particles with sizes between 30 and 60 μm were collected for the binding experiment.
Recombinant human amelogenin proteins with or without exon4 (RH189 and RH175) were expressed and purified as previously described (Liu et al. 2001). After the proteins were dissolved in H2O, the pH of the solution was titrated to pH 7.0 by the addition of 50 mM Tris-HCl, pH 9.0. The 2 proteins at a concentration of 400 ng/µL were incubated individually with 10 µg of HAP in a total volume of 250 µL, with constant shaking at 10-minute intervals, up to 60 min. After incubation, the mixtures were centrifuged at 5,000 rpm for 5 min. The unbound proteins remaining in the supernatant were measured by the Bradford assay (Bio-Rad, Hercules, CA, USA) and sodium dodecylsulfide polyacrylamide gel electrophoresis (SDS-PAGE). The amount of protein adsorbed (PA) on 1 mg of HAP crystals was calculated by the equation PA = (Ci – Cf) × V ÷ 0.1 mg, where V is the volume of the solution, Ci is the initial protein concentration, Cf is the final protein concentration, and 0.1 mg is HAP used for binding. Statistical analysis was performed by Student’s t test.
Results
AMG+4 Protein Fragments Were Identified Only in Zone 2 (Early-maturation Stage) Rat Enamel Matrix
Mass spectrometry analysis of enamel matrix proteins isolated from rat zone 1 (primarily secretory) and zone 2 (primarily early maturation) (Fig. 1A) showed that the exon4 peptide sequence (Fig. 1B) was found only in the fragments isolated from early-maturation enamel (Fig. 1C).
Immunostaining of sagittal sections of mouse incisors showed exon4 peptide as present in secretory-stage ameloblasts (Fig. 1E-a), with the immunoreaction increasing in the region of the Golgi apparatus in the transition- to early-maturation-stage ameloblasts (Fig. 1E-b and c). Immunostaining in the Golgi apparatus indicated active synthesis of AMG+4 at this stage. Similar to the mass spectrometry results, exon4 immunostaining was positive in the early-maturation enamel matrix, but was not obvious in the enamel matrix at earlier stages of enamel formation (Fig. 1E).
AMG+4 mRNA Was Detected Earlier in TgLRAP Mouse Molars as Compared with WT Molars
We used the TgLRAP mouse model, which we previously showed to have earlier ameloblast differentiation as compared with that in wild-type mice (Stahl et al. 2013), to determine whether the timing of ameloblast differentiation is linked to the expression of AMG+4. To compare the timing of expression of AMG+4 in TgLRAP and WT mice, we amplified AMG+4 mRNAs from total extracts from P0 and P5 molars. RT-PCR analysis showed that relatively more m194 was amplified in P0 TgLRAP mouse molars as compared with WT molars. Transcripts for m73 were found in P0 TgLRAP, but not in P0 WT molars. Both m194 and m73 were amplified in P5 TgLRAP and WT mice (Fig. 2).
Figure 2.
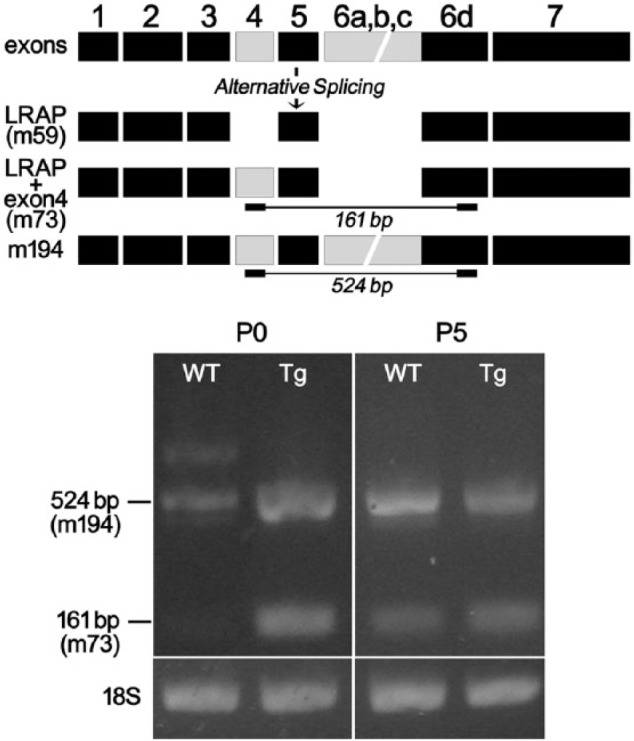
A diagram of mouse amelogenin alternatively spliced products shows the alternatively spliced isoform LRAP (m59), LRAP+ exon4 (m73), and m194 as the full-length isoform containing exon4. The locations of PCR primer sets at the beginning of exon4 and ending at exon 6d (see lines) were designed to detect amelogenin variants containing exon4. At P0, both m194 and m73 were amplified from TgLRAP mouse molars, whereas only a relatively reduced amount of m194 was amplified from P0 WT mouse molars. At P5, both m194 and m73 were amplified from TgLRAP and WT P5 molars.
