Abstract
Rolling-circle replication of streptococcal plasmid pMV158 is controlled by the concerted action of two trans-acting elements, namely transcriptional repressor CopG and antisense RNAII, which inhibit expression of the repB gene encoding the replication initiator protein. The pMV158-encoded antisense RNAII exerts its activity of replication control by inhibiting translation of the essential repB gene. RNAII is the smallest and simplest among the characterized antisense RNAs involved in control of plasmid replication. Structure analysis of RNAII revealed that it folds into an 8-bp-long stem containing a 1-nt bulge and closed by a 6-nt apical loop. This hairpin is flanked by a 17-nt-long single-stranded 5′-tail and an 8-nt-long 3′-terminal U-rich stretch. Here, the 3′ and 5′ regions of the 5′-tail of RNAII are shown to play a critical role in the binding to the target mRNA and in the inhibition of repB translation, respectively. In contrast, the apical loop of the single hairpin of RNAII plays a rather secondary role and the upper stem region hardly contributes to the binding or inhibition processes. The entire 5′-tail is required for efficient inhibition of repB translation, though only the 8-nt-long region adjacent to the hairpin seems to be essential for rapid binding to the mRNA. These results show that a “kissing” interaction involving base-pairing between complementary hairpin loops in RNAII and mRNA is not critical for efficient RNA/RNA binding or repB translation inhibition. A singular binding mechanism is envisaged whereby initial pairing between complementary single-stranded regions in the antisense and sense RNAs progresses upwards into the corresponding hairpin stems to form the intermolecular duplex.
Keywords: bacterial small non-coding RNAs, antisense RNA, target RNA, plasmid replication control, translation inhibition, RNA-RNA interaction
Introduction
Prokaryotic antisense RNAs have been defined as small, diffusible, mostly untranslated and highly structured transcripts that pair to target RNAs to control their biological function (Wagner and Simons, 1994; Brantl, 2007). More specifically, antisense RNAs are referred to as the cis-encoded group of these regulatory RNAs, i.e., those transcribed from the DNA strand opposite their target RNA (Thomason and Storz, 2010). In the early 1980s, two natural antisense RNAs were independently found to control replication of plasmids ColE1 and R1 (Stougaard et al., 1981; Tomizawa et al., 1981) and shortly afterwards, the involvement of this class of transcripts in controlling gene expression in transposons and in the bacterial chromosome was also unveiled (Simons and Kleckner, 1983; Mizuno et al., 1984; Brantl, 2012). The widespread antisense RNA control of plasmid replication works by inhibiting the function of an essential RNA through a variety of modes of action (reviewed in Wagner and Simons, 1994; del Solar and Espinosa, 2000; Brantl, 2014). In some systems, the antisense RNA inhibits maturation of the replication primer (Masukata and Tomizawa, 1986). In most cases, however, the antisense RNA inhibits transcription and/or translation of the plasmid rep gene, which encodes the replication initiator (Rep) protein (Wagner and Simons, 1994; Chai and Winans, 2005). The antisense RNA systems of plasmid replication control also exhibit a variety of RNA/RNA binding pathways, which, in the few cases where the entire process has been dissected, initiate with a “kissing” step that involves reversible base-pairing between complementary hairpin loops. This initial pairing can progress into the upper part of the corresponding stems (Malmgren et al., 1997; Wagner and Brantl, 1998; Kolb et al., 2000). Subsequent formation of the RNA/RNA stable complex starts by base-pairing between a single-stranded region of the antisense RNA and its complementary accessible region in the target RNA (Tomizawa, 1985, 1990; Persson et al., 1990; Wagner and Simons, 1994). Mutational analysis has revealed the importance of the “kissing” interaction in the stable binding of the antisense/target RNA pair (Tomizawa, 1986; Wagner and Simons, 1994). In the streptococcal theta-replicating plasmid pIP501, efficient formation of a stable complex requires the presence of both the two 3′-terminal stem-loops and the unstructured central region of the antisense RNA (Brantl and Wagner, 1994). A two-step binding pathway was proposed for this antisense/sense RNA system, where initial “kissing” between two complementary loop pairs progresses through the entire stems to form a quasi-full-length duplex (Heidrich and Brantl, 2007). Although all well-characterized antisense systems of plasmid replication control initiate the antisense/target RNA binding through loop-loop pairing, linear-linear or loop-linear recognition mechanisms can also be envisaged. Actually, a loop-linear initial pairing similar to that previously reported for the R1 hok/sok killer system has been suggested for the antisense RNA system that controls replication of the Rhizobium etli p42d plasmid (Venkova-Canova et al., 2004; Cervantes-Rivera et al., 2010). In these two systems, the 5′ single-stranded tail of the antisense molecule pairs with a hairpin loop in the target mRNA to initiate RNA binding (Thisted et al., 1994; Franch et al., 1999; Cervantes-Rivera et al., 2010). When a loop-loop initial interaction scheme is used, a YUNR motif (Y = pyrimidine; R = purine) constituting a U-turn structure has been ubiquitously found in a recognition loop of either the sense or the antisense RNA. The singular configuration of the bases in the U-turn loop structure is thought to facilitate the initial base-pairing and the progression of the intermolecular helix, causing this motif to act as a general enhancer of RNA/RNA binding rates (Franch et al., 1999).
The promiscuous streptococcal rolling-circle-replicating plasmid pMV158 is the prototype of a growing family that so far includes about 70 members isolated from a variety of bacteria belonging to the Firmicutes or Proteobacteria phyla (Ruiz-Masó et al., 2015). Replicons of the pMV158 family are characterized by a high genetic economy that relies on the smallness of the genes encoding the replication initiator and the copy-number control elements, each among the smallest in its own class (del Solar and Espinosa, 2001; del Solar et al., 2002; Boer et al., 2009). A dual mechanism based on both a transcriptional repressor protein (CopG) and an antisense RNA (RNAII) has been shown to control initiation of replication of pMV158 (del Solar and Espinosa, 1992; del Solar et al., 1995; Hernández-Arriaga et al., 2009), and is also expected to control replication in most plasmids of the family. The rnaII gene, which encodes the 48-nt long RNAII, overlaps the intergenic region of the pMV158 copG-repB operon and is transcribed in the opposite direction to it. Therefore, RNAII is a countertranscript to the sense transcript, its entire nucleotide sequence being complementary to a copG-repB mRNA target region located immediately upstream of the atypical signals involved in translation initiation of the essential replication gene, repB (del Solar et al., 1997; López-Aguilar et al., 2013). In fact, RNAII-mediated control of pMV158 replication initiation has been shown to be exerted by inhibition of repB translation (del Solar et al., 1997). Recently, we have mapped the secondary structure of RNAII by chemical and enzymatic probing (López-Aguilar and del Solar, 2013). The experimentally proven secondary structure of RNAII, which matches the computational predictions, consists of a 17-nt A-U rich single-stranded 5′-tail followed by a typical rho-independent transcriptional terminator that is in turn composed of a G-C rich hairpin and a 3′ poly(U) tail (del Solar and Espinosa, 1992; López-Aguilar and del Solar, 2013; Figures 1A,C). In vitro transcription experiments showed that Escherichia coli RNA polymerase ends synthesis of RNA at this intrinsic terminator with an efficiency of ~90% (del Solar and Espinosa, 2001). Antisense RNAII fulfills all the features regarded as essential for an efficient plasmid replication control element, i.e., it is able to inhibit plasmid replication in trans, it has a short (1–2 min) half-life (Acebo et al., unpublished data), and it determines strong incompatibility against pMV158 (del Solar and Espinosa, 1992; del Solar et al., 1995). Derivatives of pMV158 that encode a mutant copG-repB mRNA lacking the hairpin complementary to that in RNAII were still susceptible to inhibition by the wild-type (wt) antisense RNA. These findings suggested that a “kissing” step was not strictly required for the RNA pairing mechanism in pMV158 (del Solar and Espinosa, 1992), and led to the proposal that copG-repB mRNA/RNAII binding initiates via a loop-linear pairing scheme (Franch et al., 1999). With a few exceptions, the existence of antisense RNAs controlling replication of plasmids of the pMV158 family has been assumed based on what had previously been shown for the prototype plasmid, and studies on the interaction with the cognate target mRNA are still missing. In pJB01 (a pMV158-family member), the existence of an antisense RNA controlling plasmid replication was proved, and the involvement of different regions of this RNA in inhibition of rep translation was studied by mutational analysis. Copy number inspection of the mutant plasmids suggested that the entire secondary structure of the antisense RNA was important for interaction with the target mRNA (Kim et al., 2008).
