Abstract
Background
Earth’s climate is warming as a result of anthropogenic emissions of greenhouse gases from fossil fuel combustion. Bioenergy, which includes biodiesel, biohydrogen and bioethanol, has emerged as a sustainable alternative fuel source. For this reason, in recent years biodiesel production has become widespread but this industry currently generates a huge amount of glycerol as a by-product, which has become an environmental problem in its own right. A feasible possibility to solve this problem is the use of waste glycerol as a carbon source for microbial transformation into biofuels such as hydrogen and ethanol. For instance, Escherichia coli is a microorganism that can synthesize these compounds under anaerobic conditions.
Results
In this work an experimental procedure was established for screening E. coli single mutants to identify strains with enhanced ethanol and/or H2 productions compared to the wild type strain. In an initial screening of 150 single mutants, 12 novel strains (gnd, tdcE, rpiAnanE, tdcB, deoB, sucB, cpsG, frmA, glgC, fumA and gadB) were found to provide enhanced yields for at least one of the target products. The mutations, that improve most significantly the parameters evaluated (gnd and tdcE genes), were combined with other mutations in three engineered E. coli mutant strains in order to further redirect carbon flux towards the desired products.
Conclusions
This methodology can be a useful tool to disclose the metabolic pathways that are more susceptible to manipulation in order to obtain higher molar yields of hydrogen and ethanol using glycerol as main carbon source in multiple E. coli mutants.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-015-0285-6) contains supplementary material, which is available to authorized users.
Keywords: Escherichia coli, Ethanol, Glycerol, High throughput, Hydrogen
Background
Earth’s climate is warming as a result of anthropogenic emissions of greenhouse gases, particularly carbon dioxide (CO2) from fossil fuel combustion. Significant opportunities to mitigate anthropogenic emissions of these gases exist, although some will be easier to exploit than others [1]. Recently, the production and use of biofuels like biodiesel and bioethanol have become widespread because they are more sustainable, secure, renewable, and environmentally safe than the fossil fuels [2, 3]. Hydrogen (H2) is also universally recognized as an environmental friendly and safe renewable resource [4]. Therefore, the production of H2 and ethanol from biomass will probably play an important role in bioenergy generation [5]. However, although biodiesel production has emerged as a possible alternative to fossil fuels, this industry generates glycerol as a by-product in such large quantities that its market value has dropped and it has become an environmental problem in its own right. For these reasons, several practical processes for the conversion of glycerol into high-value products have been proposed [6]. In this regard, glycerol represents a cheap carbon source that has been used in several biotransformation processes for the production of added-value products [2, 7–16] including the generation of hydrogen and ethanol [17, 18].
Escherichia coli—one of the most commonly host organism used for metabolic engineering and biotechnological applications [19–21]—is a suitable species for glycerol utilization either under aerobic or anaerobic conditions [14, 22]. The fermentation of glycerol starts with its conversion to DHAP, which is mediated by a two-branch pathway: the oxidative branch by glycerol dehydrogenase and dihydroxyacetone kinase enzymes; and the reductive branch by glycerol kinase and glycerol 3-phosphate dehydrogenase enzymes. Then DHAP can be then metabolized in the glycolysis pathway to pyruvate. In this process 2 NAD+ are reduced, one of them in the assimilation of glycerol and another one in the synthesis of 1,3 bisphosphoglycerate.
Ethanol and H2 are originated in E. coli after the anaerobic mixed-acid fermentation in which pyruvate, the final product of glycolysis, is fermented to formate, the substrate for H2 synthesis, and acetyl-CoA, which after two reductive steps is transformed to ethanol in which two NADH are re-oxidized balancing in this way the NADH/NAD+ ratio during the assimilation of glycerol [14, 22].
Several factors regulate the production of hydrogen and ethanol. Among them: pH, metabolites transport from or into the cell [23, 24], and the carbon source. In this sense, it has been described that hydrogen production by the hydrogenase enzymes (Hyd-1, 2, 3 and 4) in E. coli is influenced by pH and for instance optimal pH of activity for the Hyd-3 is around pH 6.5 [2, 25, 26]. On the other hand, glycerol metabolization is enhanced at neutral pH and is highly active at alkaline pH [22]. Several relevant studies have also revealed how the hydrogen and ethanol yields can be improved by genetic engineering in E. coli [22, 27–30]. For instance it has been recently described that the deletion of transporters of precursors, such as the formic transporter FocA and FocB, can improve the production of ethanol and/or H2 [23, 24, 31]. Other strategies have consisted on the blockage of one or several pathways involved in the synthesis of competitive products [21–24, 28, 32–34] and/or the overexpression of enzymes or transcription factors. For instance, the overexpression of genes involved in the uptake and conversion of glycerol (GldA) and/or expression of hydrogenase transcriptional factors (FhlA) can help to redirect the carbon flux towards the production of these target products [22, 24, 28, 32, 33]. Both strategies have been successfully combined and, for instance, Tran et al. [23] have described recently a multiple mutant E. coli strain that increases the molar yield for hydrogen and ethanol production.
Metabolic reactions operate as a network rather than linear pathways. When a microorganism is used for the bioproduction of compounds such as H2 and ethanol, the wild type organism often renders suboptimal and unsatisfactory yields. For this reason, metabolic engineering is an emerging field whose objective is to alter the metabolic network through genetic modification in order to improve the production of biofuels such as ethanol, butanol, propanol, biodiesel and hydrogen [35]. When an experimental design in metabolic engineering is aimed towards the production of a specific product, the resulting phenotypes are often suboptimal and unsatisfactory due to the distant effects of genetic modifications or unknown regulatory interactions [36]. In contrast, genetic high-throughput screenings can reveal unexpected genetic backgrounds that are suitable for the production of a particular desired compound. These techniques also have the potential to disclose interactions between different metabolic pathways.
In order to understand the role of the protein-encoding genes in the metabolism, it is useful to study the loss of function by analysing gene knockout phenotypes. Due to the lack of available information in databases concerning growth and multi-omics data for E. coli grown under anaerobic conditions in a glycerol based-medium, in the work described here a robust and reproducible experimental design has been established for screening of E. coli single mutant strains that allowed the characterization of H2 and ethanol productions together with the glycerol consumption. In this design, the use of mini-reactors under anaerobic standardized conditions coupled to automated gas chromatography (GC) and high performance liquid chromatography (HPLC) allowed the easy and reliable measurement of hydrogen, ethanol and glycerol.
In this work an initial screening of 150 single knockout strains from the Keio and Yale Collections, was carried out and 12 novel mutants with enhanced ethanol and/or H2 production and/or glycerol consumption respect to the E. coli wild type strain were found. Moreover, based on these results, several multiple mutant strains have also been engineered in order to improve the target product yields and the consumption of glycerol.
This design could be applied in a more extensive screening, which might provide useful genetic backgrounds for the production of H2 and ethanol and will help to understand further the physiology of glycerol uptake under anaerobic conditions.
Results
Selection of mutants for analysis from the KEIO and YALE collections
In this study, 150 isogenic E. coli single knockout mutant strains were cultured under the experimental conditions described in Additional file 1: Figure S1. Each mutant strain was tested in triplicate in every set of experiments and a triplicate of the wild type strain was also analysed as a quality control (QC). The pre-selection of the mutants for analysis was based mainly on genes related to the central carbon metabolism pathways such as glycolysis (8 strains), TCA cycle (20 strains), pentose phosphate pathway (PPP) (13 strains) and intermediate metabolism pathways such as amino acid (20 strains), nucleotide (13 strains) and lipid (13 strains) pathways, although genes involved in other cellular functions were also studied. In this pre-selection some single mutant strains related to the synthesis of hydrogen and ethanol and the assimilation of glycerol were also included (13 strains) and used as experimental controls, since mutants of the same genes have previously been reported to produce higher or lower values for the target products. All of the selected knockout genes are listed in Additional file 2: Table S1.