Overexpression of LRAP and Earlier Synthesis of AMG+4 Corresponded to Earlier Matrix Maturation
To determine if earlier differentiation of ameloblasts and earlier AMG+4 production were associated with changes in mineralization of enamel matrix, we stained undecalcified molar sections with Von Kossa to detect mineral (phosphate) deposition, and with Trichrome stain to assess maturation of the enamel matrix (Duailibi et al. 2004). Both TgLRAP and WT enamel matrix showed initial Von Kossa-positive stain at similar locations, though, as previously observed, the TgLRAP mouse enamel matrix was thinner than WT enamel (Fig. 3A, B) (Stahl et al. 2013). Trichrome staining showed evidence of earlier enamel maturation (bluish grey) in P5 TgLRAP mice as compared with WT mouse molars (red). The early-secretory-stage enamel matrix was stained red in both WT (Fig. 3C, E) and TgLRAP molars (Fig. 3D, F). In contrast, though enamel matrix at the late secretory stage of WT mice (Fig. 3G) remained red, the TgLRAP enamel matrix stained bluish grey, typical of early-maturation-stage enamel matrix (Fig. 3H).
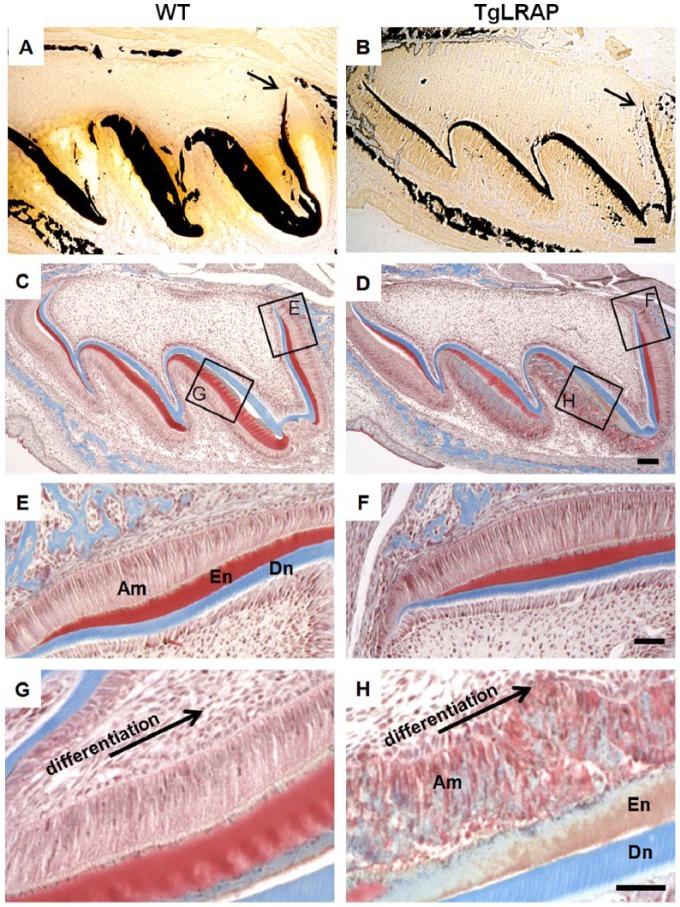
Figure 3.
Von Kossa staining showed that WT mice had a thicker mineralized matrix at P5 (A) as compared with TgLRAP mice (B), with mineralization initiating at similar locations (arrows). The enamel matrix at the cervical loop region of WT (C, E) and TgLRAP (D, F) P5 molars stained red after Trichrome staining. In the most differentiated enamel matrix (along the distal of the mesial cusp) under the late-secretory ameloblasts, the enamel matrix of WT stained red (C, G), whereas the TgLRAP enamel matrix was stained bluish gray (D, H), indicating a maturation matrix phenotype underlying the disrupted ameloblast layer. Scale bars: 100 μm for A–D; 50 μm for E–G.
More Recombinant AMG+4 (RH189) Bound to HAP as Compared with AMG-4 (RH175)
The amount of recombinant amelogenin containing exon4 transcript (rh189) that bound to HAP was significantly increased as compared with recombinant amelogenin without exon 4 (rh175). The increased binding of rh189 was significant at every time interval, up to 60 min, when the binding was fully saturated (Fig. 4) (P < 0.001). SDS-PAGE results confirmed that more of the RH189 was bound to the apatite after 60 min (data not shown).