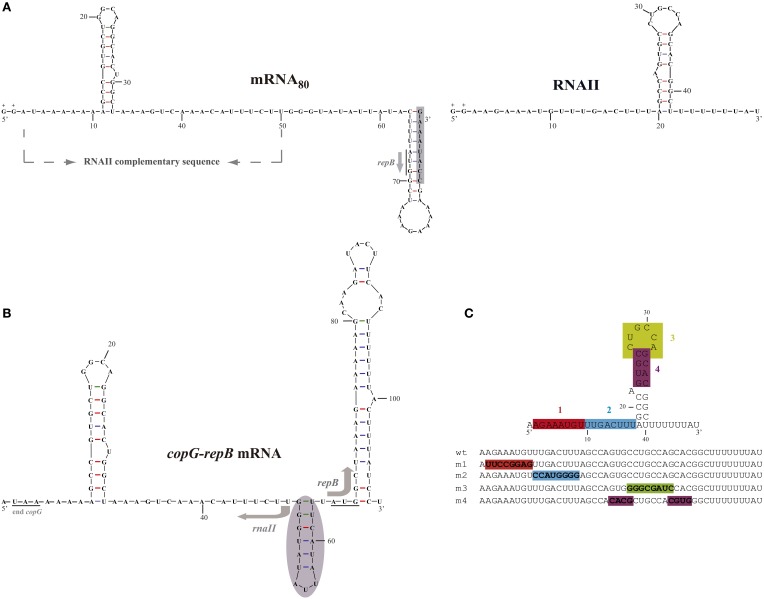
Figure 1.
Sequence and folding of the in vivo- and in vitro-synthesized antisense and target RNAs of the pMV158 replication control system. (A) Observed secondary structure of in vitro-synthesized sense mRNA80 (left) and antisense RNAII (right). The 8-nt-long 3′ overextension observed in the mRNA80 transcript is shadowed. In the mRNA80 transcript the sequence complementary to antisense RNAII is shown, and the start codon of repB is underlined and indicated by an arrow. In both transcripts, the two extra Gs added at the 5′-end of the RNA sequence for efficient in vitro transcription (López-Aguilar and del Solar, 2013) are indicated (+). (B) Predicted structure of the intergenic region of the copG-repB mRNA. The copG stop codon and the repB start codon are underlined. The shadowed region upstream of the repB start codon has been shown to be involved in translation initiation of the gene (López-Aguilar et al., 2013). The start point and direction of transcription of rnaII and repB are indicated by arrows. (C) Antisense RNA mutants. Upper, RNAII regions whose sequence was changed to generate antisense RNA mutants m1 to m4 are shown in different colors. Lower, the same color code is used to display the sequence altered in each of the antisense mutants.
In the present work, we have analyzed the role of the different secondary structure elements of the pMV158-encoded antisense RNAII both in binding to the target mRNA and in inhibition of repB translation. The 3′-half of the 5′-tail has been found to be essential for efficient binding, whereas the 5′-terminal half, although contributing only moderately to mRNA binding, has a main role in inhibition of repB translation and hence in control of plasmid replication. Unlike most antisense RNA systems controlling plasmid replication, the apical loop of the single hairpin of RNAII has a rather secondary role in binding to the copG-repB mRNA, whereas the sequence of the upper stem is not required for either binding to the target mRNA or inhibition of repB expression.
Materials and methods
Bacterial strains, plasmids and oligonucleotides
Streptococcus pneumoniae 708 (end-1, exo-2, trt-1, hex-4, malM594) was employed as the host for all the in vivo assays. Pneumococcal cells were grown and transformed as described (Lacks, 1968). The plasmids used are listed in Table 1. Selection was applied by exposure to tetracycline (Tc; 1 μg/ml, or 0.5 μg/ml when specified) or chloramphenicol (Cm; 3 μg/ml) for pneumococcal cells containing pMV158-based or pC194-based replicons, respectively.
Table 1.
List of plasmids used in this work.
| Plasmid | Size (bp) | Marker | Characteristics | References |
|---|---|---|---|---|
| pLS1 | 4408 | TcR | Δmob, ΔssoU derivative of pMV158 | Lacks et al., 1986 |
| pC194 | 2907 | CmR | Copy-up mutant of pC194 (~110 copies per chromosome equivalent in S. pneumoniae) | Ballester et al., 1990 |
| pCGA9 | 3070 | CmR | rnaII gene of pMV158 cloned in pC194 | del Solar et al., 1995 |
| pCGA9-m1 | 3070 | CmR | rnaII-m1 mutant gene cloned in pC194 | This work |
| pCGA9-m2 | 3070 | CmR | rnaII-m2 mutant gene cloned in pC194 | This work |
| pCGA9-m3 | 3070 | CmR | rnaII-m3 mutant gene cloned in pC194 | This work |
| pCGA9-m4 | 3070 | CmR | rnaII-m4 mutant gene cloned in pC194 | This work |
| pALTER-T7-copG-repB-cat | 7545 | CmR | Recombinant plasmid that carries copG, repB and cat genes of pJS3 cloned in pALTER-1 under the control of the ϕ10 promoter of phage T 7 | del Solar et al., 1997 |
Oligonucleotides (oligos) used for amplification of DNA, rnaII mutagenesis, construction of DNA templates for in vitro transcription and oligo-RNA band-shift assays were chemically synthesized and then purified from 12% polyacrylamide (PAA) sequencing gels. All oligonucleotides employed in this work are listed in Supplementary Table 1.
Plasmid DNA preparations and DNA manipulations
Total DNA preparations (crude extracts) from cells harboring plasmids were used to determine plasmid copy number as described (del Solar and Espinosa, 1992). Plasmid DNAs were prepared by the alkali denaturation method modified for S. pneumoniae (Stassi et al., 1981). DNA preparations were electrophoresed in 0.8–1% agarose gels. The nucleic acid bands were stained with ethidium bromide and visualized with the aid of a Gel-Doc documentation system. The Quantity-One software was used for the quantitative analysis of the fluorescence intensity of the different bands.
DNA fragments were purified from agarose gels by using the QIAquick Gel Extraction Kit (QIAGEN). DNA concentration was quantified using a NanoDrop™ spectrophotometer.
Enzymes for the modification of the DNA (restriction endonucleases, T4 DNA ligase and T4 polynucleotide kinase) as well as DNA polymerases were purchased from New England Biolabs and Fynnzymes, respectively, and used as specified by the suppliers.
RNA extraction from pneumococcal cells
Total RNAs were prepared from exponentially growing pneumococcal cultures harboring the different pCGA9 plasmid variants (pCGA9, pCGA9-m1, -m2, -m3, and -m4), essentially as described (López et al., 1989).
RNA secondary structure prediction
RNA structures were predicted by using the Mfold (3.2) program (Zuker, 2003)1.
Directed mutagenesis of rnaII and construction of pCGA9 derivatives harboring the mutated genes
Mutant pCGA9 plasmids (pCGA9-m1, -m2, -m3, and -m4; Table 1) were generated by directed mutagenesis of the rnaII gene of pCGA9 (a pC194-based recombinant plasmid that carries the wt rnaII gene from pMV158; del Solar and Espinosa, 1992), followed by DNA amplification, digestion of the product with HaeIII and PvuII, and cloning of the appropriate fragment in pC194. Site-directed mutagenesis of rnaII was performed by the recombinant PCR method (Higuchi, 1990). Basically, two separate PCR were prepared by using as the primer pair either the forward oligo pC1800-1818 and one of the reverse mutagenic oligos (R-RNAII-m1, -m2, -m3, or -m4), or the reverse oligo pC2798-2780 and one of the forward mutagenic oligos (F-RNAII-m1, -m2, -m3 or -m4), and pCGA9 DNA as template. Each pair of overlapping primary PCR products were denatured and allowed to reanneal together, and the resulting heteroduplex with recessed 3′ ends was extended to generate a fragment that consists of the two primary products. This fragment was amplified using oligos pC1800-1818 and pC2798-2780, and subsequently subjected to restriction with HaeIII and PvuII. Next, the HaeIII-PvuII fragment was swapped into pC194, and the resulting recombinant plasmids were used to transform competent pneumococcal cells. The existence of pCGA9 derivatives containing the desired mutations in the rnaII gene was confirmed by automated DNA sequencing.
Generation of DNA templates and in vitro synthesis of different RNA species
Double-stranded DNAs containing an improved promoter for T7 RNA polymerase (T7 RNAP)-mediated transcription (Moll et al., 2004; López-Aguilar and del Solar, 2013) followed by the sequence of the gene or gene fragment to be transcribed were used as templates to synthesize the wt and mutant antisense species, as well as the mRNA target (mRNA80) employed for the binding experiments. DNA templates encoding the different antisense RNA species were generated by extension of the common primer P-RNAII annealed to the following 83-mer oligos (Supplementary Table 1): RNAII (wt antisense RNA), RNAII-m1 (m1 mutant), RNAII-m2 (m2 mutant), RNAII-m3 (m3 mutant), or RNAII-m4 (m4 mutant). Then, the template DNAs were PCR-amplified using the primer pairs P-RNAII and P2-RNAII (for wt, m1, and m2 antisense RNAs), P-RNAII and P3-RNAII (for m3 antisense RNA), or P-RNAII and P4-RNAII (for the m4 mutant). The DNA template for in vitro synthesis of mRNA80 was generated by annealing and extension of oligos mRNA and mRNA-80 (Supplementary Table 1). The resulting double-stranded DNA was PCR-amplified using P-RNAII and mRNA-80R as the primer pair. The amplified DNA fragments were purified from PAA gels before being used as templates for synthesis of the various RNA species. The in vitro run-off transcription assays were performed using T7 RNAP from Roche, according to the manufacturer's protocol. The template DNAs were next removed by digestion with RNase-free DNase I (Roche). Samples were cleaned up by filtration through Sephadex G-25 columns (GE-Healthcare) and successive extraction with phenol/chloroform and chloroform, followed by ethanol precipitation. RNAs were quantified from their absorbance at 260 nm in a NanoDrop™ spectrophotometer.
