Summary
Cell division in Chlamydiae is poorly understood as apparent homologs to most conserved bacterial cell division proteins are lacking and presence of elongation (rod shape) associated proteins indicate non-canonical mechanisms may be employed. The rod-shape determining protein MreB has been proposed as playing a unique role in chlamydial cell division. In other organisms, MreB is part of an elongation complex that requires RodZ for proper function. A recent study reported that the protein encoded by ORF CT009 interacts with MreB despite low sequence similarity to RodZ. The studies herein expand on those observations through protein structure, mutagenesis, and cellular localization analyses. Structural analysis indicated that CT009 shares high level of structural similarity to RodZ, revealing the conserved orientation of two residues critical for MreB interaction. Substitutions eliminated MreB protein interaction and partial complementation provided by CT009 in RodZ deficient E. coli. Cellular localization analysis of CT009 showed uniform membrane staining in Chlamydia. This was in contrast to the localization of MreB, which was restricted to predicted septal planes. MreB localization to septal planes provides direct experimental observation for the role of MreB in cell division and supports the hypothesis that it serves as a functional replacement for FtsZ in Chlamydia.
Keywords: Chlamydia, RodZ, Morphogenesis, Structure
Introduction
Within the phylum of Chlamydiae, there are several species of bacteria that have significant effects on public health, causing sexually transmitted infections, blinding trachoma, and pneumonia (Corsaro & Venditti, 2004). Notwithstanding differences in the diseases they cause, all chlamydial species share a similar biphasic developmental cycle. This process begins with infection of a host cell by a metabolically inactive Elementary Body (EB). Once within the host cell, the EB differentiates into the non-infectious but metabolically active Reticulate Body (RB), which maintains itself in a parasitophorous vacuole termed an inclusion as it grows and replicates. After numerous rounds of replication, RBs begin to asynchronously convert back to EBs and are released by either host cell lysis or extrusion (Hybiske & Stephens, 2007). Despite its importance and conservation among chlamydial species, there are many aspects of this complex developmental cycle that are poorly understood, including the fundamental process of RB cell division and cell shape maintenance.
In the vast majority of bacterial species, cell division is mediated primarily by formation of the Z ring, which directs the location of the division septum (Erickson et al., 2010). The Z ring is composed of protein FtsZ which polymerizes in a GTP dependent manner and serves as a scaffold for the recruitment of all other proteins involved in septation and cytokinesis (Dajkovic & Lutkenhaus, 2006, Adams & Errington, 2009). Chlamydiae are unusual among bacteria in that no homolog to FtsZ is apparent in any Chlamydiae genome. In addition, many other well-conserved proteins that are involved in Z-ring assembly, such as FtsA and ZipA, also lack apparent homologs in chlamydial genomes. Together, this suggests that either homologs to these proteins are present in chlamydial species but are not apparent by sequence similarity or that chlamydial cell division is mediated by a novel mechanism. Along these lines, it was recently proposed that, in Chlamydia, the protein MreB functions as a central coordinator of cell division, in replacement of FtsZ (Ouellette et al., 2012). MreB is a bacterial actin homolog and is predominately associated with peptidoglycan synthesis and the regulation of non-cocci cell elongation. Chlamydia are an exception to this trend as they have coccoid morphology and yet encode MreB (CT709) (Stephens et al., 1998). Moreover, Chlamydia encode several rod-shape determining proteins, including MrdA/Pbp2 (CT682), which is involved in elongation peptidoglycan synthesis and MrdB/RodA (CT726), a predicted integral membrane lipid II flippase. These are in addition to encoding the functionally analogous but divisome-associated FtsI/Pbp3 (CT270) and FtsW (CT760). Of particular note, it was recently shown that peptidoglycan in Chlamydia can be detected and localization appeared to be restricted to likely septal planes of cell division (Liechti et al., 2014).
If MreB is utilized for cell division in Chlamydia, then it is likely that other proteins associated with MreB may also play a role in this process. In other bacteria, MreB does not function alone; rather, it is part of a large morphogenic complex and has been shown to interact with several other proteins including MreC, MreD, RodA, and RodZ (Kleinschnitz et al., 2011, Kruse et al., 2005, White et al., 2010). While RodA is predicted to be a lipid II flippase, the precise functions of MreC, MreD and RodZ are not well understood but with several other rod-shape determining proteins they are thought to play scaffolding roles in the morphogenic complex. RodZ is of particular importance because its interactions with MreB are better defined, due to a crystal structure of a protein:protein complex of these two proteins (PDB ID: 2WUS (van den Ent et al., 2010)). Importantly, while there is an annotated chlamydial RodA homolog (CT726), many of the expected MreB interaction partners are not apparent based upon sequence similarity in chlamydial species. A better understanding of the presence or absence of these MreB interaction partners in chlamydial species could provide insight into the unique cell division mechanisms employed by Chlamydia.
Open Reading Frame (ORF) CT009 is conserved in all species of Chlamydiae with sequenced genomes. Interestingly, sequence similarity searches reveals that this protein contains a predicted helix-turn-helix motif within a conserved XRE (xenobiotic response element) domain superfamily of DNA binding transcription factors. However, a recent study (Ouellette et al., 2014) using bacterial two-hybrid analysis indicated that CT009 interacts with MreB and supported a potential role as RodZ homolog. As described herein, in depth computational and experimental protein structural analyses were performed to provide more comprehensive support for CT009 as a RodZ homolog. In addition to providing structural support for CT009 as a RodZ homolog, this structural information revealed the conserved orientation of two amino acids that were shown to be critical for RodZ interaction with MreB, The functional importance of these residues were evaluated in an E. coli complementation study as well as bacterial two hybrid interaction analyses. Lastly, cellular localization of CT009 and MreB in Chlamydia was also performed to provide further support for a role in cell division.
Results
A computationally generated protein structure of the cytoplasmic domain of CT009 is most structurally similar to RodZ
Comparative sequence analyses with CT009 reveals a predicted helix-turn-helix motif within a conserved XRE (Xenobiotic Response Element)-family DNA binding domain. The Xre family (IPR001387) is expansive in regards to phylogenetic distribution and associated with many DNA regulatory functions including Lambda phage (CI/Cro) related transcription repressors (Minezaki et al., 2005) (Aggarwal et al., 1988). In contrast to many, but not all, protein sequences containing Xre DNA binding domains was the presence of a predicted carboxy-terminal transmembrane helix formed by amino acids 117-136 (Fig. 1A) in CT009, as predicted by the HMMTOP server (Tusnady & Simon, 2001).
Figure 1. Predicted domain organization of RodZ and CT009 and computational models of predicted protein structure.
A. The predicted domain architecture of RodZ from E. coli (Bendezu et al., 2009). It has an N-terminal cytoplasmic domain that contains a Cro/CI-type helix-turn-helix motif (HTH), a highly basic juxta-membrane domain (JM), a transmembrane helix (TM), a periplasmic domain rich in prolines and threonines (PT-rich) and β-strands rich domain. As predicted by HMMTOP, CT009 also possesses an N-terminal cytoplasmic domain with Cro/CI-type helix-turn-helix motif (HTH; amino acids 1-116) and a transmembrane helix (TM; amino acids 117-136). However, CT009 does not code for a basic juxta-membrane domain and its periplasmic region is only 7 amino acids long (amino acids 137-143).
B. Computational model of CT0091.116 from Chlamydia trachomatis generated by ITASSER (Roy et al., 2010) depicted in cartoon ribbon format (cyan) aligned with RodZ from T. maritima (PDB ID: 2WUS, chain R, green) with an RMSD of 0.74 Å over 63/87 Cα atoms. Conserved MreB-interacting residues, Tyr53 and Tyr57 from T. maritima RodZ and Tyr57 and Phe61 from CT009, are highlighted in purple (balls-and-sticks).
C. Computational model of CT0091.116 from C. trachomatis generated by PHYRE (Kelley & Sternberg, 2009) depicted in cartoon ribbon format (magenta) aligned with RodZ from T. maritima (PDB ID: 2WUS, chain R, green) with an RMSD of 0.31 Å over 63/87 Cα atoms. Conserved MreB-interacting residues, Tyr53 and Tyr57 from T. maritima RodZ and Tyr57 and Phe61 from CT009, are highlighted in cyan (balls-and-sticks).
In order to gain more specific information regarding the function and possible biological role of CT009, computational models were generated by I-TASSER and PHYRE. I-TASSER (Iterative Threading ASSEmbly Refinement) is a computational method that has been successful in accurately modeling protein structures (Zhang, 2014, Zhang, 2009, Roy et al., 2010). I-TASSER, as well as other protein modeling methods, is limited in the ability to accurately model membrane associated protein regions. Therefore, only the predicted amino-terminal cytoplasmic domain of CT009 was analyzed (CT0091-116). Several models were generated; the highest scored model (Fig. 1B) had a C-score (−0.92) above the recommended cutoff (C-score > −1.50) and the DALI server (Holm & Rosenstrom, 2010) was used to search for proteins with structural similarity. Unexpectedly, the top structure that aligned best with the I-TASSER model of CT0091-116 was the cytoplasmic domain of RodZ (chain R from PDB ID: 2WUS), with Z- scores of 14.5. This RodZ structure aligns well to the CT009 I-TASSER computational model with an RMSD of 0.74 Å (over 63/87 Cα atoms). The degree of structural similarity is striking considering the low level of sequence similarity between Thermotoga maritima RodZ and CT009 (BLAST E-value >10; Fig. 1B). The amino acid sequence of CT0091-116 was also submitted to PHYRE2 (Protein Homology/analogY Recognition Engine v2.0) (Kelley & Sternberg, 2009), which generated a computational model (Fig. 1C) highly similar in structure to the CT009 I-TASSER model (RMSD of 1.96 Å over 116/116 Cα atoms) as well as the previously discussed RodZ structure (Fig. 1C)., The C-terminus is predicted to adopt very different conformations within each CT0091-116 computational model (Fig. 1B and 1C), indicating this region of CT009 might be highly flexible.