Ethanol and H2 productions and glycerol consumption in the experimental control strains
An overview of the experimental control strain values for the target products (Figure 1) showed that the experimental design proposed in this work was appropriate. Thus, the strains defective in enzymes involved in ethanol and H2 synthesis and glycerol consumption—that have previously been reported to produce lower yields—namely formate hydrogenolyase (Fhl) [37], pyruvate formate lyase (PflB) [2], glyceraldehyde 3P dehydrogenase (GldABC), dihydroxyacetone kinase (DhaKL) [13], and formate dehydrogenase (FdhF) and formate hydrogenolyase transcriptional factor (FhlA) [13], consistently showed the lowest values for the specific production of the target products as well as the specific consumption of glycerol. On the other hand, the focA, ldhA, frdC [23] frdA [14] and frdB [38] mutants, which were previously described as efficient H2 and/or ethanol producers, showed higher values for these parameters than the wild type reference which is also shown in Figure 1.
Figure 1.
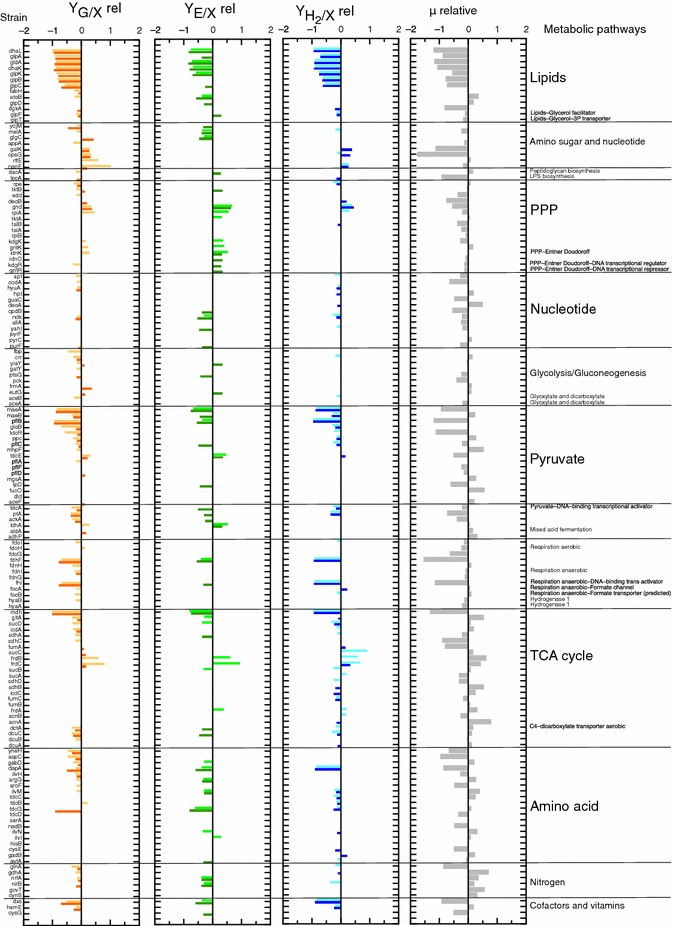
Bar charts showing relative values of the parameters evaluated respect to that of the wild type. Specific glycerol consumed (mmol/g CDW, YG/X rel) in orange; specific ethanol production (mmol ethanol/g CDW, YE/X rel) in green; specific hydrogen production (mmol hydrogen/g CDW, YH2/X rel) in blue and growth rates (µ relative) in grey. Statistically significant P < 0.05 for YH2/X and YG/X parameters and 0.01 for YE/X was used (0 denotes the wild type values). The clear colours represent the relative values at 22 h and the dark colours at 46 h. In the left-hand column are listed the mutant strains assayed in this work and in the right-hand column the metabolic pathways in which each defective mutant strain is involved. LPS lipopolysaccharide, PPP pentose phosphate pathway, TCA tricarboxylic acid.
Novel genetic backgrounds suitable for H2 and ethanol production and glycerol consumption
In order to determine which of the screened mutants showed statistically significant higher values for YE/X and/or YG/X and/or YH2/X than those of the wild type strain, the non-parametric contrast Mann–Whitney U test was applied to these parameters. From this analysis it was concluded that 45 mutants (including the experimental control strains) showed statistically significant higher values for any of the target products (Additional file 3: Table S3) and all of them showed values for growth rate (µ) > 0 (Additional file 4: Figure S2).
Nevertheless, the number of experiments carried out for each strain was too low (n = 3) to use more robust parametrical analysis such as Student’s t test. For this reason, the strains included in the 50th percentile (24 mutants) for each target product (Additional file 3: Table S3) were tested further (n ≥ 6) and the data obtained were analysed in box plots (Figure 2).
Figure 2.
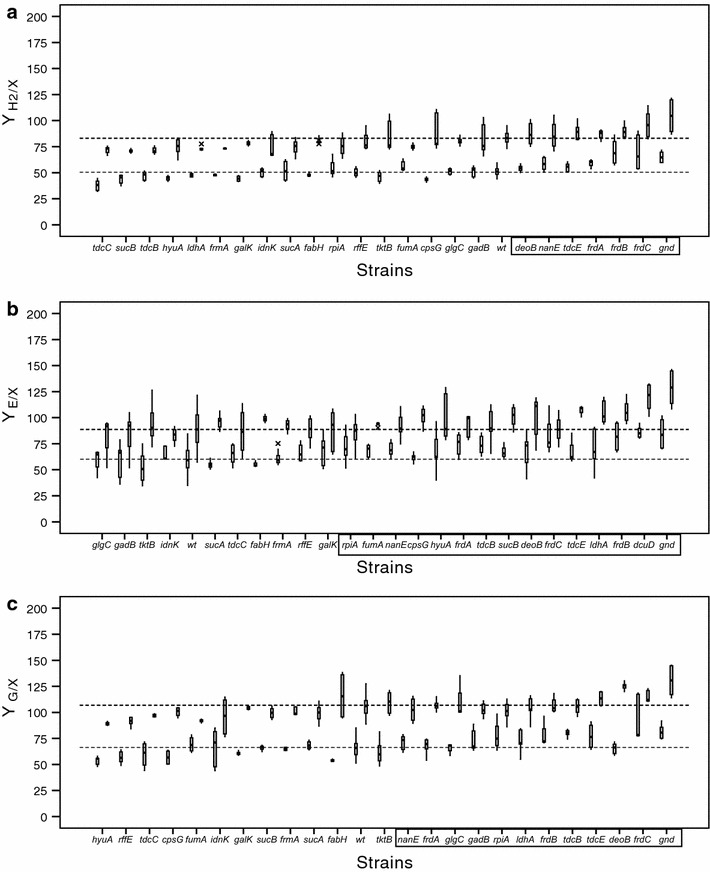
Box plots of parameters evaluated in the selected mutant and wild type strains. Specific hydrogen production (mmol hydrogen/g CDW, YH2/X) (a), specific ethanol production (mmol ethanol/g CDW, YE/X) (b) and specific glycerol consumption (mmol glycerol consumed/g CDW, YG/X) (c). In each graph the white and black boxes represent the 22 and 46 h interquartile range values respectively and bars the SD. The dashed lines in each graph indicate the wild type averages for each parameter at 22 and 46 h. In the X-axis, the strains whose average values are higher with statistical significance in comparison to that of the wild type using a P < 0.05 were framed. The wild type data was obtained from at least 75 replicates and the coefficient of variation (CV) was <11% for all parameters, except for ethanol concentration, which was lower than 21%. These results were considered to be suitable to establish a reference for comparison of the mutant strain average values with respect to those obtained for the wild type.
In order to study the interrelation of the three parameters analysed in this work, the YH2/X, YE/X and YG/X values for each of these strains and the experimental controls (frdA, frdB, frdC and ldhA mutants) were related to the wild type ones and subsequently plotted in the cumulative bar chart shown in Figure 3. These experimental control strains consistently showed similar increased values for at least one of the parameters studied in this work (Additional File 5: Table S4) as previously described in the literature [13, 23, 39, 40]. On the other hand, the remaining 12 mutants (gnd, tdcE, rpiAnanE, tdcB, deoB, sucB, cpsG, frmA, glgC, fumA and gadB) had not previously been related with the parameters studied in this work. The strains with enhanced values for the three parameters respect to the wild type strain were the gnd, tdcE, rpiA, nanE and deoB mutants.
