Abstract
Understanding how axons are guided to target locations within the brain is of fundamental importance for neuroscience, and is a widely studied area of research. Biologists have an unmet need for reliable and easily accessible methods that generate stable, soluble molecular gradients for the investigation of axon guidance. Here we developed a microfluidic device with contiguous media-filled compartments that uses gravity-driven flow to generate a stable and highly reproducible gradient within a viewing compartment only accessible to axons. This device uses high-resistance microgrooves to both direct the growth of axons into an isolated region and to generate a stable gradient within the fluidically isolated axon viewing compartment for over 22 h. Establishing a stable gradient relies on a simple and quick pipetting procedure with no external pump or tubing. Since the axons extend into the axonal compartment through aligned micro-grooves, the analysis of turning is simplified. Further, the multiple microgrooves in parallel alignment serve to increase sample sizes, improving statistical analyses. We used this method to examine growth cone turning in response to the secreted axon guidance cue netrin-1. We report the novel finding that growth cones of embryonic mouse cortical axons exhibited attractive turning in the lower concentrations of netrin-1, but were repulsed when exposed to higher concentrations. We also performed immunocytochemistry in growth cones exposed to a netrin-1 gradient within the axon viewing compartment and show that netrin receptors associated with both attraction and repulsion, DCC and UNC5H, localized to these growth cones. Together, we developed an accessible gradient chamber for higher throughput axon guidance studies and demonstrated its capabilities.
Introduction
To establish a functional neuronal network, axons synapse onto targets that are often quite distal from their cell body or soma. This is accomplished during development, when axons are guided by spatio-temporally regulated gradients of extracellular guidance cues. The motile growth cone at the tip of the extending axon contains transmembrane receptors that integrate information from numerous guidance cues to allow the axon to navigate accurately over the order of hours and days toward specific destinations. In the simplest description, when exposed to a gradient of an attractive cue, the growth cone turns up the concentration gradient. Upon encountering a gradient of a repulsive guidance cue, the growth cone turns down the gradient. In vivo, growth cones are simultaneously exposed to gradients of multiple cues, and can exhibit bimodal responses to the same cue, indicating that extracellular information is integrated by the growth cone for appropriate axon navigation. Bimodal responses are mediated by changes in the expression, localization, and activation of attractive and repulsive receptors for individual guidance cues. In instances where the axon is sufficiently long, the soma does not perceive the guidance cue and the growth cone responds independently. As such, axons orchestrate local response to soluble gradients of multiple cues to achieve proper nervous system connectivity.
Over the past two and a half decades, in vivo approaches have identified numerous axon guidance cues as well as axon guidance receptors relevant to achieving fidelity in the connectivity of the nervous system. To evaluate the function of individual cues and individual receptors in the laboratory, however, a controlled microenvironment is required. A multitude of techniques have been employed to do so. For example, explants of specific neural tissues can be embedded within a collagen matrix and exposed to a gradient of guidance cue secreted by transfected cells or diffusing from a cue-soaked agarose block.1,2 In this setup, neurite outgrowth from the side of the explant proximal to the cue is compared to outgrowth from the side distal to the cue. Although a guidance ratio is reported, changes in biased neurite outgrowth and guidance cannot be separated, particularly since the neurons are exposed to the guidance cue from the start of the experiment. This method permits quantification of the behavior of population of neurons in response to a specific cue. However the behavior of individual axons in response to the gradient differs cannot be gleaned from these experiments. Indeed, axonal turning, a key component of axon guidance is not observed in this experiment. Furthermore, as the explants and the source of guidance cues are placed by hand into the collagen gel, there is variability in the distance between the explant and the cue. Because the solidification of the collagen gel is sensitive to temperature and pH, there can be a high frequency of poor explant outgrowth or death.
A second long-used assay to interrogate axon guidance is the stripe assay.3,4 Here, linear or geometric arrays of guidance cues are adhered to glass, plastic, or membrane surfaces using micro-channels or stamps to selectively pattern cues.5 Subsequently dissociated neurons are cultured on the substrates, and changes in axonal trajectories upon reaching a stripe of substrate-bound cue can be assessed. The abrupt transition between two substrates however is unlikely to model extracellular conditions in vivo, and the behavior of an axon in this assay may reveal only a preference for one substrate over another, not necessarily substrate repulsion. Discontinuous and continuous gradients of substrate bound cues have also been established with various printing procedures, but these methods are incompatible with soluble cues or temporal control.6 Another popular assay that alternatively offers higher spatial and temporal resolution compares the turning of individual growth cones toward a micropipette source of axon guidance cue7–9 or guidance cue-coated beads.10,11 Although this offers benefits in spatial resolution, these assays suffer variable gradients, and thus difficulty in reproducibility.12 Because images of only one growth cone can be acquired per experiment, these assays are low throughput. In addition, due to the rate of growth of vertebrate central neurons, meaningful data require hours of image acquisition. The high variability in the growth and shape of axons prior to exposure to axon guidance cues complicates analysis of the growth cone turning response.
Microfluidic devices replica molded using poly(dimethylsiloxane) (PDMS) are valuable tools for studying cells because of the ability to create reproducible structures and microenvironments.13–15 In particular, a PDMS-based device developed to use passive forces for fluidic isolation that is easy to use has been well-adopted by the neuroscience community.16–21 This device uses high-resistance microgrooves embedded in barrier dividers to separate and fluidically isolate axons. Because of the high fluidic resistance of these microgrooves, hydrostatic pressure due to a differential in fluid volumes between the compartments can maintain fluidic isolation.
Microfluidic devices have been used to establish concentration gradients to study chemotaxis of cells using flow-based splitting and mixing devices22,23 and via source/sink devices,24–26 but have had limited success for studying growth cone guidance of neurons. Reasons for this include neuronal sensitivity to shear stress and the challenge of restricting the gradient to axons and their growth cones. In addition, many of the microfluidic-based gradient devices used for chemotaxis require the use of external pump equipment and tubing that are cumbersome to set up and are not amenable to repeated experimentation. Collagen gels can be used to reduce the flow effects on axons and provide a diffusion barrier,27 but also add potential molecular and imaging confounds, along with added experimental complexity and variability.