RodZ is a membrane-bound cell shape determining protein, a component of the bacterial cell morphogenic complex and an interaction partner of the bacterial actin homolog MreB (van den Ent et al., 2010, Bendezu et al., 2009, Alyahya et al., 2009, Shiomi et al., 2008). In addition to predicted structural similarity within their cytoplasmic domains, CT009 and RodZ also share a similar membrane topology; a single transmembrane helix anchors both proteins in the inner membrane (Fig. 1A; (Bendezu et al., 2009)). The only available structure of RodZ (from T. maritima) is a co-crystal with MreB (PDB 2WUS; Fig. S1). In this crystal structure, several bulky aromatic residues were identified at the interface of RodZ and MreB, two of which were required for proper interaction between RodZ and MreB (van den Ent et al., 2010). These residues are found within the fourth helix of the protein and correspond to Y53 and Y57 in RodZ encoded by T. maritima (Fig. 1B and 1C). Importantly, these residues are conserved in CT009 as Y57 and F61 and are similarly positioned in helix α4 of the CT0091-116 computational models CT009 (Fig. 1B, 1C, and 3C) Therefore, the overall structural similarity and the presence of conserved residues critical for function support that CT009 could function as a RodZ homolog within Chlamydia.
Figure 3. Crystallographic dimer of CT009 has structural similarity to RodZ from T. maritima, including conservation of key MreB-interacting residues.
A. Subdomain swap through crystallographic dimer of a CT009 (as predicted by PISA (Rocchia et al., 2002)) depicted in cartoon ribbon format and colored as in Fig. 2B (residues 1-54 and 55-103 are contributed by each protomer). Only a single “monomer” of the subdomain swap dimer is shown for clarity. Putative MreB-interacting residues (Tyr57 and Phe61) are highlighted in purple (balls-and-sticks).
B. Structural superposition of CT009 subdomain swap monomer from panel A (colored orange and cyan) with RodZ from T. maritima (PDB ID: 2WUS, chain R, lime) with an RMSD of 1.02 Å over 66/87 Cα atoms. Conserved MreB-interacting residues from both proteins are highlighted in purple (ball-and-sticks).
C. Sequence alignment of CT009 and RodZ homologs from E. coli and T. maritima highlight limited overall similarity. Alignment was generated using ClustalW (Thompson et al., 1994) and rendered with ESPRIPT (Gouet et al., 1999). Numbers above the sequences correspond to C. trachomatis CT009. Residues are colored according to conservation (cyan = identical and purple = similar) as judged by the BLOSUM62 matrix. Red stars below the sequences correspond to conserved MreB-interacting amino acid side chains.
D. Quantified β-galactosidase activity reflecting CT009 specific protein interactions with MreB and self. Full length CT009, mutant CT009 (Y57A/F61A substitution), the predicted cytoplasmic domain of CT009 (residues 1-116), and chlamydial MreB interactions were evaluated within bacterial adenylate cyclase two hybrid system as described by Kaimova et al. (Karimova et al., 2005, Karimova et al., 1998). MreB interactive clones have N-terminal T25 fusion protein (pKT25) tested with N-terminal T18 fusion (pUT18C) with CT009. CT009 self-interactions also used N-terminal T25 fusion protein (pKT25) with N-terminal T18 fusion protein (pUT18C). As a positive and negative control, self-interaction of the leucine zipper of the GCN4 protein and empty vectors was tested. Expression of the β-galactosidase reporter gene by reconstituted adenylate cyclase was assessed using a β-galactosidase assay and activity reported in Miller units. Error bars represent standard deviations calculated from assays performed in triplicate and asterisks indicated P values < 0.001 (Student's t-test) relative to vector control.
The crystal structure of CT009 shows structural similarities to RodZ
To validate the computational structure prediction of CT009, recombinant CT009 (cytoplasmic domain, residues 1-116) was expressed, purified, and screened for crystallization. Tetragonal crystals of CT0091-116 were obtained which yielded a structure that was refined using X-ray diffraction data to 1.25 Å resolution. The model could be traced to the electron density maps from Met 1 to Ser 103 including two N-terminal residues (GH) from cloning. The C-terminal residues Met 104 to Ser 116 were disordered and could not be modeled. As previously discussed, homology models of CT0091-116 were generated using several structural prediction servers (I-TASSER, PHYRE and MODELLER (Roy et al., 2010, Eswar et al., 2006, Kelley & Sternberg, 2009)). Intriguingly, only the structure generated by MODELLER produced a successful molecular replacement solution, with a single polypeptide in the asymmetric unit (Fig. 2A). Curiously, the protein appears to have crystallized in a flat or “open” conformation. The physiological relevance of this form is dubious; proteins in solution typically adopt a more compact fold, minimizing the surface area exposed to solvent. However, crystal packing analysis revealed that a symmetry mate related by a crystallographic 2-fold operator [y, x, -z], places the N-terminal (Met 1 to Leu 52) and C-terminal (Ala 59 to Ser 103) regions in close proximity (Fig. 2B). Together, these crystallographic dimer partners form a “folded” conformation where subdomains from each protomer are paired and adopt a conformation very similar to a monomeric domain (compare Fig. 2B, 3A, and 3B). This phenomenon, in which two polypeptide chains exchange subdomains, is a less common occurrence and primarily restricted to structural studies that utilize x-ray crystallography (Bennett et al., 1994). Swapping of subdomains is often an artifact of crystal packing but in certain cases the subdomain-swapped form exists in solution as well (Barrientos et al., 2002). The electron density of the “hinge” (Ile53 to Ala 58) that connects that N and C-terminal regions (Fig. 2C) demonstrates that the asymmetric unit was modeled correctly. In other words, the hinge bridges the N-terminal region of the asymmetric unit with the C-terminal region of the symmetry mate thus resulting in an asymmetric unit that is “flat”. However, a model of CT009 that adopts an expected compact fold can be constructed by considering the interactions between the N-terminal residues from amino acids 1-54 of the asymmetric unit with C-terminal region of the molecule related by crystallographic symmetry (amino acids 55-103 of its symmetry mate) (Fig. 3A). Importantly, the conserved MreB-interacting residues (Y57 and F61) are apparent in the crystal structure and face outward in a position capable of interaction with MreB (Fig. 3B and Fig. S1; highlighted in magenta).
Figure 2. 1.25 Å crystal Structure of CT009 from C. trachomatis adopts an “open” fold, resulting in a subdomain swap dimer.
A. Crystal structure of C. trachomatis CT009 (residues 1 to 103) shown in cartoon ribbon format (colored blue (N-terminus) to red (C-terminus)). A single copy of CT009 is found within the asymmetric unit. Crystal structure is rotated 90° about the horizontal axis on the right.
B. Crystallographic dimer of CT009 mediated by “open” fold is depicted in cartoon ribbon format (colored orange and cyan, respectively) and is generated by applying the following symmetry operator: [y,x,-z].
C. 2Fo-Fc electron density map (blue mesh, contoured at 3.0σ) of the CT009 “hinge” region where subdomain swap dimerization occurs (residues Ile53 – Ala58). Coloring is the same as panel B.
In addition to the subdomain-swapped crystallographic dimer, recurring positive difference (Fo-Fc) electron density at the C terminal helix, following refinement, indicated that this portion of the helix adopted two distinct conformations and was modeled as such. Structural superposition of RodZ with the predicted CT009 monomer resulted in an RMSD of 1.02 Å over 66/87 Cα atoms (Fig. 3B). The major difference between the structure of CT009 and the structure of RodZ is in the orientation of their C- terminal helices. In RodZ, helix α7 (amino acids 69-86) is orientated parallel to helix α1 thus adopting an orientation that is perpendicular to the MreB interface. However, in the structure of CT009, this helix is kinked sharply and results in two distinct helices, one short (amino acids 73-78) and the other longer (amino acids 81-103) that positions helix α7 nearly perpendicular to helix α1. The reason for this difference is not obvious, although it is possible that the final helix in CT009 is kinked so that it can make better crystal contacts with its symmetry mates. Importantly, this helix is not directly involved in contacts with MreB and the MreB-interacting surface appears to be structurally conserved in CT009. Therefore, despite the unusual crystal packing of the CT009 subdomains, the crystal structure of the cytoplasmic domain of CT009 supports the hypothesis that it is a structural homolog of RodZ and the conserved orientation of two MreB interacting residues.
CT009 interacts with MreB
As discussed previously, RodZ plays a role in rod shape maintenance in other bacteria as part of a morphogenic complex with the bacterial actin homolog MreB. Although CT009 may be a functional homolog of RodZ, this does not explain what role this protein has in Chlamydiae, a coccoid shaped bacteria. It was recently proposed that cell shape-determining proteins in Chlamydiae, including MreB, are involved in chlamydial cell division (Brown & Rockey, 2000, Ouellette et al., 2012). RodZ is thought to play a critical role in tethering MreB to the membrane through conserved amino acids at a well-defined interaction interface (van den Ent et al., 2010). Specifically, RodZ and MreB from a variety of species have been shown to interact in the context of a bacterial-two hybrid system and even co-crystallize together (Bendezu et al., 2009, Kleinschnitz et al., 2011, van den Ent et al., 2010, White et al., 2010). Given the structural similarity and conservation of these key MreB-interaction residues in CT009, we tested the hypothesis that CT009 interacts with chlamydial MreB and requires these key interactive residues using the Bacterial Adenylate Cyclase Two Hybrid (BACTH) system (Karimova et al., 2005, Karimova et al., 1998).
As it is difficult to anticipate if a certain fusion partner or orientation will effect the function of a protein, all eight different possible interactions combinations (with differing N- or C- orientations of the 25 kDa or 18 kDa adenylate cyclase as fusion proteins with CT009 or MreB) were tested to capture potential interactions. Interaction between N-terminal 25 kDa fusions to CT009 and N-terminal 18 kDa fusion to MreB proteins was detected (Fig. 3D; CT009-MreB), however, any construct in which an adenylate cyclase domain was fused to the C-terminus of full-length CT009 failed to interact with MreB. CT009 is predicted to be anchored to the inner membrane, with its N-terminus in the cytoplasm and its C-terminus in the periplasm (Fig. 1), suggesting that proper membrane localization is essential for interaction with MreB. In concert with this observation, all combinations using N-terminally tagged CT009, showed interaction between CT009 and MreB (Fig. 3D and data not shown). It is therefore likely that adenylate cyclase fusions at the carboxyl termini of CT009 either disrupts membrane anchoring of CT009 or results in the localization of the adenylate cyclase to the periplasm while cognate MreB fusion is in the cytoplasm, abrogating enzymatic restoration.
As discussed previously, RodZ from Thermatoga maritima has been shown to interact with MreB and several residues (Y53 and Y57 in T. maritima) that are critical for this process have been identified (van den Ent et al., 2010). As previously discussed, these residues are conserved in CT009 as Y57 and F61 (red stars in Fig. 3C). In order to test whether these residues are similarly required for CT009-MreB interactions, alanine substitution were introduced into CT009 (Y57A and F61A) and analyzed by BACTH. In contrast to the wild-type CT009-MreB interactions, these two alanine substitutions severely reduced protein interactions (Fig. 3D; CT009 Y57A/F61A – MreB).