Figure 3.
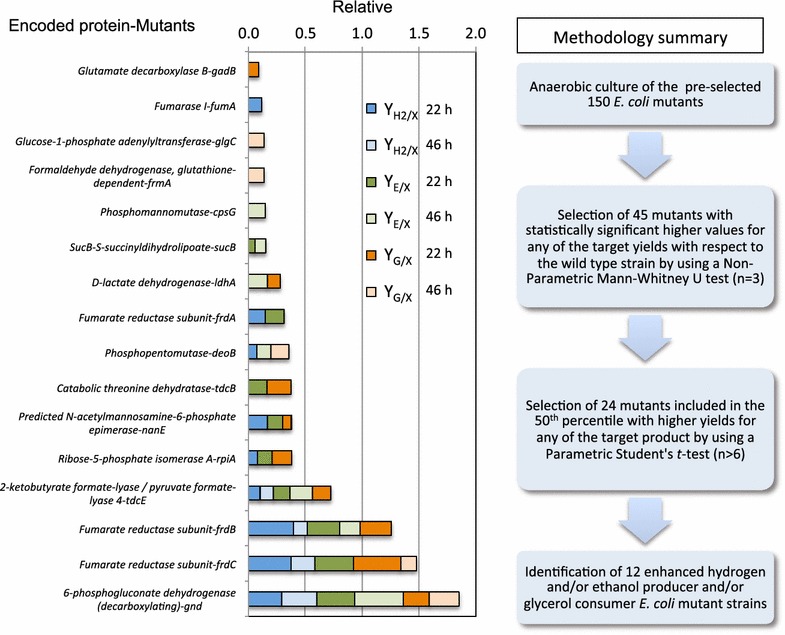
Cumulative bar charts of mutant strains with relative values respect to that of those of the wild type. Specific hydrogen production relative values (YH2/X rel) coloured in blue, specific ethanol production relative values (YE/X rel) in green and specific glycerol consumption in glycerol (YG/X rel) in orange which are significantly higher than the wild type with P < 0.05. Clear colours represent the 22 h values and the dark colours the 46 h ones.
The tdcB mutant increased the glycerol and ethanol yields. In other cases only one of the analysed parameters was improved: the sucB and cpsG mutants for ethanol, the fumA mutant for hydrogen and the gadB, frmA and glgC mutants for glycerol consumption (Figure 3).
Regarding the values of the cumulative bars, the selected mutants could be separated into three different groups: (1) the gnd, frdC and frdB mutants increased values by more than 100% with respect to the wild type (between 1.12- and 1.43-folds for any of the parameters respect to the wild type); (2) the tdcE mutant increased values by more than 50% (from 1.1- up to 1.18-folds respect to the wild type in any of the parameters) and (3) the rpiA, nanE, tdcB, deoB, frdA, ldhA, sucB, cpsG, frmA, glgC, fumA andgadB mutants showed an increase of <50% than that of the wild type (Figure 3). Consequently, the gnd mutant (1.43- and 1.31-folds in ethanol and hydrogen production respectively at 46 h) and tdcE mutant (1.19-folds at 46 h and 1.12-folds at 22 h in ethanol and hydrogen production respectively) were selected as novel potential genetic backgrounds for further studies.
Construction of multiple mutant strains by knocking out the ldhA, gnd, frdBC and tdcE genes
One of the strategies commonly used in E. coli to improve target product yields is to combine multiple mutations that may help to the redirection of carbon flux towards the synthesis of the desired products. To this aim, the following multiple mutants were constructed; ldhAgnd::kan, ldhAgndfrdBC::kan and ldhAgndfrdBCtdcE::kan denoted in this work as M2, M4 and M5 respectively. On the other hand, another variable that can be considered is the pH condition. In this sense, the same culture medium but at pH 7.5 was analysed as one of the possible variables that could increase the glycerol consumption [14, 22] in the multiple mutant strains. With this purpose, the specific productions for ethanol and hydrogen and glycerol consumption were previously evaluated in the wild type strain up to 94 h (Additional file 6: Figure S3). In these analysis, although specific hydrogen production (YH2/X) expressed in mmol of H2/g CDW values did not show significant differences between at both pH (Additional file 6: Figure S3A), the specific ethanol production (YE/X) in mmol of ethanol/g CDW and glycerol consumption (YG/X) in mmol of glycerol consumed/g CDW, were significantly higher at pH 7.5 than those obtained at pH 6.25 from 22 h on (Additional file 6: Figure S3B and C). Therefore the assays with the multiple M2, M4 and M5, the single gnd and tdcE mutants and wild type strains, were conducted at pH 7.5.
Although the YH2/X and YE/X in M4 and M5 at 94 h were significantly lower (0.9-folds for both parameters respect to the wild type) than that of the gnd, tdcE single mutants and for the wild type strain (Figure 4a, c). The molar yields of hydrogen—expressed in mmol of product per mmol of glycerol consumed—was higher for M2 1.07-fold at 22 h and for M5 1.22-fold at 70 h and 1.33-fold at 94 h respect to that of the wild type strain and were also higher than those obtained with M4, gnd, and tdcE mutants (Figure 4b). Regarding to ethanol molar yields it was found that a value for the wild type strain showed a maximum value of 1.1, which is higher than the theoretical value of 1. This effect can be explained by the fact that the culture medium used in this work is not a minimal medium and, together with glycerol, includes peptone, which can be used by the cells as C source for ethanol production. In the case of M5, ethanol molar yield values were enhanced 1.41-fold from 22 h up to 94 h respect to the wild type and they were also higher than those obtained respect to the other analysed mutant strains (Figure 4d). On the other hand, M5 showed a higher standard deviation (SD) at 70 and 94 h for hydrogen yields and at 46, 70 and 94 h for ethanol yields compared to those obtained at 22 h (Figure 4b, d). This relatively high SD is probably due to a biological variability of this mutant strain in the glycerol consumption since specific production for hydrogen and ethanol (Figure 4a, c) showed very low standard deviation.
Figure 4.
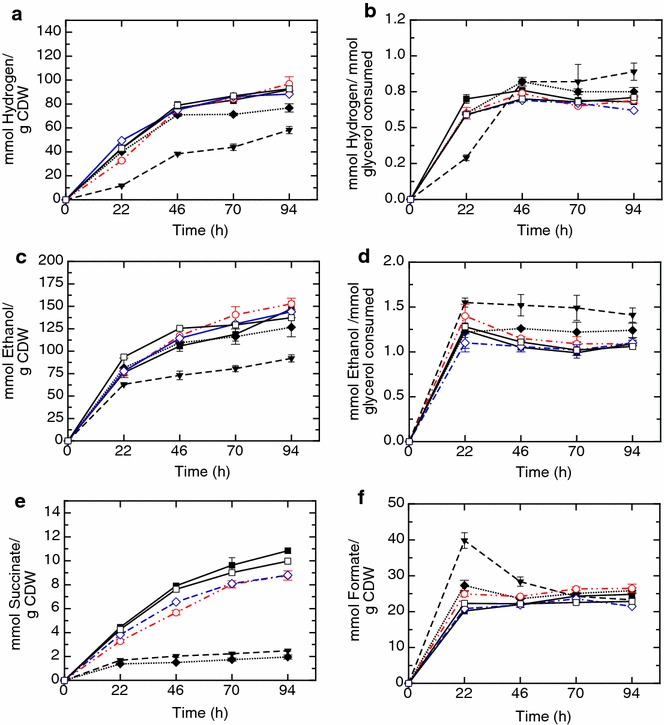
Scatter plots of mean and SD of parameters evaluated in single mutant, multiple mutant and wild type strains. Specific hydrogen production, YH2/X (a); hydrogen molar yield (b); specific ethanol production, YE/X (c); ethanol molar yield (d); specific succinate efflux, YS/X (e); and specific formate efflux, YF/X (f); graphed up to 94 h in the following mutant and wild type strains: ldhAgnd::kan (M2) (filled square); ldhAgndfrdBC::kan (M4) (filled diamonds); ldhAgndfrdBCtdcE::kan (M5) (filled inverted triangle); gnd mutant (open circle); tdcE mutant (open square) and wild type strain (open diamond). Time points evaluated were 22, 46, 70 and 94 h of experiment.