Here we designed an easy-to-use, passive microfluidic device to expose isolated axons to soluble gradients for prolonged periods of time without flow effects. This device offers several innovations and advantages. First, because axons extend through aligned microgrooves into a fluidically isolated compartment, cell bodies have negligible exposure to the axon guidance cue. Because the microgrooves align the axons prior to their exposure to the gradient, there is no likelihood of a false positive analysis of axon turning. Additionally, this device allows data acquisition from multiple pre-aligned axons in a gradient simultaneously, to improve throughput and data analyses. The axons in the device described here extend directly along the glass cover-slip; as such they are accessible to high resolution transmitted light live cell imaging as well as fixation and immunocytochemistry. Indeed these advances permitted the surprising observation of a bimodal axonal response to netrin-1 that was concentration dependent.
Results and discussion
Design of the passive microfluidic gradient chamber
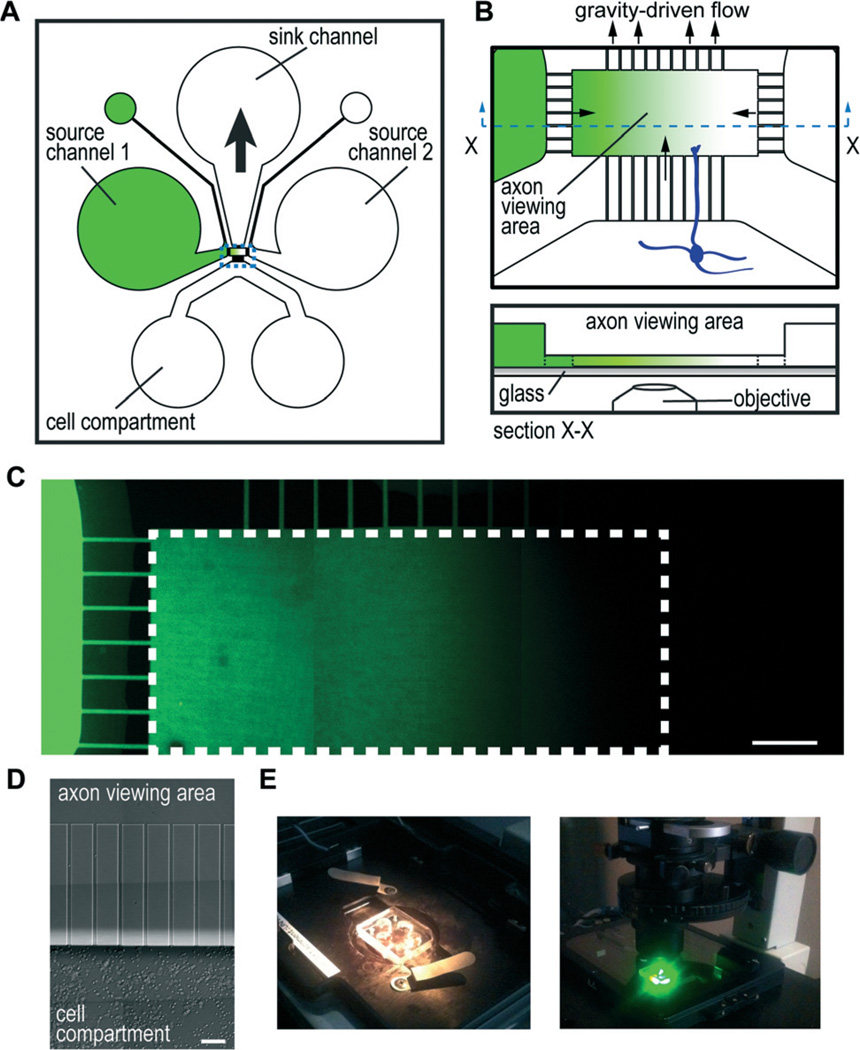
To make gradient devices user-friendly and cost effective, we focused on designing a device using passive hydrostatic forces. We designed the resulting passive microfluidic gradient chamber (or micro-pass gradient chamber), which consists of a microfluidic channel for culturing neurons (cell compartment), two source channels for adding soluble molecules to establish gradients, and a sink channel used to establish a pressure differential (Fig. 1A). These microfluidic channels are all connected to an axon viewing area (~1 mm × 0.5 mm) via microgrooves embedded within a PDMS barrier (Fig. 1B). The pressure differential induced by removing fluid from the sink well draws fluid from the source wells towards the sink to establish a gradient across the axon viewing area. The microgrooves and axon viewing area are 5 µm tall, much shorter than the other channels, which are 100 µm high; thus, the microgroove regions and axon viewing area provide high fluidic resistance such that the flow rate slows minimizing the potential influence on axon turning. The microgrooves in particular provide the most fluidic resistance due to greater surface contact with the walls of the channels. Since fluidic resistance is proportional to the length of the microgrooves, we designed the microgrooves connecting the source and sink channels to be shorter than the micro-grooves connected to the cell compartment in order to preferentially flow source solutions into the axon viewing area and not into the cell compartment when a pressure differential is established. In addition, the length of the micro-grooves allows axons to enter into the axonal viewing area after 2–4 days.
Fig. 1. Micro-pass gradient chamber.
(A) Cartoon illustration of micro-pass chamber and the channel layout. Source channels connect to a smaller outlet port to facilitate channel filling. Gradients are established by removing 30–45 µl from the sink well which draws in solution by gravitational force from the source channels/wells to create a gradient within the axon viewing area. (B) Illustration of the axon viewing area enlarged (top) and a profile view (bottom) showing that the axon viewing area and microgrooves are the same height (5 µm) and the source channels are taller (100 µm). (C) Representative fluorescence image montage of a low molecular weight dye gradient (AlexaFluor hydrazides) established within the micro-pass chamber. White dashed line outlines the axon viewing area. Scale bar, 100 µm. (D) Growth of mouse cortical neurons after 3 days within the chamber. Axons are present, but are not visible at this magnification. Scale, 50 µm. (E) Photographs of micro-pass chamber mounted on an inverted microscope. Photograph on the right shows the chamber within a humidified, temperature controlled, and CO2 adjusted stage incubator after 20 h of timelapse imaging.
The molecular gradient generated in the micro-pass chamber involves a balance of convective and diffusive forces. To evaluate the fluid velocities within the chamber, we used fluorescent microspheres applied to the solution channel and measured the travel distance over a 1 s exposure time (data not shown). We found that removing 30 µl from the sink well generated sufficient fluid velocity within the microgrooves from the source channel to the axon viewing area (>80 µm s−1) such that we would expect no diffusion into the opposing channel for a range of molecular diffusivities (40–400 µm2 s−1). The fluid velocity slowed considerably immediately after entering into the axon viewing compartment (20–30 µm s−1). In addition we found that within the center most region of the axon viewing area the fluid velocity slowed even further such that we would expect diffusion of both small and large molecular weight species, thus producing a smooth gradient. Further, we estimated that this flow-maintained gradient pattern would produce a stable gradient for longer than 24 h factoring in the time required to normalize fluid levels using our fluid velocity estimates.