Figure 4.
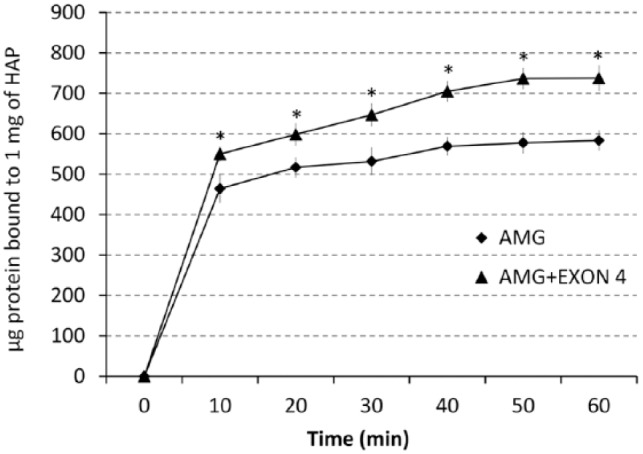
Binding of RH189 (AMG+exon4) to HAP was greater than that of RH175 (AMG). Proteins were incubated with HAP for up to 60 min, and the amount of protein absorbed onto the HAP was determined at 10-minute intervals. At all time-points, the binding of RH189 to HAP was calculated to be significantly greater (*P < 0.001).
A bioinformatics approach developed from known crystallized protein structures that bound either calcium or phosphate was used for further exploration of mineral binding by amelogenins containing the exon4-encoded peptide. This analysis predicted that amelogenins with exon4-encoded peptide had enhanced binding to both calcium and phosphate ions (see Appendix).
Discussion
Alternative splicing is a key event in eukaryotes that increases the coding capacity of the human genome, allowing for one gene to produce multiple proteins with potentially different functions. Previous studies identified AMG+4 mRNAs including exon4 (m73 and m194) transcripts in presecretory/secretory ameloblasts (P0-P1) in the mouse tooth organ (Iacob and Veis 2006). Hu and co-workers (Hu et al. 1997) amplified m194, m73, and m170 transcripts from combined extracts of P7 (maturation-stage) rat and mouse molar enamel organ and incisors (containing both secretory and maturation-stage), and identified amelogenins containing exon4 in enamel matrix extracts by Western blot. Our study is the first to show a stage-specific regulation of AMG+4 expression and secretion, and increased binding of AMG+4 to HAP.
The early maturation stage of amelogenesis is characterized by a marked decrease in organic matrix proteins and an increase in mineral content; however, ameloblasts continue to synthesize and secrete enamel matrix proteins during the early stages of maturation (Nanci et al. 1987). Our mass spectrometry analyses identified fragments containing exon4-encoded peptide only in the early-maturation enamel matrix. This finding was supported by immunohistochemistry showing increased immunostaining in the Golgi apparatus of early-transition-stage ameloblasts, and immunostaining for exon4 peptide in the early-maturation-stage enamel matrix.
We found that, in TgLRAP mice, which have earlier ameloblast differentiation, AMG+4 transcripts were also expressed earlier than in WT mice. The earlier synthesis of amelogenin mRNAs containing exon4 in TgLRAP mice was consistent with earlier transition from red to bluish grey Trichrome staining of the mineralized enamel matrix. In the TgLRAP mice, the red color, resulting from the less porous secretory enamel matrix (Duailibi et al. 2004), was lost earlier as compared with that in WT mouse enamel. These findings support the possibility that maturation of the enamel matrix results in part from increased expression and synthesis of AMG+4.
To investigate further the possible role of AMG+4 in enamel biomineralization, we used recombinant amelogenins to compare AMG+4 and AMG-4 binding to HAP. We found increased binding of RH189 as compared with RH175 to HAP in vitro. Our bioinformatics analyses of M194 as compared with M180 also predicted that exon4-encoded peptide had increased calcium and phosphate binding (see Appendix), and similar results were found with the human sequences (data not shown). Taken together, these findings suggest that AMG+4 has functional differences related to enamel biomineralization, possibly to stabilize early-formed HAP or its calcium and phosphate precursors. The functional significance of HAP stabilization at the transition and early maturation stages could be to limit mineral growth just prior to maturation, allowing for the rapid removal of matrix proteins that occurs at this stage.