The DNA template for T7 RNAP-mediated in vitro synthesis of the copG-repB mRNA employed in the experiments of repB translation inhibition was prepared by linearizing the recombinant plasmid pALTER-T7-copG-repB-cat (Table 1) at its single NcoI site located within the cat gene, as previously described (del Solar et al., 1997). The transcription assays were also carried out as described (del Solar et al., 1997). The resulting transcript specified full-length CopG and RepB proteins, and a truncated chloramphenicol acetyltransferase (T-Cat).
5′-end labeling of RNAs and oligonucleotides
In vitro synthesized antisense RNAs were dephosphorylated with alkaline phosphatase (Promega) following the manufacturer's indications. Then, the full-length RNA molecules were isolated from 12% PAA sequencing gels, extracted with phenol/chloroform and chloroform, and ethanol-precipitated, previous to the labeling of their 5′-end with T4 polynucleotide kinase (New England Biolabs) and [γ−32P] ATP. Labeled RNAs were re-isolated from 12% PAA sequencing gels as above.
The three 17-mer oligos (BS, BS-m3, and BS-m4, see Supplementary Table 1) employed in the oligo-RNA band-shift experiments were labeled at their 5′-end as described (del Solar et al., 1995).
RNA-RNA binding assays
Previous to the binding reaction, the antisense and target RNAs were incubated separately at 65°C for 10 min and then at 37°C for 10 min, in order to obtain their native conformation. The binding mixtures (10 μl) contained 0.2 nM of any of the 5′-end-labeled antisense RNA variants and various concentrations (0–400 nM) of the unlabeled target RNA (mRNA80) in TMN buffer (100 mM NaCl, 20 mM Tris-OAc, 10 mM Mg(OAc)2. Binding was allowed for 30 min at 37°C, and terminated by adding an equal volume of loading buffer (glycerol 30%, tRNA 4%, xylene cyanol 0.4%, and bromophenol blue 0.4% in TMN buffer) and placing the mixture on ice. The samples were applied to non-denaturing 10% PAA gels. Gels were run at 300 V for 2 h.
Radioactive bands were visualized and quantified from fixed and dried gels using the storage phosphor technology, with the aid of a FLA-3000 (FUJIFILM) imaging system and the Quantity One software (Bio-Rad). The mRNA concentration required for half-maximal binding under the described experimental conditions (Kd) was obtained with the Sigmaplot 9.0 software from data point fitting to the equation:
where y is the fraction of complexed antisense RNA, Bmax is the amplitude of the reaction, and x is the concentration of the target RNA.
Time-course experiments of the RNA-RNA association
Standard binding mixtures containing 0.2 nM of the 5′-end-labeled antisense RNA and the indicated concentration, always in molar excess, of the unlabeled target RNA in TMN buffer were prepared as above and incubated at 37°C. Samples were withdrawn at different times and the reaction was stopped by diluting the samples into an equal volume of gel application buffer (94% formamide, 17 mM EDTA, 0.025% xylene cyanol, and 0.025% bromophenol blue) on ethanol-dry ice. Sample electrophoresis as well as visualization and quantification of the bands were performed as above. Least-squares analysis of the data from a log-linear plot of the fraction of free antisense RNA as a function of the incubation time yielded a straight line, at least until ~10% of the antisense RNA remained unbound. The slope of the curve was used to obtain the pseudo-first-order rate constant (b). The second-order rate constant (ka) was calculated as follows (Persson et al., 1988):
where the concentration of the target RNA may be considered constant, due to the large molar excess of the mRNA relative to the antisense RNA.
Antisense RNA-mediated inhibition of in vitro repB translation
E. coli S30 Extract System for Linear Templates (Promega) was employed to provide the host machinery needed for protein synthesis. Previous to the addition of the antisense and messenger RNAs, translation mixtures were prepared as directed by the manufacturers, maintained at 0°C, and treated with rifampicin (25 μg/ml) to prevent subsequent transcription from DNA traces that might be present in the mRNA preparation. Then, mRNA (2.5 μg) synthesized from pALTER-T7-copG-repB-cat (see above) and one of the antisense RNA variants (at a 20-Fold molar excess over the mRNA) were added, and the translation mixtures were incubated at 37°C for 90 min. De novo-synthesized proteins, labeled with [35S]-methionine, were separated on 15% SDS-PAA gels. Radioactivity in the bands was detected and quantified as described above. Values of the radioactivity in RepB were normalized with respect to those in CopG, whose synthesis is not regulated by the antisense RNA. Similar results were obtained when the values were normalized with respect to the levels of T-Cat.
Incompatibility tests and copy number determinations
To test whether wt and mutant antisense RNAs exert incompatibility toward the pMV158 replicon, type I quantitative incompatibility tests (Nordström et al., 1980) were performed using as donor plasmids the pC194-based recombinants. Competent pneumococcal cultures (500 μl) harboring pLS1 (a pMV158 derivative, see Table 1) as the resident plasmid were transformed with 0.25 μg of DNA of the donor plasmid (pCGA9, pCGA9-m1, pCGA9-m2, pCGA9-m3, or pCGA9-m4). After allowing for phenotypic expression (70 min), cultures were induced with 0.5 μg/ml of Cm for 20 min, and transformants were selected in plates containing 3 μg/ml of Cm. Ten clones from each transformation were picked and grown in the presence of selective pressure for only the donor plasmid, and their total DNA content was analyzed by preparation of crude extracts and electrophoresis in agarose gels, as previously described (del Solar and Espinosa, 1992). The copy number of pLS1 in the heteroplasmid strains was compared with that in the homoplasmid strain. The percentage of pLS1-containing cells was also estimated at this stage (after ~37 generations without selective pressure for the resident plasmid) as the % of cells resistant to 0.5 μg/ml Tc, which is the lowest amount of antibiotic that provides selection (del Solar and Espinosa, 1992).
Whenever the presence in trans of a mutant antisense RNA did not result in clear incompatibility against pLS1 after ~37 generations of bacterial growth in the absence of Tc, three out of the transformant clones were grown for 60 more generations in the presence of only Cm, and the plasmid content as well as the percentage of Tc resistant (TcR) cells was analyzed as above after each 10 generations.
Oligo-RNA band-shifts assays
To determine the relative intracellular amount of the different antisense RNA variants, “oligo-RNA band-shifts” experiments were performed basically as described (del Solar et al., 1995). Total RNA (4 μg) from pneumococcal cultures harboring any of the recombinant pCGA9-type plasmids (wt or m1 to m4 mutants) was mixed with 0.25 pmol of the appropriate 5′-end-labeled oligo (BS for RNA extracted from pneumococcal cells containing pCGA9, pCGA9-m1 or –m2; BS-m3 for RNA obtained from cells containing pCGA9-m3; and BS-m4 for RNA extracted from cells harboring pCGA9-m4) in a final volume of 5 μl hybridization buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 1 mM EDTA). The mixture was heated at 75°C for 5 min and then slowly allowed to cool down to 42°C over a period of 120 min. Then the mixture was kept at 42°C for 30 more min before adding 1.5 μl of loading buffer (60% glicerol, 0.075% bromophenol blue, 0.25% xylene cyanol, 10 mM EDTA). Oligo-RNA hybrids were run on non-denaturing 15% PAA gels in Tris-acetate buffer (40 mM Tris-acetate, 1 mM EDTA) at 4°C, 110 V for 3 h. Labeled bands were visualized and quantified as described above. Each experiment was repeated at least twice for each of two RNA independent preparations. To estimate the amount of antisense RNA per μg of total RNA isolated from pneumococcal cells harboring a given pCGA9 plasmid variant, 0.025 and 0.0025 pmol of the corresponding in vitro synthesized antisense RNA variant were hybridized with the appropriate 5′-end-labeled oligo, and the samples were processed and analyzed in the same way as those containing the cellular RNA.
Results
Features of the antisense and sense RNAs synthesized in vivo and in vitro
The 5′-end of pMV158-encoded RNAII synthesized in pneumococcal cells has been determined by primer extension experiments (del Solar et al., 1997). Also, the 3′-end of this antisense RNA has been mapped on the basis of the increased size of the in vitro synthesized full-length transcript relative to run-off transcripts generated from the rnaII gene truncated at different positions (del Solar and Espinosa, 2001). Hence, antisense RNAII is a 48-nt-long transcript.