In addition to interacting with MreB, RodZ has been shown to interact with itself, although the requirement of this self-interaction for function has not been explored (Bendezu et al., 2009, Kleinschnitz et al., 2011, White et al., 2010). Analysis of the soluble cytoplasmic domain of CT009 (residues 1-116) by analytical ultracentrifugation indicated that stable higher order structures were not formed (Fig. S2). In order to further assess the ability of full length CT009 to interact with itself and the possible importance of membrane localization, all eight possible combinations of full-length CT009 self-interaction (with differing orientation of the fusion proteins) were tested using BACTH Only a single combination resulted in a positive interaction (Fig. 3D; CT009 WT-Self) and, similar to the CT009-MreB interaction, required that the T25 and T18 domains were fused to CT009 at its N terminus and localized in the cytoplasm. This supports that full length CT009 interacts with itself in concert with other RodZ homologs. Similarly, the two alanine substitutions (Y57A and F61A) in CT009 had little effect on self-interactions (Fig. 3D; CT009 Y57/F61A - self), supporting that these substitutions likely have not altered the overall structure of the protein and provides validating support for their role in providing specificity to MreB interaction.
As mentioned previously, the CT009 cytoplasmic domain did not form higher order structures with itself in solution (Fig. S2). This observation strengthens the likelihood that the crystallographic CT009 dimer (Fig. 2B) is likely an artifact of crystallization and may not be physiologically relevant. To further evaluate if cytoplasmic domains interact, self-interactions with the cytoplasmic domain of CT009 were tested using BACTH (Fig. 3D, CT009 cyt-CT009 cyt). In contrast to the full-length protein, the cytoplasmic domain of CT009 did not interact with itself. Again, all possible interaction combinations were tested, but no combination lead to a positively identified interaction (data not shown). Importantly, the possibility that the self-interactions observed with full-length CT009 are a result of cytoplasmic regions adopting a domain-swap dimer conformation, similar to what was observed in the crystal structure, cannot be excluded. Interaction between the cytoplasmic domain of CT009 and MreB was also tested (Fig. 3D, CT009 cyt-MreB). No interactions between these two proteins were observed (data not shown). This suggests that the transmembrane helix and/or the short periplasmic tail may be involved in mediating or strengthening dimerization of CT009 and that dimerization and/or localization to the membrane may be essential for MreB interaction.
CT009 is able to partially complement rod shaped morphology of RodZ deficient E. coli
With the hypothesis that CT009 is a chlamydial RodZ homolog, its ability to complement the function of RodZ in the heterologous system of E. coli was tested. RodZ is involved in directing cell shape and in rodZ knockout strains of E. coli there is a well-defined phenotype: cells grow to be spherical instead of rod-shaped (Bendezu et al., 2009, Shiomi et al., 2008). rodZ knockout cells (FB60) were transformed with plasmids that either encoded E. coli RodZ or chlamydial CT009. In addition, a plasmid containing modified CT009 (CT009 Y57A/F61A) was also transformed into FB60 cells. To test for phenotype complementation in RodZ deficient cells, plasmid-containing FB60 cells were grown to exponential phase, fixed, stained with the dye malachite green and visualized by fluorescence microscopy (Fig. 4). As expected for K-12 E. coli, control MC1000 cells are rod shaped (Fig. 4). rodZ deficient E. coli cells (FB60) with an empty vector have an aberrant morphology and are more spheroid with less of a defined rod shape (Bendezu et al., 2009) (Fig. 4). When FB60 cells are complemented with a plasmid expressing E. coli RodZ, wild type phenotype is restored and cells have rod morphology (Bendezu et al., 2009) (Fig. 4). When FB60 cells are complemented with a plasmid expressing CT009, rod morphology is partially restored (Fig. 4). Although cells supplemented with CT009 are longer and more rod-like than RodZ knockout cells, they are also wider than cells complemented with RodZ. This may be due to the fact that CT009 lacks a periplasmic domain; a previous report in which RodZ knockout cells complemented with a truncated RodZ (lacking its periplasmic domain) showed complementation accompanied by widening of cells (Bendezu et al., 2009). Additionally, when FB60 cells express a CT009 that contains mutations in the two conserved residues that mediate the interacting between RodZ and MreB (Y57A/F61A), cells fail to restore wild-type phenotype (Fig. 4).
Figure 4. CT009 is able to partially complement morphology of rodZ deficient E. coli.
Representative images of E. coli cells that were fixed, stained with malachite green, and imaged by epifluorescent microscopy for analysis of cellular morphology. E. coli strains analyzed with specific genetic complementation are indicated above each panel and include wild type E. coli (MC1000), FB60 rodZ deficient E. coli (Bendezu et al., 2009) containing empty vector (pMLB1113), FB60 cells complemented with plasmid expressing E. coli RodZ (pFB290), FB60 cells complemented with plasmid expressing chlamydial CT009, and FB60 cells complemented with plasmid expressing CT009 Y57A/F61A mutant. Morphology of individual E. coli cells was quantitatively assessed by measuring major and minor axis using ImageJ. Ratio of major to minor axis were calculated for at least 100 cells per sample and then averaged. Averages and standard deviation were calculated from triplicate experiments. Asterisks indicate significant difference (p<0.05) as compared to FB60 sample.
To provide a quantitative assessment of functional complementation by CT009, the width and length (major/minor axis) of over 100 individual bacterial cells per sample were measured using ImageJ. Normal E. coli cells are about 2.5 to 3 times longer than they are wide as reflected by wild-type (MC1000) and RodZ complemented bacteria (FB60-RodZ; Fig. 4). In contrast, RodZ deficient cells (FB60) are almost equidistant at any axis. When CT009 was introduced into a RodZ deficient strain (FB60-CT009), cells were 2 times longer than wide on average supporting a partial restoration. Substitutions in two MreB interacting aromatic amino acids (Y571/F61A) of CT009 (FB60-CT009 mut) resulted in a decrease to approximately 1.5 times long as wide. Interestingly, statistical analysis of complemented CT009 support significant difference (p<0.05) from rodZ deficient (FB60) E. coli, however, CT009-FB60 complementation was not significantly different (p>0.05) from FB60-CT009 mutant. This could indicate that mutated CT009 retained interactions with some elongation proteins, other than MreB, that are involved with RodZ function and provided minimal, but more than rodZ deficient (FB60), function in E. coli elongation. Lastly, as a previous study with GFP tagged CT009 observed no functional complementation in E. coli with differential interference contrast used to view cells, this technique was also employed. This analysis (Fig. S3) revealed a very similar overall observation of partial morphologic complementation with CT009 as seen with malachite green stained cells. Together, these data support that CT009 is able to partially serve as a functional homolog to RodZ in E. coli.
CT009 and mreB transcript expression patterns
The role of RodZ, in most other bacteria, appears to be for peptidoglycan synthesis during elongation of rod shaped bacteria (Alyahya et al., 2009, Bendezu et al., 2009). This is typically in concert with MreB and other peptidoglycan machinery. Given the observations herein that CT009 interacts with MreB and is likely participating in a shared cellular role (cell division), it is expected that their expression patterns would be similar. Moreover, if the role of CT009 and MreB is exclusively for cell division, the gene expression patterns should be similar to other cell growth and replication components.
Relative gene expression analysis was performed on Chlamydia trachomatis (strain LGV) infected mouse fibroblasts throughout a 36-hour infection. RNA was isolated at 6-hour increments in order to study the transcription patterns of candidate genes throughout the developmental cycle. For LGV strains of Chlamydia trachomatis, EBs have converted into RBs within 6 hours post infection and continue to replicate throughout the developmental cycle and between 18-24 hpi, RBs begin asynchronously converting into EBs. To distinguish between transcripts associated with RB cell division/replication processes to those associated with RB to EB conversion and later developmental processes, transcript levels are normalized to a general secretory component (secY) at each time point and each gene expression levels is compared to cognate 6 hpi expression levels. As figure 5 indicates, transcription patterns of DNA replication components gyrB, dnaN, and dnaE and transcription machinery rpoA are fairly equal and constitutive throughout the developmental cycle. In contrast, transcription of the RB to EB associated gene hctB (Histone like protein B) is strongly upregulated (~256 fold) between 18-24 hpi. Interestingly, patterns of expression for CT009 and mreB are very similar exhibiting a continual step-wise increase in levels until about 30 hpi. Expression of two other cell division encoding genes (rodA and ftsK) was analyzed. RodA is typically associated with peptidoglycan cell elongation for rod shaped organisms and FtsK is a DNA translocase that is associated with cell division and chromosome segregation. Transcript expression patterns for rodA showed a minimal but apparent step-wise increase through 24 hpi whereas expression of ftsK had a two-fold increase between 12-18-24 hpi. Overall, transcript expression patterns and relative levels of CT009 and mreB are very similar, supporting a shared biological role of the encoded proteins. However, the expression patterns are not paired with RB replication associated processes like DNA and RNA polymerase components. Furthermore, expression patterns of CT009 and mreB are more similar to other cell division components. This suggests that the role of cell division components, including CT009 and MreB, may extend beyond bacterial cell replication and may include RB to EB conversion events. Importantly, MreB or CT009 were not detected in purified EB samples (data not shown) indicating that that any role beyond cell division is likely early in RB to EB transition and concluded well before final formation of EB.
Figure 5. Quantitative gene expression analysis of CT009, cell replication, and cell division encoding components.
Total RNA was isolated at 6, 12, 18, 24, 30 and 36 hours post-infection from L929 cells infected with C. trachomatis. Relative gene expression ratios for each gene was normalized to secY from matching time point and charted as log2 transformed expression ratio (compared to 6 hpi). As comparative controls for RB replication, gene expression for DNA replication, gyrB (DNA gyrase), dnaN (DNA polymerase clamp), dnaE (DNA polymerase helicase) and transcription rpoA (RNA polymerase alpha subunit) gene were analyzed. Additionally, an indicator for RB to EB conversion was also included (hctB). Error bars represent the standard deviation from triplicate experimental samples.