In conclusion, these studies show that AMG+4 are temporally and spatially synthesized and secreted at the early maturation stage of enamel matrix formation. In vitro and in silico analyses suggest that AMG+4 modulates mineralization through its increased mineral-binding capacity. Future studies will further elucidate the role of AMG+4 in enamel matrix formation, and the possibility that this peptide can be used to stabilize early HAP formation to modulate enamel biomineralization.
Author Contributions
J. Stahl, Y. Nakano, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; J. Horst, L. Zhu, contributed to design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; M. Le, contributed to data acquisition and interpretation, drafted and critically revised the manuscript; Y. Zhang, contributed to data acquisition, drafted and critically revised the manuscript; H. Liu, contributed to data analysis, drafted and critically revised the manuscript; W. Li, contributed to design, data analysis, and interpretation, drafted and critically revised the manuscript; P.K. Den Besten, contributed to conception, design, data analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Acknowledgments
The UCSF Sandler-Moore Mass Spectrometry Core Facility acknowledges the support of the Sandler Family and the Gordon and Betty Moore Family Foundations. We thank the laboratory of Dr. Carolyn Gibson at the University of Pennsylvania for sharing the LRAP transgenic mouse line.
Footnotes
This study was supported by the U.S. Navy (J.S.), Department of Orofacial Sciences, and National Institute of Dental and Craniofacial Research (NIDCR) grants 2R01DE015821 and 3R01DE015281S1 to W.L.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, or the U.S. Government.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Copyright Statement: J.S. is a member of the military, and this work was prepared as part of his official duties. Title 17 U.S.C. §105 provides that “Copyright protection under this title is not available for any work of the United States Government.” Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Bleicher F, Couble ML, Farges JC, Couble P, Magloire H. 1999. Sequential expression of matrix protein genes in developing rat teeth. Matrix Biol. 18(2):133–143. [DOI] [PubMed] [Google Scholar]
- Boabaid F, Gibson CW, Kuehl MA, Berry JE, Snead ML, Nociti FH, Jr, Katchburian E, Somerman MJ. 2004. Leucine-rich amelogenin peptide: a candidate signaling molecule during cementogenesis. J Periodontol. 75(8):1126–1136. [DOI] [PubMed] [Google Scholar]
- Chen E, Yuan ZA, Wright JT, Hong SP, Li Y, Collier PM, Hall B, D’Angelo M, Decker S, Piddington R, et al. 2003. The small bovine amelogenin LRAP fails to rescue the amelogenin null phenotype. Calcif Tissue Int. 73(5):487–495. [DOI] [PubMed] [Google Scholar]
- Cho ES, Kim KJ, Lee KE, Lee EJ, Yun CY, Lee MJ, Shin TJ, Hyun HK, Kim YJ, Lee SH, et al. 2014. Alteration of conserved alternative splicing in AMELX causes enamel defects. J Dent Res. 93(10):980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Besten PK. 1986. Effects of fluoride on protein secretion and removal during enamel development in the rat. J Dent Res. 65(10):1272–1277. [DOI] [PubMed] [Google Scholar]
- Duailibi MT, Duailibi SE, Young CS, Bartlett JD, Vacanti JP, Yelick PC. 2004. Bioengineered teeth from cultured rat tooth bud cells. J Dent Res. 83(7):523–528. [DOI] [PubMed] [Google Scholar]
- Eastoe JE. 1964. The chemical composition of bone and teeth. In: JL Hardwick, editor. Advance in Fluorine Research Dental Caries Prevent. Oxford (UK): Pergamon Press; p. 5–17. [PubMed] [Google Scholar]
- Fincham AG, Belcourt AB, Termine JD, Butler WT, Cothran WC. 1981. Dental enamel matrix: sequences of two amelogenin polypeptides. Biosci Rep. 1(10):771–-778. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Simmer JP, Sarte P, Lau EC, Diekwisch T, Slavkin HC. 1994. Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J Struct Biol. 112(2):103–109. [DOI] [PubMed] [Google Scholar]
- Gibson CW, Golub E, Ding WD, Shimokawa H, Young M, Termine J, Rosenbloom J. 1991. Identification of the leucine-rich amelogenin peptide (LRAP) as the translation product of an alternatively spliced transcript. Biochem Biophys Res Commun. 174(3):1306–1312. [DOI] [PubMed] [Google Scholar]
- Gibson CW, Golub EE, Abrams WR, Shen G, Ding W, Rosenbloom J. 1992. Bovine amelogenin message heterogeneity: alternative splicing and Y-chromosomal gene transcription. Biochemistry. 31(35):8384–8388. [DOI] [PubMed] [Google Scholar]