To experimentally verify that we could generate a gradient within the axon viewing area of this chamber, we used a low molecular weight fluorescent dye introduced into a source channel to enable visualization of the resulting gradient (Fig. 1C). Because only simple pipetting is required, the procedure to initiate a gradient required less than 5 min. We found negligible fluorescence in the opposing source channel (Fig. 1C) and in the cell compartment (data not shown), showing that the concentration gradient established was specifically isolated within the axon viewing compartment. These results were reproduced >20 times. We also found equivalent results for the higher molecular weight fluorescein conjugated dextran (70 kDa).
Because flow rate and flow pattern are critical to establish a gradient, the fluid levels within the wells must be precise. For example, if the fluid level is too high in the cell compartments, the flow pattern will skew the gradient such that it will not be orthogonal to the axons entering into the axon viewing area. We found that 150 µl in the source wells and 85–100 µl in the cell compartments produced a consistently orthogonal gradient.
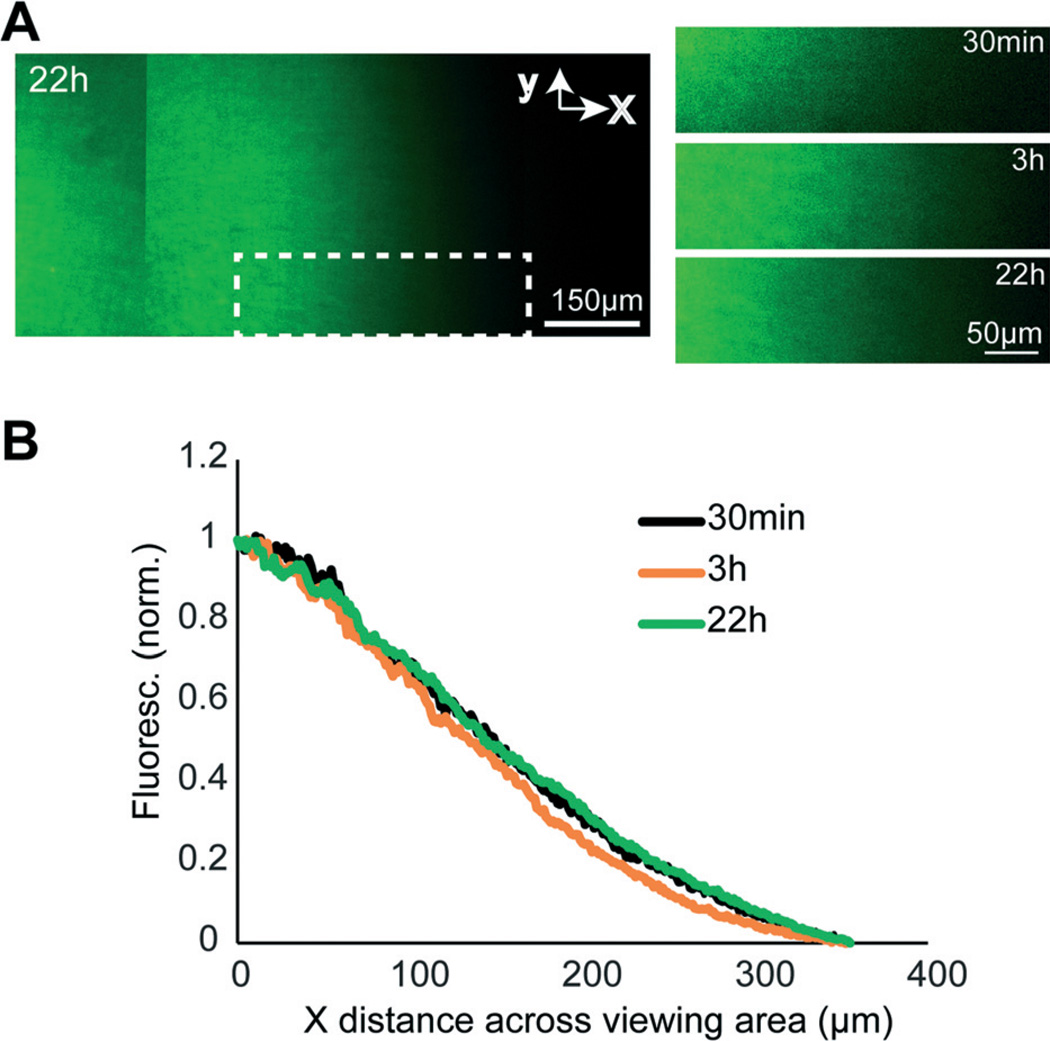
Gradient stability
To test whether gradients established within the micro-pass gradient chamber were stable over a prolonged period, we used a low molecular weight fluorescent dye and examined the gradient profile across the axon viewing area between 30 min and 22 h. A representative chamber is shown in Fig. 2. We found that the gradient was extremely stable; the normalized fluorescence throughout the axonal viewing area was equivalent between 30 min and 22 h.
Fig. 2. Gradient stability.
(A) Montaged fluorescence images of the axon viewing area within a representative micro-pass gradient chamber 22 h after initiating a gradient using AlexaFluor dye (1 µM). The graphs on the right show the stability of gradient within the same white dashed region at different timepoints. (B) Gradient profiles within the white dashed region highlighted in (A) at 30 min, 3 h, and 24 h. Fluorescence intensity values were normalized to the maximum and minimum within the boxed region.