Chemical and enzymatic probing of in vitro synthesized RNAII (López-Aguilar and del Solar, 2013) has confirmed the previous computational predictions (del Solar and Espinosa, 1992) of the very simple secondary structure of this antisense RNA that comprises a single hairpin. RNAII consists of a 17-nt-long single-stranded 5′-tail followed by a hairpin structure (with a bulge and an apical loop) and a 3′-terminal U-stretch (Figure 1A). The hairpin and the U-stretch have been shown to be required in vitro for rho-independent transcription termination (del Solar and Espinosa, 2001). While performing this work, we observed that severe changes in the RNAII hairpin seemed also to impair transcriptional termination in vivo, leading to read-through transcription that, in the pCGA9-derivative plasmids encoding these extended RNAs, converged into the cat gene (see map of pCGA9 in del Solar and Espinosa, 1992) and prevented its expression and hence the Cm-resistance phenotype (not shown). Therefore, feasible selection of the pCGA9-type recombinants for Cm resistance implies that the cloned rnaII mutants encode an efficient intrinsic terminator.
In this work, wild-type RNAII and four antisense RNA mutants (m1, m2, m3, and m4) harboring 8-nt sequence substitutions in different elements of the RNAII secondary structure have been synthesized both in vivo and in vitro in order to compare their ability to bind to the target RNA and to inhibit translation of the essential repB gene. The mutated regions (Figure 1C) corresponded, respectively, to the 5′- (region-1, nt 2–9) and 3′- (region-2, nt 10–17) halves of the 5′-tail; to the hairpin loop (region-3, nt 26–33), and to the upper stem (region-4, nt 23–26 and 33–36). To change the sequence of region-4 without modifying the stability of the hairpin, the left and right arms of the upper stem were swapped, thus maintaining the base pairing (Figure 1C). All the antisense RNA mutants were predicted to keep a similar transcription terminator hairpin whose existence is also indicated by preliminary RNA structure probing experiments (not shown). Moreover, the intrinsic terminators of the antisense RNA mutants appear to be functional in pneumococcal cells, as inferred from the feasibility of cloning the corresponding genes in pC194 and selecting the recombinants for Cm resistance (see previous paragraph).
Cis-encoded antisense RNAII is complementary to the untranslated intergenic region of the bicistronic copG-repB mRNA (Figure 1B). The mRNA target is predicted to consist of secondary structure elements (a stem-loop flanked by single-stranded regions) of complementary sequence to their corresponding counterparts in antisense RNAII (Figure 1B). Previously, we produced in vitro two different targets of the antisense RNA (mRNA60 and mRNA80) by T7 RNAP-mediated run-off transcription of the intergenic region of the copG-repB operon (López-Aguilar and del Solar, 2013). Both target RNAs, which comprised the entire sequence complementary to antisense RNAII and lacked any intrinsic terminator, started at the position 5′ adjacent to the copG stop codon and spanned supposedly 60 nt (mRNA60) or 80 nt (mRNA80) on the copG-repB mRNA. However, run-off target products were longer-than-expected and contained 3′ extensions that arose most likely from the RNA-dependent polymerase activity of T7 RNAP and were involved in terminal hairpins (López-Aguilar and del Solar, 2013). Sequence and structure probing of both target RNAs showed that the mRNA80 mimicked quite well the structure predicted for the intergenic region of the copG-repB mRNA since it exhibited the sequences complementary to the 5′ and 3′ tails of RNAII in single-stranded conformation (Figures 1A,B). In mRNA60, by contrast, part of the sequence complementary to the RNAII region-1 was sequestered in the stem of the quite stable 3′-terminal hairpin (López-Aguilar and del Solar, 2013). The mRNA80 transcript was found to be 2–3 times more efficient in binding to RNAII than mRNA60 (López-Aguilar and del Solar, 2013). This different binding ability might be due to the distinct conformation of the region complementary to the 5′ terminus of antisense RNAII in both target transcripts, as it has been reported that the structural accessibility of the target RNA constitutes an important factor for antisense activity (Kittle et al., 1989; Wagner and Simons, 1994; Shao et al., 2006). The mRNA80 run-off transcript was chosen as the target RNA for analyzing the binding of the different antisense RNA species in view of the close resemblance between its structure and the structure predicted for the antisense RNA target region in the copG-repB mRNA.
Region-2 of antisense RNAII plays a critical role in binding to the target mRNA
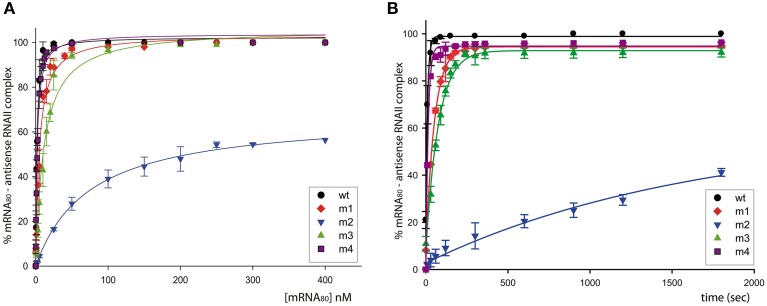
To know the contribution of the intermolecular pairing through different RNAII regions to the binding with the target RNA (mRNA80), we studied the efficiency of formation of RNA-RNA complexes that persist throughout electrophoresis on non-denaturing PAA gels. Two parameters were determined to analyze the efficiency of RNA-RNA binding: the concentration of free target RNA for half maximal binding (Kd) and the association rate constant (ka).
RNA-RNA binding efficiency was most impaired by substitution of the RNAII region-2 (antisense RNA m2), which resulted in a 45-Fold increase in the concentration of free mRNA for half-maximal binding, and a 200-Fold decrease in the association rate constant (Figure 2 and Table 2). In contrast, substitution of region-1 (m1 mutant) or region-3 (m3 mutant) of RNAII caused only a 3.5- or 5.5-Fold increase, respectively, in the concentration of target RNA required for half-maximal binding, accompanied by a similar decrease in the association rate constant. Finally, the efficiency of binding was only slightly impaired in the antisense RNA m4, which has an altered sequence in region-4 (see Figure 2 and Table 2). These results show that the single-stranded regions of RNAII (regions 1, 2, and 3) contribute significantly to the binding to the target RNA, whereas the upper stem of the hairpin does not. In addition, region-2 is shown to have the greatest contribution.
Figure 2.
Binding between the antisense RNA (wt or its variants) and the mRNA80 target. (A) mRNA concentration dependence of the RNA-RNA binding. The binding mixtures were incubated for 30 min at 37°C. (B) Kinetic of the RNA-RNA association. Time-course experiments were performed in the presence of 150 nM of mRNA80.
Table 2.
Efficiency of binding of the different antisense RNA variants to mRNA80.
| mRNA80 | |||||
|---|---|---|---|---|---|
| Antisense RNA | Binding rate constant (ka) (105 M−1s−1) | Relative ka | 1/K*d (nM−1) | Bmax ± s.d. (%) | Relative 1/Kd |
| wt | 3.75 ± 0.50 | 1 | 0.56 ± 0.05 | 102.39 ± 0.68 | 1 |
| m1 | 1.08 ± 0.11 | 0.29 | 0.17 ± 0.01 | 103.56 ± 0.64 | 0.30 |
| m2 | 0.02 ± 0.001 | 0.005 | 0.013 ± 0.001 | 67.58 ± 1.46 | 0.023 |
| m3 | 0.66 ± 0.09 | 0.18 | 0.10 ± 0.01 | 106.26 ± 1.19 | 0.18 |
| m4 | 2.29 ± 0.06 | 0.61 | 0.43 ± 0.02 | 103.89 ± 0.62 | 0.77 |
Values are the mean of at least three independent assays.
Kd, concentration of free mRNA for half-maximal binding.