Cellular Localization of MreB and CT009 in Chlamydia
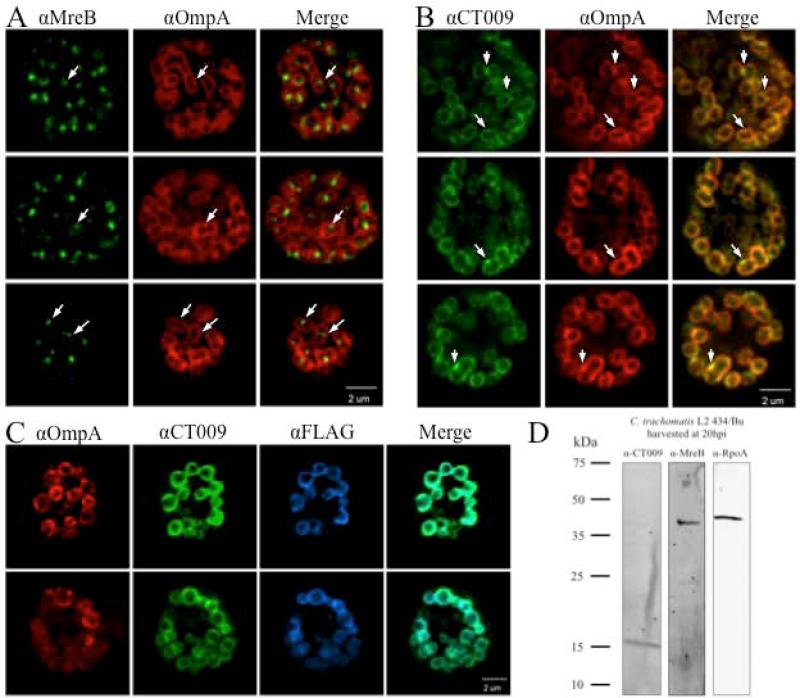
Recent studies using fluorescently labeled peptidoglycan have shown localization predominately restricted to cell division septal planes (Liechti et al., 2014). These observations, and those linking MreB and RodZ with peptidoglycan production, support the hypothesis that MreB and CT009 are localized similarly to chlamydial division septal planes. To test this hypothesis, affinity purified antibodies to CT009 and MreB were used for immunofluorescent labeling of Chlamydia infected cells. At 16 hpi, replicating RBs are easily identifiable in which division septae can be visually predicted based upon membrane invaginations between elongated dividing cells (Fig. 6A; white arrows and red OmpA labeling). MreB was distinctly localized to relatively small foci near predicted division sites or more complete localization across the division plane (Fig. 6A and Sup. Movie 1). In stark contrast to MreB localization, CT009 was almost uniformly localized to the membrane, supporting the accurate prediction of the transmembrane domain and maintenance of full-length CT009 protein. However, there were rare unique membrane localization patterns where CT009 appeared to be enriched near division sites relative to OmpA labeling (Fig. 6B; note merged image and more pronounced green labeling). While these data support the role of MreB in cell division and association with peptidoglycan, the observed unrestricted membrane localization suggests that CT009 would require other components to limit MreB localization to division site at this stage in the developmental cycle. It is possible that the uniform localization observed for CT009 is due to cross-reactivity with another Chlamydia membrane-localized protein. Immunoblot analysis of Chlamydia infected cellular lysates revealed a single band at approximately 16 kDa (Fig. 6D). In order to provide orthogonal support for the cellular localization of CT009, C. trachomatis was transformed with a plasmid expressing CT009 with FLAG epitope encoded at the N-terminus. IFA analysis of transformed Chlamydia infected cells using antibodies to the FLAG tag also showed a uniform, albeit more intense, labeled RB membrane (Fig. 6C) providing very strong evidence for the previous specificity of the antibodies and overall observation regarding CT009 localization.
Figure 6. Localization of CT009 or MreB within C. trachomatis.
L929 cells infected with C. trachomatis L2 were fixed at 16 hpi and stained with affinity-purified antibodies and images acquired by confocal microscopy with 150× objective. Two or three representative inclusions are shown for
A. Anti-MreB (green), anti-OmpA (red), and merged image
B. Anti-CT009 (green), anti-OmpA (red), and merged image White arrows are highlighting potential sites for bacterial cell division.
C. Anti-OmpA (red), anti-CT009 (green), anti-FLAG (blue), and merged image of CT009 and FLAG
D. Monospecificity of CT009 and MreB antibodies. Expression levels of CT009, MreB and RpoA from C. trachomatis L2 434/Bu were analyzed at 20 hpi. Western blot was performed with affinity purified rabbit anti-CT009, rabbit anti-MreB, or rat anti-RpoA. Calculated MW of each protein are as follows: CT009, 16.2 kDa; MreB, 39.5 kDa; and RpoA, 41.8 kDa.
Protein association of CT009 with other cell division components
Oullette et al. (Ouellette et al., 2012) demonstrated that MreB interacts with FtsK using the same bacterial two-hybrid approach described herein. FtsK is a septal protein and DNA translocase that facilitates chromosomal dimer segregation during cell division (Yu et al., 1998, Wang & Lutkenhaus, 1998) flippase and is essential for peptidoglycan synthesis typically during elongation. Additionally, RodA is an integral membrane lipid II. RodA has previously been shown to interact with RodZ in C. crescentus, E. coli, and B. subtilis (White et al., 2010). To continue the observations of proteins that interact with CT009 and extend support for a role in cell division, bacterial two-hybrid analyses were performed. As with other bacterial-two hybrid experiments described herein, all eight combinations of fusion protein and complementary transformations were tested. Two amino-terminal fusions of T25 or T18 fragments to CT009 (Fig. 7; pKT25-CT009 and pUT18C-CT009) displayed positive protein interactions with complementary carboxy fusions with RodA (pUT18-RodA and pKNT25-RodA, respectively). A carboxyl fusion of T18 fragment to CT009 also showed interaction with amino-fusion of T25 to RodA. This was unexpected as carboxyl fusions to CT009 are likely to effect membrane localization suggesting that this (membrane localization of CT009) may not be critical for interaction with RodA. FtsK also demonstrated interaction with CT009, albeit with only one combination of fusion partners (Fig. 7; pKT25-FtsK and pUT18C-CT009). The RodA interactions provide further support that CT009 is functioning as a RodZ homolog and interactions with FtsK strengthen the association of CT009 with cell division processes in Chlamydia.
Figure 7. Qualitative β-galactosidase activity reflecting CT009 protein interactions with FtsK and RodA.
Full length CT009, FtsK, and RodA interactions were evaluated within the bacterial adenylate cyclase two hybrid system as described by Kaimova et al. (Karimova et al., 2005, Karimova et al., 1998). As a positive and negative control, self-interaction of the leucine zipper of the GCN4 protein and empty vectors were tested, respectively. Amino (N) or carboxyl (C) terminal fusions of the T25 or T18 fragment to CT009 (indicated on the top of each box of spotted bacteria) were evaluated for interaction against C or N terminal fusion of T18 or T25 fragment to RodA or FtsK (indicated to the side of each set of boxes of spotted bacteria). Expression of the β-galactosidase reporter gene by reconstituted adenylate cyclase was assessed using a enzymatic cleavage of X-galactoside in M63 agar plates after 48 hours at 30°C. Results are representative of observations from duplicate experiments.
Computational analyses identify potential RodZ homologs in other bacterial species
At the time of its original annotation, CT009 contained a predicted helix-turn-helix motif as well as a predicted XRE family DNA binding domain. For this reason, it was annotated as a transcription factor. However, our structural and functional studies on this protein now show that it has properties of a RodZ homolog, interacting with MreB and is likely to be involved in chlamydial cell division. Support for our initial exploration of CT009 as a homolog to RodZ, rather than a transcription factor came from structural similarity and its possession of conserved residues involved in MreB interaction. Another key feature of RodZ that is conserved in CT009 is the presence of a C-terminal transmembrane helix. Given the large number of proteins with high sequence similarity to CT009 that are annotated as Xre family DNA binding domain or not annotated functionally (hypothetical), we hypothesized that many of these are also RodZ homologs.
To support this hypothesis, the protein sequence of CT009 was used in a BLAST search (Altschul et al., 1997). The top 20 proteins of similar sequence to CT009 (nonchlamydial, from non-redundant genera) were first analyzed by the HMMTOP server, which predicts the presence of transmembrane helices and protein topology (Tusnady & Simon, 2001). Fourteen of these proteins were found to have a single predicted transmembrane helix, with the N-terminus of the proteins oriented in the cytoplasm (like RodZ; Table II). These fourteen proteins were then analyzed further. All of these proteins are either annotated as hypothetical proteins (HP) or as transcription factors, helix-turn-helix containing proteins (HTH) or XRE-family members (XRE). All of the species that produce these proteins are rod shaped in morphology and all encode an MreB homolog (except for Carboxydibrachium pacificum), suggesting a need for a RodZ homolog. Importantly, none of these species have an annotated homolog to RodZ.
Table II.
Putative RodZ functional homologs based on sequence similarity to CT009
| CT009 | MreB-interacting | ||||
|---|---|---|---|---|---|
| Name | Annotation | Organism | Transmembrane Helix | E value | Residues Present |
| RodZ | RodZ | Escherichia coli | 112-131 | 0.004 | Yes, F60, Y64 |
| CT009 | HTH | Chlamydia trachomatis | 117-136 | N/A | Yes, Y57, F61 |
| ALO_19967 | XRE | Acetonema longum | 109-128 | 3e-14 | Yes, F49, F53 |
| CYB_0195 | HP | Synechococcus sp. | 110-129 | 1e-10 | Yes, Y56, F60 |
| Moth_1073 | XRE | Moorella thermoacetica | 100-117 | 1e-10 | Yes, Y49, F53 |
| Thewi_1795 | HP | Thermoanaerobacter wiegelii | 109-127 | 2e-10 | Yes, Y49, F53 |
| Btus_1597 | XRE | Kyrpidia tusciae | 107-125 | 2e-10 | Yes, Y49, F53 |
| Thit_1638 | HP | Thermoanaerobacter italicus | 110-128 | 4e-10 | Yes, Y49, F53 |
| CDSM653_1192 | HP | Carboxydibrachium pacificum | 107-126 | 5e-10 | Yes, Y49, F53 |
| Daes_1731 | HTH | Desulfovibrio aespoeensis | 115-133 | 6e-10 | Yes, Y51, F55 |
| TC41_1408 | XRE | Alicyclobacillus acidocaldarius | 157-181 | 7e-10 | Yes, Y65, F53 |
| syc0492_c | HP | Synechococcus elongatus | 112-134 | 1e-9 | Yes, F57, F61 |
| ANT_03850 | XRE | Anaerolinea thermophila | 241-260 | 2e-9 | No, Q181, M185 |
| PTH_1295 | HP | Pelotomaculum thermopropionicum | 105-122 | 3e-9 | Yes, Y48, F52 |
| TM_1864 | HP | Thermotoga maritima | 105-122 | 4e-9 | Yes, Y53, Y57 |
| Thexy_0625 | XRE | Thermoanaerobacterium xylanolyticum | 112-129 | 7e-9 | Yes, Y49, F53 |
HP - Hypothetical Protein; XRE – Xenobiotic response element
Finally, the amino acid sequences of these proteins were aligned to that of E. coli RodZ and C. trachomatis CT009 using ClustalW2 (Larkin et al., 2007). As mentioned previously, structural studies of RodZ and MreB revealed two aromatic residues that are crucial for mediating interactions between RodZ and MreB (red stars in Fig. 3C). Conservation of these MreB-interacting residues was examined and all CT009 homologs, except for one, possess these key amino acids. While the protein ANT_03850 from the filamentous thermophile Anaerolinea thermophile lacks these residues, its sequence homology to CT009 along with its predicted transmembrane helix suggest that it may be a RodZ homolog that mediates interactions with MreB through a novel mechanism. While this is not an exhaustive exploration of potential RodZ homologs among all bacterial species, it does showcase the power of going beyond sequence similarity to identify proteins of homologous function in other organisms.