Growth cones exposed to a control gradient do not exhibit consistent turning responses
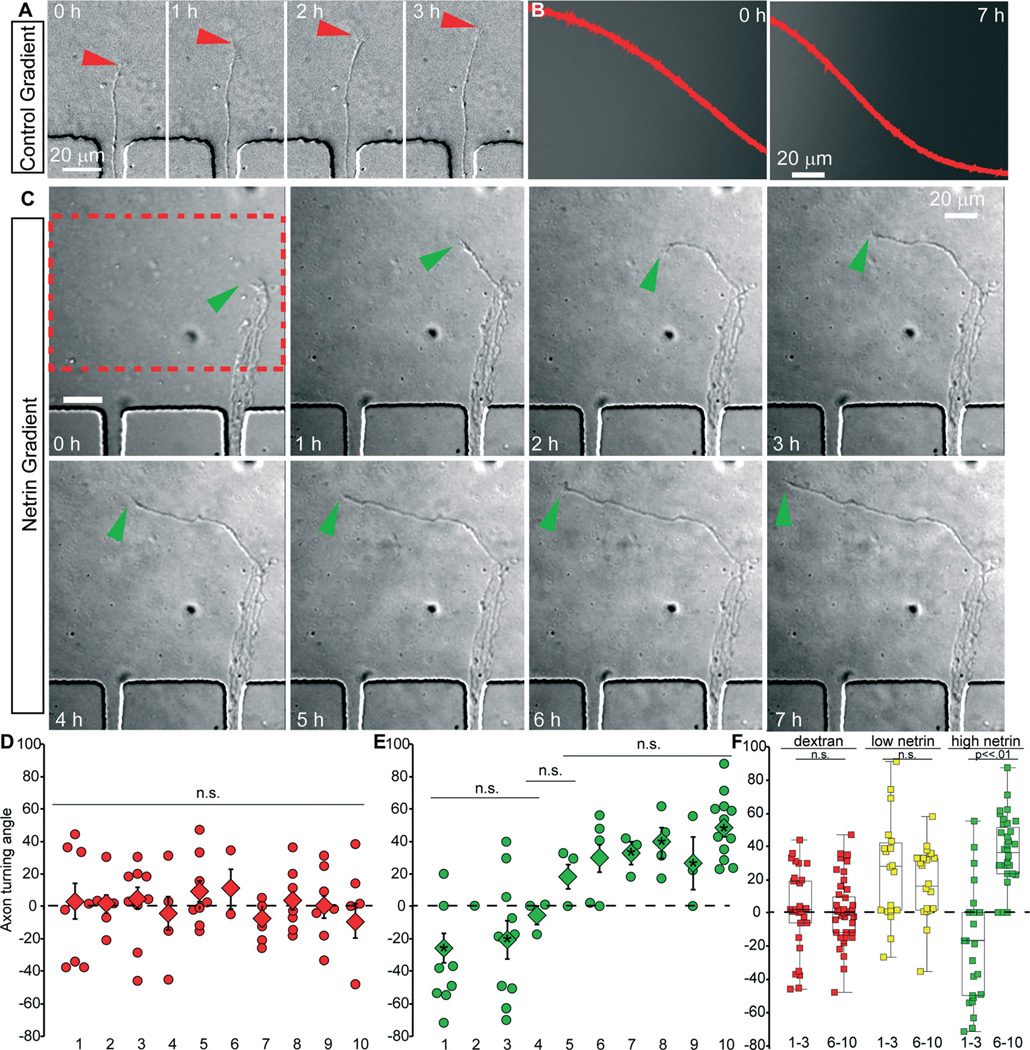
To demonstrate the functionality of the micro-pass gradient chamber, we first confirmed that axons of embryonic mouse neocortical neurons did not exhibit significant turning in response to fluid flow within the axon viewing area upon establishing a dextran gradient (Fig. 3A, red arrowhead). The micro-pass gradient chambers were seeded with cortical neurons isolated from embryonic day (E)15.5 mice. By approximately three to four days in vitro, cortical axons extended from one or more of the 10 microgrooves into the axon viewing compartment. At this time, media from one source well was replaced with growth media containing 1 µM fluorescent dextran (70 kDa). The viability of neurons within the micro-pass gradient chamber was excellent, both within the cell culture incubator and on the microscope (Fig. 3A, C). Images of axons (DIC) and the gradient (Epifluorescence) were acquired every 5 minutes for 8–18 hours. To measure axon turning, we calculated the angle between the initial trajectory of the axon, and the trajectory following exposure to the gradient. Angles were measured for axons that were within the viewing area before initiation of the gradient, as well as those that exited a microgroove into the viewing area over the course of the experiment. The mean angle of axon turning for all axons (n = 18 devices, 79 axons) was 1.5° with a 95% confidence interval of 2.3°, indicating there was only a random turning response. Furthermore, for each microgroove the mean turning angle was close to zero and not significantly different between any microgroove (Fig. 3D, within 95% confidence interval of the mean, nonparametric Kruskal–Wallis significance test). Importantly, these results clearly demonstrate that the low levels of induced flow caused by removal of media from the sink well in the absence of netrin-1 in a source well was not sufficient to induce axon turning within this device, as no consistent turning response was observed.
Fig. 3. The micro-pass chamber reveals the response of neocortical axons to netrin is concentration dependent.
(A) Example DIC images from time-lapse of cortical axon entering the axon viewing area containing only a dextran gradient from µgroove 8. This axon continues to extend in the same direction and does not turn. Red arrowhead denotes growth cone, time in hours. (B) Epifluorescence images at indicated time (hours) of dextran/netrin gradient from region of interest in C. The overlay red plot denotes quantification of dextran/netrin gradient across the ROI. X/Y scales are constant between images. (C) Example DIC images from time-lapse of cortical axons entering the axon viewing area containing a netrin-1 gradient from µgroove 8. This axon turns up the netrin-1 gradient, indicative of attraction. The growth cone tip is denoted by green arrowhead, time in hours. (D) Angle of turning in a dextran gradient of individual axons (diamonds) and average angle of turning (square) reported for each µgroove. (E) Angle of turning in a netrin-1 gradient (starting at 600 ng ml−1) of individual axons (diamonds) and average angle of turning (square) reported for each µgroove. (F) Pooled axon turning angles for µgrooves 1–3 and 6–10 in a dextran gradient (red), and netrin-1 gradients starting at 250 ng ml−1 (yellow) and 600 ng ml−1 (green).