- Gibson CW, Li Y, Suggs C, Kuehl MA, Pugach MK, Kulkarni AB, Wright JT. 2011. Rescue of the murine amelogenin null phenotype with two amelogenin transgenes. Eur J Oral Sci. 119(Suppl 1):70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomori G. 1950. A rapid one-step trichrome stain. Am J Clin Pathol. 20(7):661–664. [DOI] [PubMed] [Google Scholar]
- Hu CC, Ryu OH, Qian Q, Zhang CH, Simmer JP. 1997. Cloning, characterization, and heterologous expression of exon-4-containing amelogenin mRNAs. J Dent Res. 76(2):641–647. [DOI] [PubMed] [Google Scholar]
- Iacob S, Veis A. 2006. Identification of temporal and spatial expression patterns of amelogenin isoforms during mouse molar development. Eur J Oral Sci. 114(Suppl 1):194–200. [DOI] [PubMed] [Google Scholar]
- Le TQ, Zhang Y, Li W, Denbesten PK. 2007. The effect of LRAP on enamel organ epithelial cell differentiation. J Dent Res. 86(11):1095–1099. [DOI] [PubMed] [Google Scholar]
- Le Norcy E, Kwak SY, Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. 2011. Potential role of the amelogenin N-terminus in the regulation of calcium phosphate formation in vitro. Cells Tissues Organs. 194(2-4):188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li W, DenBesten PK. 1995. Alternative splicing of amelogenin mRNA from rat incisor ameloblasts. J Dent Res. 74(12):1880–1885. [DOI] [PubMed] [Google Scholar]
- Li W, Mathews C, Gao C, DenBesten PK. 1998. Identification of two additional exons at the 3′ end of the amelogenin gene. Arch Oral Biol. 43(6):497–504. [DOI] [PubMed] [Google Scholar]
- Liu HX, Oei PT, Mitchell EA, McGaughran JM. 2001. Interstitial deletion of 3p22.2-p24.2: the first reported case. J Med Genet. 38(5):349–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngstadaas SP, Risnes S, Sproat BS, Thrane PS, Prydz HP. 1995. A synthetic, chemically modified ribozyme eliminates amelogenin, the major translation product in developing mouse enamel in vivo. EMBO J. 14(21):5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Beertsen W, van den Bos T, Kawamoto T, Oda K, Takano Y. 2004. Site-specific localization of two distinct phosphatases along the osteoblast plasma membrane: tissue non-specific alkaline phosphatase and plasma membrane calcium ATPase. Bone. 35(5):1077–1085. [DOI] [PubMed] [Google Scholar]
- Nanci A, Slavkin HC, Smith CE. 1987. Immunocytochemical and radioautographic evidence for secretion and intracellular degradation of enamel proteins by ameloblasts during the maturation stage of amelogenesis in rat incisors. Anat Rec. 217(2):107–123. [DOI] [PubMed] [Google Scholar]
- Salido EC, Yen PH, Koprivnikar K, Yu LC, Shapiro LJ. 1992. The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes. Am J Hum Genet. 50(2):303–316. [PMC free article] [PubMed] [Google Scholar]
- Sire JY, Huang Y, Li W, Delgado S, Goldberg M, Denbesten PK. 2012. Evolutionary story of mammalian-specific amelogenin exons 4, “4b”, 8, and 9. J Dent Res. 91(1):84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Nanci A. 1989. A method for sampling the stages of amelogenesis on mandibular rat incisors using the molars as a reference for dissection. Anat Rec. 225(3):257–266. [DOI] [PubMed] [Google Scholar]
- Stahl J, Nakano Y, Kim SO, Gibson CW, Le T, Denbesten P. 2013. Leucine rich amelogenin peptide alters ameloblast differentiation in vivo. Matrix Biol. 32(7-8):432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K, Le T, Zhu L, Chen J, Featherstone JD, Li W, DenBesten P. 2008. Effects of fluoride on the interactions between amelogenin and apatite crystals. J Dent Res. 87(1):39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. 1980. Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem. 255(20):9760–9768. [PubMed] [Google Scholar]
- Warotayanont R, Frenkel B, Snead ML, Zhou Y. 2009. Leucine-rich amelogenin peptide induces osteogenesis by activation of the Wnt pathway. Biochem Biophys Res Commun. 387(3):558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakoshi Y. 2011. Porcine amelogenin: alternative splicing, proteolytic processing, protein–protein interactions, and possible functions. J Oral Biosci. 53(3):275–283. [PMC free article] [PubMed] [Google Scholar]
- Ye L, Le TQ, Zhu L, Butcher K, Schneider RA, Li W, Den Besten PK. 2006. Amelogenins in human developing and mature dental pulp. J Dent Res. 85(9):814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.