Discussion
A combination of protein structure, bacterial two hybrid analyses, mutagenesis, and heterologous complementation provide comprehensive support that CT009 is a RodZ homolog in Chlamydia. Protein structure revealed that CT009 displays a high degree of structural similarity to RodZ. While the experimentally determined protein structure has adopted a domain swap dimer conformation, this does not negate nor depreciate the structural value. Evidence for this is the maintained orientation of the two key MreB interacting residues as well as the high amount of similarity to the previously solved RodZ protein structure. Furthermore, rod shaped morphology of rodZ deficient E. coli was partially restored with complementation of CT009 supporting structural observations and providing evidence that CT009 functions as a homolog to RodZ. Bacterial two-hybrid analyses demonstrated interaction with the key morphogenic protein MreB, similar to previous report (Ouellette et al., 2014). Mutagenesis of two amino acids in CT009 that are predicted to be central to interactions with MreB provided additional level of specificity for these interactions. Interestingly, localization studies of native and ectopically expressed epitope-tagged CT009 in Chlamydia, revealed localization uniformly around RB membranes. This localization was in contrast to MreB localization, which appears to be restricted to septal planes of dividing Chlamydia. Septal localization of MreB provides direct observational support to the hypothesis that MreB is serving as a functional FtsZ homolog in cell division. Overall, these observations provide comprehensive support for the functional annotation of CT009 as a RodZ homolog, which interacts with septal localized MreB protein.
The benefits of utilizing computational structure prediction for insight into protein function/role
An important observation to highlight from this report is the accuracy at which the I-TASSER server modeled the structure of CT009 and the importance of this structural information in the ultimate identification of this protein's role. The model generated was similar to the structure of RodZ with a RMSD value of 0.74 Å over 63/87 Cα atoms; this model lead to the initial hypothesis that CT009 is a RodZ homolog. The accuracy of this modeling is of note because of the extremely low sequence similarity between CT009 and RodZ protein sequences (BLAST E value >0.004) and particularly challenging category (i.e. ab initio) of computational protein structure modeling. It should also be noted that initial models of CT009, using the full-length sequence, were more similar to the transcription factors CI and Cro (data not shown). This predicted structure similarity, and the predominant association of helix-turn-helix domain with nucleic acid interactions, supported a hypothesis that CT009 functions as a transcription factor. As highlighted previously, accurate modeling of membrane-associated regions of proteins is very difficult for many structural modeling servers (I-TASSER included). Only after the putative trans-membrane region of CT009 was excluded was a confidently scored model of the protein generated, leading to the proposed hypothesis that CT009 functions as a RodZ homolog. These observations with computational structural modeling of CT009 therefore demonstrate some of the strengths, and weaknesses, of a structure-based annotation of protein function. However, it is clear that structural modeling can provide very valuable information regarding the function of a protein with otherwise ambiguous or unclear function.
Absence of periplasmic tail in CT009 could provide explanation for incomplete restoration of rod shaped phenotype in E. coli
Although the cytoplasmic domain of CT009 is structurally similar to RodZ and this membrane localized domain partially complements the function of RodZ in E. coli, the cells were wider than wild type (Fig. 4). This phenotype may be explained by the absence of a periplasmic domain in CT009. CT009 itself is significantly shorter than RodZ (Fig. 1) although the cytoplasmic domains are of very similar size. The major difference is the presence of a large periplasmic domain in RodZ (199 amino acids) while the predicted periplasmic domain of CT009 is only 7 amino acids long. Deletion studies on the different domains of RodZ have demonstrated that the periplasmic domain of RodZ is not strictly required for the maintenance of cell shape. Importantly, deletion of this domain led to a widening of cells (Bendezu et al., 2009). Therefore, it is unsurprising that, in the absence of this domain, CT009 is still able to functionally complement RodZ by rendering rod shaped cells that are also wider than normal. Partial complementation of CT009 in E. coli in this study is in contrast to those in a recent report (Ouellette et al., 2014). One of the more notable differences between the experiment performed herein is the use of an amino GFP-tagged CT009 in the previous study. While the GFP fusion did not affect membrane localization of CT009 or the function of RodZ in E. coli, given the sequence differences it seems plausible that CT009 interactions with critical morphologic proteins may be more sensitive to GFP perturbations as compared to native RodZ protein.
The precise function of the periplasmic portion of RodZ has not been defined, but there have been 16 RodZ homologs identified that lack this domain (Alyahya et al., 2009). These RodZ proteins with a periplasmic domain shorter than 10 amino acids were typically found in bacteria with reduced genomes or with round morphology (including but not limited to Rickettsia spp., Xanthamonas campestris and Staphylococcus spp.), suggesting dispensability for this domain. Unfortunately, RodZ homologs in these species have not, to date, been experimentally analyzed. Being both coccoid in morphology and possessing a reduced genome, it is unsurprising that CT009 also lacks a periplasmic domain.
CT009 Helix-turn-helix motif is likely for protein interaction and not transcriptional regulation
Original annotation of CT009 was a helix-turn-helix transcriptional regulator with the gene nomenclature yfgA (Stephens et al., 1998). However, current BLAST analysis indicates a conserved helix-turn-helix domain of the XRE superfamily. This functional assignment is supported by similar annotation of virtually all other Chlamydia genomes. Furthermore, sequence similarity to YfgA is sparse with E values greater than 0.004. While the gene annotation of CT009 as yfgA is somewhat enigmatic, the function or role of YfgA in any organism was unknown until genome wide analysis of single gene disruptions in E. coli suggested a role in integrity of outer membrane (Niba et al., 2007). This report was followed by an elegant analysis of YfgA being required for proper assembly of MreB cytoskeleton and cell shape in E. coli and being named RodZ (Bendezu et al., 2009). These observations, combined with those herein and from recent publication (Ouellette et al., 2014), provide a strong compliment of support for functional annotation. Another recent analysis searching for additional transcription factors in chlamydial species using a bioinformatics approach identified CT009 as the number one strongest predicted DNA-binding protein (Akers et al., 2011). This is not surprising, as we have now experimentally shown that CT009 possesses a helix-turn-helix motif. Although this motif is most frequently utilized for DNA binding, there are examples of this motif functioning as an interface for protein-protein interactions (Aravind et al., 2005). Importantly, RodZ also contains a helix-turn-helix motif; however, it utilizes this motif to stabilize protein-protein interactions, specifically with MreB (van den Ent et al., 2010). Therefore, the helix-turn-helix motif of CT009 is also likely involved in interactions with MreB, based on its structural and functional homology to RodZ. Interestingly, chlamydial species already encode a relatively low number of identified transcription factors and these data support exclusion of CT009 as a candidate. Similarly, structural and functional studies have supported that CT296, originally described as a divalent cation repressor, should also be removed as a candidate transcription factor (Kemege et al., 2011). This places a greater importance on the search for additional factors involved in regulation of chlamydial gene expression and the virulence defining developmental cycle as well as the expansion of the role of known transcription factors.
Gene expression patterns of CT009 and mreB and potential dual role in Chlamydia replication and development
An unexpected finding was the contrasting transcription expression patterns of CT009 and mreB relative to other cell replication components (Fig. 6). CT009 and mreB were continually upregulated throughout 30 hpi, whereas transcription for genes encoding gyrB, which is a part of the DNA replication machinery; dnaN, which is a part of the DNA polymerase III; and dnaE, a catalytic α-subunit of DNA polymerase III acting as a DNA helicase, were constitutively expressed throughout the developmental cycle. Additionally, rpoA encodes the α-subunit of RNA polymerase involved in transcription and was expressed constitutively as well. Not only did CT009 and mreB display increasing transcription levels during the developmental cycle, but so did rodA and ftsK. These data suggest the possibility that CT009 and mreB, and other cell division components might have dual roles, such as cellular processes involved in later stages of the chlamydial developmental cycle (e.g. RB to EB conversion). In support of this hypothesis, a recent report suggested that lateral transfer of peptidoglycan synthesis genes occurred between Chlamydiae and Streptomyces (Griffiths & Gupta, 2002). Interestingly, Streptomyces coelicolor use MreB to generate infectious particles (i.e. spores) (Mazza et al., 2006) and also interacts with RodZ as part of the cell morphogenic complex involved in spore cell wall formation. While this is intriguing consideration, clearly spore composition and formation in Streptomyces is very contrasting from those of chlamydial EBs.
Transcription expression patterns reported herein may appear to be in contrast to a previous study (Ouellette et al., 2014), however, it is important to note that these analyses were normalized differently. In this study, gene expression comparisons were made to replicating RB and the associated transcription of numerous genes encoding DNA replication, secretion, and RNA polymerase components. Comparison to transcription of genes associated with replicating RB enables a wider dynamic analysis and focuses on transcriptional changes that occur as RB convert to EB and other later developmental stage events. This is in contrast to the previous study which used genomic DNA for normalization and places an emphasis on relative transcriptional shut-off that occurs for most genes as more and more transcriptionally inert EBs are generated during the middle and late stages of the developmental cycle. Despite this difference in normalization, it is of interest to note from this previous report that the transcription levels of ct009 and mreB (~ 1 log decrease) do not appear to decrease as rapidly as those for the cell division component ftsI (~ 2 log decrease). These qualitative observations of those data roughly match the expression pattern for ct009, mreB, rodA, and ftsK reported herein showing a relative enhancement of transcription compared to bacterial replication encoding genes.