In order to better understand the redirection of the carbon rewiring, formate—a precursor of hydrogen synthesis—and succinate—the final product of TCA reductive branch—were analysed in the culture medium. These products are actively exported out from the cell in anaerobic conditions. As can be expected, succinate efflux (YS/X) (mmol/g CDW) was significantly lower for M4 and M5 0.22- and 0.28-fold respectively compare to that of the wild type at 94 h, which are defective in the fumarate reductase enzyme. On the other hand, the M2 showed higher values, 1.23-fold respect to those of the gnd mutant and wild type strain at 94 h (Figure 4e). In the case of extracellular formate (YF/X) expressed in mmol/g CDW, the M5 strain showed significantly higher values, up to twofold, at 22 h than those obtained for the M2, and wild type strains and 1.5-fold respect to M4, tdcE and gnd mutants. These values gradually decreased and eventually reached similar values to that of the gnd mutant at 94 h (Figure 4f).
Discussion
The recent development of two knockout E. coli collections—the Keio Collection and Yale University CGSC Stock Center [41, 42]—has allowed the systematic search of phenotypes in diverse conditions. Recent advances in high-throughput omic technologies have led to the possibility of deciphering an organism’s genotype-to-phenotype relationships [43]. However, obtaining useful biological knowledge from a single type of omic data—for example, DNA microarray only—is not an easy task. In this work we propose a design for a high-throughput methodology for the systematic analysis of E. coli knockout strains for the study of hydrogen and ethanol synthesis and glycerol consumption in order to identify novel E. coli phenotypes with enhanced yields for these parameters (Figure 5). In order to test this design, a preliminary screening of 150 single mutants (n = 3) from collection strains (Additional file 2: Table S1) was carried out in this work. This methodological design was considered suitable for our purpose due to its reproducibility. In order to validate the experimental procedure, several mutants used as experimental controls (Figure 1), consistently showed statistically significant differences with respect to the reference strain for the same parameters, which validated this methodology. In addition to the control strains, 45 mutants showed statistically significant enhanced parameters (non-parametric test) for one or more of the target products (Figure 1). Those mutants in the 50th percentile of these 45 strains, for any of the parameters evaluated, were selected for further analysis (Additional file 3: Table S3). These 19 selected mutants, together with the experimental control strains (ldhA, frdA, frdB and frdC mutants), were further tested (n ≥ 6) by the parametrical t test. In this new analysis the experimental control strains consistently showed increased YH/X, YE/X and YG/X (Figure 2) and also confirmed that 12 out of the 19 mutant strains showed an enhanced ethanol and/or hydrogen production and/or glycerol consumption (gnd, tdcE, rpiAnanE, tdcB, deoB, sucB, cpsG, frmA, glgC, fumA and gadB). These mutated genes are mainly related to the metabolism of amino acids, PPP, TCA, gluconeogenesis, amino sugar and nucleotides.
Figure 5.
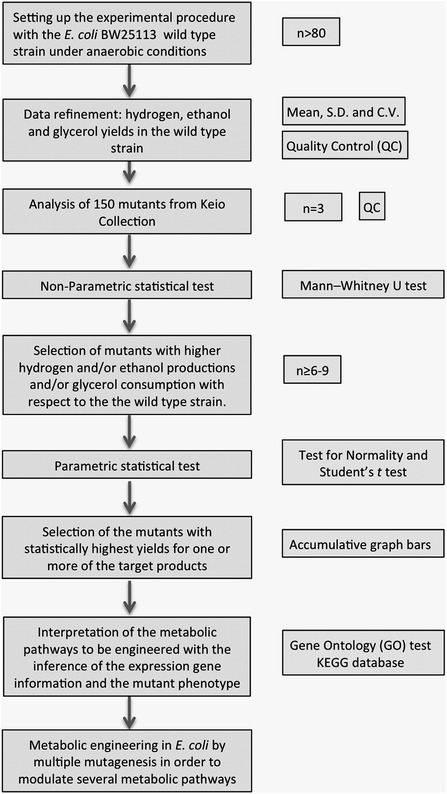
Methodology for a high throughput screening of E. coli mutant strains.
It is remarkable that 2 out of the 12 selected mutant strains were defective in enzymes involved in the PPP, including the gnd mutant—defective for the 6-phosphogluconate dehydrogenase enzyme—for which YH/X, YE/X and YG/X parameters were significantly enhanced at the two times post inoculum studied in this work. A possible explanation for this phenotype may be that the lack of activity of this enzyme can increase the pool of 6-PG, which would lead to an increase in glyceraldehyde 3-P (G3P) and phosphoenolpyruvate (PEP) by the Entner–Doudoroff pathway, which feeds the ‘bottom half’ of glycolysis [44]. On the other hand, the rpiA mutant (defective in ribulose-5P-isomerase) also increased the three parameters analysed in this work, albeit to a lesser extent than the gnd mutant. The enzyme codified by this gene is involved in the interconversion of ribose-5P to ribulose-5P. The lack of this enzyme could also have a similar effect to that of the gnd mutant, i.e., the increase of glyceraldehyde 3-phosphate (GA3P) and fructose-6P from pentose sources. Another mutant that is related to the metabolism of pentoses was also found in the 12 selected mutants, namely deoB phosphopentomutase mutant, which showed a similar phenotype to that of the rpiA mutant. This enzyme is involved in the catabolism of nucleotides and deoxynucleotides and promotes the interconversion of ribose-1P or deoxyribose-1P to their corresponding 5P form. It can therefore be concluded from these results that the reorganization of the metabolism through the blocking of several enzymes involving the PPP is a promising possibility for the rewiring of the metabolism in the use of glycerol as the main C source towards pyruvate and, subsequently synthesising acetyl-CoA and formate which are transformed respectively into ethanol and hydrogen.
In this screening 39 mutants that involve the nitrogen metabolites were also tested. From these mutants only the tdcE (2-ketobutyrate formate-lyase/pyruvate formate lyase) and the tdcB (threonine dehydratase) mutants, both of which codify enzymes involved in threonine degradation [45], showed enhanced hydrogen yield and glycerol consumption and the tdcE mutant also showed a higher ethanol yield. In this regard, the mutation of any of these enzymes may increase the acetyl-CoA pool and, consequently, the ethanol production. Although not all of the enzymes related to amino acid metabolism were tested, it is significant that two of the selected mutants were related to threonine catabolism. Nevertheless, more in-depth studies are needed to investigate the possibility of rewiring the amino acid metabolism and the rest of the mutants (nanE, deoB, sucB, cpsG, frmA, glgC, fumA, gadB) found in this work.
In order to gain a deeper understanding of which biological functions could be depleted to avoid redundant or deleterious mutations, with the ultimate aim of constructing a multiple mutant strain, these genes were analysed in the Gene Ontology (GO) database [46]. Although several of the GO matches proved to be redundant, the most significant biological processes that were affected were the fermentation, energy derivation by oxidation of organic compounds, generation or precursor metabolites and energy, cell projection, generation of precursor metabolites and energy, oxidation–reduction process and flagella motility among others (Table 1). The results of this analysis seem to indicate that the cells try to rewire the metabolic process to maintain the energy balance, which is translated to the alteration in the synthesis of precursors for the anabolic metabolic processes that cause a decrease in the cell division rate, as can be observed by the relative µ values for the mutants studied in this work (Additional file 4: Figure S2). In addition, the oxidation–reduction process involving the NAD(P)H/NAD(P)+ balance within the cell is fundamental in the equilibrium of glycolysis and fermentation pathways in order to synthesis ATP by level-substrate phosphorylation and also in thousands of energetic-dependent reactions.