The entire 5′-tail of antisense RNAII is required for efficient inhibition of in vitro repB translation
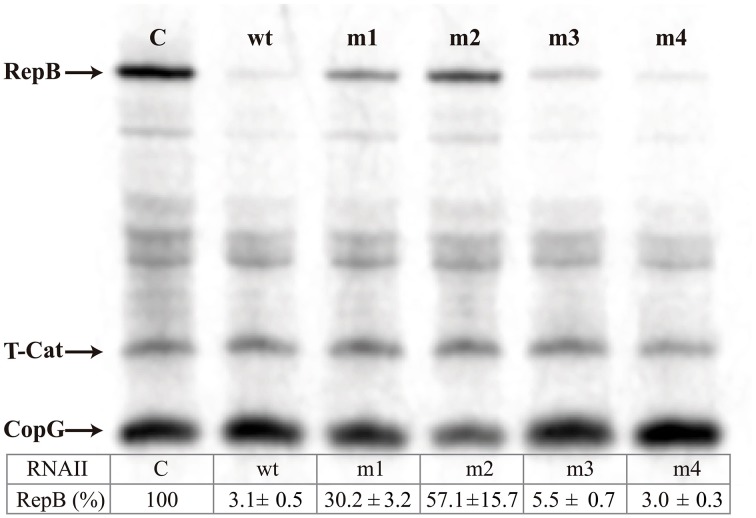
The ability of the antisense RNA variants to inhibit translation of repB from an in vitro synthesized copG-repB-truncated cat mRNA was tested in E. coli S30 extracts. Quantification of the results (inserted Table in Figure 3) shows the percentage of RepB synthesized in the presence of the different antisense RNA variants, relative to the amount of repB translation in the absence of antisense RNA. The highest inhibitory activity was exhibited by the wt and m4 mutant antisense RNAs, which reduced repB translation to ~3%. Substitution of the RNAII hairpin loop (m3 mutant) only slightly impaired the inhibitory ability of the antisense RNA. In contrast, significant levels of RepB were synthesized in the presence of the m1 or m2 mutant RNAs, indicating that substitution of region-1 or, particularly, of region-2 resulted in a severe decrease of the inhibitory activity of the antisense RNA (Figure 3).
Figure 3.
Specific inhibition of RepB synthesis by antisense RNA variants. In vitro translation of the copG-repB-truncated cat mRNA synthesized in vitro was performed either in the absence (C) or in the presence of the indicated antisense RNA variants. The table below shows the percentage of residual RepB synthesis. The numbers indicate the mean and standard deviation values obtained from at least four independent experiments.
The entire 5′-tail of antisense RNAII is required for efficient inhibition of pMV158 replication in vivo
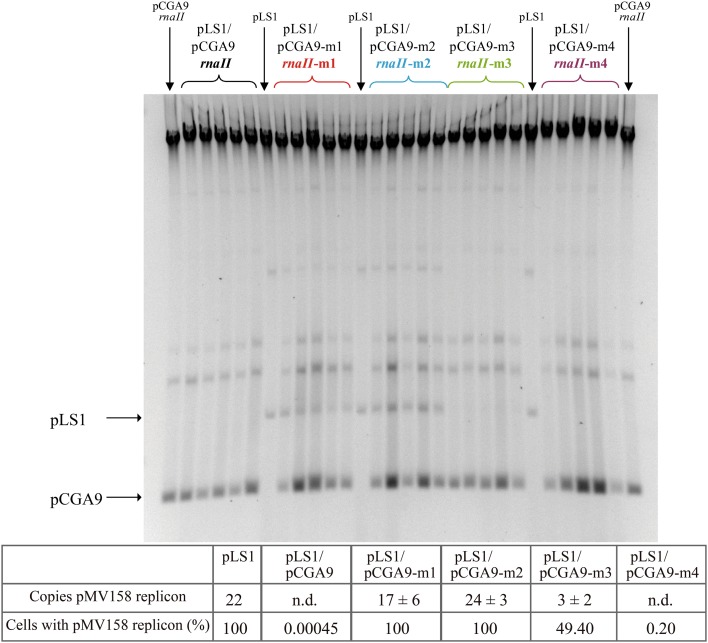
Incompatibility tests were performed to analyze the effect of the different antisense RNA variants on the replication of pLS1, a plasmid derivative carrying the pMV158 replicon (Lacks et al., 1986). Genes encoding the RNAII variants (wt, m1, m2, m3, and m4) were cloned in pC194, a plasmid that has ~100 copies per chromosome equivalent in S. pneumoniae (Table 1) and is totally compatible with pLS1 (del Solar and Espinosa, 1992; del Solar et al., 1995). The resulting recombinant pCGA9 variants (see Materials and Methods) were transformed separately into competent pneumococcal cells containing pLS1. Ten individual clones from each transformation were grown for ~37 generations in the absence of selection for the resident plasmid, and the percentage of pLS1-containing cells as well as the average pLS1 copy number were then determined in all of them. In vivo replication of pLS1 was most impaired in the presence of wt RNAII synthesized from pCGA9: at the time of the analysis, only 0.5 × 10−3% of the cells contained pLS1 as an average (values ranged from <0.001 × 10−3 to 6.5 × 10−3% in the different colonies), and the DNA of this plasmid could not be detected in any of the clones (Figure 4). Antisense RNA m4 encoded by pCGA9-m4 also produced a strong incompatibility against the pMV158 replicon, which resulted in an average 0.20% (with values ranging from <0.001 × 10−3% to 0.95% in the different colonies) of cells containing the resident plasmid after ~37 generations, and again no detectable pLS1 DNA in the bacterial extracts (Figure 4). This indicates that pairing between RNAII and copG-repB mRNA through the bases located in the upper stems of the complementary hairpins is not required for efficient inhibition of pLS1 replication. RNA m3 still inhibited the pMV158 replicon though with a severely reduced efficiency compared to the wt RNAII, since at the time of analysis, plasmid pLS1 was present in about 50% (the percentage ranged from 2 to 76% in the different clones) of the cells containing pCGA9-m3, and its average copy number was estimated to be ~3 (Figure 4). Therefore, preventing formation of a “kissing complex” does not abolish the ability of the antisense RNA to inhibit in vivo pMV158 replication. In contrast, the m1 and m2 antisense RNA mutants lacked the in vivo inhibitory ability, so that plasmid pLS1 maintained the same copy number as in the homoplasmid strain in virtually all cells containing pCGA9-m1 or pCGA9-m2 (Figure 4). Total segregational stability of pLS1 in the presence of m1 or m2 antisense RNAs was confirmed after growing several heteroplasmid clones for 60 more generations in the absence of Tc (not shown). These results demonstrated that pairing with the copG-repB mRNA through the entire 5′-tail of antisense RNAII is required for in vivo inhibition of repB translation.
Figure 4.
In vivo inhibition of the pMV158 replicon in the presence of the antisense RNA variants. Incompatibility tests were performed as indicated in Materials and Methods. Pneumococcal cultures harboring plasmid pLS1, which contains the pMV158 replicon, were transformed with the indicated pCGA9 recombinant plasmid variants. The gel shows the total DNA content of several transformant clones. Homoplasmid strains harboring pLS1 or pCGA9 were used as controls. Supercoiled monomeric forms of both plasmids are indicated in the gel. The table below shows the average copy number of pLS1 and the % of cells retaining this plasmid, as estimated from at least 10 independent colonies. n.d.: not detectable.
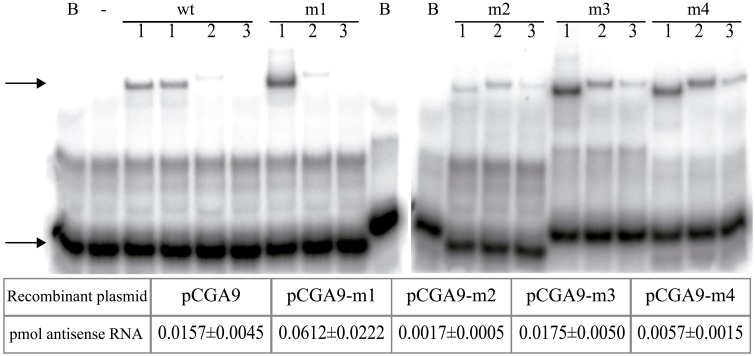
Determination of the relative amount of antisense RNA in pneumococcal cells containing different pCGA9 plasmid variants
Mutations in the rnaII gene may alter the turnover, and hence the intracellular concentration, of the mutant antisense RNAs. Since the intracellular concentration of RNAII has been shown to correlate with the level of incompatibility against the pMV158 replicon (del Solar et al., 1995), unbiased analysis of the in vivo inhibitory ability of the antisense RNA variants required estimation of the relative amount of these RNAs in pneumococcal cells containing the corresponding pCGA9 plasmid variant. To this end, “oligo-RNA band-shift” assays (Figure 5) were performed by using total RNA from each homoplasmid strain and a radioactively-labeled complementary oligo (BS, BS-m3, or BS-m4, see Materials and Methods). As controls, known amounts of the in vitro synthesized antisense RNAs were employed in these experiments. Table in Figure 5 shows the estimated amount of antisense RNA per μg of total RNA in cells bearing each recombinant pCGA9 variant. Only the mutation affecting the hairpin loop seemed not to alter significantly the turnover of the antisense RNA, as the intracellular concentration of the m3 mutant was similar to that of the wt RNA. Mutation in the 5′-end (region-1) resulted in a 4-Fold increase of the intracellular amount of the antisense RNA, whereas mutation in region-2 or in the upper stem (region-4) of RNAII caused a 9- or 3-Fold decrease, respectively, in the relative concentration of the mutant antisense RNAs.
Figure 5.