Cell shape and division in chlamydial species
One of the more significant observations in this report is the localization of MreB to predicted cell division sites (Fig. 6). An early report indicating that peptidoglycan in Chlamydia may be restricted to cell division planes was provided by Brown and Rockey (Brown & Rockey, 2000) using antisera raised against a mixture of mycobacterial cell wall skeleton. This original observation has been solidified through the elegant use of click chemistry and peptidoglycan labeling to reveal a clear localization at cell division sites (Liechti et al., 2014). The directed orientation of peptidoglycan synthesis to cell division sites through MreB was also recently proposed and based upon studies using chemical inhibitors of MreB that led to a non-dividing state in chlamydial cells (Ouellette et al., 2012). This proposed function of MreB in cell division is also in the context of an absence of FtsZ and possibly serving to, in some form, replace the function of the FtsZ during cell division. The observations provided herein, regarding localization of MreB at division sites, provide direct evidence and serve to strengthen the proposed role of MreB in cell division and functioning to direct peptidoglycan synthesis at this site.
Potentially disconcerting is the observation that CT009 was relatively unrestricted throughout the membrane of dividing Chlamydia (Fig. 6). One explanation may be that CT009 continues to interact with MreB and assist in tethering it to the membrane, as reported for other bacteria, but MreB requires other cell division proteins to direct and restrict localization to cell division planes. This would also suggest that CT009 has other functions associated with the observed broad membrane localization. The observation that CT009 interacts with FtsK and RodA provides further support for a role in cell division, especially as FtsK is associated with chromosome segregation during replication. It is important to consider that these observations were from bacterial two-hybrid analysis only and that subsequent in vivo analyses are needed to strengthen this biologic association. Related to these observations are those made by Jacquier et al. (Jacquier et al., 2014) of which they report on the localization of a RodZ homolog in the obligate intracellular bacteria Waddlia chondrophila. In this report, localization of RodZ was observed as the mid-cell during early division or upon treatment with penicillin. In contrast to the division site localization of MreB observed herein, Waddlia MreB appears to be enriched near division sites but also present throughout the membrane of the organism. A direct explanation for the contrasting observations is not readily apparent, however it is worth noting that while Waddlia is within the Chlamydiales order it does belong to a different family (Waddliaceae). As such there are many differences from Chlamydia genera including encoded resistance to β-lactam and encoding a much larger genetic repertoire (over 2 Mb vs ~1 Mb for C. trachomatis) (Bertelli 2010). Future studies employing the recent genetic advances in Chlamydia to directly monitor protein localization (fluorescent protein fusion) or evaluate close proximity protein partners (e.g. FRET) are expected to be influential in clarifying discrepancies such as these and providing greater insight to the basic cell biology of Chlamydia.
Experimental Procedures
I-TASSER Protein Structure Modeling
The I-TASSER server has been described previously (Roy et al., 2010, Zhang, 2009) and has its application in the modeling of chlamydial proteins (Kemege et al., 2011). The following amino acid sequence from the genome of Chlamydia trachomatis serovar L2 strain 434/Bu (L2/434/Bu; NC_010287.1) was selected as the target: GHMSEHVHKELLHLGEVFRSQREERALSLKDVEAATSIRLSALEAIEAGHLGKLI SPVYAQGFMKKYAAFLDMDGDRLLKEHPYVLKIFQEFSDQNMDMLLDLESMG GRNSPEKAIRS. This proteins sequence represents amino acids 1-116 of CT009, the predicted cytoplasmic domain. In addition, there are two extra amino acids added to the N-terminus of the sequence. In our purification strategy for recombinant protein, these amino acids are the remnant of an N-terminal solubility tag. In order to make a comparison between the modeled structure and the experimentally determined structure, the exact sequence of the recombinant protein used for crystallography was used for computational modeling. In the nomenclature of the 434/Bu genome, this protein is named CTL0264. As these two homologs share 100% sequence identity, the more widely utilized serovar D/UW-3 genome nomenclature is used and the protein is referred to as CT009.
Cloning, Protein Expression and Purification of CT009 and MreB
The putative cytoplasmic domain of CT009 (residues 1-116) and truncated MreB (residues 163-367 to eliminate predicted N-terminal amphipathic helix) were amplified using Chlamydia trachomatis (L2/434/Bu) genomic DNA and cloned into the NdeI and BamHI sites of pDZ1b (generous gift from Dr. Susan Egan, University of Kansas). The vector pDZ1b encodes a His6-tagged GB1 protein (the B1 immunoglobulin domain of Protein G from Streptococcus sp., used as a solubility tag (Cheng & Patel, 2004)) and a Tobacco Etch Virus (TEV) protease cleavage site upstream of the cloning site (resulting in an N-terminally tagged protein) (Chatterjee et al., 2011). It has been previously reported that three amino acids changes can be made to the GB1 protein (resulting in a protein termed “GB1basic”) that increase its pI to 8.0 but do not alter its efficacy as a solubility tag (Zhou & Wagner, 2010). These alterations increased the solubility of the protein and separation of desired protein from GB1 tag. Ligation reactions were then transformed into E. coli BL21-derivative Acella cells (Edge BioSystems). Cells were grown at 37°C with shaking at 220 rpm in LB medium supplemented with 100 μg mL−1 ampicillin to an optical density (OD600) of 0.6. Expression of recombinant protein was induced by the addition of 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) followed by shaking incubation at 15°C and 220 rpm for ~16 hours. Cells were harvested by centrifugation (10,000 × g, 20 min, 4°C) and frozen at −20°C.
For CT009, cell pellets were suspended in lysis/wash buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl), lysed by sonication and centrifuged at 16,000 × g, 20 min, 4°C. The soluble fraction was filtered through a 0.45 μm filter before it was applied to TALON metal (Co2+) affinity resin (Clontech) as per the manufacturer's guidelines. The beads were washed with lysis/wash buffer and GB1basic-CT0091-116 was eluted with lysis/wash buffer supplemented with 500 mM imidazole. Peak fractions were pooled and purity was assessed by SDS-PAGE, followed by staining with IRDye Blue protein stain (LI-COR) and visualization by an Odyssey infrared imaging system (LI-COR).
Purified GB1basic-CT0091-116 was treated with purified recombinant TEV protease as it was dialyzed into 50 mM Tris-HCl pH 8.0 at room temperature for ~16 hours. Protein digest was filtered through a 0.22 μm filter and NaCl was added to a final concentration of 500 mM. Protein solution was then applied to fresh TALON metal (Co2+) affinity resin. Flowthrough and washes (with lysis/wash buffer) were collected and assessed for purity by immunoblot against polyhistidine and CT009. Fractions with CT0091-116 but without the His6-tagged GB1basic were pooled and concentrated to 30.1 mg ml−1, using an Amicon 3,000 molecular weight cut-off centrifugal filtration device (Millipore). Purity of CT0091-116 was assessed by SDS-PAGE, followed by stain and visualization, as described above. Protein was further purified through gel filtration chromatography. Purified CT009 was concentrated for further use as antigen for generating rabbit anti-serum (Proteintech, Chicago, IL) and in crystallization studies.
For MreB163-367, protein inductions were performed the same as for CT009, however protein remained localized to inclusion bodies following sonication and centrifugation. Pellets were suspended in 6 M guanidine hydrochloride (GdnHCl) and incubated on ice for an hour before centrifugation at 39,000 × g for 20 min. The soluble fraction was filtered through a 0.45 μm filter before being applied to TALON metal (Co2+) affinity resin (Clontech) as per the manufacturer's guidelines. Flow through and washes (with lysis/wash buffer) were collected and assessed for purity by immunoblot against polyhistidine MreB. Elution fractions enriched with MreB163-367 were pooled prior to renaturation procedure. Renaturation was performed by dialysis at 4°C with a stepwise reduction of GdnHCl from 6 M, 3 M, 2 M, 0.5 M, and 0 M (30 mM Tris-HCl, pH 7.6, 200 mM KCl, 1 mM EDTA, 5 mM DTT, and 10% v/v glycerol) with a minimum of 4 hrs for each buffer exchange. Resulting protein sample was centrifuged at 16,000 × g for 30 minutes at 4°C before application of soluble protein to gel filtration chromatography. Purified MreB163-367 fractions were pooled, concentrated, and used as antigen for generating rabbit anti-serum (Proteintech, Chicago, IL).
Crystallization and Data Collection
A purified sample of CT009 was concentrated to 43.5 mg ml−1 in 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, pH 8.0 buffer for crystallization screening. All crystallization experiments were conducted using Compact Jr. (Emerald Biosystems) sitting drop vapor diffusion plates at 20 °C using equal volumes of protein and crystallization solution equilibrated against 75 μL of the latter. Initial crystals, which formed as clusters of needles in 1-2 days, were obtained from the Crystal Screen HT screen (Hampton Research) condition A6 (30% w/v PEG 4K, 100 mM Tris-HCl pH 8.5, 200 mM MgCl2). For X-ray data collection, samples were transferred to a fresh drop composed of 80% crystallization solution and 20% PEG 400 before flash freezing in liquid nitrogen. Initial X-ray diffraction data were collected in-house at 93K using a Rigaku RU-H3R rotating anode generator (Cu-Kα) equipped with Osmic Blue focusing mirrors and an R-Axis IV++ image plate detector. Subsequent crystals, which were used for high-resolution data collection, were obtained from a custom PEG Screen, with the final buffer composition being 100 mM Tris-HCl (pH 8.5), 200 mM MgCl2, 30% w/v PEG 5K, pH 8.5. Data were collected at the Advanced Photon Source beamline 17-ID using a Dectris Pilatus 6M pixel array detector.
Structure Solution and Refinement
Intensities for the in-house diffraction data were integrated using XDS (Kabsch, 2010). Synchrotron data were integrated using XDS via the XDSAPP interface (Krug et al., 2012). The Laue class check and data scaling were performed with Aimless (Evans, 2011). The highest probability Laue class was 4/mmm and space groups P41212 or P43212. The Matthew's coefficient (Matthews, 1968) (Vm =2.5 Å3/Dalton and 50.8% solvent content) indicated that there was a single CT009 molecule in the asymmetric unit. Structure solution was conducted by molecular replacement with Phaser (McCoy, 2007) via the Phenix (Adams et al., 2010) interface using the in-house diffraction data scaled to 2.6Å resolution. All space groups in the Laue class 4/mmm were tested. The search model for molecular replacement was generated using the protein structure prediction program MODELLER (Eswar et al., 2006). The top solution was obtained in the space group P41212, which was used from this point forward. The model was improved using the Autobuild suite via the Phenix interface which converged at Rfactor/Rfree = 25.0/33.0 following refinement. This model was used for subsequent molecular replacement searches against synchrotron data that was scaled to a resolution of 1.25Å. Structure refinement using anisotropic atomic displacement parameters and manual model building were conducted with Phenix and Coot (Emsley et al., 2010) respectively. Structure validation was conducted with Molprobity (Chen et al., 2010). Figures were prepared using PyMol (DeLano, 2002).