Table 1.
Gene-Ontology (GO) database search in EcoCyc of the knockout mutants selected in based on the statistically significant results of the parameters evaluated shown in Figure 3
| Gene-Ontology-terms | p-valuesa | Matches (mutant strains) |
|---|---|---|
| Fermentation | 2.22E−08 | ldhA//“frdB”//“frmA”//“frdA”//“frdC” |
| Generation of precursor metabolites and energy | 1.58E−07 | frdB//“tdcE”//“frdA”//“frdC”//“glgC”//“ldhA”//“frmA”//“fumA” |
| Energy derivation by oxidation of organic compounds | 5.45E−07 | frdB//“tdcE”//“frdA”//“frdC”//“glgC”//“ldhA”//“frmA” |
| Aspartate family amino acid catabolic process | 2.02E−05 | sucB//“tdcB”//“tdcE” |
| Bacterial-type flagellum assembly | 2.57E−05 | frdA//“frdB”//“frdC” |
| Ethanol metabolic process | 5.57E−05 | ldhA//“frmA” |
| Oxidation–reduction process | 6.95E−05 | gnd//“rpiA”//“frdB”//“tdcE”//“frdA”//“frdC”//“glgC”//“ldhA”//“frmA” |
| Single-organism metabolic process | 9.44E−05 | rpiA//“gnd”//“deoB”//“gadB”//“tdcB”//“frdB”//“tdcE”//“frdA”//“frdC”//“glgC”//“frmA”//“cpsG”//“ldhA”//“sucB”//“nanE” |
| Single-organism catabolic process | 1.52E−04 | ldhA//“gnd”//“rpiA”//“deoB”//“tdcB”//“sucB”//“tdcE”//“nanE”//“frmA” |
| Tricarboxylic acid cycle | 1.57E−04 | fumA//“frdB”//“sucB” |
| l-threonine catabolic process to propionate | 3.86E−04 | tdcB//“tdcE” |
| Anaerobic respiration | 4.10E−04 | frdB//“tdcE”//“frdA”//“frdC” |
| Organic substance catabolic process | 6.34E−04 | frmA//“tdcB”//“sucB”//“tdcE”//“ldhA”//“gnd”//“rpiA”//“nanE”//“deoB” |
| Glucose catabolic process | 6.49E−04 | ldhA//“gnd”//“rpiA” |
| Cell motility | 8.04E−04 | frdA//“frdB”//“frdC” |
| Carbohydrate metabolic process | 8.48E−04 | rpiA//“gnd”//“ldhA”//“glgC”//“cpsG”//“nanE”//“tdcE” |
| Small molecule metabolic process | 8.52E−04 | frmA//“cpsG”//“deoB”//“gnd”//“rpiA”//“ldhA”//“gadB”//“tdcB”//“sucB”//“tdcE”//“nanE” |
| Cellular metabolic process | 8.65E−04 | deoB//“frdB”//“tdcE”//“frdA”//“frdC”//“frmA”//“gnd”//“rpiA”//“cpsG”//“glgC”//“ldhA”//“gadB”//“tdcB”//“nanE”//“sucB”//“fumA” |
| Cellular respiration | 9.59E−04 | frdB//“tdcE”//“frdA”//“frdC” |
aThe matches were found using a P < 0.001 which are denoted as scientific notation (E).
Once established the main metabolic pathways, in which these mutants were involved, the gnd and tdcE mutants were selected for further analysis because they showed the higher values in all of the parameters evaluated respect to the wild type strain. One of the strategies commonly used to enhance the efficiency of the biotransformation processes carried out by E. coli is the redirection of C-flux by multiple mutations. In this work the mutation of gnd gene (defective in the synthesis of d-Ribulose 5-P) was combined with the deletion of lactate synthesis (ldhAgnd::kan, M2), the succinate synthesis (ldhAgndfrdBC::kan, M4) and the threonine degradation (ldhAgndfrdBCtdcE::kan, M5). These mutant strains together with the reference strain were also evaluated at pH 7.5 due to it has been previously described that glycerol metabolization is favored at this pH [22]. In the case of M4 and M5 mutants, the blockage of succinate synthesis was observed because in both strains the succinate efflux was substantially decreased (Figure 4e) as can be expected from the mutation of frdBC genes [7, 47]. The hydrogen and ethanol molar yields obtained from these engineered strains, indicates that metabolism of C source is shunt towards formate, in the case of gnd and tdcE single mutants, M4 and specially M5 mutant in which formate is increased twofold respect wild type strain and 1.5-fold respect to gnd mutant (Figure 4f).
This effect correlated with an enhanced ethanol molar yield in the M5 mutant (1.41-fold respect to that of the wild type) (Figure 4d). This improvement is higher than that obtained in the septuplet mutant (BW25113frdC ldhA fdnG ppc narG mgsA, hycA) reported by Tran et al. [23] (0.67-fold respect to the wild type at 24 h). The only difference between M4 and M5 strains is the deletion of TdcE enzyme gene, which is involved in catabolism of threonine into propanoate [45]. This deletion in M5 may provoke a significant metabolic redirection—through unknown metabolic regulation mechanisms—towards pyruvate and subsequently to formate—which is exported out the cell- and acetyl-CoA that is converted more efficiently into ethanol (Figure 4d). The excess of formate is exported out the cell due to accumulation of this molecule in the cell to toxic levels, although after 22 h could be imported [31, 48] and then converted to hydrogen (Figure 4a). In fact, the M5 strain growth was significantly depleted after 22 h (Additional file 7: Figure S4) and specific productions were very low respect to all the analysed strains (Figure 4a, c). In this mutant the higher molar yield measured for hydrogen and ethanol could mean that glycerol consumption was lower, although the metabolization of this product is more efficient in M5 mutant than in the wild type. In this sense M5 mutant could be an interesting genetic background for further studies in which the excess of formate may be converted into hydrogen.
Conclusion
In conclusion, the systematic analysis of the target products in E. coli mutant strains proposed in this work is a feasible methodology to identify novel suitable genetic backgrounds to enhance the synthesis of hydrogen and/or ethanol in cells cultured in a glycerol-based medium. In this work we identified several mutants (chiefly gnd, and the tdcE) that could be combined in multiple mutant strains in order to enhance the yields of the desired products by metabolic engineering. This kind of studies can also help to understand the metabolic rewiring to reveal the pathways that are most susceptible to genetic modification, which could in turn facilitate the design of more efficient strategies to engineer E. coli strains.
Methods
Bacterial strains and chemicals
The strain BW25113 was used as the wild type strain in this work. Isogenic single-gene knock out derivatives [48] were obtained from the National Bioresource Project, Keio Collection (NIG, Japan) and from the Coli Genetic Stock Center (CGSC) (Yale University, USA) [41] and they are listed in Additional file 2: Table S1.
Molecular and functional information for the metabolic pathways used in this work was compiled from the Kyoto Encyclopaedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) [49] and EcoCyc (http://ecocyc.org/) [50].
Kanamycin (Kan) was purchased from Gibco™ (Invitrogen, UK) and was used for pre-culturing the isogenic knockouts with chromosomal kan resistance markers (KanR) at a concentration of 50 µg mL−1. The chemicals used for the culture media were as follows: peptone, yeast extracts, agar–agar were obtained from Panreac (PANREAC QUIMICA S.A., Spain) and KH2PO4, Na2HPO4 (extra pure), Na2SO4, NaCl, MgSO∙47H2O and glycerol (food grade 99% extra pure) were obtained from Scharlau (Scharlab S.L., Spain).