Growth cones exposed to a netrin-1 gradient show concentration-dependent responses
We next tested the response of axons to a gradient of netrin-1. Netrin-1 is a secreted axon guidance cue28 of approximately 80 kDa, that can be either attractive or repulsive, based upon the netrin receptors present.29 DCC is typically an attractive netrin receptor, whereas UNC5H alone or in collaboration with DCC mediates repulsion.29–31 Cortical neurons are a mixed population composed of multiple neuronal subtypes that extend axons to various destinations. This includes netrin-sensitive projections that cross the corpus callosum or extend to the thalamus.28,32–34 We chose to add 600 ng ml−1 of netrin-1 to the source well, as this concentration increases filopodia number,35 a harbinger of axon turning. Lower concentrations of netrin that have been used in micropipette assays and axon branching assays would then occur within the axon viewing area. After removal of 30–45 µl of media from the sink, a stable gradient of netrin-1 was established (Fig. 3B). Axons emerged from the microgrooves into the gradient, facilitating the quantification of turning angle. Axon growth was not significantly altered by the presence of netrin-1 (n = 62 axons, 12 devices for netrin-1, 19.5 ± 4.3 µm per hour with netrin-1 versus 13 ± 2.5 µm per hour, p > .05) or the microgroove from which the axon emerged (not shown). This is consistent with previous findings that netrin-1 does not increase axon length in cortical neurons.36,37 Although axon growth was unaffected, the turning of axons was modulated by the netrin gradient. Axons extending from microgrooves 1 and 3, which were exposed to the highest concentrations of netrin, were repulsed and turned down the netrin-1 gradient (Fig. 3E). This response was significantly different from axons extending from micro-grooves 1 and 3 in devices exposed to only a fluorescent dextran gradient (Kruskal Wallis ANOVA with Bonferonni posthoc test, p < .03). In microgrooves 4 and 5, axon turning was not significantly different between the two conditions (Kruskal Wallis ANOVA with Bonferonni posthoc test, p = .6, and p = .4). Axons extending from microgrooves 7–10, which were exposed to the lowest netrin concentrations, turned up the gradient toward higher netrin-1 concentrations, in a response significantly different from observed in the control gradient (Kruskal Wallis ANOVA with Bonferonni posthoc test, p < .05 in each condition). Statistical comparison of netrin-dependent axon turning angles between microgrooves revealed that axons extending from microgrooves 1–3 and 6–10 were distinct populations (Kruskal Wallis ANOVA with Bonferonni posthoc test, p > .18 between microgrooves within a population, p < .01 between populations). To increase the statistical power of our analysis, we pooled the axon turning angles for microgrooves 1–3 (n = 26 netrin axons, 34 dextran axons) and 6–10 (n = 36 netrin axons, 45 dextran axons). With increased sampling and normal distribution, we performed ANOVA with Bonferroni post-hoc analysis of these populations (Fig. 3F). The two populations were significantly different from each other and both dextran populations (p < .0001). In contrast the two dextran populations were not different. These surprising results suggested that the turning response of cortical neurons to a gradient of netrin-1 was concentration dependent, while axon growth was not affected.
Although netrin-1 has both attractive and repulsive properties, to our knowledge, this is the first evidence of a concentration-dependent switch in axonal response to netrin-1. Since the gradients formed in the micro-pass chamber are reproducible and stable (Fig. 2), we curve fit the fluorescence data averaged from 3 micro-pass chambers to a polynomial equation to estimate the concentration of netrin at each microgroove. This analysis revealed that the axons of cortical neurons were extremely sensitive to netrin-1 and were attracted to netrin-1 at low concentrations (<15 ng ml−1). This has not been observed previously, likely due to axon guidance assays typically performed on the order of <2 hours. Furthermore, this analysis demonstrated that netrin-1 concentrations >300 ng ml−1, which are typically used to promote filopodia formation and axon turning acutely, are repulsive over longer time courses. To confirm this surprising observation, we lowered the netrin-1 concentration in the source well to 250 ng ml−1. Under this paradigm, repulsive axon turning in microgrooves 1–3 was no longer observed and axons turned up the gradient (Fig. 3F, average turning angle 29.8 ± 10, n = 28 axons and 5 devices, significantly different from grooves 1–3 in high netrin-1 starting concentrations, p < .002).
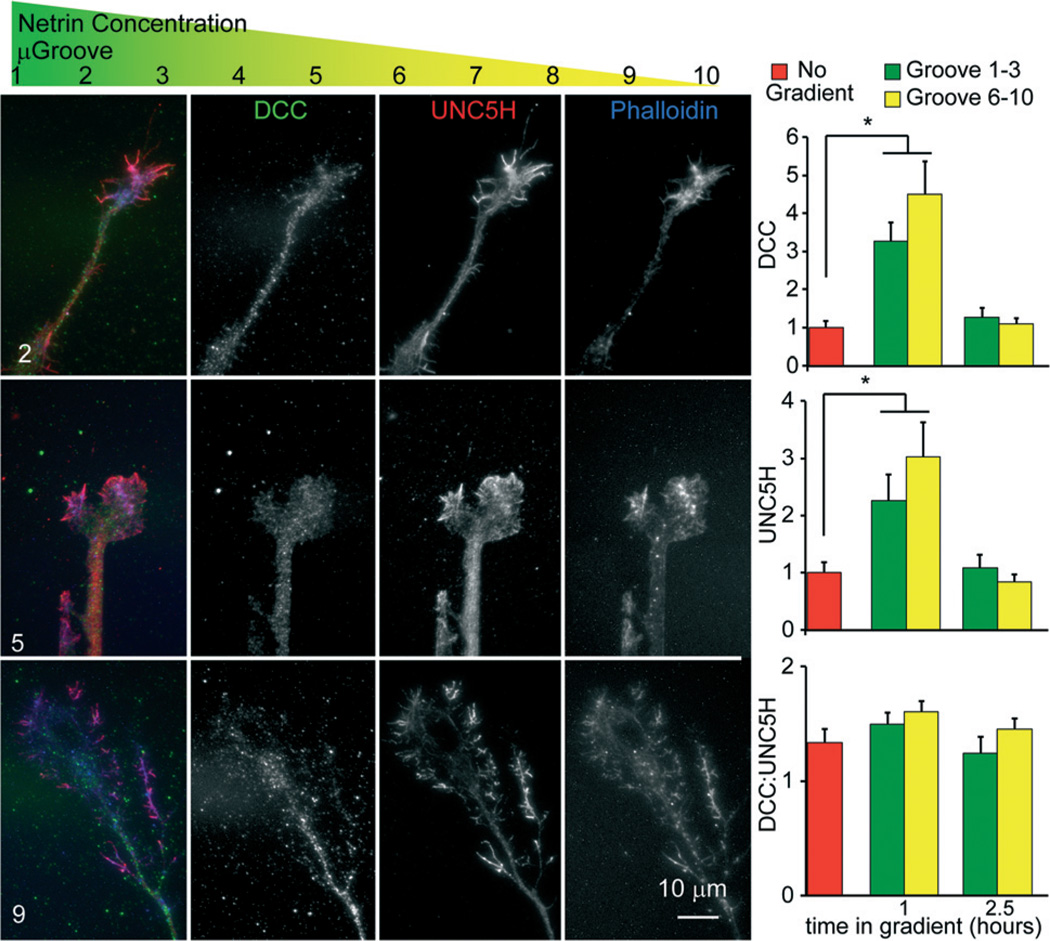
Growth cones within the axon viewing compartment contain netrin receptors