Relative intracellular amounts of the antisense RNA variants. Oligo-RNA band-shifts assays were performed with total RNA extracted from pneumococcal cells containing the different pCGA9 recombinant plasmid variants. Arrows point to the position of the free oligonucleotide (lower band) and that of the oligo-RNAII hybrids (upper band). B, sample lacking any RNA; -, sample containing in vitro synthesized mRNA80 and lacking antisense RNA;. Lane 1, samples containing 4 μg total RNA from cells that encode the indicated antisense RNA. Samples prepared with two independent extractions of RNAII wt are shown. Lanes 2 and 3, samples containing 0.025 and 0.0025 pmol, respectively, of the indicated in vitro synthesized antisense RNAs. The table below shows the amount of antisense RNA (pmol) per μg of total RNA. The values displayed are the mean of at least four independent assays.
Discussion
In this communication we report the analysis of the RNAII-mRNA interaction pathway leading to the formation of the inhibitory complex that controls replication of plasmid pMV158. As highlighted in a recent review, RNAII is the first and still only extrachromosomal element-encoded antisense RNA characterized in pneumococci (Wilton et al., 2015). Moreover, RNAII and, by extension, the equivalent RNAs encoded by plasmids of the pMV158 replicon family are among the smallest and simplest bacterial antisense RNAs described so far (Thomason and Storz, 2010). Hence, a detailed study on the pMV158 antisense control system can help to understand the mechanisms by which these small RNAs cope with base-pairing with, and regulation of, their target RNAs. Here we have analyzed the role of different regions and secondary structure elements of pMV158 RNAII in the binding to the target RNA and in the inhibition of repB translation. To this end, we mutated the nucleotide sequence of several elements of RNAII (Figure 1C) that might be involved in the recognition of the target mRNA, namely each of the two 8-nt-long halves of the 5′-tail (region-1 and region-2), as well as the apical loop (region-3) and the upper stem region (region-4) of the hairpin. As we wanted to analyze the effect of these changes on the in vitro binding to the mRNA and on in vitro and in vivo inhibition of repB expression, preservation of a functional intrinsic terminator (a stable hairpin followed by a 3′ U-tract), was a goal. Therefore, investigation on the participation of the RNAII 3′ U-tail is missing in the present work, although no indications of a main role of the equivalent region have been reported for other antisense RNA systems (Wagner and Simons, 1994; Cervantes-Rivera et al., 2010). Our results show that binding to mRNA involves mainly the RNAII region-2, located adjacent to the single hairpin of the antisense RNA (Figure 1C). In addition, pairing of the 5′-terminal region-1 of RNAII with its complementary sequence in the mRNA is shown to be essential for the efficient inhibition of repB translation initiation in vitro and in vivo. Taken together, these data reveal that the entire 5′-tail of RNAII plays a dual critical role, being required both for the RNAII/mRNA binding and for the mechanism of repB translation inhibition.
In vitro analysis of the interaction between antisense RNAII and its target region in mRNA80 yields binding-rate constants (Table 2) that are well within the range reported for other antisense-RNA control systems (1–50 × 105 M−1 s−1; Wagner and Simons, 1994; Wagner and Brantl, 1998; Franch et al., 1999). These studies also show that region-2 of RNAII has a critical role in the RNAII/mRNA binding, as indicated by the fact that antisense RNA m2 binds to the mRNA by far with the lowest efficiency (Figure 2 and Table 2). In contrast, intermolecular pairing through the upper stem region of RNAII is dispensable for the rapid binding to the mRNA, whereas the loop and the 5′-terminal region-1 contribute moderately to the binding (Figure 2 and Table 2). Therefore, only single-stranded regions of RNAII seem to have a significant role in initiating binding to the target mRNA, as was expected.
Based on these data, we propose that binding of the antisense/sense RNAs of the pMV158 replication control system initiates by pairing of the single-stranded 5′-tail of RNAII with its complementary sequence in the mRNA, and is facilitated by formation of a “kissing complex” between antisense/sense cognate hairpin loops. Moreover, progression of the “kissing” pairing into the upper stems of the complementary hairpins is not required for efficient formation of an RNAII/mRNA stable complex. This antisense/sense RNA interaction pathway based on a linear-linear scheme contrasts with the loop-loop model proved for other plasmid replication control systems, where the “kissing” interaction is crucial for binding of both RNAs, and the initial loop/loop base-pairing is propagated into the upper part (as in R1) or even into the lower portion (as in pIP501) of the stems (Hjalt and Wagner, 1995; Wagner and Brantl, 1998; Kolb et al., 2000; Heidrich and Brantl, 2007). Mutations in the recognition loops of the antisense RNAs of these well-known systems lead to new incompatibility groups (Tomizawa and Itoh, 1981; Wagner and Simons, 1994; Brantl and Wagner, 1996), which highlight the importance of the “kissing” interaction for RNA-RNA binding. Contrarily, changing the entire sequence of the apical loop of the single RNAII hairpin does not abolish the ability of the mutant antisense RNA (RNA m3) to produce incompatibility against pMV158 (Figure 4). In fact, only changes in the 5′-tail of RNAII (RNAs m1 and m2) suppress the inhibitory activity of the antisense RNA and are, therefore, predicted to lead to new incompatibility groups (Figure 4). The rather secondary role played by the apical loop of pMV158 RNAII (and hence by the corresponding “kissing”) in antisense/sense RNA binding is consistent with previous observations that repB mRNA variants lacking the complementary target hairpin were still susceptible to RNAII-mediated inhibition of repB translation (del Solar and Espinosa, 1992). These results led to the suggestion that the pMV158 system was unique among the plasmid replication control systems mediated by antisense RNA in that the initial interaction with the mRNA was based in base-pairing through the 5′-tail of RNAII (del Solar and Espinosa, 1992; Franch et al., 1999). In the linear-linear model proposed here, the entire 5′-tail and the hairpin loop of RNAII would participate in the recognition of the mRNA, although the 3′ half of the 5′-tail would play a further critical role in formation of a stable complex. Dispensability of the putative “kissing complex” in the pMV158 system has been related to the lack of U-turn motifs in the complementary hairpin loops of the inhibitor and target RNAs (Kim et al., 2008). It is worth noting that, although initiation of the RNAII/mRNA binding pathway seems to differ notably from the general U-turn-dependent “kissing” reported for other plasmid replication antisense-control systems, the RNA/RNA association rate constant obtained for the pMV158 system is in the same order of magnitude as those reported for the other plasmids. Thereby, the lack of a U-turn motif in both of the complementary hairpin loops of the pMV158 antisense/sense RNAs appears to be overcome by an efficient bi-partite interaction involving both the hairpin loop and the single-stranded 5′-tail of RNAII, as well as their corresponding complementary sequences in the mRNA.
The fact that sequence substitution of region-2 impairs binding to the mRNA much more severely than substitution of region-1 is rather intriguingly as both RNAII regions share features theoretically related to the efficiency of interaction with the target RNA (Matveeva et al., 2003; Shao et al., 2006). Namely, region-1 and region-2 are located in the 5′-tail of RNAII and have the same high A-U content (6 A/U out of 8 nt), and hence they would have been expected to contribute similarly to RNAII/mRNA binding. It should be noted that the RNA/RNA binding experiments performed in this work do only allow the observation of complexes persistent to gel electrophoresis. Hence, the extraordinaire impairment in binding efficiency observed for antisense RNA m2 might arise from the participation of region-2 not only in the initial interaction with the mRNA, but also in a subsequent step of intermolecular pairing progression leading to the formation of a stable complex.
We have previously proposed that the different folding of mRNA60 and mRNA80 (with the latter exposing the entire sequence complementary to the 5′-tail of RNAII as single strand) might account for the ~2-3-Fold higher efficiency of binding of the antisense RNA to mRNA80 relative to mRNA60 (López-Aguilar and del Solar, 2013). The differential binding of RNAII to mRNA60 and mRNA80 suggests that the efficiency of the RNAII/mRNA interaction might vary as the structure of the nascent mRNA changes. However, timing does not seem to be critical for RNAII-mediated repB translation inhibition, since this occurs efficiently irrespective of whether RNAII binds to growing mRNA molecules (in vivo incompatibility tests) or to full-length copG-repB mRNA (in vitro translation experiments). These findings contrast with those reported for the antisense control system of ColE1, where inhibitory binding of the antisense RNA occurs when nascent pre-primer RNA is 100-150 nt long. Once the pre-primer has grown longer, binding of the antisense RNA is still possible, but it does not prevent folding of the pre-primer RNA into the set of secondary and tertiary structures that are crucial for primer maturation (Masukata and Tomizawa, 1986; Wagner and Simons, 1994). The existence of a time window for inhibition of ColE1 replication by the antisense RNA arises from the indirect control mechanism used in this system, where pairing between the antisense and target RNAs blocks the formation of essential distal structures in the untranslated pre-primer RNA. On the contrary, the apparent lack of a time window for inhibition mediated by the pMV158 antisense RNA is consistent with direct hindrance of the access of the ribosomes to the repB translation initiation signals upon binding of RNAII to its complementary region in the mRNA. Although the RNAII target does not include actually any sequence element required for efficient repB translation, formation of a stable RNA-RNA duplex may prevent binding of the ribosomes to the immediately downstream region and hence expression of the essential gene (López-Aguilar et al., 2013). Moreover, the tight coupling of transcription and translation in prokaryotes likely constrains the folding of the mRNA within local regions (Shao et al., 2006), so that the structure of the target of RNAII, which lies entirely in the copG-repB intergenic portion of the mRNA, may remain rather invariable throughout the biosynthesis of the messenger.