Culture conditions
Escherichia coli knock out strains listed in Additional file 2: Table S1 were initially streaked from −80°C glycerol stocks on LB agar plates containing Kan. Knock out mutations were checked by PCR [50]. Cultures for all of the experiments were incubated in an orbital incubator shaker at 200 rpm and 37°C.
For the screening of 150 single mutant strains, a fresh single colony was inoculated in 2 mL Luria–Bertani (LB) medium supplemented with Kan and the colony was cultured overnight. This aerobic pre-culture was used to inoculate the bacteria under microaerobic conditions in LB-glycerol medium [11] in 50 mL Falcom tubes (VWR International Eurolab S.L., Spain). These cells were centrifuged at 4,900×g for 15 min at 4°C (Sigma 2K15, Laborzentrifugen GmbH, Germany). Inside an anaerobic glove box, previously purged with Ar to diminish the oxygen level to below 1%, the pellet obtained was suspended in approximately 40 mL glycerol-based medium [11], (KH2PO4, 7.19 g L−1; Na2HPO4, 1.98 g L−1; Na2SO4, 0.0806 g L−1; NaCl, 0.0152 g L−1; MgSO∙47H2O, 0.031 g L−1; glycerol 10 g L−1 (109 mM) and peptone 4.25 g L−1, pH 6.25) previously sparged with Ar for 5 min, in order to obtain an OD600nm of 0.83 ± 0.025. These cells were poured into 12 mL crimp-top vials and sealed with a butyl rubber septum and aluminium caps. Both LB-glycerol and glycerol-based medium were previously sparged with argon (Ar) gas (99.9%) for 5 min to ensure that they were completely deprived of O2. The wild type and the gnd and the multiple mutant strains were also assayed in the experimental conditions described previously except for the glycerol-based medium pH, which was adjusted at pH 7.5 by using the following salt buffer concentrations: KH2PO4, 1.78 g L−1 and Na2HPO4, 7.65 g L−1. Triplicates of each E. coli strains were incubated for 22 and 46 h.
Construction of knock out strains
The multiple mutants were constructed using the ldhA::kan single mutant as the parental strain following the homologous recombination method described by Datsenko and Wanner [51]. Firstly, the Kan resistance marker inserted in the ldhA mutant strain (ΔldhA::kan) was removed after transformation with the pCP20 plasmid. Clones were selected by replica plating in LB agar plates supplemented with Cm or Kan. For plasmid curing, several clones were randomly selected; replica plated in LB agar plates with no antibiotic and incubated at 42°C. For the multiple strain constructions, gnd, frdBC and tdcE genes, pairs of primers were designed in order to have short 5′ (H1) or 3′ (H2) homology sequences of the target genes (in capital letters) that were flanking P4 or P1 priming sequences of the pKD13 vector (in lower case letters) (Additional file 8: Table S2). Using these primers and the pKD13 vector as the template, PCR products were performed containing the KanR gene and flanked by FLP recognition targets (FRTs). PCR was carried out with Velocity™ DNA polymerase (Bioline Reagents Ltd., London, UK). The PCR-generated products were transformed by electroporation in the strains previously transformed with the pKD46 plasmid. The transformants were then selected in LB agar plates supplemented with Kan. Gene disruptions of the generated mutant strains; were confirmed by PCR amplification by using external and internal primers (Additional file 8: Table S2).
All DNA manipulations were performed according to standard methods [52, 53], plasmid isolation was achieved using the NucleoSpin® Plasmid Kit (Macherey–Nagel, Düren, Germany GmbH & Co.) and PCR Clean-up was performed with the QIAquick PCR Purification Kit (QUIAGEN, Hilden, Germany).
Analytical techniques and methods
The volume of gas generated (H2 and CO2) in the headspace was measured by inserting a needle, which was connected to a water column manometer, into the rubber septum. Hydrogen quantification in the headspace was measured by injecting 100 µL aliquots into a Bruker 450-Gas Chromatograph (GC) equipped with a Poraplot Q Plot FS 25 × 53 column and a thermal conductivity detector (TCD) (Bruker Daltonik GmbH, Fahrenheitstr, Germany). The injector and detector were maintained at 250 and 150°C, respectively and the Ar carrier gas flow rate was maintained at 10 mL min−1.
Cell growth was estimated by measuring OD600 nm (1 OD = 0.31 g of cell dry weight (CDW)/L) according to standard procedures [54] on a Spectroquant® Pharo 100 spectrophotometer (© Merck KGaA, Darmstadt, Germany).
Glycerol, ethanol, succinate and formate efflux, were measured from the supernatant of the samples, filtered through 0.22 µm nylon filters, and quantified by HPLC as previously described [11].
Calculation of parameters and statistical analysis
The raw data for H2 (%), concentration of ethanol and glycerol consumed (g L−1) and the volume of gas generated (mL) were analysed and were used to calculate the following parameters for the wild type and mutant strains: specific hydrogen (YH2/X) and ethanol productions (YE/X), specific glycerol consumption (YG/X) values referred to the biomass (X) (g of CDW). For the multiple mutant, gnd and tdcE single mutants and wild type strains, succinate (YS/X) and formate efflux (YF/X) were also calculated and referred to the biomass.
For each parameter the average (m), standard deviation (SD) and the coefficient of variation (CV) were calculated using at least three biological replicates for the mutant strains and at least 75 replicates for the wild type strain. For the experiments with the pre-selected mutant, the statistical analysis for each mutant parameter was considered to be significantly different based on the non-parametric contrast of the Mann–Whitney U test. The P < 0.01 for YE/X and <0.05 for YH2/X, CDW, and YG/X parameters were used. The statistically significant values of the YH2/X, YE/X, YG/X, and µ parameters (Y rel.) were used to relativize the mutant values with the wild type ones (Ymut–Ywt/Ywt) (wild type denoted as 0). The molar yields for H2 and ethanol (mmol product/mmol glycerol consumed) were also calculated in the multiple mutant, gnd and tdcE single mutants, and wild type strains.
For mutant selection on the basis of the best parameter (YH2/X, YG/X and YE/X results) obtained at 22 and/or 46 h, the distribution of continuous variables was evaluated by the Shapiro–Wilk’s normality test and Levene’s test for homogeneity of variances was employed to inform the choice of the appropriate statistical test. As conditions for the application of parametric tests, Student’s t test was used to evaluate the statistical significance of differences the parameters between the groups. IBM® SPSS® Statistics 20 software was used for statistical analysis.
Gene Ontology (GO) database information on gene expression was used to provide additional information about the selected mutant phenotypes (Table 1). GO is a database of standardized biological terms used to annotate gene products and it comprises several thousand terms divided into three branches: molecular function, biological process and cellular component. This analysis of putative biological processes was applied to the selected mutants and the assignment of a biological process was restricted to a P < 0.001 in the Fisher’s test was used in this database.
Authors’ contributions
AV carried out the experimental procedure and the data analysis. AV, JB, DC and GC conceived of the study and participated in its design. JB and AV drafted the manuscript and GC and DC reviewed it. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the Consejería de Innovación, Ciencia y Empresa de la Junta de Andalucía through the Project for Excellence P09-TEP-4830 (2009) co-funded by FEDER Programme 2007-2013 and the PAIDI groups CTS-569 and TEP-105.
Compliance with ethical guidelines
Competing interest The authors declare that they have no competing interests.
Abbreviations
- GLDA
glycerol dehydrogenase
- PflB
pyruvate formate lyase
- GldABC
glyceraldehyde 3P dehydrogenase
- DhaKL
dihydroxyacetone kinase
- FdhF
formate dehydrogenase-H
- FhlA
formate hydrogenolyase transcriptional factor
- PPP
pentose phosphate pathway
- TCA
tricarboxilyc acid cycle
- GA3P
glyceraldehyde 3-phosphate
- PEP
phosphoenolpyruvate
- GO
gene ontology
- QC
quality control
- µ
growth rate
Additional files
Figure S1. In the screening carried out in this work, the culture conditions were first normalized and the procedures for the measurement of the ethanol and hydrogen production and the glycerol consumption were established by a gradual adaptation of the bacteria to anaerobic growth and the use of glycerol as the carbon source. This adaptation process is summarized in four steps; the overnight aerobic pre-culture grown in LB medium (A), 1% of microaerobic inoculum (E) cultured in LB-glycerol containing a negligible oxygen concentration culture up to 4 h (B); the anaerobic inoculum, in which the LB-glycerol medium is removed and replaced by the glycerol-based medium (C); and the anaerobic mini-reactors (D); with 60% headspace was chosen for higher hydrogen production and two time points at 22 and 46 h post-inoculum due to were representative of the LP and SP for the analysis of the target products (F).