Protein-Protein Interactions by Bacterial Two-Hybrid
Genes encoding CT009 (full-length and residues 1-116), mreB (CT709), the rodA (CT726) and ftsK (CT739) were amplified by PCR using Chlamydia trachomatis LGV (L2/434/Bu) genomic DNA (Supplemental Table I). The gene encoding the CT009 Y57A, F61A mutant was amplified by PCR using pMLB1113-CT009 Y57A, F61A plasmid as a template. Vector plasmids (pKT25, pKNT25, pUT18 and pUT18C; Euromedex) were digested with BamHI and KpnI enzymes. Vector and inserts were processed using the In-Fusion HD ligation-independent cloning system (Clontech) and were transformed into α-Select Gold Efficiency cells (Bioline). Plasmids were isolated from overnight culture and then co-transformed into chemically competent BTH101 cells (Euromedex).
Interactions between proteins were assessed using the Bacterial Adenylate Cyclase Two Hybrid (BACTH) system, as described previously (Karimova et al., 1998). Briefly, BTH101 cells (a Δcya strain of E. coli) freshly transformed with compatible plasmids, each encoding a different fragment of the adenylate cyclase gene fused to a chlamydial protein of interest, and plated onto LB plates containing 100 μg ml−1 ampicillin, 50 μg ml−1 kanamycin, 0.5 mM IPTG and 50 μg ml−1 X-gal. Plates were incubated at 30°C for up to 48 hours and single colonies were picked and grown in liquid culture at 30°C. Expression of the β-galactosidase reporter gene by reconstituted adenylate cyclase was assessed using a β-galactosidase assay using mid-logarithmic phase growth cultures (1:100 dilutions from overnight growth cultures) containing 0.5 mM IPTG, as described previously (Miller, 1972). Assays were performed in triplicate and Student's t-test performed for statistical analysis. For qualitative analysis, 10 μl of turbid culture was plated on M63 media supplemented with Amp (50 μg ml−1), Kan (25 μg ml−1), IPTG (0.5 mM) and X-Gal (40 μg ml−1) and incubated for 2-5 days at 30 °C or RT prior to analysis.
RodZ Functional Complementation
Full length CT009 was amplified by PCR using C. trachomatis (L2/434/Bu) genomic DNA and specific primers (Table S1). Amplicons for CT009 were directionally cloned in the vector pMLB1113 (Bendezu et al., 2009)) at the EcoRI/SmaI sites to place expression under the control of Plac. The ligation reaction was transformed into E. coli BL21-derivative Acella cells (Edge BioSystems), successful cloning was verified by sequencing of the CT009 gene and finally purified pMLB1113-CT009 plasmid was transformed into FB60 cells. FB60 is an E. coli cell line, derived from the TB28 cell line, which lacks the RodZ gene (Bendezu et al., 2009). pFB290 is pMLB1113 with E. coli RodZ also under expression control of Plac and was used previously to demonstrate functional restoration in FB60. The pMLB1113 plasmid, pFB290, and the FB60 cell line were generous gifts from the laboratory of Piet de Boer (Case Western University). A plasmid encoding full length CT009 Y57A, F61A was generated by subjecting the pMLB1113-CT009 plasmid to site directed mutagenesis using the QuikChange II XL site-directed mutagenesis kit according to the manufacturer's protocol (Agilent Technologies) with specific primers (Table S1).
For functional complementation assays, cells were grown overnight in flasks of FB60 media (M9 minimal media with 0.2% maltose, 0.2% casamino acids and 50 mM thiamine-HCl) at 30 °C to an OD600 of 0.2-0.3 with 250 μM IPTG. Cells were prepared for fluorescence microscopy by suspending the cells from 3 ml of culture (harvested by centrifugation for 1 minute at 16,000 × g in a benchtop centrifuge) in 1 ml of Fixative Solution (0.05 mg ml−1 malachite green and 2.5% v/v glutaraldehyde). Cells were fixed for 1 hour at room temperature and then were washed twice with 1 ml of 1× PBS before resuspension in 100 μl of 1× PBS. Fixed cells were mounted on glass coverslips and visualized using an Axioplan-2 upright fluorescent microscope (Zeiss) and a 40X/1.3 oil Plan-Neofluor objective. Images were captured with an ORCA-ER digital camera (Hamamatsu). ImageJ was used to evaluate bacterial cell morphology by measuring major versus minor length of individual cells and calculating ratios of major/minor length. For each sample of bacterial strain analyzed, the average of major length versus width was calculated for more than 100 cells. The average from the three experimental averages and standard deviation were calculated. Student's test used to determine p values.
Quantitative gene expression
Total RNA was isolated from C. trachomatis (L2/434/Bu) infected L929 cells every 6 hpi through 36 hpi using TRIzol® Reagent (Life Technologies, Grand Island, NY). Isolated RNA was treated with DNase using the TURBO DNA-free kit (Life Technologies) according to the manufacturer's protocol and cleaned further using RNAeasy columns (Qiagen, Valencia, CA). Concentrations of RNA samples were assessed by spectrophotometry. For each RNA sample, cDNA was synthesized using the High Capacity RNA-to-cDNA kit (Applied Biosystems, Carlsbad, CA) following the manufacturer's instruction. Primer pairs for each gene target were verified for efficiency using serially diluted cDNA as a template for PCR. Resulting CT values were used to generate standard curves using the StepOnePlus software and confirmed that primer efficiency was suitable for relative quantification using the comparative CT method (2−ΔΔCT) (Schmittgen & Livak, 2008). Applied Biosystem's StepOnePlus Real-time PCR system was used for relative quantification of target genes. To verify that RNA samples were free of contaminating DNA, real-time PCR was performed using respective RT-negative reactions for each RNA sample. Relative quantification was performed for each gene target and for each time point using the comparative CT method (Schmittgen & Livak, 2008). Reactions were set up in duplicate with each mixture containing 500 nmol of each primer and Fast SYBR Green Master Mix (Applied Biosystems). Endogenous control, secY (CT510), was used as the internal sample normalizing control for all target genes and to reveal patterns of expression throughout the developmental cycle, relative quantification values were calibrated against the 6-hpi time point for each gene target.
Confocal microscopy and imaging
L929 cells in 8-well μ-Slide plates (Ibidi, Germany) were infected with wild-type C. trachomatis (L2/434/Bu). At 16 hpi, the cells were fixed with methanol for 10 minutes. For immunofluorescence staining, the primary antibodies were goat anti-MOMP (Virostat, Portland, ME) at 1:500 dilution, custom generated (Proteintech, Chicago, IL) affinity-purified rabbit anti-CT009 at 1:500 dilution and anti-MreB at 1:500 dilution, and mouse monoclonal anti-FLAG M2 (Sigma, St. Louis, MO) at 1:1000. Affinity purification of rabbit anti-CT009 and MreB was performed using AminoLink Plus coupling resin according to manufacture's protocol (Pierce-ThermoScientific, Rockford, IL). The secondary antibodies for the anti-CT009 images were Alexa Fluor 568 donkey anti-goat IgG (H+L) and Alexa Fluor 647 goat anti-rabbit IgG (H+L) (Invitrogen). The two secondary antibodies were incubated sequentially (anti-goat first, then goat anti- rabbit) with several washes in between to avoid cross-reactivity. To evaluate the specificity of the secondary antibodies, control wells were tested just the individual secondary antibodies, and the primary antibodies were tested separately with the non-matching secondary antibody. In each case, no cross-reactivity was observed. The secondary antibodies for the anti-MreB images were Alexa Fluor 568 Donkey Anti-Goat IgG (H+L) and Alexa Fluor 647 Donkey Anti-Rabbit IgG (H+L) (Invitrogen). For IFA staining of FLAG tagged CT009 samples, secondary antibodies were Alexa Fluor 568 donkey anti-mouse IgG (H&L), Alexa Fluor 488 donkey anti-goat IgG (H&L), and Alexa Fluor 647 donkey anti-rabbit. All secondary antibodies were used at a 1:750 dilution. DAPI was also used in staining, but not included in images. Wells were washed with PBS before 90% glycerol (in 0.1 M Tris-HCl, pH 9.0) was added to minimize photobleaching during imaging. Images were captured by spinning disk confocal microscopy with SlideBook (Intelligent Imaging Innovations, Inc., Denver, CO) software. Select images were pseudo-colored to maintain continuity with previous images. An Olympus IX81 inverted fluorescence microscope (Center Valley, PA) with a 150× objective (numerical aperture of 1.45) and a Hamamatsu EM-CCD camera was used. 3D Volume View feature in Slidebook was used with confocal images with Z-plane to stacks of 0.1 μm increments generate the 3D view.
For expression of Flag-tagged CT009 in C. trachomatis, a shuttle vector was constructed similar to previously described plasmid (Wickstrum et al., 2013) by combining a modified version of the E. coli expression plasmid pASK-IBA33plus (IBA, Göttingen) with the plasmid from C. trachomatis serovar L2 (434/Bu). The multiple cloning site in pASK was replaced with a single AgeI site such that any insert would provide the start codon immediately downstream of the AgeI junction. Briefly, the two plasmids were separately amplified by PCR that added compatible restriction endonuclease sites for subsequent digestion and ligation, and the resulting shuttle vector was named pTL2. Additionally, a variation of this shuttle vector was made by inserting a FLAG tag-encoding sequence at the AgeI site in pTL2. The inserted sequence also contained a Strep-tag II-encoding sequence as part of a tandem affinity purification (TAP) tag, although that was not used for this study. The AgeI site from pTL2 was recreated at the 5’ end of the FLAG tag-encoding sequence and an NheI site was created at the 3’ end so that genes inserted into the NheI site will have an N-terminal FLAG tag. For this study, the CT009 gene was inserted into the NheI site using the In-Fusion HD cloning kit (Clontech) to express CT009 with a N-terminal TAP tag. The NheI site was recreated at the 3’ end of the CT009 gene, but not at the 5’ end. See Figure S4 for the sequence of the FLAG-tagged CT009. Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA) and all PCR was done using the Phusion High Fidelity Polymerase kit (Thermo Scientific). Plasmid constructs were confirmed by DNA sequencing. Transformation into C. trachomatis L2 (434/Bu) and selection of stable shuttle vector was accomplished as described previously (Wickstrum et al., 2013)
Immunoblot analysis
L929 cells were infected with C. trachomatis L2 434/Bu and harvested at 20 hpi. Media was aspirated from cells and washed three times with PBS before 50 μl SDS-PAGE Laemmli buffer was added per 10 cm2 well surface area. Wells were scraped with a pipet tip until the cell layer detached (~10 seconds). An equal volume of PBS was added in order to pipet the detached cells, which were then boiled at 100 °C for 5 min prior to analysis (~10% of total lysate was analyzed per lane). Affinity purified antibodies to CT009 (rabbit), MreB (rabbit), or RpoA (rat) and IR tagged secondary antibodies (Rockland Immunochemicals, Limerick, PA) were used at 1:5000 dilution.