The novel finding of a differential response of cortical axons to a netrin-1 gradient dependent upon the microgroove and thus netrin-1 concentration was surprising. Attractive and repulsive axonal responses to netrin-1 depend upon the receptors DCC and UNC5H, respectively,29,33,38 but a role for the concentration of netrin has not been described. Our results suggested that cortical axons must contain the machinery to be both attracted and repulsed by netrin-1, and that their activity may be concentration dependent. To determine if both receptors were expressed and localized to the axonal growth cones of cortical neurons, we established a netrin-1 gradient within the micro-pass gradient device for 4 h, before fixing and staining for netrin-1 receptors using immunocytochemistry (Fig. 4). This revealed that both DCC and UNC5H were expressed and localized to the growth cones of cortical neurons exposed to a netrin gradient within micro-pass gradient devices, consistent with the ability of cortical axons to respond both positively and negatively to a netrin gradient. However there were no differences in the amount of DCC ( p = .19) or UNC5H ( p = .18) within the growth cone. In response to netrin-1 treatment, DCC accumulates at the cell surface and is subsequently endocytosed and degraded by the proteasome.39 Surface levels of UNC5H are also regulated by endocytosis.40 To determine if high and low netrin-1 concentrations had differential effects on the localization of netrin receptors to the growth cone surface, we probed non-permeabilized growth cones with antibodies that recognize the receptor extracellular domains. The localization patterns of DCC and UNC5H in the growth cone were indistinguishable from those of permeabilized cells shown in Fig. 4. As previously observed, surface levels of DCC increased and subsequently decreased following exposure to netrin-1. However, there were no significant differences between axons extending into attractive and repulsive netrin-1 concentrations ( p = .46). The surface levels of UNC5H behaved similarly. Furthermore the ratio of DCC:UNC5H on the growth cone surface was not different under any condition. These findings indicate that additional factors are involved in switching growth cone response to netrin-1 between attraction and repulsion. This could include differential dimerization and/or dynamics of the receptors or differential activation of downstream signalling pathways.
Fig. 4. Attractive and repulsive netrin receptors localize to the tip of neocortical axons extending in a netrin-1 gradient.
Schematic showing where each µgroove sits within the netrin-1 gradient. Images of neo-cortical axons within the axon viewing area from the indicated µgroove. Axons were stained with phalloidin (blue), an antibody against DCC (green), and an antibody against UNC5H (red). Both DCC and UNC5H localized to the growth cones and axon tips of growth cones in the low netrin concentrations/attractive regime (µgroove 6–10), and the higher netrin concentrations, repulsive regime (µgroove 1–3). Graphs show quantification of surface levels of the netrin receptors in growth cones extending from indicating µgrooves at indicated times in a netrin-1 gradient (starting concentration 600 ng ml−1).
Experimental
Device fabrication
SU-8 masters were fabricated using photolithography and replica molding was performed using poly(dimethylsiloxane) (PDMS) as described previously.16 Briefly, SU-8 2005 (Microchem) was spun onto a clean silicon wafer to achieve a thickness of 5 µm and then exposed through a chrome mask (Photo Sciences Inc.) to pattern the microgrooves and axon viewing area. A second layer of SU-8 2150 (Microchem) was then spun onto the wafer to achieve a thickness of 100 µm and then a second chrome mask containing the boundaries of the cell compartment, source wells, and sink well was aligned to the wafer, exposed and developed. The developed SU-8 master was then placed into a petri dish for PDMS replica molding.
Device assembly and preparation
Either 30 × 22 mm 1.5 glass coverslips or 24 × 50 mm 1.0 glass coverslips (Carolina Glass) were sonicated in 100% EtOH for 30 minutes, and coated with PDL (40 mg L−1; BD Biosciences 354210) at 37 °C overnight, rinsed, and dried as described previously. PDMS chambers were cleaned, sterilized with 70% EtOH and attached to cover glass.16,17,41
Animals
All mice were on a C57BL/6 background and were bred at UNC with approval from the Institutional Animal Care and Use Committee. Timed pregnant females were obtained by placing male and female mice together overnight; the following day was designated as E0.5 if the female had a vaginal plug.
Neuronal culture
Neurons were dissociated from E15.5 mouse cortex as previously described.36 Briefly, dams were sacrificed by CO2 followed by cervical dislocation. Embryonic neocortex was removed and dissociated with trypsin for 20 minutes at 37 °C. After dissociation, neurons were collected by centrifugation, and resuspended to a concentration of 15000 cells per µl in Neurobasal media supplemented with B27 (Invitrogen). To load the micro-pass gradient chamber, 150 µl of Neurobasal media supplemented with B27 was added to the sink channel, allowing media to fill the axon viewing area and micro-grooves. Then, approximately 150 000 cells were added into the cell compartment, and allowed to adhere for five minutes.16 Following adherence, we added media to the remaining channels. Devices were incubated at 37 °C, 5% CO2 until axons had extended into the axon viewing area.
Establishing gradients
For the gradient stability experiments, we filled the micro-pass gradient chamber with buffer and incubated the chamber overnight. We added fluid to the sink channel, ensuring that the fluid fills the axon viewing area and microgrooves and then filled the remaining channels (150 µl per well for all wells except for the cell compartment wells which received 85 µl per well because they are 6 mm in diameter instead of 8 mm). Air occasionally became trapped initially within the sink channel closest to the microgroove region, but released through the PDMS within 1–2 days after filling. After equilibration, we induced a concentration gradient by replacing the source channels with 150 µl of fluorescent dye solution: 1 µM of AlexaFluor 488 hydrazide (570 MW; Invitrogen) or 1 µM AlexaFluor 568 hydrazide (731 MW; Invitrogen). We then immediately removed 30–45 µL from the sink channel and added 10 µL of mineral oil to each well of the cell compartment.