Table S1. List of E. coli knock out mutant strains purchased from Keio and Yale collections.
Table S3. E. coli mutant strains with significantly higher values of specific hydrogen production (YH2/X) in blue, specific ethanol production (YE/X) in green and specific glycerol consumption (YG/X) in orange with respect to the wild type values. Intensity of colours indicates higher relative values. The 50th percentile of the values are: 14% and 13% for the YH2/X at 22 and 46 h, respectively, 47% and 33% for the YE/X at 22 and 46 h respectively, and 32% and 20% for the YG/X at 22 and 46 h respectively. The mutant strains selected according to the 50th percentile are marked in bold
Figure S2. Growth rates (μ) calculated for the mutants and wild type strain (in frame) ordered from top to bottom in ascendant order.
Table S4. Relative hydrogen and ethanol production in E. coli mutant strains using different carbon sources and pH.
Figure S3. Scatter plots of mean and SD of specific hydrogen production, YH2/X (A); specific ethanol production, YE/X (B); and specific glycerol consumption, YG/X (C) of wild type strain with culture medium at pH 6.25 (filled inverted triangle) and pH 7.5 (open circle).
Figure S4. Cell dry weight (CDW) curves in g/L of the multiple mutants: ldhAgnd::kan (M2) (filled square); ldhAgndfrdBC::kan (M4) (filled diamond); ldhAgndfrdBCtdcE::kan (M5) (filled inverted triangle); the single mutants: gnd (open circle) and tdcE (open square) and the wild type strain (open diamond).
Table S2. The primers from 1 to 6 were used to obtain multiple mutants; ldhAgnd:kan (M2), ldhAgndfrdBC::kan (M4) and ldhAgndfrdBCtdcE::kan (M5). Capital letters indicate the homologous sequences of the target genes and in lower case letters indicate the priming sequences of the pKD13 vector. In the names of the primers, H2P1 indicates the forward primers and H1P4 the reverse primers. Primers from 7 to 15 were used for PCR verification of the deletion of the targeted genes.
Contributor Information
Antonio Valle, Email: antonio.valle@uca.es.
Gema Cabrera, Email: gema.cabrera@uca.es.
Domingo Cantero, Email: domingo.cantero@uca.es.
Jorge Bolivar, Email: jorge.bolivar@uca.es.
References
- 1.Montzka SA, Dlugokencky EJ, Butler JH. Non-CO2 greenhouse gases and climate change. Nature. 2011;476(7358):43–50. doi: 10.1038/nature10322. [DOI] [PubMed] [Google Scholar]
- 2.Dharmadi Y, Murarka A, Gonzalez R. Anaerobic fermentation of glycerol by Escherichia coli: a new platform for metabolic engineering. Biotechnol Bioeng. 2006;94(5):821–829. doi: 10.1002/bit.21025. [DOI] [PubMed] [Google Scholar]
- 3.Hansen AC, Zhang Q, Lyne PWL. Ethanol–diesel fuel blends—a review. Bioresour Technol. 2005;96(3):277–285. doi: 10.1016/j.biortech.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Das D, Veziroglu TN. Advances in biological hydrogen production processes. Int J Hydrog Energ. 2008;33(21):6046–6057. doi: 10.1016/j.ijhydene.2008.07.098. [DOI] [Google Scholar]
- 5.Cai G, Jin B, Monis P, Saint C. Metabolic flux network and analysis of fermentative hydrogen production. Biotechnol Adv. 2011;29(4):375–387. doi: 10.1016/j.biotechadv.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Johnson DT, Taconi KA. The glycerin glut: options for the value-added conversion of crude glycerol resulting from biodiesel production. Environ Prog. 2007;26(4):338–348. doi: 10.1002/ep.10225. [DOI] [Google Scholar]
- 7.Hu H, Wood TK. An evolved Escherichia coli strain for producing hydrogen and ethanol from glycerol. Biochem Biophys Res Commun. 2010;391(1):1033–1038. doi: 10.1016/j.bbrc.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 8.Ito T, Nakashimada Y, Senba K, Matsui T, Nishio N. Hydrogen and ethanol production from glycerol-containing wastes discharged after biodiesel manufacturing process. J Biosci Bioeng. 2005;100(3):260–265. doi: 10.1263/jbb.100.260. [DOI] [PubMed] [Google Scholar]
- 9.Dasilva G, Mack M, Contiero J. Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol Adv. 2009;27(1):30–39. doi: 10.1016/j.biotechadv.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 10.Yuwa-amornpitak T. Bio-hydrogen production from biodiesel glycerol waste from used oil by bacterium isolated from waste water sludge. J Environ Sci Technol. 2012;5(5):373–380. doi: 10.3923/jest.2012.373.380. [DOI] [Google Scholar]
- 11.Cofré O, Ramírez M, Gómez JM, Cantero D. Optimization of culture media for ethanol production from glycerol by Escherichia coli. Biomass Bioenergy. 2012;37:275–281. doi: 10.1016/j.biombioe.2011.12.002. [DOI] [Google Scholar]
- 12.Durnin G, Clomburg J, Yeates Z, Alvarez PJJ, Zygourakis K, Campbell P, et al. Understanding and harnessing the microaerobic metabolism of glycerol in Escherichia coli. Biotechnol Bioeng. 2009;103(1):148–161. doi: 10.1002/bit.22246. [DOI] [PubMed] [Google Scholar]
- 13.Yazdani SS, Gonzalez R. Engineering Escherichia coli for the efficient conversion of glycerol to ethanol and co-products. Metab Eng. 2008;10(6):340–351. doi: 10.1016/j.ymben.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Murarka A, Dharmadi Y, Yazdani SS, Gonzalez R. Fermentative utilization of glycerol by Escherichia coli and its implications for the production of fuels and chemicals. Appl Environ Microbiol. 2008;74(4):1124–1135. doi: 10.1128/AEM.02192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trchounian K, Trchounian A. Escherichia coli hydrogenase 4 (hyf) and hydrogenase 2 (hyb) contribution in H2 production during mixed carbon (glucose and glycerol) fermentation at pH 7.5 and pH 5.5. Int J Hydrog Energy. 2013;38(10):3921–3929. doi: 10.1016/j.ijhydene.2013.01.138. [DOI] [Google Scholar]
- 16.Lee S, Kim S, Kang S, Han S, Park C, Kim S. Effect of crude glycerol-derived inhibitors on ethanol production by Enterobacter aerogenes. Bioprocess Biosyst Eng. 2012;35(1):85–92. doi: 10.1007/s00449-011-0607-y. [DOI] [PubMed] [Google Scholar]
- 17.Kim JH, Choi KH, Choi YS. Hydrogen generation from solid NaBH4 with catalytic solution for planar air-breathing proton exchange membrane fuel cells. Int J Hydrog Energy. 2010;35(9):4015–4019. doi: 10.1016/j.ijhydene.2010.02.021. [DOI] [Google Scholar]