Inhibition of repB translation by RNAII does not require formation of an antisense RNA/mRNA complete duplex, since swapping the complementary bases at the upper stem of the RNAII hairpin hardly decreases the inhibitory ability of the antisense RNA in vivo or in vitro (Figures 3, 4). In contrast, efficient inhibition by RNAII depends on pairing to the target mRNA through the 5′-tail of the antisense RNA.
Since stable binding to its target mRNA might be a prerequisite for RNAII to inhibit repB translation, we cannot rule out that the almost null inhibitory ability of antisense RNA m2 results solely from the severe impairment that substitution of region-2 causes on the efficiency of binding to the mRNA (Figures 2–4 and Table 2). Therefore, no information on the participation of region-2 in the mechanism of inhibition itself could be inferred from either the in vivo incompatibility tests or the in vitro translation inhibition assays. Contrarily, region-1 of RNAII is shown to have a main role in the mechanism of inhibition of repB translation since antisense RNA m1, while still retaining a moderate efficiency of binding to the mRNA (Figures 2–4 and Table 2), lacks the ability to produce incompatibility against pMV158 (Figure 4) and only slightly reduces repB translation (Figure 3). The failure of antisense RNA m1 to inhibit in vivo replication of pMV158 occurs despite its increased intracellular concentration, which is 4 times as high as that of the wt RNAII (Figure 5). On the other hand, base-pairing with the mRNA target through the apical loop and upper stem of the RNAII hairpin does not seem to contribute to the inhibitory mechanism itself. Although, compared to the wt RNAII, the m3 mutant shows a 5-Fold decreased efficiency of binding to the mRNA, it still causes a significant level of incompatibility against pMV158 and reduces repB translation by ~20-Fold. Swapping the base pairs at the upper stem hardly impaired the binding efficiency of the antisense RNA or its ability to inhibit repB translation in vivo or in vitro (Figures 2–4 and Table 2). The fact that inhibition of repB expression requires intermolecular pairing through region-1 of RNAII is consistent with the target of this region on the mRNA lying just upstream of the sequences involved in binding of the ribosomes (Figure 1B; López-Aguilar et al., 2013). Hence, whenever the antisense RNA/mRNA complex, although efficiently formed, allows the access of the ribosomes to the repB translation initiation signals, inhibition is avoided and synthesis of the plasmid replication initiator protein takes place.
Concluding remarks
Taking into account the inhibition and binding results obtained with the antisense RNA variants, we can conclude that the 5′-tail of RNAII plays critical roles in both processes. The 3′ half of this tail is required for generation of a stable RNA-RNA complex, predictably because its location adjacent to the RNAII hairpin renders this region essential for propagation of intermolecular pairing into the G-C rich sequence of the antisense RNA. On the other hand, the 5′ half of the tail is essential for inhibiting repB translation.
The pMV158 antisense system studied in the present work is singular among the plasmid replication control systems in that: (i) antisense RNAII (48-nt long) is the smallest characterized so far; (ii) contrarily to other systems that also involve pairing of complementary sequences located on cognate secondary structure elements of the inhibitor and target RNAs, formation and progression of a “kissing complex” is not a critical step in this plasmid; (iii) efficient binding to the mRNA requires intermolecular pairing through the RNAII region-2 5′-adjacent to the hairpin; (iv) in contrast to what has been reported for other systems (Wagner and Simons, 1994), efficient RNA-RNA binding does not ensure efficient in vivo or in vitro inhibition of repB translation; instead, two distinct regions of the 5′-tail of RNAII have critical roles in binding to the mRNA target and in preventing repB translation.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank J.A. Ruiz-Masó and M. Sanz for fruitful discussions and help. This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (Grants BFU2010-19597 and Complementary Action BFU2008-00179-E-REDEEX to GS; BFU2009-08137 to AB; CSD2008/00013-INTERMODS to ME); CTS-5077 from Junta de Andalucía and FEDER funds from EU to AB.
Footnotes
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fgene.2015.00225
References
- Ballester S., Alonso J. C., López P., Espinosa M. (1990). Comparative expression of the pC194-cat gene in Streptococcus pneumoniae, Bacillus subtilis and Escherichia coli. Genetics 86, 71–79. 10.1016/0378-1119(90)90115-8 [DOI] [PubMed] [Google Scholar]
- Boer D. R., Ruiz-Masó J. A., López-Blanco J. R., Blanco A. G., Vives-Llàcer M., Chacón P., et al. (2009). Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains. EMBO J. 28, 1666–1678. 10.1038/emboj.2009.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S. (2007). Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 10, 102–109. 10.1016/j.mib.2007.03.012 [DOI] [PubMed] [Google Scholar]
- Brantl S. (2012). Acting antisense: plasmid- and chromosome-encoded small regulatory RNAs (sRNAs) from Gram-positive bacteria. Future Microbiol. 7, 853–871. 10.2217/fmb.12.59 [DOI] [PubMed] [Google Scholar]
- Brantl S. (2014). Plasmid replication control by antisense RNAs. Microbiol. Spectrum 2:PLAS-0001-2013. 2. 10.1128/microbiolspec.PLAS-0001-2013 [DOI] [PubMed] [Google Scholar]
- Brantl S., Wagner E. G. H. (1994). Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: an in vitro study of plasmid pIP501. EMBO J. 13, 3599–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S., Wagner E. G. H. (1996). An unusually long-lived antisense RNA in plasmid copy number control: in vivo RNAs encoded by the streptococcal plasmid pIP501. J. Mol. Biol. 255, 275–288. 10.1006/jmbi.1996.0023 [DOI] [PubMed] [Google Scholar]
- Cervantes-Rivera R., Romero-López C., Berzal-Herranz A., Cevallos M. A. (2010). Analysis of the mechanism of action of the antisense RNA that controls the replication of the repABC plasmid p42d. J. Bacteriol. 192, 3268–3278. 10.1128/JB.00118-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Winans S. C. (2005). A small antisense RNA downregulates expression of an essential replicase protein of an Agrobacterium tumefaciens Ti plasmid. Mol. Microbiol. 56, 1574–1585. 10.1111/j.1365-2958.2005.04636.x [DOI] [PubMed] [Google Scholar]
- del Solar G., Acebo P., Espinosa M. (1995). Replication control of plasmid pLS1: efficient regulation of plasmid copy number is exerted by the combined action of two plasmid components, CopG and RNA II. Mol. Microbiol. 18, 913–924. 10.1111/j.1365-2958.1995.18050913.x [DOI] [PubMed] [Google Scholar]
- del Solar G., Acebo P., Espinosa M. (1997). Replication control of plasmid pLS1: the antisense RNA II and the compact rnaII region are involved in translational regulation of the initiator RepB synthesis. Mol. Microbiol. 23, 95–108. 10.1046/j.1365-2958.1997.1981561.x [DOI] [PubMed] [Google Scholar]
- del Solar G., Espinosa M. (1992). The copy number of plasmid pLS1 is regulated by two trans-acting plasmid products: the antisense RNA II and the repressor protein, RepA. Mol. Microbiol. 6, 83–94. 10.1111/j.1365-2958.1992.tb00840.x [DOI] [PubMed] [Google Scholar]
- del Solar G., Espinosa M. (2000). Plasmid copy number control: an ever-growing story. Mol. Microbiol. 37, 492–500. 10.1046/j.1365-2958.2000.02005.x [DOI] [PubMed] [Google Scholar]
- del Solar G., Espinosa M. (2001). In vitro analysis of the terminator TII of the inhibitor antisense rna II gene from plasmid pMV158. Plasmid 45, 75–87. 10.1006/plas.2000.1503 [DOI] [PubMed] [Google Scholar]
- del Solar G., Hernández-Arriaga A. M., Gomis-Rüth F. X., Coll M., Espinosa M. (2002). A genetically economical family of plasmid-encoded transcriptional repressors involved in control of plasmid copy number. J. Bacteriol. 184, 4943–4951. 10.1128/JB.184.18.4943-4951.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franch T., Petersen M., Wagner E. G. H., Jacobsen J. P., Gerdes K. (1999). Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol. 294, 1115–1125. 10.1006/jmbi.1999.3306 [DOI] [PubMed] [Google Scholar]