Recombinant netrin-1 was concentrated as previously described.28 To establish a gradient of netrin-1 and fluorescent dextran (or fluorescent dextran alone), media was removed from the source channel wells, and replaced with media supplemented with fluorescent dextran or with media supplemented with both fluorescent dextran and 600 ng ml−1 or 250 ng ml−1 netrin-1. Subsequently 30 µl of media was removed from the sink channel, and the formation of a gradient was monitored by widefield epifluorescence of fluorescent dextran. If a gradient failed to form, an additional 15 µl was removed from the sink channel.
Imaging and analysis
To assess gradient stability using fluorescent dyes, we used a spinning disk confocal imaging system (Yokogawa CSU-X1) configured for an Olympus IX81 zero-drift microscope (Andor Revolution XD system). Light excitation was provided by 50 mW 488 nm and 50 mW 561 nm lasers. The following band pass emission filters (Semrock brightline) were used 525–30 nm (TR-F525-030) and 607–36 nm (TR-F607-036). We used Andor iQ software to acquire montage images using an automated stage (Ludl Bioprecision2) and 10× objective (0.25 NA). Images were stitched using Andor iQ.
Time-lapse imaging of cells was performed on an inverted microscope (IX81-ZDC2) with an automated XYZ stage (Prior) with MetaMorph acquisition software, a 20×, 1.4 NA Olympus Plan Apo-chromat objective, an electron-multiplying charge-coupled device (iXon), and an imaging chamber (Stage Top Incubator INUG2-FSX; Tokai Hit), which maintained humidity, 37 °C and 5% CO2. Images of axons and the gradient were acquired every 5 minutes for 8–18 hours. The change in axon length and angle of axon turning over this time course was analyzed using ImageJ analysis software. Briefly, the change in length was calculated for all growing axons within the axon viewing area. The angle of axon turning was measured from the initial trajectory of the axon. Positive turning angles indicate turning up the gradient, whereas negative angles indicate turning down the gradient. We did not analyze axons that had deviated more than 15° from normal before a gradient was established.
Immunocytochemistry
Once axons extended into the axon viewing compartment, a netrin-1 gradient was established as described above for 0–4 hours within the cell culture incubator, using a starting netrin-1 concentration of 600 ng ml−1. Subsequently, the devices were removed from the coverslips, and cells were fixed with 4% paraformaldehyde. For the images shown in Fig. 4, cells were subsequently washed in PBS, permeabilized in 0.1% TritonX100, and blocked in 10% BSA. A monoclonal antibody against the extracellular domain DCC (A-20, Santa Cruz Biotechnology, Inc.) and a goat polyclonal against extracellular domain of UNC5H1 (E-15, Santa Cruz Biotechnology, Inc.), Alexa Fluor secondary antibodies and Alexa Fluor phalloidin were used for immunocytochemistry. To measure surface levels of the two receptors, the same procedure was performed, without permeabilization with TritonX100. For immunocytochemistry, images were acquired with an Olympus 100× 1.49 NA Plan Apo-chromat TIRF objective. All imaging parameters were maintained over the experiment (exposure time/camera gain) to allow quantitative comparison of fluorescence intensities. Total and surface levels of DCC and UNC5 were measured in growth cones and normalized to values of non-stimulated growth cones stained simultaneously.
Statistical analysis
At least 2 independent experiments were performed for each assay. Data distribution normality was determined using the Shapiro–Wilk test. Normally distributed data were compared by unpaired t-test, for two independent samples, or ANOVA with Tukey post-hoc correction, for >2 samples. For non-normal data, the Kruskal–Wallis nonparametric ANOVA with Bonferonni posthoc correction for >2 samples. All data are presented as means ± standard error of the mean, unless where 95% confidence interval (CI) was reported. Statistical significance is represented as such: *p < .05, **p < .01, ***p < .005).
Conclusions
We have established a novel technology that uses passive fluid flow to produce stable, reproducible gradients of soluble extracellular cues in a fluidically isolated axon viewing compartment. This device permits high viability, high spatial and temporal resolution imaging, and simple analysis of axon growth and turning within the gradient. Using this device, we found a concentration dependent turning response of embryonic cortical axons within a gradient of netrin; at high concentrations of netrin-1, axons are repelled, at lower concentrations are attracted to netrin-1.
Acknowledgements
We thank the Taylor and Gupton laboratories for thoughtful critique of the manuscript. We thank Haeijin Song, Joyce Kamande, Naucika Desouza, Cassie Meeker, and Asif Khan for their contributions and assistance. This work was supported by the National Institutes of Health grants: GM108970 (S.L.G), R41MH097377 (A.M.T.), K12HD073945 (A.M.T.) and a University Research Council grant provided by the University of North Carolina at Chapel Hill School of Medicine (S.L.G), and an American Heart Association fellowship 14POST20450085 (S.M.). A.M.T. is an Alfred P. Sloan Research Fellow.
Footnotes
Yes there is potential competing interest. A.M.T. is an inventor of microfluidic chambers to compartmentalize neurons (US 7419822 B2) and has financial interest in Xona Microfluidics, LLC. S.M. and S.L.G. declare no competing financial interests.