Supplementary Material
Table I.
Crystallographic data for CT009 refined to 1.25Å resolution.
| CT009 | |
|---|---|
| Data Collection | |
| Unit-cell parameters (Å, °) | a=55.17, c=88.12 |
| Space group | P41212 |
| Resolution (Å)1 | 46.76-1.25 (1.27-1.25) |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 100 |
| Observed reflections | 318,621 |
| Unique reflections | 38,420 |
| <I/σ(I)>1 | 25.1 (2.5) |
| Completeness1 | 99.9 (98.5) |
| Multiplicity1 | 8.3 (7.5) |
| Rmerge (%)1, 2 | 3.6 (82.4) |
| Rmeas (%)1, 4 | 3.8 (88.5) |
| Rpim (%) 1, 4 | 1.3 (32.0) |
| CC½1, 5 | 0.999 (0.816) |
| Refinement | |
| Resolution (Å) | 29.21-1.25 |
| Reflections (working/test) | 36,417/1,921 |
| Rfactor / Rfree (%)3 | 15.9/17.3 |
| No. of atoms (Protein/Water) | 1,007/111 |
| Model Quality | |
| R.m.s deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.437 |
| Average B-factor (Å2) | |
| All Atoms | 20.2 |
| Protein | 18.8 |
| Water | 32.9 |
| Coordinate error (maximum) likelihood) (Å) | 0.13 |
| Ramachandran Plot | |
| Most favored (%) | 99.2 |
| Additionally allowed (%) | 0.8 |
| PDB ID | 4GQM |
Values in parenthesis are for the highest resolution shell.
Rmerge = ∑hkl ∑ i |Ii(hkl) - <I(hkl)>| / ∑ hkl ∑ i Ii(hkl), where Ii(hkl) is the intensity measured for the ith reflection and <I(hkl)> is the average intensity of all reflections with indices hkl.
Rfactor = ∑ hkl ∥Fobs (hkl) | - |Fcalc (hkl) ∥ / ∑hkl |Fobs (hkl)|; Rfree is calculated in an identical manner using 5% of randomly selected reflections that were not included in the refinement.
Rmeas = redundancy-independent (multiplicity-weighted) Rmerge (Evans, 2006, Evans, 2011). Rpim = precision-indicating (multiplicity-weighted) Rmerge (Weiss, 2001, Diederichs & Karplus, 1997).
CC½ is the correlation coefficient of the mean intensities between two random half-sets of data (Evans, 2012, Karplus & Diederichs, 2012).
Acknowledgements
This research was supported by NIH (R03AI081099). KK was supported partially by a National Institutes of Health T32 Training Grant (AI070089). PSH, MLB, JMH, and NB were supported by National Institutes of Health (AI079083). JW was supported by NIH (AI103711). Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the University of Kansas Protein Structure Laboratory was supported by grants from National Center for Research Resources (5P20RR017708-10) and the National Institute of General Medical Sciences (8P20GM103420-10). We would also like to thank Lei Hu and Patricia Braun for technical assistance. We would also like to thank Dr. Piet de Boer (Case Western Reserve University) for strains and plasmids used in the RodZ functional complementation assays. Lastly, Shauna Moore for purification of MreB recombinant protein and Katie Summers for critical reading of manuscript.
References
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nature reviews. Microbiology. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal AK, Rodgers DW, Drottar M, Ptashne M, Harrison SC. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988;242:899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Akers JC, HoDac H, Lathrop RH, Tan M. Identification and functional analysis of CT069 as a novel transcriptional regulator in Chlamydia. Journal of bacteriology. 2011;193:6123–6131. doi: 10.1128/JB.05976-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C. RodZ, a component of the bacterial core morphogenic apparatus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1239–1244. doi: 10.1073/pnas.0810794106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Barrientos LG, Louis JM, Botos I, Mori T, Han Z, O'Keefe BR, Boyd MR, Wlodawer A, Gronenborn AM. The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures. Structure. 2002;10:673–686. doi: 10.1016/s0969-2126(02)00758-x. [DOI] [PubMed] [Google Scholar]
- Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Choe S, Eisenberg D. Domain swapping: entangling alliances between proteins. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WJ, Rockey DD. Identification of an antigen localized to an apparent septum within dividing chlamydiae. Infect Immun. 2000;68:708–715. doi: 10.1128/iai.68.2.708-715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Zhong D, Nordhues BA, Battaile KP, Lovell S, De Guzman RN. The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci. 2011;20:75–86. doi: 10.1002/pro.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D. Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Patel DJ. An efficient system for small protein expression and refolding. Biochemical and biophysical research communications. 2004;317:401–405. doi: 10.1016/j.bbrc.2004.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsaro D, Venditti D. Emerging chlamydial infections. Crit Rev Microbiol. 2004;30:75–106. doi: 10.1080/10408410490435106. [DOI] [PubMed] [Google Scholar]
- Dajkovic A, Lutkenhaus J. Z ring as executor of bacterial cell division. Journal of molecular microbiology and biotechnology. 2006;11:140–151. doi: 10.1159/000094050. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System, 2009. 2002 [Google Scholar]
- Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nature structural biology. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiology and molecular biology reviews : MMBR. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Andreas D, Baxevanis, editors. Comparative protein structure modeling using Modeller. Current protocols in bioinformatics / editoral board. 2006 doi: 10.1002/0471250953.bi0506s15. [et al.] Chapter 5: Unit 5 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta crystallographica. Section D, Biological crystallography. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Evans P. Biochemistry. Resolving some old problems in protein crystallography. Science. 2012;336:986–987. doi: 10.1126/science.1222162. [DOI] [PubMed] [Google Scholar]
- Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta crystallographica. Section D, Biological crystallography. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Griffiths E, Gupta RS. Protein signatures distinctive of chlamydial species: horizontal transfers of cell wall biosynthesis genes glmU from archaea to chlamydiae and murA between chlamydiae and Streptomyces. Microbiology. 2002;148:2541–2549. doi: 10.1099/00221287-148-8-2541. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic acids research. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Frandi A, Pillonel T, Viollier P, Greub G. Cell wall precursors are required to organize the chlamydial division septum. Nature communications. 2014;5:3578. doi: 10.1038/ncomms4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta crystallographica. Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two5 hybrid analysis. Journal of bacteriology. 2005;187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kemege KE, Hickey JM, Lovell S, Battaile KP, Zhang Y, Hefty PS. Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes. Journal of bacteriology. 2011;193:6517–6528. doi: 10.1128/JB.05488-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschnitz EM, Heichlinger A, Schirner K, Winkler J, Latus A, Maldener I, Wohlleben W, Muth G. Proteins encoded by the mre gene cluster in Streptomyces coelicolor A3(2) cooperate in spore wall synthesis. Molecular microbiology. 2011;79:1367–1379. doi: 10.1111/j.1365-2958.2010.07529.x. [DOI] [PubMed] [Google Scholar]
- Krug M, Weiss MS, Heinemann U, Mueller U. XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. Journal of Applied Crystallography. 2012;45:568–572. [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Molecular microbiology. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature. 2014;506:507–510. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. Journal of molecular biology. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Mazza P, Noens EE, Schirner K, Grantcharova N, Mommaas AM, Koerten HK, Muth G, Flardh K, van Wezel GP, Wohlleben W. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Molecular microbiology. 2006;60:838–852. doi: 10.1111/j.1365-2958.2006.05134.x. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta crystallographica. Section D, Biological crystallography. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press; Plainview, NY.: 1972. p. 466. [Google Scholar]
- Minezaki Y, Homma K, Nishikawa K. Genome-wide survey of transcription factors in prokaryotes reveals many bacteria-specific families not found in archaea. DNA research : an international journal for rapid publication of reports on genes and genomes. 2005;12:269–280. doi: 10.1093/dnares/dsi016. [DOI] [PubMed] [Google Scholar]
- Niba ET, Naka Y, Nagase M, Mori H, Kitakawa M. A genome-wide approach to identify the genes involved in biofilm formation in E. coli. DNA research : an international journal for rapid publication of reports on genes and genomes. 2007;14:237–246. doi: 10.1093/dnares/dsm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette SP, Karimova G, Subtil A, Ladant D. Chlamydia Co-Opts the Rod-Shape Determining Proteins MreB and Pbp2 for Cell Division. Molecular microbiology. 2012 doi: 10.1111/j.1365-2958.2012.08100.x. [DOI] [PubMed] [Google Scholar]
- Ouellette SP, Rueden KJ, Gauliard E, Persons L, de Boer PA, Ladant D. Analysis of MreB interactors in Chlamydia reveals a RodZ homolog but fails to detect an interaction with MraY. Frontiers in microbiology. 2014;5:279. doi: 10.3389/fmicb.2014.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. Journal of computational chemistry. 2002;23:128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schuck P, Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers. 2000;54:328–341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Shiomi D, Sakai M, Niki H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 2008;27:3081–3091. doi: 10.1038/emboj.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Johnson CM, Persons L, de Boer P, Lowe J. Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J. 2010;29:1081–1090. doi: 10.1038/emboj.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Molecular microbiology. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- Weiss MS. Global indicators of X-ray data quality. Blackwell; Oxford: 2001. ROYAUME-UNI. [Google Scholar]
- White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Molecular microbiology. 2010;76:616–633. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- Wickstrum J, Sammons LR, Restivo KN, Hefty PS. Conditional gene expression in Chlamydia trachomatis using the tet system. PloS one. 2013;8:e76743. doi: 10.1371/journal.pone.0076743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Weihe EK, Margolin W. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. Journal of bacteriology. 1998;180:6424–6428. doi: 10.1128/jb.180.23.6424-6428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;77(Suppl 9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Interplay of I-TASSER and QUARK for template-based and ab initio protein structure prediction in CASP10. Proteins 82 Suppl. 2014;2:175–187. doi: 10.1002/prot.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Wagner G. Overcoming the solubility limit with solubility17 enhancement tags: successful applications in biomolecular NMR studies. J Biomol NMR. 2010;46:23–31. doi: 10.1007/s10858-009-9371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.