- 18.Panagiotopoulos IA, Bakker RR, Budde MAW, de Vrije T, Claassen PAM, Koukios EG. Fermentative hydrogen production from pretreated biomass: a comparative study. Bioresour Technol. 2009;100(24):6331–6338. doi: 10.1016/j.biortech.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Maeda T, Sanchez-Torres V, Wood TK. Metabolic engineering to enhance bacterial hydrogen production. Microb Biotechnol. 2008;1(1):30–39. doi: 10.1111/j.1751-7915.2007.00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valle A, Le Borgne S, Bolivar J, Cabrera G, Cantero D. Study of the role played by NfsA, NfsB nitroreductase and NemA flavin reductase from Escherichia coli in the conversion of ethyl 2-(2′-nitrophenoxy)acetate to 4-hydroxy-(2H)-1,4-benzoxazin-3(4H)-one (D-DIBOA), a benzohydroxamic acid with interesting biological properties. Appl Microbiol Biotechnol. 2012;94(1):163–171. doi: 10.1007/s00253-011-3787-0. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Zhou L, Tian K, Kumar A, Singh S, Prior BA, et al. Metabolic engineering of Escherichia coli: a sustainable industrial platform for bio-based chemical production. Biotechnol Adv. 2013;31(8):1200–1223. doi: 10.1016/j.biotechadv.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez R, Murarka A, Dharmadi Y, Yazdani SS. A new model for the anaerobic fermentation of glycerol in enteric bacteria: trunk and auxiliary pathways in Escherichia coli. Metab Eng. 2008;10(5):234–245. doi: 10.1016/j.ymben.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Tran KT, Maeda T, Wood TK. Metabolic engineering of Escherichia coli to enhance hydrogen production from glycerol. Appl Microbiol Biotechnol. 2014;98(10):4757–4770. doi: 10.1007/s00253-014-5600-3. [DOI] [PubMed] [Google Scholar]
- 24.Maeda T, Sanchez-Torres V, Wood TK. Hydrogen production by recombinant Escherichia coli strains. Microb Biotechnol. 2012;5(2):214–225. doi: 10.1111/j.1751-7915.2011.00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trchounian K. Transcriptional control of hydrogen production during mixed carbon fermentation by hydrogenases 4 (hyf) and 3 (hyc) in Escherichia coli. Gene. 2012;506(1):156–160. doi: 10.1016/j.gene.2012.06.084. [DOI] [PubMed] [Google Scholar]
- 26.Trchounian K, Sanchez-Torres V, Wood TK, Trchounian A. Escherichia coli hydrogenase activity and H2 production under glycerol fermentation at a low pH. Int J Hydrog Energy. 2011;36(7):4323–4331. doi: 10.1016/j.ijhydene.2010.12.128. [DOI] [Google Scholar]
- 27.Trchounian K, Poladyan A, Trchounian A. Relation of potassium uptake to proton transport and activity of hydrogenases in Escherichia coli grown at low pH. Biol Membr. 2009;26(2):111–118. [Google Scholar]
- 28.Cintolesi A, Clomburg JM, Rigou V, Zygourakis K, Gonzalez R. Quantitative analysis of the fermentative metabolism of glycerol in Escherichia coli. Biotechnol Bioeng. 2012;109(1):187–198. doi: 10.1002/bit.23309. [DOI] [PubMed] [Google Scholar]
- 29.Khanna S, Goyal A, Moholkar VS. Microbial conversion of glycerol: present status and future prospects. Crit Rev Biotechnol. 2012;32(3):235–262. doi: 10.3109/07388551.2011.604839. [DOI] [PubMed] [Google Scholar]
- 30.Trchounian K, Soboh B, Sawers RG, Trchounian A. Contribution of hydrogenase 2 to stationary phase H2 production by Escherichia coli during fermentation of glycerol. Cell Biochem Biophys. 2013;66(1):103–108. doi: 10.1007/s12013-012-9458-7. [DOI] [PubMed] [Google Scholar]
- 31.Trchounian K, Trchounian A. Different role of focA and focB encoding formate channels for hydrogen production by Escherichia coli during glucose or glycerol fermentation. Int J Hydrog Energy. 2014;39(36):20987–20991. doi: 10.1016/j.ijhydene.2014.10.074. [DOI] [Google Scholar]
- 32.Gonzalez R, Campbell P, Wong M. Production of ethanol from thin stillage by metabolically engineered Escherichia coli. Biotechnol Lett. 2010;32(3):405–411. doi: 10.1007/s10529-009-0159-2. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Torres V, Mohd Yusoff MZ, Nakano C, Maeda T, Ogawa HI, Wood TK. Influence of Escherichia coli hydrogenases on hydrogen fermentation from glycerol. Int J Hydrog Energy. 2013;38(10):3905–3912. doi: 10.1016/j.ijhydene.2013.01.031. [DOI] [Google Scholar]
- 34.Trchounian K, Trchounian A. Escherichia coli multiple [Ni–Fe]-hydrogenases are sensitive to osmotic stress during glycerol fermentation but at different pHs. FEBS Lett. 2013;587(21):3562–3566. doi: 10.1016/j.febslet.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu K. 2.30—metabolic regulation analysis and metabolic engineering. In: Moo-Young M, editor. Comprehensive biotechnology. 2. Burlington: Academic Press; 2011. pp. 407–420. [Google Scholar]
- 36.Garrett RH, Grisham CM (2013) Metabolism: an overview. In: Biochemistry, 5th edn. Brooks/Cole, Cengage Learning, pp 551–573
- 37.Maeda T, Sanchez-Torres V, Wood TK. Escherichia coli hydrogenase 3 is a reversible enzyme possessing hydrogen uptake and synthesis activities. Appl Microbiol Biotechnol. 2007;76(5):1035–1042. doi: 10.1007/s00253-007-1086-6. [DOI] [PubMed] [Google Scholar]
- 38.Agapakis CM, Ducat DC, Boyle PM, Wintermute EH, Way JC, Silver PA. Insulation of a synthetic hydrogen metabolism circuit in bacteria. J Biol Eng. 2010;4(1):3. doi: 10.1186/1754-1611-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikel PI, Ramirez MC, Pettinari MJ, Mendez BS, Galvagno MA. Ethanol synthesis from glycerol by Escherichia coli redox mutants expressing adhE from Leuconostoc mesenteroides. J Appl Microbiol. 2010;109(2):492–504. doi: 10.1111/j.1365-2672.2010.04668.x. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida A, Nishimura T, Kawaguchi H, Inui M, Yukawa H. Enhanced hydrogen production from glucose using ldh- and frd-inactivated Escherichia coli strains. Appl Microbiol Biotechnol. 2006;73(1):67–72. doi: 10.1007/s00253-006-0456-9. [DOI] [PubMed] [Google Scholar]
- 41.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, Touda Y, et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol Syst Biol. 2009;5:335. doi: 10.1038/msb.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yup LS. Systems biology and biotechnology of Escherichia coli. The Netherlands: Springer; 2009. pp. 6–7. [Google Scholar]
- 44.Peekhaus N, Conway T. What’s for dinner? Entner–Doudoroff metabolism in Escherichia coli. J Bacteriol. 1998;180(14):3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hesslinger C, Fairhurst SA, Sawers G. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades l-threonine to propionate. Mol Microbiol. 1998;27(2):477–492. doi: 10.1046/j.1365-2958.1998.00696.x. [DOI] [PubMed] [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, Botstein D, Butler HJ, Cherry M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–59. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maeda T, Sanchez-Torres V, Wood TK. Enhanced hydrogen production from glucose by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol. 2007;77(4):879–890. doi: 10.1007/s00253-007-1217-0. [DOI] [PubMed] [Google Scholar]
- 48.Trchounian A, Gary Sawers R. Novel insights into the bioenergetics of mixed-acid fermentation: can hydrogen and proton cycles combine to help maintain a proton motive force? IUBMB Life. 2014;66(1):1–7. doi: 10.1002/iub.1236. [DOI] [PubMed] [Google Scholar]
- 49.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, et al. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33(Database issue):D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(6):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Miller JH. Experiments in molecular genetics. New York: Cold Spring Harbor Laboratory Cold Spring Harbor; 1972. [Google Scholar]
- 54.Greenberg AE, Clesceri LS, Eaton AD (1992) Standard methods for the examination of water and wastewater, vol 13, 18th edn. APHA, Washington, p 936