- Heidrich N., Brantl S. (2007). Antisense RNA-mediated transcriptional attenuation in plasmid pIP501: the simultaneous interaction between two complementary loop pairs is required for efficient inhibition by the antisense RNA. Microbiology. 153, 420–427. 10.1099/mic.0.2006/002329-0 [DOI] [PubMed] [Google Scholar]
- Hernández-Arriaga A. M., Rubio-Lepe T. S., Espinosa M., del Solar G. (2009). Repressor CopG prevents access of RNA polymerase to promoter and actively disociates open complexes. Nucleic Acids Res. 37, 4799–4811. 10.1093/nar/gkp503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R. (1990). Recombinant PCR, in PCR Protocols: a Guide to Methods and Applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (San Diego, CA: Academic Press; ), 177–183. [Google Scholar]
- Hjalt T., Wagner E. G. H. (1995). Bulged-out nucleotides in an antisense RNA are required for rapid target RNA binding in vitro and inhibition in vivo. Nucleic Acids Res. 23, 580–587. 10.1093/nar/23.4.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. W., Jeong I. S., Jeong E. J., Tak J. I., Lee J. H., Eo S. K., et al. (2008). The terminal and internal hairpin loops of the ctRNA of plasmid pJB01 play critical roles in regulating copy number. Mol. Cells. 26, 26–33. [PubMed] [Google Scholar]
- Kittle J. D., Simons R. W., Lee J., Kleckner N. (1989). Insertion sequence IS10 antisense pairing initiates by an interaction between the 5′ end of the target RNA and a loop in the anti-sense RNA. J. Mol. Biol. 210, 561–572. 10.1016/0022-2836(89)90132-0 [DOI] [PubMed] [Google Scholar]
- Kolb F. A., Engdahl H. M., Slagter-Jäger J. G., Ehresmann B., Ehresmann C., Westhof E., et al. (2000). Progression of a loop-loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. EMBO J. 19, 5905–5915. 10.1093/emboj/19.21.5905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S. A. (1968). Genetic regulation of maltosaccharide utilization in Pneumococcus. Genetics 60, 685–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S. A., López P., Greenberg B., Espinosa M. (1986). Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J. Mol. Biol. 192, 753–765. 10.1016/0022-2836(86)90026-4 [DOI] [PubMed] [Google Scholar]
- López P., Martínez S., Díaz A., Espinosa M., Lacks S. A. (1989). Characterization of the polA gene of Streptococcus pneumoniae and comparison of the DNA polymerase I it encodes to homologous enzymes from Escherichia coli and phage T7. J. Biol. Chem. 264, 4255–4263. [PubMed] [Google Scholar]
- López-Aguilar C., del Solar G. (2013). Probing the sequence and structure of in vitro synthesized antisense and target RNAs from the replication control system of plasmid pMV158. Plasmid 70, 94–103. 10.1016/j.plasmid.2013.02.005 [DOI] [PubMed] [Google Scholar]
- López-Aguilar C., Ruiz-Masó J. A., Rubio-Lepe T. S., Sanz M., del Solar G. (2013). Translation initiation of the replication initiator repB gene of promiscuous plasmid pMV158 is led by an extended non-SD sequence. Plasmid 70, 69–77. 10.1016/j.plasmid.2013.01.011 [DOI] [PubMed] [Google Scholar]
- Malmgren C., Wagner E. G. H., Ehresmann C., Ehresmann B., Romby P. (1997). Antisense RNA control of plasmid R1 replication: the dominant product of the antisense RNA-mRNA binding is not a full RNA duplex. J. Biol. Chem. 272, 12508–12512. 10.1074/jbc.272.19.12508 [DOI] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. (1986). Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell 44, 125–136. 10.1016/0092-8674(86)90491-5 [DOI] [PubMed] [Google Scholar]
- Matveeva O. V., Mathews D. H., Tsodikov A. D., Shabalina S. A., Gesteland R. F., Atkins J. F., et al. (2003). Thermodynamic criteria for high hit rate antisense oligonucleotide design. Nucleic Acids Res. 31, 4989–4994. 10.1093/nar/gkg710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Chou M.-Y., Inouye M. (1984). A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (mic-RNA). Proc. Natl. Acad. Sci. U.S.A. 81, 1966–1970. 10.1073/pnas.81.7.1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll P. R., Duschl J., Richter K. (2004). Optimized RNA amplification using T7-RNA-polymerase based in vitro transcription. Anal. Biochem. 334, 164–174. 10.1016/j.ab.2004.07.013 [DOI] [PubMed] [Google Scholar]
- Nordström K., Molin S., Aagaard-Hansen H. (1980). Partitioning of plasmid R1 in Escherichia coli: II. Incompatibility properties of the partitioning system. Plasmid 4, 332–349. 10.1016/0147-619X(80)90071-2 [DOI] [PubMed] [Google Scholar]
- Persson C., Wagner E. G. H., Nordström K. (1988). Control of replication of plasmid R1: kinetics of in vitro interaction between the antisense RNA, CopA, and its target, CopT. EMBO J. 7, 3279–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Wagner E. G. H., Nordström K. (1990). Control of replication of plasmid R1: structures and sequences of the antisense RNA, CopA, required for its binding to the target RNA, CopT. EMBO J. 9, 3767–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Masó J. A., Machón C., Bordanaba-Ruiseco L., Espinosa M., Coll M., del Solar G. (2015). Plasmid rolling-circle replication. Microbiol. Spectrum 3, PLAS-0035-2014 10.1128/microbiolspec.PLAS-0035-2014 [DOI] [PubMed] [Google Scholar]
- Shao Y., Wu Y., Chan C. Y., McDonough K., Ding Y. (2006). Rational design and rapid screening of antisense oligonucleotides for prokaryotic gene modulation. Nucleic Acids Res. 34, 5660–5669. 10.1093/nar/gkl715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. (1983). Translational control of IS10 transposition. Cell 34, 683–691. 10.1016/0092-8674(83)90401-4 [DOI] [PubMed] [Google Scholar]
- Stassi D. L., López P., Espinosa M., Lacks S. A. (1981). Cloning of chromosomal genes in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 78, 7028–7032. 10.1073/pnas.78.11.7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard P., Molin S., Nordström K. (1981). RNAs involved in copy-number control and incompatibility of plasmid R1. Proc. Natl. Acad. Sci. U.S.A. 78, 6008–6012. 10.1073/pnas.78.10.6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisted T., Sorensen N. A., Wagner E. G. H., Gerdes K. (1994). Mechanism of post-segregational killing: sok antisense RNA interacts with Hok mRNA via its 5′-end single-stranded leader and competes with the 3′-end of Hok mRNA for binding to the mok translation initiation region. EMBO J. 13, 1960–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason M. K., Storz G. (2010). Bacterial antisense RNAs: how many are there and what are they doing? Annu. Rev. Genet. 44, 167–188. 10.1146/annurev-genet-102209-163523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. (1985). Control of ColE1 plasmid replication: initial interaction of RNA I and the primer is reversible. Cell 40, 527–535. 10.1016/0092-8674(85)90201-6 [DOI] [PubMed] [Google Scholar]
- Tomizawa J. (1986). Control of ColE1 plasmid replication: binding of RNA I to RNA II and inhibition of primer formation. Cell 47, 89–97. 10.1016/0092-8674(86)90369-7 [DOI] [PubMed] [Google Scholar]
- Tomizawa J. (1990). Control of ColE1 plasmid replication: intermediates in the binding of RNA I and RNA II. J. Mol. Biol. 212, 683–694. 10.1016/0022-2836(90)90230-J [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T. (1981). Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc. Natl. Acad. Sci. U.S.A. 78, 6096–6100. 10.1073/pnas.78.10.6096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T., Selzer G., Som T. (1981). Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc. Natl. Acad. Sci. U.S.A. 78, 1421–1425. 10.1073/pnas.78.3.1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkova-Canova T., Soberón N. E., Ramírez-Romero M. A., Cevallos M. A. (2004). Two discrete elements are required for the replication of a repABC plasmid: an antisense RNA and a stem-loop structure. Mol. Microbiol. 54, 1431–1444. 10.1111/j.1365-2958.2004.04366.x [DOI] [PubMed] [Google Scholar]
- Wagner E. G. H., Brantl S. (1998). Kissing and RNA stability in antisense control of plasmid replication. Trends Biochem. Sci. 23, 451–454. 10.1016/S0968-0004(98)01322-X [DOI] [PubMed] [Google Scholar]
- Wagner E. G. H., Simons R. W. (1994). Antisense RNA control in bacteria, phages and plasmids. Annu. Rev. Microbiol. 48, 713–742. 10.1146/annurev.mi.48.100194.003433 [DOI] [PubMed] [Google Scholar]
- Wilton J., Acebo P., Herranz C., Gomez A., Amblar M. (2015). Small regulatory RNAs in Streptococcus pneumoniae: discovery and biological functions. Front. Genet. 6:126. 10.3389/fgene.2015.00126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.