Contributor Information
A. M. Taylor, Email: amtaylor@unc.edu.
S. L. Gupton, Email: sgupton@email.unc.edu.
Notes and references
- 1.Tessier-Lavigne M, Placzek M, Lumsden AG, Dodd J, Jessell TM. Nature. 1988;336:775–778. doi: 10.1038/336775a0. [DOI] [PubMed] [Google Scholar]
- 2.Serafini T, Colamarino SA, Leonardo ED, Wang H, Beddington R, Skarnes WC, Tessier-Lavigne M. Cell. 1996;87:1001–1014. doi: 10.1016/s0092-8674(00)81795-x. [DOI] [PubMed] [Google Scholar]
- 3.Walter J, Henke-Fahle S, Bonhoeffer F. Development. 1987;101:909–913. doi: 10.1242/dev.101.4.909. [DOI] [PubMed] [Google Scholar]
- 4.Walter J, Kern-Veits B, Huf J, Stolze B, Bonhoeffer F. Development. 1987;101:685–696. doi: 10.1242/dev.101.4.685. [DOI] [PubMed] [Google Scholar]
- 5.Knoll B, Weinl C, Nordheim A, Bonhoeffer F. Nat. Protoc. 2007;2:1216–1224. doi: 10.1038/nprot.2007.157. [DOI] [PubMed] [Google Scholar]
- 6.Dupin I, Dahan M, Studer V. J. Neurosci. 2013;33:17647–17655. doi: 10.1523/JNEUROSCI.3277-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ming GL, Song HJ, Berninger B, Holt CE, Tessier-Lavigne M, Poo MM. Neuron. 1997;19:1225–1235. doi: 10.1016/s0896-6273(00)80414-6. [DOI] [PubMed] [Google Scholar]
- 8.Song HJ, Ming GL, Poo MM. Nature. 1997;388:275–279. doi: 10.1038/40864. [DOI] [PubMed] [Google Scholar]
- 9.Song H, Ming G, He Z, Lehmann M, McKerracher L, Tessier-Lavigne M, Poo M. Science. 1998;281:1515–1518. doi: 10.1126/science.281.5382.1515. [DOI] [PubMed] [Google Scholar]
- 10.Lin CH, Forscher P. Neuron. 1995;14:763–771. doi: 10.1016/0896-6273(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 11.Suter DM, Forscher P. J. Neurobiol. 2000;44:97–113. [PubMed] [Google Scholar]
- 12.Pujic Z, Giacomantonio CE, Unni D, Rosoff WJ, Goodhill GJ. J. Neurosci. Methods. 2008;170:220–228. doi: 10.1016/j.jneumeth.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Chiu DT, Jeon NL, Huang S, Kane RS, Wargo CJ, Choi IS, Ingber DE, Whitesides GM. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2408–2413. doi: 10.1073/pnas.040562297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Annu. Rev. Biomed. Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 15.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. Nat. Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor AM, Rhee SW, Tu CH, Cribbs DH, Cotman CW, Jeon NL. Langmuir. 2003;19:1551–1556. doi: 10.1021/la026417v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor AM, Dieterich DC, Ito HT, Kim SA, Schuman EM. Neuron. 2010;66:57–68. doi: 10.1016/j.neuron.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baleriola J, Walker CA, Jean YY, Crary JF, Troy CM, Nagy PL, Hengst U. Cell. 2014;158:1159–1172. doi: 10.1016/j.cell.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castle MJ, Perlson E, Holzbaur EL, Wolfe JH. Mol. Ther. 2014;22:554–566. doi: 10.1038/mt.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor AM, Wu J, Tai HC, Schuman EM. J. Neurosci. 2013;33:5584–5589. doi: 10.1523/JNEUROSCI.2944-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeon NL, Baskaran H, Dertinger SK, Whitesides GM, Van de Water L, Toner M. Nat. Biotechnol. 2002;20:826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 23.Lin F, Saadi W, Rhee SW, Wang SJ, Mittal S, Jeon NL. Lab Chip. 2004;4:164–167. doi: 10.1039/b313600k. [DOI] [PubMed] [Google Scholar]
- 24.Diao J, Young L, Kim S, Fogarty EA, Heilman SM, Zhou P, Shuler ML, Wu M, DeLisa MP. Lab Chip. 2006;6:381–388. doi: 10.1039/b511958h. [DOI] [PubMed] [Google Scholar]
- 25.Irimia D, Charras G, Agrawal N, Mitchison T, Toner M. Lab Chip. 2007;7:1783–1790. doi: 10.1039/b710524j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng SY, Heilman S, Wasserman M, Archer S, Shuler ML, Wu M. Lab Chip. 2007;7:763–769. doi: 10.1039/b618463d. [DOI] [PubMed] [Google Scholar]
- 27.Kothapalli CR, van Veen E, de Valence S, Chung S, Zervantonakis IK, Gertler FB, Kamm RD. Lab Chip. 2011;11:497–507. doi: 10.1039/c0lc00240b. [DOI] [PubMed] [Google Scholar]
- 28.Serafini T, Kennedy TE, Galko MJ, Mirzayan C, Jessell TM, Tessier-Lavigne M. Cell. 1994;78:409–424. doi: 10.1016/0092-8674(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 29.Hong K, Hinck L, Nishiyama M, Poo MM, Tessier-Lavigne M, Stein E. Cell. 1999;97:927–941. doi: 10.1016/s0092-8674(00)80804-1. [DOI] [PubMed] [Google Scholar]
- 30.Moore SW, Tessier-Lavigne M, Kennedy TE. Adv. Exp. Med. Biol. 2007;621:17–31. doi: 10.1007/978-0-387-76715-4_2. [DOI] [PubMed] [Google Scholar]
- 31.Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS, Culotti JG, Tessier-Lavigne M. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- 32.Deck M, Lokmane L, Chauvet S, Mailhes C, Keita M, Niquille M, Yoshida M, Yoshida Y, Lebrand C, Mann F, Grove EA, Garel S. Neuron. 2013;77:472–484. doi: 10.1016/j.neuron.2012.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fazeli A, Dickinson SL, Hermiston ML, Tighe RV, Steen RG, Small CG, Stoeckli ET, Keino-Masu K, Masu M, Rayburn H, Simons J, Bronson RT, Gordon JI, Tessier-Lavigne M, Weinberg RA. Nature. 1997;386:796–804. doi: 10.1038/386796a0. [DOI] [PubMed] [Google Scholar]
- 34.Metin C, Deleglise D, Serafini T, Kennedy TE, Tessier-Lavigne M. Development. 1997;124:5063–5074. doi: 10.1242/dev.124.24.5063. [DOI] [PubMed] [Google Scholar]
- 35.Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, Gertler FB. Neuron. 2004;42:37–49. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- 36.Winkle CC, McClain LM, Valtschanoff JG, Park CS, Maglione C, Gupton SL. J. Cell Biol. 2014;205:217–232. doi: 10.1083/jcb.201311003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dent EW, Barnes AM, Tang F, Kalil K. J. Neurosci. 2004;24:3002–3012. doi: 10.1523/JNEUROSCI.4963-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leonardo ED, Hinck L, Masu M, Keino-Masu K, Ackerman SL, Tessier-Lavigne M. Nature. 1997;386:833–838. doi: 10.1038/386833a0. [DOI] [PubMed] [Google Scholar]
- 39.Kim TH, Lee HK, Seo IA, Bae HR, Suh DJ, Wu J, Rao Y, Hwang KG, Park HT. J. Neurochem. 2005;95:1–8. doi: 10.1111/j.1471-4159.2005.03314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams ME, Wu SC, McKenna WL, Hinck L. J. Neurosci. 2003;23:11279–11288. doi: 10.1523/JNEUROSCI.23-36-11279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor AM, Rhee SW, Jeon NL. Methods Mol. Biol. 2006;321:167–177. doi: 10.1385/1-59259-997-4:167. [DOI] [PubMed] [Google Scholar]